Abstract
Acid sensing ion channels (ASICs) are voltage-independent, amiloride-sensitive channels implicated in diverse physiological processes ranging from nociception to taste. Despite the importance of ASICs in physiology, we know little about the mechanism of channel activation. Here we show that psalmotoxin activates non- and sodium-selective currents in chicken ASIC1a at pH 7.25 and 5.5, respectively. Crystal structures of ASIC1a – psalmotoxin complexes map the toxin binding site to the extracellular domain and illuminate how toxin binding triggers an expansion of the extracellular vestibule and stabilization of the open channel pore. At pH 7.25 the pore is ~10 Å in diameter whereas at pH 5.5 the pore is largely hydrophobic and elliptical in cross section with dimensions of ~5 by ~7 Å, consistent with a barrier mechanism for ion selectivity. These studies define mechanisms for activation of ASICs, illuminate the basis for dynamic ion selectivity and provide the blueprints for new therapeutic agents.
Acid-sensing ion channels (ASICs)1, members of the epithelial sodium channel/degenerin (ENaC/DEG) superfamily of cation channels2, 3, open a transmembrane pore upon exposure to low pH4. Primarily found in the central and peripheral nervous systems5–7, ASICs occupy diverse physiological roles that include nociception8, 9, mechanosensation8, synaptic plasticity, learning and memory7, and fear conditioning10. The ASIC subfamily is coded by four genes which give rise to seven isoforms11, of which ASIC1a is permeable to Na+ and Ca2+ and is implicated in ischemic neuronal injury12, 13. The ENaC channel3, found throughout the human body, is crucial to the regulation of blood pressure14 and is directly involved in Liddle’s syndrome15 and pseudohypoaldosteronism16.
ASICs and ENaCs are trimeric17, voltage-independent and sodium-selective ion channels sensitive to the classic ENaC blocker amiloride1, 3. Whereas ASICs display a selectivity of Na+:K+ ranging from 3 to 30:1 and are inhibited by micromolar concentrations of amiloride, ENaCs harbor a preference for Na+:K+ of >100:1 and are blocked by nanomolar concentrations of amiloride2, 18. For both ASICs and ENaCs, Li+ permeability is similar to that of Na+ and monovalent ions larger than K+, such as Cs+, are generally impermeable19. However, the ‘peak’ and ‘sustained’ or ‘steady state’ ionic currents carried by ASICs display variable ion selectivity and blocker sensitivity20–25, properties reminiscent of the dynamic ion selectivity of trimeric P2X receptors26. At present there is no understanding of how ASICs adopt sodium-selective and non-selective conformations with differential sensitivity to the blocker amiloride.
Activation of the ion channel pore in ASICs is classically conditioned by drops in extracellular pH from ~7.5 to pH 4–64 with the currents of ASIC1a exhibiting rapid and nearly complete desensitization27. Psalmotoxin (PcTx1), classified as an inhibitor cystine knot toxin from a South American tarantula28, 29, acts potently on ASIC1a, increasing the channel’s affinity for protons30 and, contingent on the species and splice variant of the channel, acts as an agonist, eliciting steady state current or as an antagonist, diminishing ion channel activation31, 32. The action of PcTx1 as an antagonist confers both analgesic33 and neuroprotective12 properties.
Here we report crystallographic and electrophysiological studies of the action of PcTx1 on chicken ASIC1a, showing the determinants of toxin binding, the mechanism by which toxin binding opens the ion channel, and the architecture of non- and Na+-selective conformations of the ion channel pore. Our studies inform mechanisms of gating and permeation in ASICs and ENaC/DEG channels and lay a foundation for development of new molecules for modulation of ion channel activity.
Function and architecture of ASIC1a – PcTx1 complex
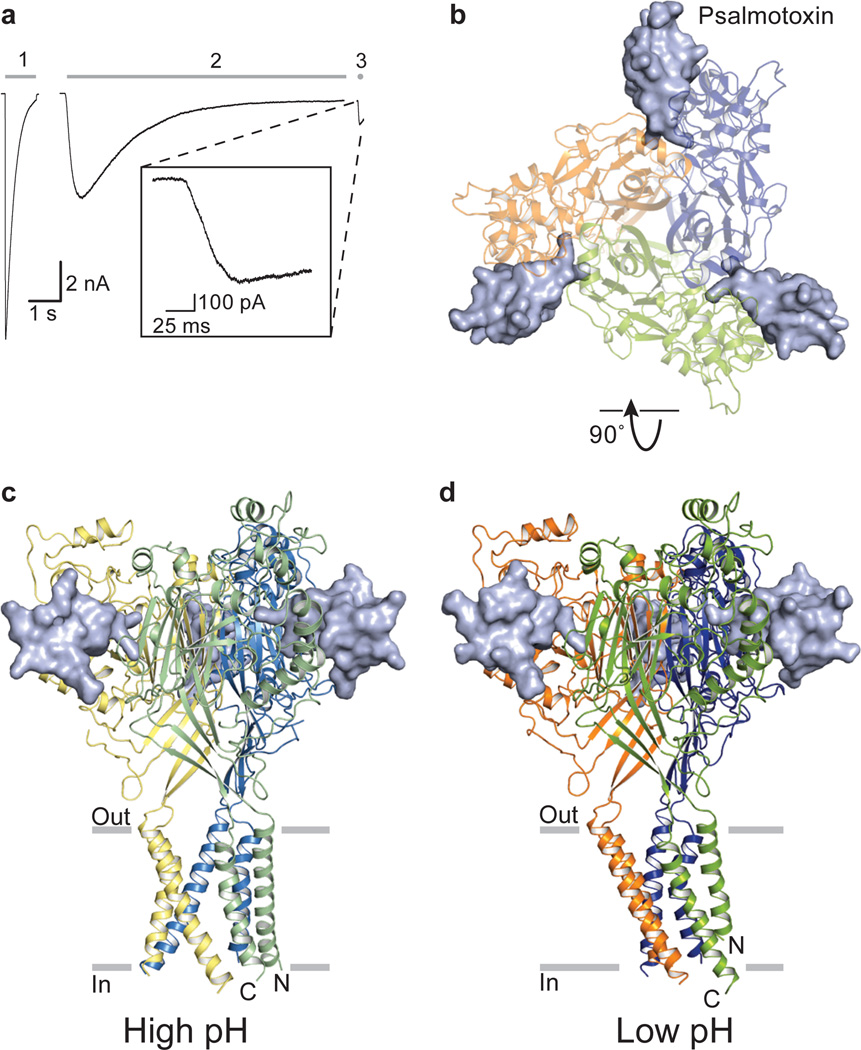
PcTx1 slows desensitization of ASIC and yields substantial steady state current when applied to ASIC1a/1b chimeras, and thus we asked whether PcTx1 stabilizes open channel states of cASIC1a31. We generated a cASIC1a construct for structural studies by removing 13 and 63 residues from the N- and C-termini respectively, yielding a channel with wild type-like electrophysiological properties (Δ13; Supplementary Figs. 1 and 2). Application of a pH 5.5 solution to Δ13 gives rapidly activating and desensitizing inward current while perfusion of a saturating PcTx1 solution at pH 7.25 elicits a current that activates and decays over a time scale of seconds to yield a steady state current (Fig.1a and Supplementary Fig. 3). Subsequent application of saturating PcTx1 at pH 5.5 further activates peak current and steady state currents.
