Abstract
Purpose
Hypospadias is one of the most frequent genital malformations in the male newborn, and results from abnormal penile and urethral development. The etiology of hypospadias remains largely unknown despite intensive investigations. Fetal androgens have a crucial role in genital differentiation. Recent studies have suggested that molecular mechanisms that underlie the effects of androgens on the fetus may involve disruption of epigenetic programming of gene expression during development. We assessed whether epigenetic modification of DNA methylation is associated with hypospadias in a case-control study of 12 hypospadias and 8 control subjects.
Materials and Methods
Genome-wide DNA methylation profiling was performed on the study subjects using the Illumina Infinium® HumanMethylation450 Bead-Chip, which enables the direct investigation of methylation status of more than 485,000 individual CpG sites throughout the genome. The methylation level at each CpG site was compared between cases and controls using the t test and logistic regression.
Results
We identified 14 CpG sites that were associated with hypospadias with p <0.00001. These CpG sites were in or near the SCARB1, MYBPH, SORBS1, LAMA4, HOXD11, MYO1D, EGFL7, C10orf41, LMAN1L and SULF1 genes. Two CpG sites in SCARB1 and MYBPH genes remained statistically significant after correction for multiple testing (p = 2.61×10−09, pcorrected = 0.008; p = 3.06×10−08, pcorrected = 0.02, respectively).
Conclusions
To our knowledge this is the first study to investigate hypospadias using a unique and novel epigenetic approach. Our findings suggest DNA methylation patterns are useful in identifying new genes such as SCARB1 and MYBPH that may be involved in the etiology of hypospadias.
Keywords: hypospadias, DNA methylation, epigenomics, case-control studies, growth and development
Hypospadias is one of the most frequent genital malformations in the male newborn, and results from abnormal penile and urethral development. The prevalence of hypospadias in the United States is approximately 2 to 6 per 1,000 live male births, and has been increasing during the last few decades.1–4 The cause of hypospadias remains largely unknown. However, current epidemiology and laboratory studies have shed new light on the etiology of hypospadias. It has been postulated that changes in the concentrations of sex hormones during the critical period of penile and urethral development (weeks 8 to 14), caused by endogenous or exogenous factors, may have a part in the development of hypospadias. 3,5–7 Since certain endogenous or exogenous factors such as hormone levels, diet and exposure to chemical substances can cause epigenetic alterations, particularly maternal exposure during fetal development, it is likely that epigenetic alterations are involved in the etiology of hypospadias.8–10
Male sexual differentiation required the production of testosterone in the testes, its activation to dihydrotestosterone in the genital target tissues and the expression of a normal androgen receptor.11,12 In a recent study Vottero et al found evidence of epigenetic abnormalities of the AR gene in foreskin from children with hypospadias.13 They found that the AR gene in target tissues from patients with hypospadias was more methylated than in control children, resulting in decreased expression of AR. In another study Bens et al compared the DNA methylation of labia majora fibroblast strains from 46,XY patients with androgen insensitivity syndrome to scrotal fibroblast strains from normal 46,XY males to evaluate whether epigenetic mechanisms translate AR mutation into androgen dependent gene expression patterns.14 They found frequently higher HOXA5 DNA methylation levels in labia majora fibroblast strains in androgen insensitivity syndrome compared to normal male scrotal fibroblast strains. This indicates that androgen programming in sexual differentiation is not restricted to global gene transcription, but that it also occurs at the epigenetic level.
Endocrine disrupting chemicals such as phthalates that behave similarly to biological hormones can interfere with the physiological functions of the endogenous hormones. In humans a recent multigenerational nationwide cohort study of 529 families of women exposed to diethylstilbestrol (a synthetic nonsteroidal estrogen) showed that a significant proportion of boys born to diethylstilbestrol daughters exhibited hypospadias, suggesting a transgenerational effect of EDCs.15 Wu et al analyzed global DNA methylation status and examined expression levels of the DNA methyltransferases in fetal testes of mice after maternal exposure to diethylhexyl phthalate, an EDC used for producing flexible polyvinyl chloride.16 Fetal testes exposed to diethylhexyl phthalate significantly had a more than 10% relative increase in global DNA methylation as well as increased DNA methyltransferase expression.16 These findings suggest that the molecular mechanisms that underlie the effects of EDCs on the fetus and in subsequent generations may involve changes in the epigenetic background and alteration of DNA methylation.
