SUMMARY
While the chromatin state of pluripotency genes has been extensively studied in embryonic stem cells (ESCs) and differentiated cells, their potential interactions with other parts of the genome remain largely unexplored. Here, we identified a genome-wide, pluripotency-specific interaction network around the Nanog promoter by adapting circular chromosome conformation capture-sequencing (4C-seq). This network was rearranged during differentiation and restored in induced pluripotent stem cells. A large fraction of Nanog-interacting loci were bound by Mediator or cohesin in pluripotent cells. Depletion of these proteins from ESCs resulted in a disruption of contacts and the acquisition of a differentiation-specific interaction pattern prior to obvious transcriptional and phenotypic changes. Similarly, the establishment of Nanog interactions during reprogramming often preceded transcriptional upregulation of associated genes, suggesting a causative link. Our results document a complex, pluripotencyspecific chromatin "interactome" for Nanog and suggest a functional role for longrange genomic interactions in the maintenance and induction of pluripotency.
INTRODUCTION
The three-dimensional (3D) chromatin architecture is important for many biological processes including transcriptional regulation. Looping between promoter and enhancer or insulator elements controls the transcriptional activation or repression of genes, respectively (Engel and Tanimoto, 2000; Ling et al., 2006; Zhao et al., 2006). Although long-range chromatin interactions have been observed mostly in cis along the same chromosome (Schoenfelder et al., 2010), they can also occur in trans between different chromosomes. Trans interactions are associated with co-regulation of imprinted genes (Zhao et al., 2006) or genes associated with erythropoiesis (Schoenfelder et al., 2010), with stochastic selection for monoallelic activation of the IFN-β locus (Apostolou and Thanos, 2008) and olfactory genes (Clowney et al., 2012; Lomvardas et al., 2006) and with AID-mediated translocations (Klein et al., 2011; Rocha et al., 2012). Although the organization of chromosomes into defined territories was shown three decades ago (Schardin et al., 1985), the molecular principles of global chromatin architecture have only recently been explored with high-throughput technologies such as the Hi-C method (Dixon et al., 2012; Duan et al., 2010; Lieberman-Aiden et al., 2009; Sexton et al., 2012; Zhang et al., 2012).
Chromatin organization also plays a role in the control of pluripotency and cellular differentiation. For instance, pluripotency-associated genes such as Sox2, Nanog and Klf4 relocate from the nuclear center to the periphery upon differentiation of mouse embryonic stem cells (ESCs)(Peric-Hupkes et al., 2010). Moreover, the loss of promoter-enhancer interactions at key pluripotency genes, including Nanog and Oct4, during ESC differentiation has been associated with silencing of these genes (Kagey et al., 2010; Levasseur et al., 2008). Proteins involved in chromatin looping, comprising CTCF, cohesin and Mediator, co-occupy many genomic targets of pluripotency factors (Kagey et al., 2010; Nitzsche et al., 2011) or directly interact with them (Donohoe et al., 2009; Tutter et al., 2009). These molecules might therefore cooperate to arrange a higher-order chromatin structure that maintains pluripotency. Indeed, depletion of Mediator and cohesin subunits from ESCs results in unscheduled differentiation (Kagey et al., 2010). A more recent study using the Hi-C technology in mouse and human ESCs and differentiated cells identified a network of local chromatin interactions domains, so-called topological domains, with conserved boundaries among different species and cell types (Dixon et al., 2012). Although that report documented important general principles of chromatin organization in pluripotent and differentiated cells, a high-resolution map of genome-wide interactions of pluripotency genes in ESCs is lacking. It also remains unclear which molecules might be involved in establishing such putative connections and if and how these patterns change upon differentiation.
Forced expression of the transcription factors Oct4, Sox2, Klf4 and c-Myc is sufficient to endow somatic cells with pluripotency, giving rise to induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006). In-depth molecular analysis of reprogramming intermediates has been achieved only recently with improved technologies to study rare and defined cell populations (Buganim et al., 2012; Golipour et al., 2012; Polo et al., 2012; Soufi et al., 2012). In addition, molecular characterization of stable partially reprogrammed iPSC (piPSC) lines shed light on the earliest events in cellular reprogramming (Mikkelsen et al., 2008; Sridharan et al., 2009). Although these studies reported the reestablishment of an ESC-like transcriptional and epigenetic state, it remains unclear whether, when and how 3D chromatin structure is reset during cellular reprogramming into iPSCs.