Figure 1. PcTx1 activates the chicken ASIC1a Δ13 construct.
a, Whole-cell, patch-clamp current traces of activation by steps into pH 5.5 (1), pH 7.25 and 1 µM PcTx1 (2), and pH 5.5 and 1 µM PcTx1 (3). Inset, current trace of step into pH 5.5 and 1 µM PcTx1. b, Structure of low pH Δ13-PcTx1 complex viewed from the extracellular side. c, d, High pH (c) and low pH (d) complexes viewed parallel to the membrane. Each subunit is in a different color and toxin is in solvent-accessible surface representation.
The Δ13-PcTx1 complex forms crystals at pH 7.25 (high pH) that belong to the R3 space group, diffract to ~3.3 Å resolution, and have a single ASIC subunit-toxin complex positioned on the 3-fold axis of crystallographic symmetry. Crystals grown at pH 5.5 (low pH) belong to the C2 space group with an ASIC trimer and three toxin molecules in the asymmetric unit and a diffraction limit of ~2.8 Å resolution (Figs. 1b, c, d, Supplementary Fig. 4). Structures of both crystal forms were solved by molecular replacement and refined to good crystallographic statistics (Supplementary Table 1).
PcTx1 binds at subunit interfaces
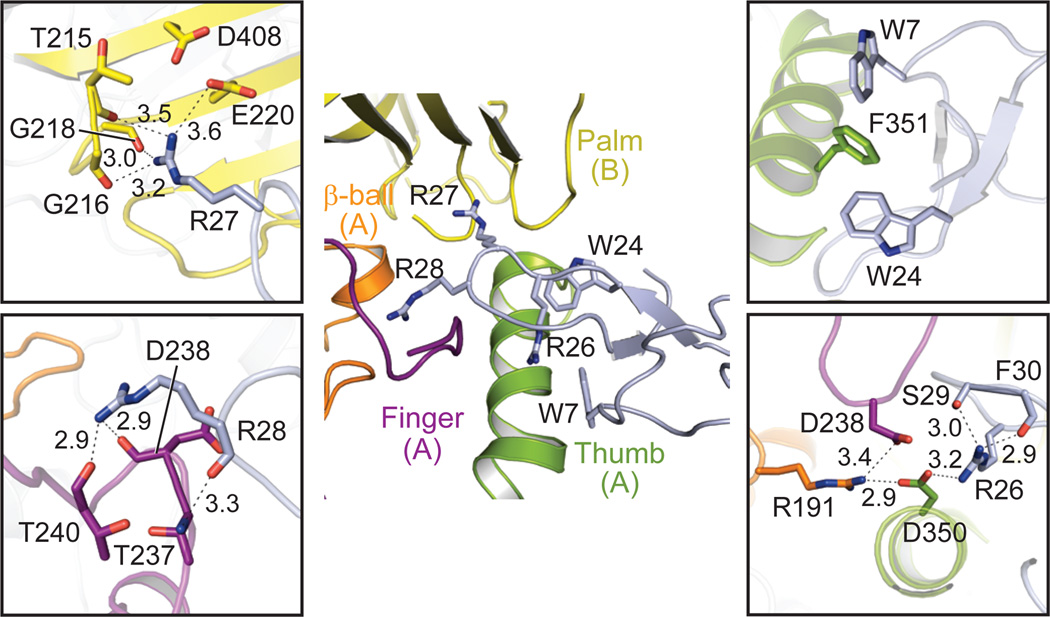
The high and low pH channel-toxin complexes show three toxin molecules bound to the extracellular domain of each trimer at similar subunit interfaces ~45 Å from the transmembrane domain (Fig. 1). PcTx1 molecules bury ~900 Å of solvent accessible surface area and make multiple ionic, polar and hydrophobic contacts consistent with studies mapping sites of channel-toxin interaction34 (Supplementary Fig. 5) yet distinct from computational modeling of the channel-toxin complex35, 36. Docked on the thumb domain, toxin molecules form nonpolar interactions mediated by aromatic residues, and they nestle an arginine-rich hairpin between adjacent subunits, making polar interactions in the acidic pocket (Fig. 2). Together, these interactions bridge the finger, β-ball, and thumb domains of one subunit and the palm domain of the adjacent subunit. Arg 26 of PcTx1 tunnels under the toxin β-sheet to hydrogen bond with the side chain carboxylate of Asp 350 of the thumb domain, which in turn interacts with Asp 238 and Arg 191 of the finger domain. In contrast, Arg 27 is oriented in the opposite direction, forming hydrogen bonds with backbone oxygens of Thr 215, Gly 216, and Gly 218 and also lying near the Glu 220-Asp 408 acidic pair, all on the palm domain of an adjacent subunit. Like Arg 27, Arg 28 interacts with backbone oxygens of Asp 238 and Thr 240 in the finger domain.
Figure 2. Extensive interactions adhere PcTx1 to the Δ13 ion channel.
Close-up views of toxin-binding site of the low pH complex. Dashed lines indicate possible hydrogen bonds.
PcTx1 is further anchored on the thumb domain by an aromatic ‘embrace’ between Trp 7 and Trp 24 of PcTx1 and Phe 351 (Fig. 2), a residue that when mutated in human ASIC1a to leucine renders the channel insensitive to PcTx137. Phe 351 is conserved in ASIC1 orthologs17 and appears crucial to the specificity of PcTx1 to ASIC1 channels. Site-directed mutagenesis of PcTx135 previously implicated Trp 24, Arg 26, and Arg 27 as central to toxin specificity, consistent with our structures of the channel-toxin complex. In addition, a recent structure of PcTx1 in complex with a non functional construct of cASIC1a supports our definition of the toxin binding site38.
Conformational changes in the extracellular domain
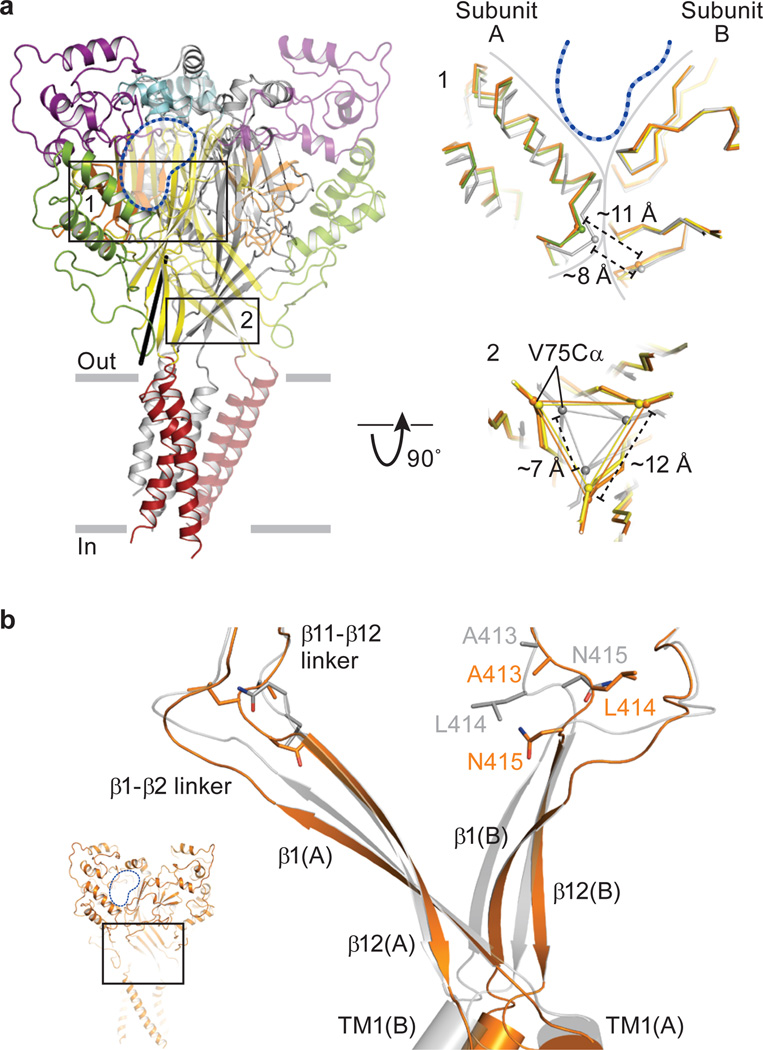
Structural comparisons of the high and low pH PcTx1-bound states with the desensitized state (PDB code: 3HGC) show that the upper palm β-strands and knuckle domains define a structurally conserved scaffold (Supplementary Fig. 6), reminiscent of the scaffold of P2X receptors39. Relative to the desensitized state, the lower palm domain and the wrist of the high pH Δ13-PcTx1 complex rotate by as much as ~13° around an axis positioned below the scaffold (Fig. 3a). This rotation shifts subunit-subunit interfaces compared to those of the desensitized state structure, separating the thumb and palm domains of adjacent subunits by 2–3 Å and displacing the finger domains of the high and low pH complexes (Fig. 3a).