Based on these previous studies we hypothesized that changes in the DNA methylation level caused by changes in endogenous sex hormones or exposure to EDCs can affect gene expression and, consequently, may contribute to abnormal male sexual differentiation and hypospadias. To test our hypothesis we compared the genome-wide DNA methylation profile in bisulphite converted genomic DNA obtained from preputial tissue samples of 20 subjects with hypospadias and healthy controls.
MATERIALS AND METHODS
Study Participants
A total of 12 patients with isolated hypospadias and 8 healthy controls were enrolled as part of a Committee on Human Research approved study at University of California, San Francisco. Patients with an undescended testis, disorders of sex development or known endocrine abnormalities were excluded from the study. The position of the urethral meatus, associated anomalies and family history of hypospadias were assessed by history and physical examination. Of the 12 patients 8 had mild hypospadias, defined as ectopic urethral meatus between the corona and midshaft of the penis, while 4 had severe hypospadias, defined as ectopic urethral meatus between the proximal penile shaft and perineum. The age of the patients ranged from 13 to 78 months (median 26). No patients received preoperative testosterone treatment. All 12 patients were screened for and did not have any known mutations in AR, MAMLD1, SF1 and SRD5A2 genes. Controls consisted of subjects undergoing elective circumcision. For patients, excess preputial tissue not used for reconstruction at the time of hypospadias surgery was obtained. No periurethral tissue was taken. For control subjects, excess preputial tissue was obtained during elective circumcision.
Genome-wide DNA Methylation Analysis
Genomic DNA was extracted from the preputial tissue samples using QIAamp® DNeasy tissue kit. Genome-wide DNA methylation profiling was performed using Illumina Infinium HumanMethylation450 BeadChip arrays and kit. The BeadChip arrays enable the direct investigation of more than 485,000 methylation sites per sample across the genome at single nucleotide resolution. The BeadChip covers 99% of RefSeq genes, with an average of 17 CpG sites per gene region.
We first performed bisulphite conversion of 1 µg genomic DNA using the EZ-96 DNA Methylation-Gold™ Kit, followed by Illumina Infinium methylation assay according to manufacturer instructions. Bisulphite converted and unconverted (ie methylated) sites were simultaneously evaluated by hybridization of DNA to site specific probes attached to beads on the chip, one for unmethylated and the other for methylated sites, followed by allele specific base extension that included a fluorescent label. Two different labels were used, and fluorescent signals were specific for the T (unmethylated) or C (methylated) alleles. Methylation scores were represented as β scores, and were computed based on the ratio of methylated-to-methylated plus unmethylated signal outputs.
Statistical Analysis
Initial array results were visualized using Illumina® GenomeStudio®. All statistical analyses were performed using the R package (http://www.r-project.org),17 Bioconductor18 and Stata® 11. For quality control of the methylation data, samples were monitored for coverage (fraction of CpGs with detectable intensity values above background) and BSCE using the controls provided on the Illumina BeadChip. All the samples passed cutoff for BSCE or coverage. We restricted the analysis a priori to CpGs with average methylation β score ≥0.05 in control subjects. The methylation level at each CpG site was compared between patients with hypospadias and controls using the t test. Mutually adjusted logistic regression models were built for CpG sites significantly associated with the risk of hypospadias in univariate analyses. The covariates included in the multivariate regression model were age at hypospadias surgery or circumcision, race/ethnicity, and factors for chip and BSCE. To correct the p values for multiple comparisons we estimated the FDR using the q-value framework.19 It has been shown that the analytical q-value estimates are similar to those obtained using permutations of sample labels preserving the potential correlative structure between CpGs.20
RESULTS
Methylation Profile of Arrays
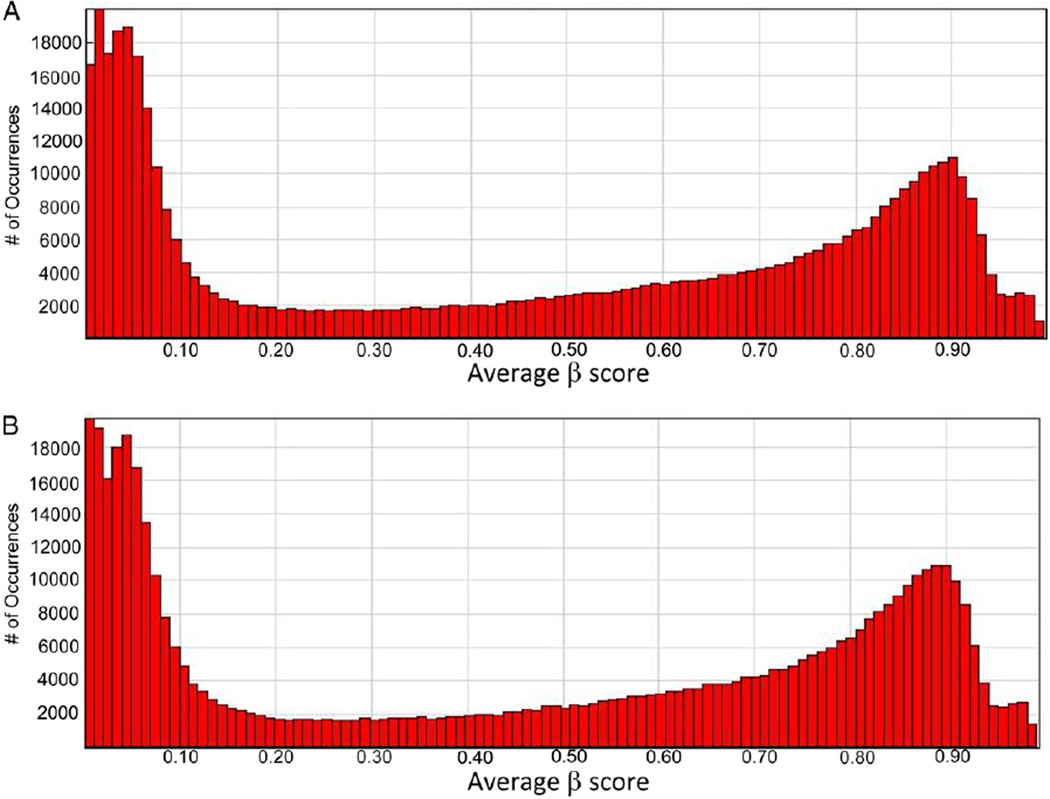
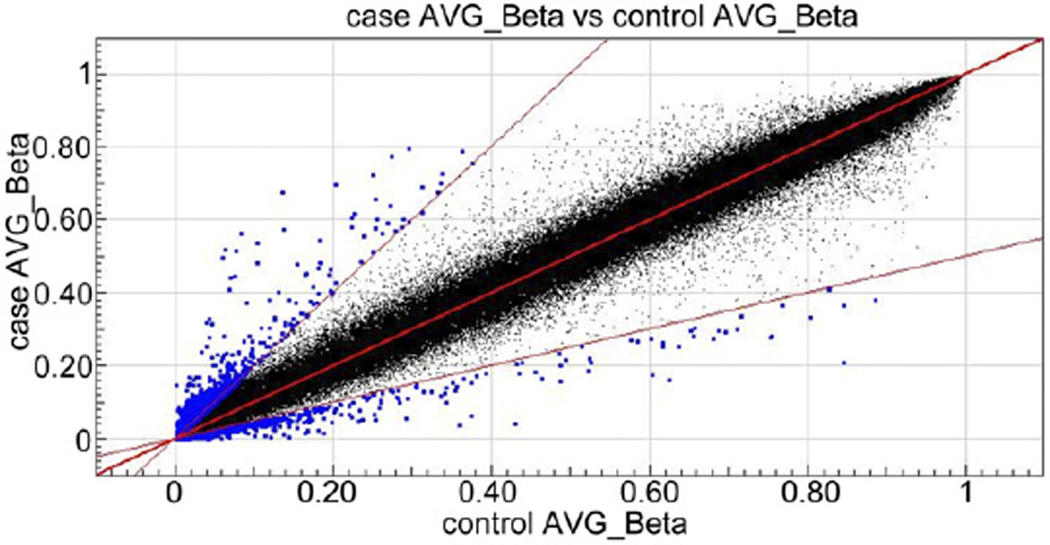
The methylation values, as defined by β scores ranging between 1 and 0 on a continuous scale, or fully to nonmethylated, are displayed for all 485,577 CpGs queried as histograms in figure 1, for controls and patients with hypospadias, respectively. Both groups show a similar pattern with a high peak of hypomethylated loci and a low peak in the hypermethylated loci which is the expected U shape of methylation pattern of the genome. Of 485,577 CpG sites assayed 98.0% had complete data, and 99.1% and 99.5% had 1 or less and 2 or less missing values, respectively. The scatterplot in figure 2 shows highly equivalent average β scores for CpG sites between cases and controls (r2 = 0.994).
Figure 1.
Average β scores across array in healthy controls (A) and patients with hypospadias (B)
Figure 2.
Scatterplot showing highly equivalent average β scores between patients with hypospadias and healthy controls. CpG sites with twofold or greater change in methylation level in patients compared to controls are shown in blue.