Figure 3. Conformational changes in the extracellular domain.
a, Low pH Δ13-PcTx1 structure is in cartoon representation and colored as in Fig. 2. Black line indicates the axis around which the lower palm domain and the wrist region rotate following superpositions of the desensitized and open state structures. Close-up views of selected regions are boxed. The low pH complex is colored by domain, the high pH complex is orange, and the desensitized state (PDB code: 3HGC) is gray. Approximate position of PcTx1 is indicated by blue dashed lines and the boundaries between adjacent subunits is shown by solid gray lines. Measured Cα distances are between residues Asn 357 (A) and Arg 85 (B) in box 1. In box 2 the distances are between Val 75 Cα atoms on adjacent subunits. b, Close-up view of strands β1 and β12, the β1- β2/β11- β12 linkers and the extracellular boundary of the TM domains from two subunits of the high pH Δ13-PcTx1 complex (orange) and the desensitized state (gray) following superposition of the respective scaffold domains. Inset shows location of close-up view in the context of the entire channel.
The consequences of toxin binding are manifested in the flexing of a blanket of β-sheets encapsulating the negatively charged central vestibule40, a cavity composed of the lower palm domains, poised between the toxin binding site and the wrist (Fig. 3a and Supplementary Fig. 7). With the Cα atom of Val 75 as a landmark, the distances between adjacent subunits are ~11 Å and ~12 Å in the low and high pH structures, respectively; upon formation of the desensitized state, the distance diminishes to ~7 Å. Chemical modification of the E79C mutant of ASIC341, a residue equivalent to Glu 80 in cASIC1a and predicted to face the interior of the central vestibule, slows the rate and extent of channel desensitization, consistent with the notion that the central vestibule contracts upon transition from the PcTx1-bound states to the desensitized state. Small molecules that activate23 or potentiate the steady state current of ASIC322, 42, respectively, bind within the central vestibule23, or at the subunit interface near Ala 82, and stabilize the central vestibule in an expanded conformation, recalling the ATP-dependent expansion of the extracellular vestibule of P2X receptors39.
A striking conformational change in the palm domain, common to both PcTx1 complexes and located within the β1-β2 linker, is a ~180o flip of the Thr 84-Arg 85 peptide bond (Supplementary Fig. 8), inducing a shift of β1 away from the equivalent β-sheet of the adjacent subunit. In addition, Ala 413, Leu 414, and Asn 415 in the β11-β12 linker in the high pH structure adopt conformations in which the side chains of Leu 414 and Asn 415 have effectively swapped positions (Fig. 3b and Supplementary Fig. 9). Indeed, the Cα atoms of residues Ala 82 and Ala 413 are farther apart in the high pH state (8.1 Å), in comparison to the low pH state (6.4 Å), consistent with the notion that a disulfide can form between the equivalent residues in shark ASIC1b, stabilizing the channel in the desensitized state25 (Supplementary Fig. 10). Studies at sites equivalent to Leu 86, a residue that interacts with Leu414 in the high pH Δ13-PcTx1 state, and Asn 415 reinforce the conclusion that the β1-β2 and β11-β12 linkers are crucial to gating42–44.
The structures of the high and low pH Δ13-PcTx1 complexes suggest molecular mechanisms underlying the pH dependent conformations of the β1-β2 and β11-β12 linkers. We speculate that at high pH, the carboxyl group of Glu 80 is ionized, favoring an expanded conformation of the central cavity (Supplementary Fig. 7). As the pH drops, acidic residues in the vestibule bind protons, allowing Glu 80 and adjacent residues to adopt a ‘contracted’ central vestibule conformation. In fact, the distance between Cα of Glu 80 and Glu 412, residues that flank both linkers, diminishes from 14.1 Å to 11.3 Å in the high and low pH states, respectively. Neutralization of Glu 80 by mutation to Ala results in a channel that desensitizes ~40-fold more rapidly than the parent construct and that does not yield measureable steady state current upon application of PcTx1 at pH 7.25 (Supplementary Fig. 11), thus underscoring the role that titratable residues play in pH dependent conformational changes of the central vestibule.
A highly conserved non polar residue in the β11-β12 linker, Leu 414, is also important to the conformational transitions of the central vestibule. In the high pH Δ13-PcTx1complex, Leu 414 interacts with Leu 86 of the β1-β2 linker. Upon transition to the low pH Δ13-PcTx1 complex and to the desensitized state, however, Leu 414 forms multiple hydrophobic contacts involving Leu 281, Ile 306, and Val 368 of the adjacent subunit. In addition, Asn 415 hydrogen bonds with Tyr 416 and the backbone nitrogen of Ala 83 in the β1-β2 linker (Supplementary Fig. 9c, d). Together, these rearrangements highlight the importance of the β1-β2 and β11-β12 linkers and nearby subunit interfaces to conformational transitions of the central vestibule.
Ion channel at high pH
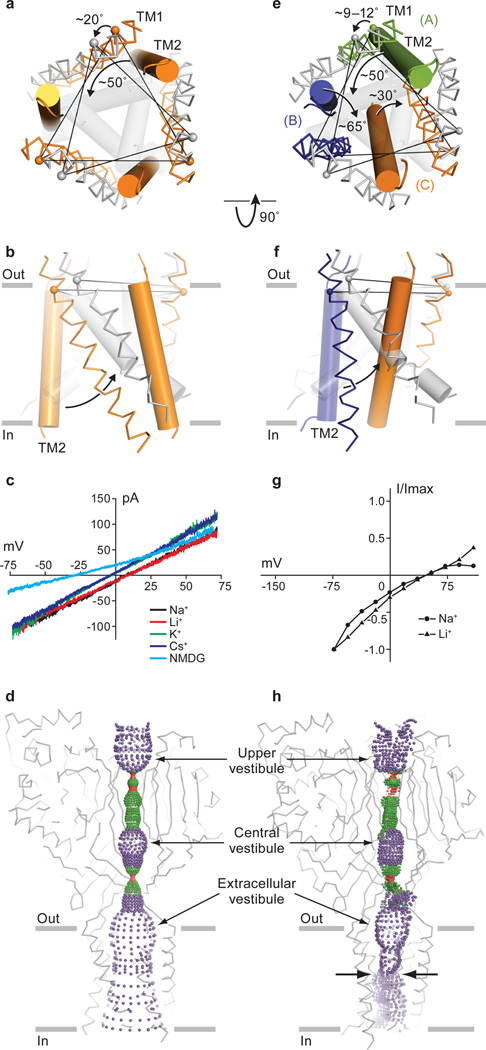
The high pH Δ13-PcTx1 complex harbors a cavernous, 3-fold symmetric pore in which TM2 resides on the periphery of the pore and lines the ion channel together with TM1 (Fig. 4a, b). In comparison to the desensitized state, a kink in TM2 at Asp 433 results in an effective rotation of the cytoplasmic end of TM2 by ~90° around the 3-fold axis and a tilting of TM2 by ~50°; movements in TM1 are smaller, characterized by a ~20° rotation around the 3-fold axis. Collectively, these movements rupture intrasubunit interactions between TM1 and TM2 and completely disrupt intersubunit contacts between TM2 segments that define the closed, desensitized channel gate, yielding sparse intersubunit and hydrophobic contacts between TM1 and TM2 mediated primarily by Ile 66 and Ile 434, and Val 46 and Ile 446 in the high pH complex (Supplementary Fig. 12).
Figure 4. Structural rearrangements and ion selectivity of the transmembrane pores.
a, b, Comparison of transmembrane domains from the high pH (orange) and desensitized (gray) state structures with TM1 in ribbon and TM2 in cylinder representation. Transmembrane domains are viewed from the extracellular side (a) and parallel to the membrane (b). c, Current/voltage experiment showing that at neutral pH the Δ13-PcTx1 complex forms a non selective cation channel. d, Mapping of solvent accessible pathway along the 3-fold axis shows that the high pH complex has a large, transmembrane pore. The occluded pathway along the 3-fold in the extracellular domain suggests that ions access the pore by way of lateral fenestrations. e, f. A comparison of the transmembrane domains from the low pH complex, where each subunit is in a different color. The desensitized state is gray. TM1 and TM2 segments are in ribbon and cylinder representations, respectively. Transmembrane domains are viewed from the extracellular side (e) and parallel to the membrane (f). g, Current/voltage experiment demonstrating that the ion channel of the low pH Δ13-PcTx1 complex is sodium selective. h, Mapping of a solvent accessible pathway along the pseudo 3-fold axis of the low pH complex shows that it has an asymmetric ion channel pore and a constriction (opposing arrows) halfway across the bilayer. Maps of solvent-accessible pathways (d and h) were generated using the HOLE software (red < 1.4 Å < green < 2.3 Å < purple).
The large TM pore of the pH 7.25 PcTx1 complex, with a diameter of ~10 Å near Asp 433 (Supplementary Fig. 13), led us to probe the selectivity of the Δ13-PcTx1 complex using bi-ionic patch clamp electrophysiology (Fig. 4c, d). At neutral pH, the channel-toxin complex does not discriminate between Li+, Na+, K+, and Cs+, and we suggest that these ions permeate through the pore in a fully hydrated state. Furthermore, N-methyl-D-glucamine (NMDG), with a radius of ~4.0 Å, permeates through the ion channel and application of 500 µM amiloride only blocks 10% of the steady-state current (Supplementary Fig. 14). This non selective behavior of the Δ13-PcTx1 complex is reminiscent of the non selective behavior previously observed in steady state currents of wild type ASIC3 and in the degenerin mutants of ASICs20, 21, and of the pore-dilated behavior of trimeric P2X receptors26.
Architecture of low pH ion channel pore
The TM helices of each subunit in the low pH Δ13-PcTx1 complex not only adopt different conformations but they also occupy distinct positions relative to the pore axis (Fig. 4e, f). TM2 segments of subunits A and B bend and ‘stretch’ at residues 433–435, similar to conformations observed in the high pH structure. In comparison to the desensitized state, the TM1 helices of the low pH complex rotate by 9–12° around the pore axis and the TM2 helices of subunits A, B and C tilt by ~47o, 65o and 30o, respectively. Thus the TM2 helices adopt an orientation nearly perpendicular to the membrane plane, reminiscent of the TM conformation of the low pH ΔASIC1 structure17.
TM2 of the C subunit adopts a straight α-helix, resulting in a ~4 residue displacement along the pore axis relative to subunits A and B, shifting subunit C toward the extracellular side of the membrane, thus conferring axial asymmetry onto the pore. This asymmetry has precedent in chemical modification studies of ENaC which were interpreted in terms of a model in which the β subunit is displaced along the pore axis by about 1 turn of an α-helix45. The axial displacement of the TM’s gives rise to a striking constellation of hydrophobic contacts mediated by the staggered arrangement of Leu 440 and Leu 447 of subunits B and C that resembles a leucine zipper motif (Supplementary Fig. 15a, b). The extensive contacts between the TM2 domains of the B and C subunits give rise to TM1 of subunit B residing on the periphery of the ion channel domain, while the proximity of TM2 of subunits A and C shields TM1 of subunit C from exposure to the pore. The remaining 4 TM segments line the pore and participate in intersubunit interactions that include contacts between Val 46 (C) and Ile 446 (A), as observed in the high pH structure (Supplementary Fig. 15c, d). The exposure of portions of TM1 from subunit A to the pore concurs with the results from accessibility studies of TM1 residues of FaNaC46, a peptide-gated channel, but stands in contrast to cysteine-directed chemical modification studies of lamprey ASIC47, which suggest that TM2 primarily lines the pore. Determining how the conformation of the asymmetric pore in the low pH Δ13-PcTx1 complex is related to the conductive pore of the Δ13 construct upon activation by protons will require additional studies. Nevertheless, we note that ENaCs likely harbor an asymmetric pore based on variable reactivity of cysteine residues at equivalent sites on different subunits45 and that the extensive intra- and intersubunit interactions between TM1 and TM2 seen in the low pH Δ13-PcTx1 complex renders asymmetric pore formation favorable.
Low pH pore is sodium selective
The low pH complex harbors an elliptical pore with a region of constriction spanning a helical turn and located approximately halfway along the transmembrane domain, primarily lined by hydrophobic side chains of Leu 440 (Supplementary Fig. 16). The position of the constriction exposes the putative amiloride binding site, Gly 439 and the ‘GAS’ selectivity tract, to the extracellular side of the membrane, consistent with the inhibitory mechanism of amiloride as an open channel blocker48. Strikingly, ion channel blockers of ASICs and ENaCs, including amiloride, are characteristically planar, non-symmetric molecules that roughly mirror the shape of the extracellular portion of the low pH pore (Supplementary Fig. 16).