DNA Methylation Analysis
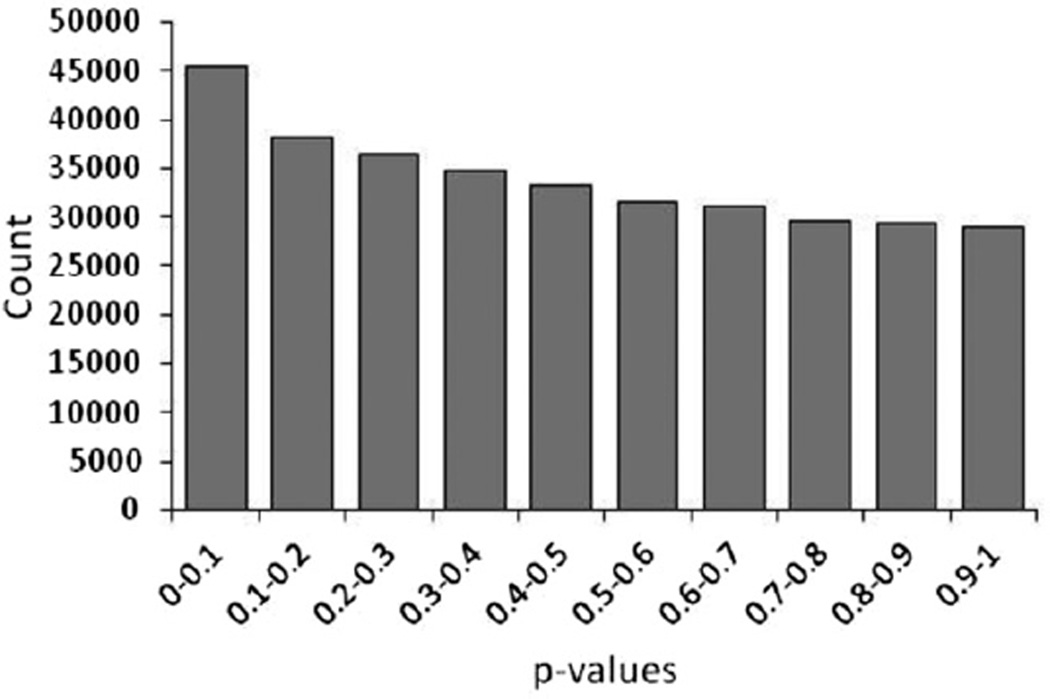
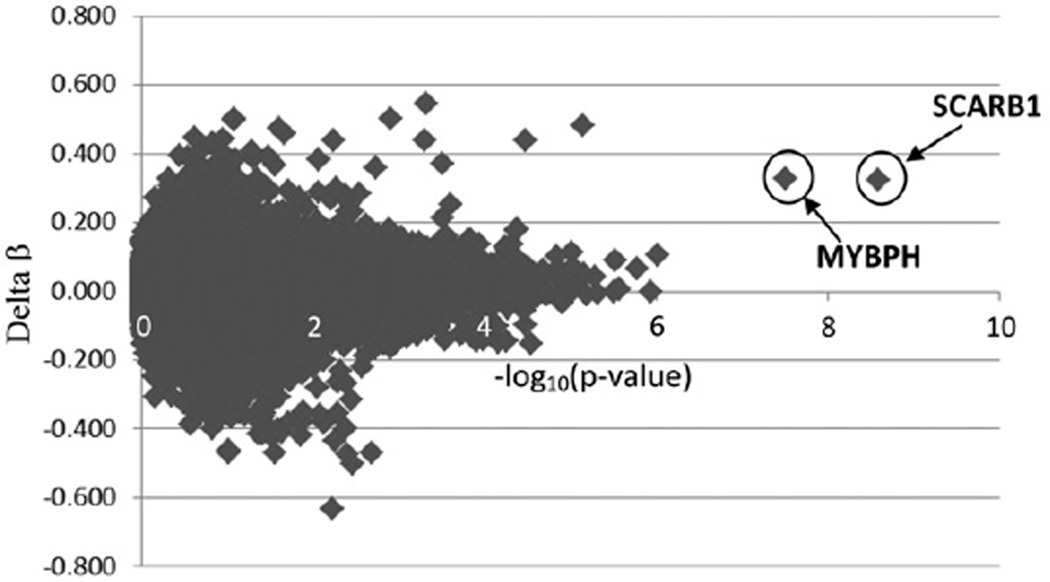
Of 485,577 CpG sites interrogated on the array further analysis was performed only for 393,704 that had an average β score of at least 0.05 in controls. A total of 887 CpG sites were differentially methylated between patients with hypospadias and healthy controls. There were 542 CpG sites that showed a two-fold or greater increase and 345 that showed a two-fold or greater decrease in methylation level among patients vs controls. We performed the t test to compare the methylation level at each CpG site between patients and controls. The resulting p value distribution from the t test of 393,704 CpG sites was significantly different from a uniform distribution with an excess of p values close to 0, suggesting that a significant number of CpGs are correlated with hypospadias (fig. 3). Figure 4 shows the distribution of difference in β scores between patients and controls for each CpG site (delta β) and their −log10 (p value) for association with hypospadias. Two loci clearly showed a highly significant p value and large delta β (cg23556238 in SCARB1, p = 2.61×10−09 and cg04083966 in MYBPH, p = 3.06×10−08). Note that a conservative adjustment for multiple testing using the Bonferroni method and a standard type I error rate of 0.05 would result in a cutoff for genome-wide significance of p value 0.05/393,704 CpG sites = 1.27×10−7.