To assess the properties of the Δ13-PcTx1 ion channel pore at low pH, we determined its ion selectivity properties and found that the complex remains selective for Na+ and Li+ with a selectively for Na+ over K+ of 10:1 (Fig. 4g). Because the pore is primarily lined by hydrophobic residues at the constriction point (Fig. 4h), we assume that Na+ ions are hydrated when traversing the pore. We suggest that the pore discriminates ions by the size of a fully or partially hydrated ion and that the mechanism underlying ion selectivity is best described by a barrier model. Indeed, Hille proposed that sodium selective pores have a rectangular cross section with dimensions of 3.2 Å by ~5.2 Å, large enough to allow one sodium ion and one water molecule to percolate but too small to allow passage of hydrated potassium ions49. The elliptical outline of the constriction in the low pH Δ13-PcTx1 structure is in general accord with the proposed geometry of a Na+-selective pore albeit larger, with constrictions of ~5 Å by ~7 Å at Leu 440 of subunit C and of ~4 Å by 10 Å at Leu 440 of subunits A and B (Supplementary Fig. 16). The extent to which the mechanism of Na+-selectivity upon proton activation, in the absence of PcTx1, hinges on Leu 440 and on the low pH Δ13-PcTx1 conformation requires further experimentation. Nevertheless, mutation of Leu 440 to Ala or Ser (Supplementary Fig. 17) demonstrates that Leu 440 is crucial to the formation and function of the ion channel pore based on the reduced activity of the mutants compared to the parent construct.
Ion binding sites
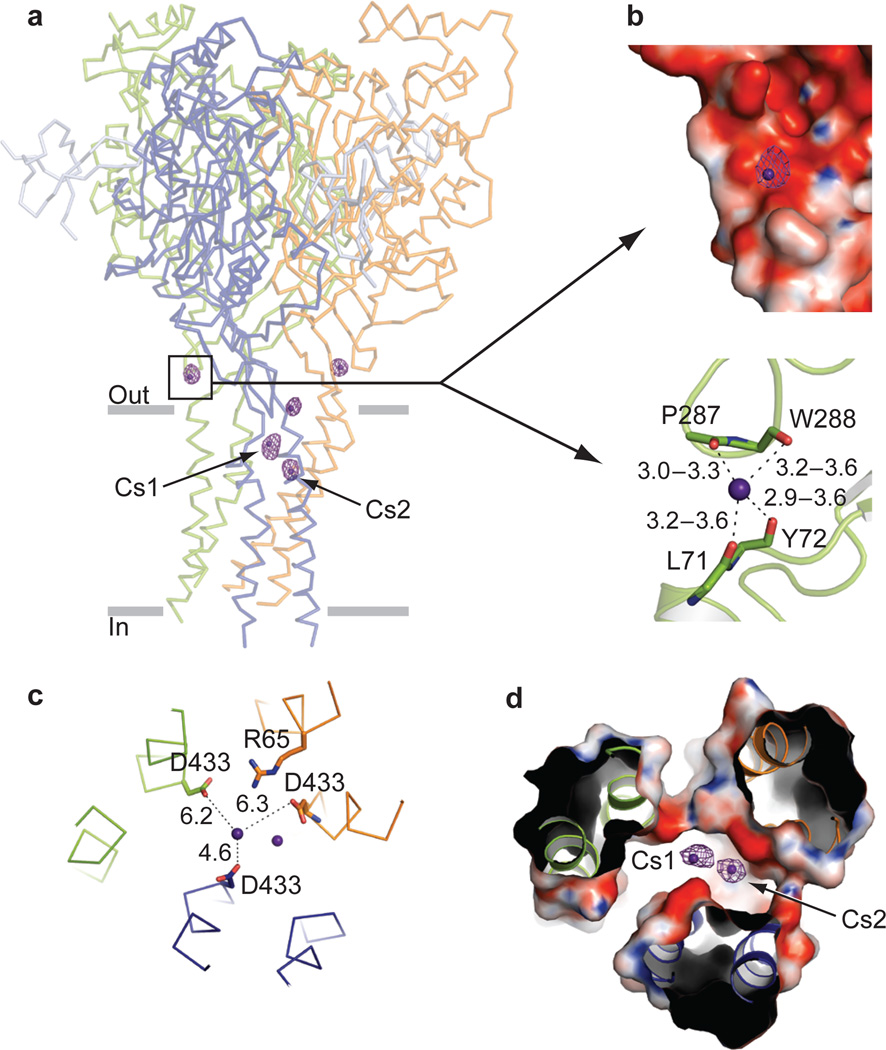
To map ion binding sites we soaked crystals in solutions containing Cs+, a voltage-dependent, open channel blocker of the Δ13 construct (Supplementary Fig. 18). Inspection of anomalous difference electron density maps revealed a site (6–12 σ) in each subunit, common to the high and low pH Δ13-PcTx1 crystal forms and located at the interface between the wrist and the extracellular end of TM1 (Fig. 5a and Supplementary Fig. 19). In both crystal forms, the backbone carbonyl oxygens of Leu 71, Tyr 72, Pro 287, and Trp 288 coordinate Cs+ (Fig. 5b). Interestingly, the three crystallographically independent Cs+ sites in the C2 structure vary in strength, with subunit C having the weakest signal, thus suggesting that the binding of ions to these sites is influenced by the variation in TM1/wrist conformation between subunits. Because Cs+ soaking experiments with crystals of the desensitized state failed to reveal ion binding to this site, and TM1 (C) in low pH Δ13-PcTx1 state harbors the weakest Cs+ site, we suggest the occupancy of the site is state dependent, with ion binding favored when the pore is in an open state, augmenting interactions between Tyr 72 and Trp 288.
Figure 5. Cs+ binding sites.
a, Anomalous difference electron density map contoured at 3.5 σ. The low pH complex is in ribbon representation. Cesium sites in the outer pore region are labeled Cs1 and Cs2. b, Close-up view of the Cs+ site in the wrist region. Top, electrostatic potential contoured from −20kT (red) to +20kT (blue). Bottom, Cs+ coordination by backbone carbonyl oxygens. The main chain is drawn as sticks. c,d, Close-up view of Cs+ sites near Asp 433 shown in ribbon and stick representation (c) and solvent accessible surface colored by electrostatic potential (d) viewed from the extracellular side.
We identified two Cs+ sites in the electrostatically negative mouth of the pore, near Asp 433 in the C2 structure (Fig 5c, d), a residue implicated in stabilization of the open state yet not crucial to ion selectivity50. Ions at these sites are ~5 Å from the closest protein residues consistent with the notion that they are low affinity, transiently occupied cation sites bound by water-mediated contacts. We did not observe anomalous difference density features deeper into the pore, perhaps because the hydrophobic pore is devoid of favorable binding sites and the structural analysis was carried out in the absence of a membrane potential.
Mechanism
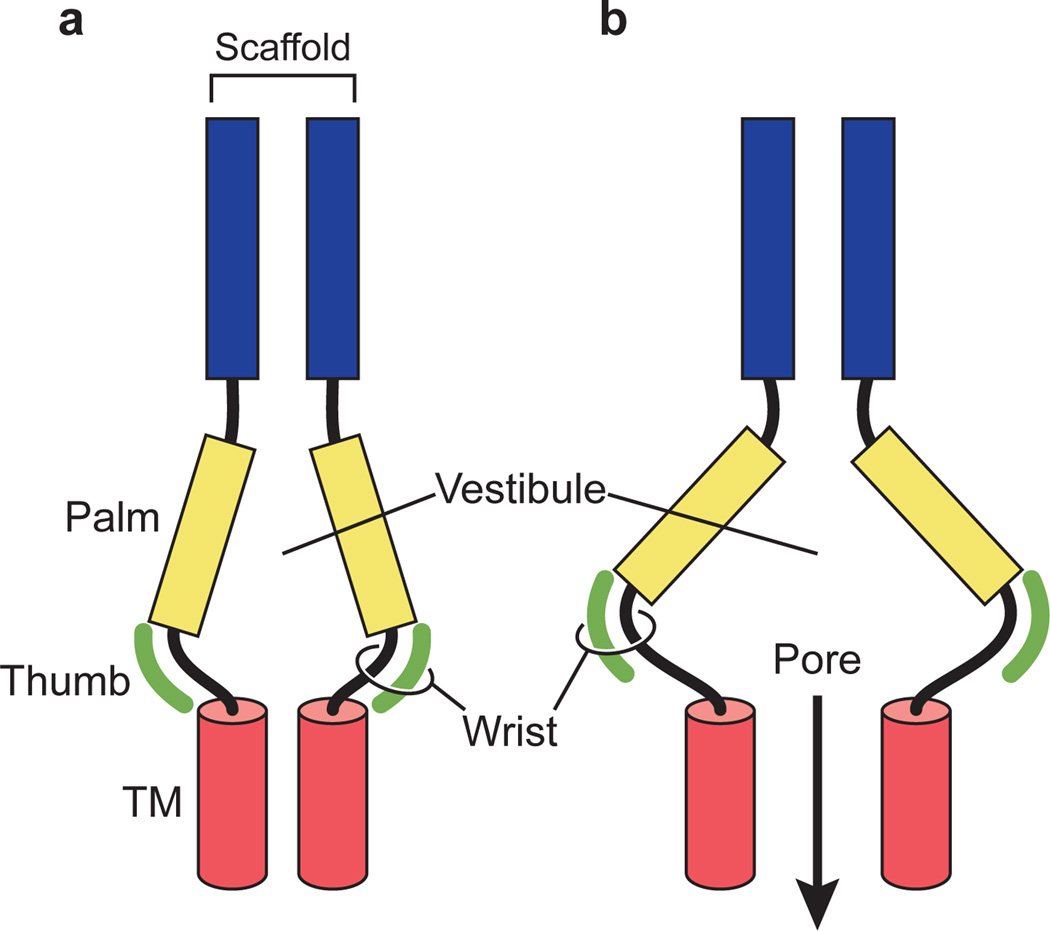
We used PcTx1 to stabilize open states of the Δ13 construct at high and low pH. Comparison of the open state and desensitized state structures defines the upper palm and knuckle as a structural scaffold and the lower palm as a conformationally flexible, proton-sensitive domain at the core of ion channel gating (Supplementary Fig. 7). The finger and thumb domains flank the palm domain, harbor binding sites for protons and PcTx1 and modulate movements of the lower palm domain by alterations in intersubunit contacts. In the open conformations, the subunit interface between the thumb and the palm domain separates while the extracellular and transmembrane domain interface forming the extracellular vestibule expands. The presence of a modulator, such as PcTx1, precludes ‘collapse’ of the thumb – palm subunit interface to a desensitized state conformation. The motions of the lower palm domain converge at the wrist region, inducing radial and rotational movements of the transmembrane domains, gating the ion channel (Fig. 6 and Supplementary Fig. 20).
Figure 6. Schematic representation of gating.
a, The extracellular vestibule in the closed, desensitized state structure adopts a contracted conformation. b, In the open pore conformation the vestibule occupies an expanded conformation, stabilized by the thumb domain. The wrist region, in turn, couples the conformational changes of the extracellular domain to the transmembrane domains (red cylinder) of the ion channel pore.
Conclusion
Our functional and structural studies of the chicken ASIC1a complex with PcTx1 at two proton concentrations provide new insights into how the movements of the multiple domains of ASICs are coupled to ligand and pH dependent gating. Channel opening is correlated to the expansion of the extracellular vestibule and to rearrangements at subunit interfaces, movements coupled to the ion channel pore by the direct connections of extracellular β-strands to the transmembrane α-helices and also by the non covalent contacts between the thumb and wrist region, a previously unrecognized site of cation binding in the open state. The non selective and sodium selective states of the ion channel pore illustrate how transmembrane helices can rearrange to form a large, non selective pore at high pH and a small, asymmetric and sodium selective channel at low pH Together, these studies illuminate mechanisms of gating and selectivity in ASIC/ENaC/DEG channels.
Methods Summary
The Δ13 construct was expressed in insect or mammalian cells using baculovirus mediated expression and purified by metal ion affinity and size-exclusion chromatography. The high and low pH crystal forms were obtained in conditions containing 20 mM Tris (pH 7.25) and 14–18% PEG 550 MME, and 100 mM sodium acetate (pH 5.5) and 9–12% PEG 2000 MME, respectively. Data processing, model building, and refinement were performed using the HKL2000, COOT, and PHENIX computer programs. The structures were solved by molecular replacement and subjected to crystallographic refinement. Whole-cell recordings were performed using CHO-K1 cells transfected with plasmid DNA encoding the Δ13 construct.
Methods
Expression and purification
The construct Δ13 was derived from the chicken ASIC1 gene and was expressed as N-terminal fusion with octa-histidine-tagged GFP using baculovirus expression systems in insect cells and mammalian cells51 with two thrombin sites and one thrombin site, respectively, encoded upstream of Δ13 generating a final protein sequence containing residues Gly 14 to Arg 463, verified by N-terminal amino acid sequencing. Protein expressed in insect cells was purified as described40, while the Δ13 expressed in mammalian cells were collected by centrifugation (1000g) and sonicated in the presence of 150 mM NaCl, 20 mM Tris (pH 8.0) and protease inhibitors, then subsequently solubilized in 40 mM n-dodecyl β-D-maltoside (DDM) for 1 hour at 4°C. The solubilized material was clarified by centrifugation (19,000g) for 1 hour at 4°C and the supernatant was incubated with TALON resin for 1.5 hours at 4°C. Bound protein was then eluted with 150 mM NaCl, 20 mM Tris (pH 8.0), 250 mM imidazole, and 1 mM DDM. Cleavage of the histidine-tagged GFP was achieved by thrombin. The resulting Δ13 protein was further purified by size-exclusion chromatography using a mobile phase containing 150 mM NaCl, 20 mM Tris (pH 7.4), 1 mM DDM, 1mm DTT, and 1 mM EDTA. Peak fractions were collected and concentrated to 1.60 mg/mL based on A280 measurement. Synthetic psalmotoxin (Peptides International) was immediately added in a 6:1 molar ratio of toxin to channel prior to crystallization.
Crystallization
Crystals of the high pH form of the Δ13-PcTx1 complex were grown with a protein solution supplemented with 100 µM 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) by way of vapor diffusion using a reservoir solution composed of 20 mM Tris (pH 7.25), 14–18% PEG 550 MME and drops composed of a ratio of 1.5:1 and 2:1, reservoir to protein, respectively, at 4 °C. For cryoprotection, crystals were soaked in reservoir solution supplemented with increasing concentrations of glycerol (15% v/v final concentration), 1 mM DDM, and 1 mM DMPC. For Cs+ anomalous diffraction experiments, crystals were soaked in reservoir solutions supplemented with 300 mM CsCl, 500 uM DMPC and 1 mM DDM.