Figure 3.
Histogram of p values from t test reveals excess of significant p values correlated with hypospadias.
Figure 4.
Distribution of difference in β scores (delta β) between patients and controls for each CpG site and −log10(p value) for association with hypospadias. Most significant associations were observed for CpG sites in SCARB1 and MYBPH genes.
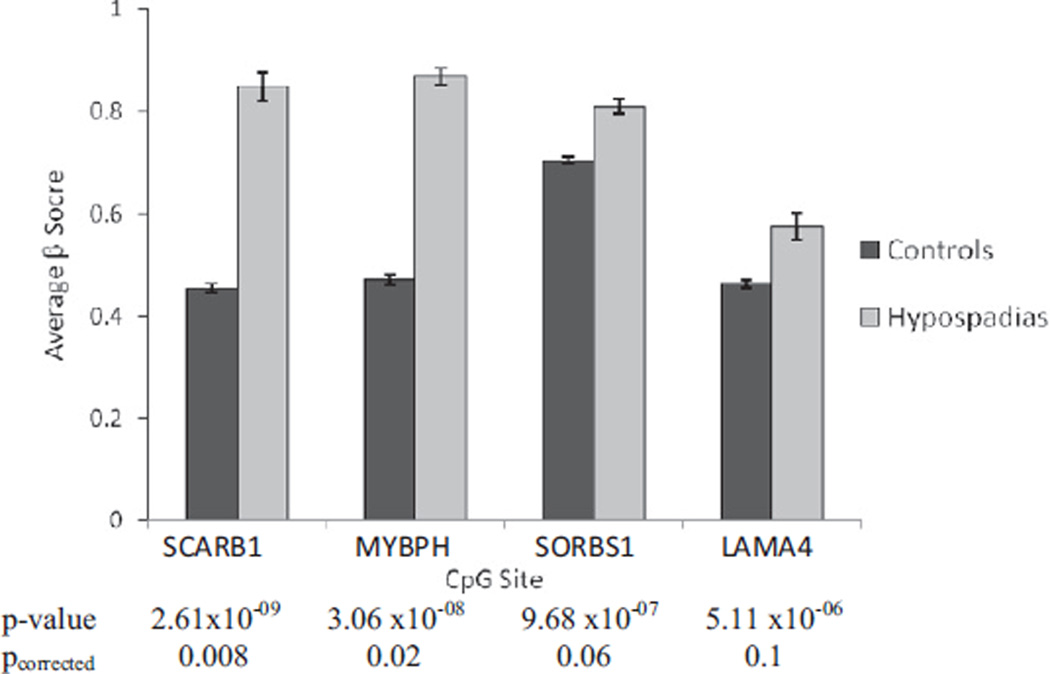
The table lists the 14 CpG sites that showed significantly different methylation scores between patients and controls at a less stringent p $0.00001. As age and race/ethnicity could confound the analysis, we also performed multivariate logistic regression analysis for the 14 top CpG sites including the confounding factors as covariates. At none of the 14 CpG sites was the methylation level associated with subject age or race/ethnicity and, therefore, the association of CpG and hypospadias did not change substantially after adjusting for covariates. Of the 14 sites 11 showed a higher level of methylation in patients than controls (hypermethylation), whereas CpG sites cg15182635 in the HOX11D gene, cg17769084 in c10orf41 and cg01708273 on chromosome 3 showed hypomethylation in patients. We further applied the FDR method to correct for multiple comparisons. At a FDR of 0.05 the association between hypospadias and methylation level remained statistically significant only for CpG sites in SCARB1 (pcorrected = 0.008) and MYBPH genes (pcorrected = 0.02), and borderline significant for SORBS1 (pcorrected = 0.06) and LAMA4 genes (pcorrected = 0.1). Figure 5 shows the average (SD) β scores for these 4 CpG sites in SCARB1, MYBPH, SORBS1 and LAMA4 genes were 0.455 (0.008), 0.47 (0.01), 0.704 (0.007) and 0.463 (0.007) in controls, and 0.848 (0.003), 0.868 (0.003), 0.811 (0.002) and 0.575 (0.002) in patients with hypospadias, respectively. We further examined whether the association varied by severity of hypospadias for the 14 top associated CpG sites. The direction of association and average β score were similar in subjects with mild and severe hypospadias for all 14 sites except cg09301498 on chromosome 7. This site had a significantly higher methylation level in patients with severe hypospadias (β score 0.698) compared to mild hypospadias (β score 0.610) and controls (β score 0.422).