Crystals of the low pH form of the Δ13-PcTx1 complex were obtained by vapor diffusion in hanging drop configuration using a 1:2 reservoir to protein ratio at 4°C. The reservoir solution typically contained 100 mM sodium acetate (pH 5.5) and 9–12% PEG 2000 MME. Cryoprotection was accomplished using reservoir solution supplemented with increasing concentrations of glycerol (20% v/v final concentration). For Cs+ anomalous diffraction experiments, a modified reservoir solution was employed in which the sodium acetate was replaced by cesium acetate, and the solution was supplemented with 500 mM CsCl and 1 mM DDM.
Structure determination
X-ray diffraction data sets from crystals grown at pH 7.25 and belonging to the R3 space group were collected at the Advanced Light Source (beamline 5.0.2) and diffraction was measured to ~3.35 Å resolution. The best x-ray diffraction data sets from the C2 form, pH 5.5 crystals, were collected at the Advanced Photon Source (beamline 24ID-C) and scaled to ~2.80 Å resolution. Diffraction data for crystals soaked in cesium containing solutions were measured using low energy x-rays (9000 eV) at the Advanced Light Source. Diffraction data were indexed, integrated, and scaled using the HKL2000 software52. The structures were solved by molecular replacement using the program PHASER53, with the extracellular domain coordinates of the ΔASIC1 structure17 (PDB code: 2QTS) as a search probe. Resulting maps after molecular replacement showed strong density of psalmotoxin, and thus the toxin was fitted into the density at the subunit interface using the solution structure29. Iterative model building and refinement were performed using COOT54 and PHENIX55 package. Crystals in the R3 space group contained one subunit and one toxin in the asymmetric unit; consequently, the channel is built using the subunits and toxins related by crystallographic symmetry. Alternatively, crystals in the C2 space group contained one trimer and three toxins in the asymmetric unit. Manual building of the transmembrane domains was guided by electron density maps calculated using the crystallographically refined coordinates of the extracellular domain - toxin complex. “Omit” electron density maps were calculated to validate residue registration, particularly with the transmembrane domains. Subsequent iterative cycles of model building and refinement were performed after building the transmembrane regions. Non-crystallographic symmetry (NCS) restraints were implemented in the extracellular domain and toxin for the low pH structure containing regions defined automatically by the PHENIX package to improve maps. These regions consist of residues 72–135, 138–153, 155–291, 303–331, 333–360, 362–386, and 388–427 in the extracellular domain and residues 2–38 in the toxin. As for the high pH structure, TLS parameters were refined with 7 TLS groups defined by PHENIX, which comprised of 5 groups in the ASIC subunit and 2 groups in the toxin. Structure validation was performed using MolProbity56. The low pH final model contains channel subunits with residues 50–454, 45–450 and 42–454 of A, B, and C chains, respectively and toxins with residues 2–38, 2–37, and 1–38 of M, N, and O chains, respectively. The high pH model contains residues 41–450 of the ASIC subunit, and 2–13 and 16–38 of the toxin. A region in the thumb domain of the high pH structure lacked electron density consisting of residues 298–301 and was omitted in the final structure. Pore surface and dimension were determined using the software HOLE57, while the rotation axis shown in Figure 3a was analyzed using Dyndom58.
Electrophysiology
Whole-cell recordings were carried out with CHO-K1 cells 24–48 hours after transfection by plasmid DNA encoding the Δ13 construct and GFP expressed from an internal ribosome entry site. Pipettes were pulled and polished to 2–3 MΩ resistance and filled with internal solution containing (in mM): 150 KCl, 2 MgCl2, 5 EGTA and 10 HEPES (pH 7.35). External solution contained (in mM): 150 NaCl, 2 MgCl2, 2 CaCl2, 8 Tris and 4 MES. For ion selectivity experiments, NaCl was substituted with equimolar concentrations of LiCl, KCl, CsCl, or NMDG in the external solution. Proton concentration-response profiles were recorded and normalized to maximal current generated by application of external solution buffered at pH 5.8, while the PcTx1 concentration-response curve was generated by normalizing to maximal current generated by application of 1 µM of toxin. All concentration-response curves were fitted to the Hill equation.
Supplementary Material
Acknowledgements
We appreciate assistance in the initial characterization of the action of PcTx1 on chicken ASIC1a by D. Samways and T. Egan, mass spectrometry analysis by D. King, together with comments from C. Jahr. We are grateful to K.C. Garcia (Stanford), C. Lee and A. Goehring for assistance with protein expression in mammalian cells. We thank L. Vaskalis for assistance with figures, H. Owen for help with manuscript preparation, H. Krishnamurthy for assistance in initial data collection, and Gouaux lab members for helpful discussion. This work was supported by an individual National Research Service Award from the National Institute of Neurological Disorders and Stroke (I.B.) and the NIH (E.G.). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Author Contributions
I.B. and E.G. designed the project. I.B. performed protein purification, crystallography, and electrophysiology. I.B. and E.G. wrote the manuscript.
References
- 1.Gründer S, Chen X. Structure, function, and pharmacology of acid-sensing ion channels (ASICs): focus on ASIC1a. Int J Physiol Pathophysiol Pharmacol. 2010;2:73–94. [PMC free article] [PubMed] [Google Scholar]
- 2.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol. Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 3.Kashlan OB, Kleyman TR. ENaC structure and function in the wake of a resolved structure of a family member. Am. J. Physiol. Renal. Physiol. 2011;301:F684–F696. doi: 10.1152/ajprenal.00259.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishtal OA, Pidoplichko VI. A receptor for protons in the nerve cell membrane. Neuroscience. 1980;5:2325–2327. doi: 10.1016/0306-4522(80)90149-9. [DOI] [PubMed] [Google Scholar]
- 5.Waldmann R. Proton-gated cation channels--neuronal acid sensors in the central and peripheral nervous system. Adv Exp Med Biol. 2001;502:293–304. doi: 10.1007/978-1-4757-3401-0_19. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez de la Rosa D, et al. Distribution, subcellular localization and ontogeny of ASIC1 in the mammalian central nervous system. J. Physiol. 2003;546:77–87. doi: 10.1113/jphysiol.2002.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wemmie JA, et al. Acid-sensing ion channel 1 is localized in brain regions with high synaptic density and contributes to fear conditioning. J. Neurosci. 2003;23:5496–5502. doi: 10.1523/JNEUROSCI.23-13-05496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deval E, et al. Acid-sensing ion channels (ASICs): pharmacology and implication in pain. Pharmacol Ther. 2010;128:549–558. doi: 10.1016/j.pharmthera.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Bohlen CJ, et al. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coryell MW, et al. Restoring Acid-sensing ion channel-1a in the amygdala of knock-out mice rescues fear memory but not unconditioned fear responses. J Neurosci. 2008;28:13738–13741. doi: 10.1523/JNEUROSCI.3907-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishtal OA. The ASICs: Signaling molecules? Modulators? Trends Neurosci. 2003;26:477–483. doi: 10.1016/S0166-2236(03)00210-8. [DOI] [PubMed] [Google Scholar]