CpG sites most significantly associated with hypospadias
| Illumina ID | Gene Name | Chromosome | Enhancer | Av β Score in Controls |
Av β Score in Pts |
Difference in β Pts vs Controls |
p Value (t test) |
|---|---|---|---|---|---|---|---|
| cg23556238 | SCARB1 | 12 | 0.455 | 0.848 | 0.394 | 2.61 × 10−09 | |
| cg04083966 | MYBPH | 1 | 0.471 | 0.868 | 0.397 | 3.06 × 10−08 | |
| cg22640982 | SORBS1 | 10 | 0.704 | 0.811 | 0.106 | 9.68 × 10−07 | |
| cg04204246 | LAMA4 | 6 | True | 0.463 | 0.575 | 0.112 | 5.11 × 10−06 |
| cg01708273 | HOXD11 | 2 | 0.615 | 0.507 | −0.108 | 2.92 × 10−05 | |
| cg06900514 | 2 | True | 0.447 | 0.552 | 0.105 | 3.28 × 10−05 | |
| cg18200150 | MYO1D | 17 | 0.492 | 0.604 | 0.113 | 3.87 × 10−05 | |
| cg09301498 | 7 | 0.422 | 0.639 | 0.217 | 4.24 × 10−05 | ||
| cg14197156 | EGFL7 | 9 | 0.648 | 0.812 | 0.165 | 4.31 × 10−05 | |
| cg08979191 | 5 | 0.119 | 0.251 | 0.132 | 4.91 × 10−05 | ||
| cg15182635 | C10orf41 | 10 | 0.389 | 0.273 | −0.116 | 5.52 × 10−05 | |
| cg21948564 | LMAN1L | 15 | 0.448 | 0.557 | 0.110 | 5.52 × 10−05 | |
| cg05355411 | SULF1 | 8 | True | 0.523 | 0.626 | 0.103 | 5.82 × 10−05 |
| cg17769084 | 3 | True | 0.493 | 0.363 | −0.130 | 6.87 × 10−05 |
Figure 5.
Average β score of 4 most significantly associated CpG sites in patients with hypospadias and healthy controls. CpG sites cg23556238, cg04083966, cg22640982 and cg04204246 were in SCARB1, MYBPH, SORBS1 and LAMA4 genes, respectively.
DISCUSSION
We investigated the methylation state of approximately 485,000 CpG sites spread throughout the genome in a study sample of 20 subjects with hypospadias and healthy controls. The analysis identified an excess of CpG sites observed to have a change in methylation status that could be correlated with hypospadias with a FDR of 0.2. Using a more stringent FDR cutoff of 0.05, 2 CpGs in SCARB1 (scavenger receptor class B, member 1) and MYBPH (myosin binding protein H) genes were identified as associated with hypospadias disease status.
In several studies the down-regulation of SCARB1 together with other genes has been shown to have a role in di(n-butyl) phthalate induced suppression of steroidogenesis.21,22 Levels of methylation for CpG site cg23556238 in SCARB1 almost doubled in patients with hypospadias vs controls, suggesting the down-regulation of SCARB1 in hypospadias. Although this CpG site is present in the body of the SCARB1 gene near a CpG island, methylation in CpG island shores has recently been suggested as being of particular biological importance.23 The other significant CpG site identified in our study is located in the first exon of the MYBPH gene. To date, little is known about the functions of MYBPH, even in regard to its interaction with muscle myosin with virtually nothing reported regarding its roles in non-muscle cells.24,25 The H isoform is believed to be associated with the assembly of myofibrils containing myofilaments of appropriate length and composition for effective contractile function.26 However, in the absence of pertinent data on the role of MYBPH in male urogenital development, it remains unclear how or to what extent the rather localized methylation difference described here might affect gene regulation. Future studies of SCARB1 and MYBPH are needed to corroborate the functionality and clinical importance of the differential methylation described here in context of male penile and urethral development.
Although our study identified 2 significantly associated CpG sites at the genome-wide level, we may have missed others due to the low power of our study because of the small sample size. Therefore, we also compared methylation status at candidate gene level for known hypospadias genes WT1, SF1, BMP4, BMP7, HOXA4, HOXB6, AR, FGF8, FGFR2, HSD3B2, SRD5A2, ATF3, MAMLD1, MID1, BNC2, ESR1, ESR2, ATF3, DGKK, CYP1A1, GSTM1, GSTT1, CTGF, CYR61 and EGF.27 Two CpG sites in AR, 1 in BMP4 and 2 in CTGF genes were significantly associated with hypospadias (data not shown) at a candidate gene level, suggesting that these genes may have a role in the etiology of hypospadias via epigenetic mechanisms. Consistent with our findings, Vottero et al also recently found higher AR gene methylation in foreskin tissues from patients with hypospadias than in normal children.13
Like all genome-wide association studies, this investigation was designed to find statistically significant associations, in this case with DNA methylation variation, but not to identify the underlying mechanism(s) for the cause or function of the observed variation. Potential mechanisms that may drive changes in DNA methylation level in hypospadias could be underlying genetic variations or external factors such as EDCs. Since this is a post hoc analysis in which we examined changes in methylation patterns in subjects at approximately age 12 to 36 months, whereas hypospadias occurs in utero during first trimester of pregnancy, it is difficult to explain whether the observed methylation changes represent the cause of hypospadias or are secondary to an altered fetal steroid milieu. In addition, we do not know if the temporal changes in methylation at the time of occurrence of hypospadias would persist after birth. However, Kalfa et al support transmission of EDC induced changes in DNA methylation in subsequent generations.15
Another limitation of our study is that DNA methylation is tissue specific. We examined methylation patterns in preputial tissue from the dorsal aspect of the penis, whereas hypospadias is a developmental defect of the ventral aspect. In humans periureteral tissue is difficult to sample since it is precious during surgery. Therefore, studies in humans are limited to analyzing preputial tissue as performed in this and other previous studies.13
CONCLUSIONS
This study is the first to our knowledge to investigate hypospadias using this cutting-edge, high-throughput DNA methylation 450K array based assay. Significant associations were observed between methylation levels of individual CpG sites and hypospadias. With the examination of 20 subjects with hypospadias and controls, our study supports the utility of a genome-wide DNA methylation approach, and the potential prospects for larger and subsequent replication studies of epigenomic factors in the etiology of hypospadias.
ACKNOWLEDGMENTS
Drs. Mark Segal and Henrik Bengtsson provided advice on data analysis.
Study received approval from the Committee on Human Research.
Supported by the National Institutes of Health through the UCSF K12 Urological Research (KURe) Career Development Program sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (5K12DK083021) (SC) and by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases R01DK058105 (LSB).