- 12.Xiong ZG, et al. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 13.Yermolaieva O, Leonard AS, Schnizler MK, Abboud FM, Welsh MJ. Extracellular acidosis increases neuronal cell calcium by activating acid-sensing ion channel 1a. Proc Natl Acad Sci U S A. 2004;101:6752–6757. doi: 10.1073/pnas.0308636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schild L. The epithelial sodium channel and the control of sodium balance. Biochim Biophys Acta. 2010;1802:1159–1165. doi: 10.1016/j.bbadis.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Snyder PM, et al. Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel. Cell. 1995;83:969–978. doi: 10.1016/0092-8674(95)90212-0. [DOI] [PubMed] [Google Scholar]
- 16.Chang SS, et al. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1. Nat Genet. 1996;12:248–253. doi: 10.1038/ng0396-248. [DOI] [PubMed] [Google Scholar]
- 17.Jasti J, Furukawa H, Gonzales E, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 18.Canessa CM, Horisberger J-D, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- 19.Palmer LG. Ion selectivity of the apical membrane Na channel in the toad urinary bladder. J. Membr. Biol. 1982;67:91–98. doi: 10.1007/BF01868651. [DOI] [PubMed] [Google Scholar]
- 20.Lingueglia E, et al. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J. Biol. Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 21.de Weille JR, Bassilana F, Lazdunski M, Waldmann R. Identification, functional expression and chromosomal localization of a sustained human proton-gated cation channel. FEBS Lett. 1998;433:257–260. doi: 10.1016/s0014-5793(98)00916-8. [DOI] [PubMed] [Google Scholar]
- 22.Yagi J, Wenk HN, Naves LA, McCleskey EW. Sustained currents through ASIC3 ion channels at the modest pH changes that occur during myocardial ischemia. Circ Res. 2006;99:501–509. doi: 10.1161/01.RES.0000238388.79295.4c. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, et al. A nonproton ligand sensor in the acid-sensing ion channel. Neuron. 2010;68:61–72. doi: 10.1016/j.neuron.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Li WG, Yu Y, Huang C, Cao H, Xu TL. Nonproton Ligand Sensing Domain Is Required for Paradoxical Stimulation of Acid-sensing Ion Channel 3 (ASIC3) Channels by Amiloride. J Biol Chem. 2011;286:42635–42646. doi: 10.1074/jbc.M111.289058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Springauf A, Bresenitz P, Grunder S. The interaction between two extracellular linker regions controls sustained opening of acid-sensing ion channel 1. J Biol Chem. 2011;286:24374–24384. doi: 10.1074/jbc.M111.230797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khakh BS, Lester HA. Dynamic selectivity filters in ion channels. Neuron. 1999;23:653–658. doi: 10.1016/s0896-6273(01)80025-8. [DOI] [PubMed] [Google Scholar]
- 27.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 28.Escoubas P, et al. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J. Biol. Chem. 2000;275:25116–25121. doi: 10.1074/jbc.M003643200. [DOI] [PubMed] [Google Scholar]
- 29.Escoubas P, Bernard C, Lambeau G, Lazdunski M, Darbon H. Recombinant production and solution structure of PcTx1, the specific peptide inhibitor of ASIC1a proton-gated cation channels. Protein Sci. 2003;12:1332–1343. doi: 10.1110/ps.0307003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X, Kalbacher H, Grunder S. The tarantula toxin psalmotoxin 1 inhibits acid-sensing ion channel (ASIC) 1a by increasing its apparent H+ affinity. J Gen Physiol. 2005;126:71–79. doi: 10.1085/jgp.200509303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Kalbacher H, Grunder S. Interaction of acid-sensing ion channel (ASIC) 1 with the tarantula toxin psalmotoxin 1 is state dependent. J Gen Physiol. 2006;127:267–276. doi: 10.1085/jgp.200509409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samways DS, Harkins AB, Egan TM. Native and recombinant ASIC1a receptors conduct negligible Ca2+ entry. Cell Calcium. 2009;45:319–325. doi: 10.1016/j.ceca.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazzuca M, et al. A tarantula peptide against pain via ASIC1a channels and opioid mechanisms. Nat Neurosci. 2007;10:943–945. doi: 10.1038/nn1940. [DOI] [PubMed] [Google Scholar]
- 34.Salinas M, et al. The receptor site of the spider toxin PcTx1 on the proton-gated cation channel ASIC1a. J. Physiol. 2006;570:339–354. doi: 10.1113/jphysiol.2005.095810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saez NJ, et al. A dynamic pharmacophore drives the interaction between Psalmotoxin-1 and the putative drug target acid-sensing ion channel 1a. Mol Pharmacol. 2011;80:796–808. doi: 10.1124/mol.111.072207. [DOI] [PubMed] [Google Scholar]
- 36.Pietra F. Docking and MD simulations of the interaction of the tarantula peptide psalmotoxin-1 with ASIC1a channels using a homology model. J. Chem. Inf. Model. 2009;49:972–977. doi: 10.1021/ci800463h. [DOI] [PubMed] [Google Scholar]
- 37.Sherwood T, et al. Identification of protein domains that control proton and calcium sensitivity of ASIC1a. J Biol Chem. 2009;284:27899–27907. doi: 10.1074/jbc.M109.029009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson RJP, et al. Structure of the acid-sensin ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nature Commun. 2012;3 doi: 10.1038/ncomms1917. [DOI] [PubMed] [Google Scholar]
- 39.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485:207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cushman KA, Marsh-Haffner J, Adelman J, McCleskey EW. A conformational change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J. Gen. Physiol. 2007;129:345–350. doi: 10.1085/jgp.200709757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T, Yang Y, Canessa CM. Asn415 in the beta11-beta12 linker decreases proton-dependent desensitization of ASIC1. J Biol Chem. 2010;285:31285–31291. doi: 10.1074/jbc.M110.160382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T, Yang Y, Canessa CM. Leu85 in the beta1-beta2 linker of ASIC1 slows activation and decreases the apparent proton affinity by stabilizing a closed conformation. J Biol Chem. 2010;285:22706–22712. doi: 10.1074/jbc.M110.134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T, Yang Y, Canessa CM. Two residues in the extracellular domain convert a nonfunctional ASIC1 into a proton-activated channel. Am J Physiol Cell Physiol. 2010;299:C66–C73. doi: 10.1152/ajpcell.00100.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Sheng S, Perry CJ, Kleyman TR. Asymmetric organization of the pore region of the epithelial sodium channel. J Biol Chem. 2003;278:13867–13874. doi: 10.1074/jbc.M300149200. [DOI] [PubMed] [Google Scholar]
- 46.Pöet M, et al. Exploration of the pore structure of a peptide-gated Na+ channel. EMBO. J. 2001;20:5595–5602. doi: 10.1093/emboj/20.20.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T, Yang Y, Canessa CM. Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat Commun. 2011;2:399. doi: 10.1038/ncomms1409. [DOI] [PubMed] [Google Scholar]
- 48.Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M. Molecular cloning and functional expression of a novel amiloride-sensitive Na+ channel. J Biol Chem. 1995;270:27411–27414. doi: 10.1074/jbc.270.46.27411. [DOI] [PubMed] [Google Scholar]
- 49.Hille B. Ion channels of excitable membranes. Sinauer Associates, Inc.; Sunderland, MA: 2001. [Google Scholar]
- 50.Li T, Yang Y, Canessa CM. Asp433 in the closing gate of ASIC1 determines stability of the open state without changing properties of the selectivity filter or Ca2+ block. J. Gen. Physiol. 2011:289–297. doi: 10.1085/jgp.201010576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 56.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smart OS, Neduvelil JG, Wang X, Wallace BA, Samsom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 58.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J. Mol. Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.