Abbreviations and Acronyms
- AR
androgen receptor
- BSCE
bisulfite conversion efficiency
- EDC
endocrine disrupting chemical
- FDR
false discovery rate
REFERENCES
- 1.Aho M, Koivisto AM, Tammela TL, et al. Is the incidence of hypospadias increasing? Analysis of Finnish hospital discharge data 1970–1994. Environ Health Perspect. 2000;108:463. doi: 10.1289/ehp.00108463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canning DA. Hypospadias trends in two US surveillance systems. Rise in prevalence of hypospadias. J Urol. 1999;161:366. [PubMed] [Google Scholar]
- 3.Dolk H. Rise in prevalence of hypospadias. Lancet. 1998;351:770. doi: 10.1016/S0140-6736(05)78924-8. [DOI] [PubMed] [Google Scholar]
- 4.Paulozzi LJ. International trends in rates of hypospadias and cryptorchidism. Environ Health Perspect. 1999;107:297. doi: 10.1289/ehp.99107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akre O, Lipworth L, Cnattingius S, et al. Risk factor patterns for cryptorchidism and hypospadias. Epidemiology. 1999;10:364. [PubMed] [Google Scholar]
- 6.Baskin LS. Hypospadias and urethral development. J Urol. 2000;163:951. [PubMed] [Google Scholar]
- 7.Silver RI, Rodriguez R, Chang TS, et al. In vitro fertilization is associated with an increased risk of hypospadias. J Urol. 1999;161:1954. [PubMed] [Google Scholar]
- 8.Aagaard-Tillery KM, Grove K, Bishop J, et al. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. J Mol Endocrinol. 2008;41:91. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin DI, Cropley JE, Suter CM. Environmental influence on epigenetic inheritance at the Avy allele. Nutr. Rev., suppl. 2008;66:S12. doi: 10.1111/j.1753-4887.2008.00057.x. [DOI] [PubMed] [Google Scholar]
- 10.Zheng YG, Wu J, Chen Z, et al. Chemical regulation of epigenetic modifications: opportunities for new cancer therapy. Med Res Rev. 2008;28:645. doi: 10.1002/med.20120. [DOI] [PubMed] [Google Scholar]
- 11.Jia L, Shen HC, Wantroba M, et al. Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol Cell Biol. 2006;26:7331. doi: 10.1128/MCB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEwan IJ. Molecular mechanisms of androgen receptor-mediated gene regulation: structure-function analysis of the AF-1 domain. Endocr Relat Cancer. 2004;11:281. doi: 10.1677/erc.0.0110281. [DOI] [PubMed] [Google Scholar]
- 13.Vottero A, Minari R, Viani I, et al. Evidence for epigenetic abnormalities of the androgen receptor gene in foreskin from children with hypospadias. J Clin Endocrinol Metab. 2011;96:E1953. doi: 10.1210/jc.2011-0511. [DOI] [PubMed] [Google Scholar]
- 14.Bens S, Ammerpohl O, Martin-Subero JI, et al. Androgen receptor mutations are associated with altered epigenomic programming as evidenced by HOXA5 methylation. Sex Dev. 2011;5:70. doi: 10.1159/000323807. [DOI] [PubMed] [Google Scholar]
- 15.Kalfa N, Paris F, Soyer-Gobillard MO, et al. Prevalence of hypospadias in grandsons of women exposed to diethylstilbestrol during pregnancy: a multigenerational national cohort study. Fertil Steril. 2011;95:2574. doi: 10.1016/j.fertnstert.2011.02.047. [DOI] [PubMed] [Google Scholar]
- 16.Wu S, Zhu J, Li Y, et al. Dynamic epigenetic changes involved in testicular toxicity induced by di-2-(ethylhexyl) phthalate in mice. Basic Clin Pharmacol Toxicol. 2009;106:118. doi: 10.1111/j.1742-7843.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- 17.Ihaka R, Gentleman RC. R: A Language for Data Analysis and Graphics. J Comput Graph Stat. 1996;5:299. [Google Scholar]
- 18.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barlow NJ, Phillips SL, Wallace DG, et al. Quantitative changes in gene expression in fetal rat testes following exposure to di(n-butyl) phthalate. Toxicol Sci. 2003;73:431. doi: 10.1093/toxsci/kfg087. [DOI] [PubMed] [Google Scholar]
- 22.Shultz VD, Phillips S, Sar M, et al. Altered gene profiles in fetal rat testes after in utero exposure to di(n-butyl) phthalate. Toxicol Sci. 2001;64:233. doi: 10.1093/toxsci/64.2.233. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welikson RE, Fischman DA. The C-terminal IgI domains of myosin-binding proteins C and H (MyBP-C and MyBP-H) are both necessary and sufficient for the intracellular crosslinking of sarcomeric myosin in transfected non-muscle cells. J Cell Sci. 2002;115:3517. doi: 10.1242/jcs.115.17.3517. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto K. Effect of H-protein on the formation of myosin filaments and light meromyosin paracrystals. J Biochem. 1988;103:274. doi: 10.1093/oxfordjournals.jbchem.a122260. [DOI] [PubMed] [Google Scholar]
- 26.Vaughan KT, Weber FE, Ried T, et al. Human myosin-binding protein H (MyBP-H): complete primary sequence, genomic organization, and chromosomal localization. Genomics. 1993;16:34. doi: 10.1006/geno.1993.1136. [DOI] [PubMed] [Google Scholar]
- 27.van der Zanden LF, van Rooij IA, Feitz WF, et al. Aetiology of hypospadias: a systematic review of genes and environment. Hum Reprod Update. 2012;18:260. doi: 10.1093/humupd/dms002. [DOI] [PubMed] [Google Scholar]