In this study, we have investigated the genome-wide interaction network of the Nanog gene, which is indispensable for development as well as the derivation of ESCs (Mitsui et al., 2003) and iPSCs (Chambers et al., 2003; Silva et al., 2009). We developed a modified version of circular chromosome conformation capture-sequencing (m4C-seq) to determine the genome-wide interaction partners of the Nanog locus in ESCs, iPSCs and mouse embryonic fibroblasts (MEFs) at high resolution. Our study provides the first detailed chromatin interaction map of a key pluripotency locus on a genomic scale and offers novel mechanistic insights into how chromatin architecture is regulated during the acquisition and maintenance of pluripotency.
RESULTS
The Nanog locus engages in distinct genome-wide interactions in pluripotent and differentiated cells
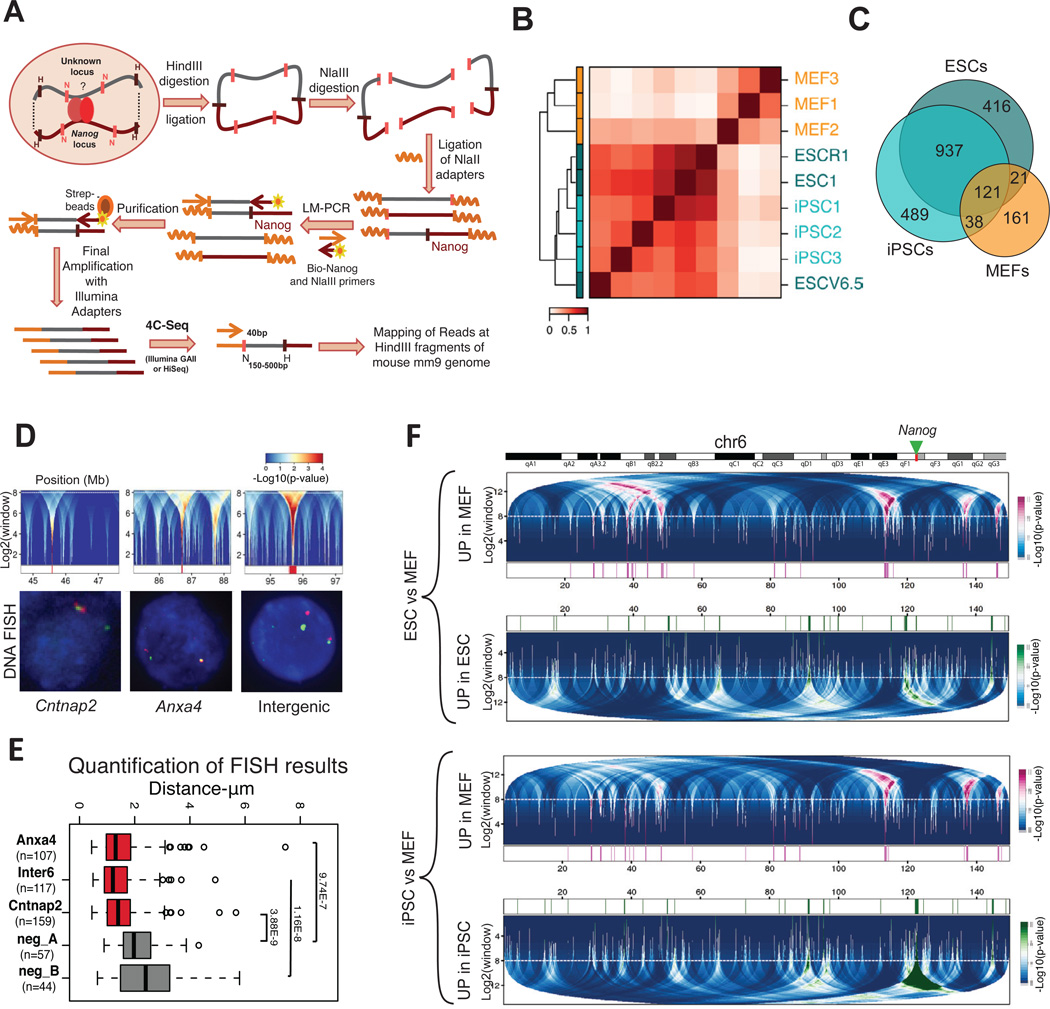
We developed a modified version of 4C-seq (m4-seq) for genome-wide unbiased capture of Nanog’s interactions in pluripotent and differentiated cells (Figure 1A; see Experimental Procedures). Briefly, 4C technology is based on the proximity-ligation principle, in which unknown chromatin loci that interact with a known “bait” locus (e.g., Nanog) are ligated into chimeric DNA molecules and then identified by deep sequencing (Dekker et al., 2002). m4C-seq involves ligation of universal adapters to the linearized hybrid molecules, followed by ligation-mediated PCR with an adapter-specific oligonucleotide and a biotinylated primer recognizing the Nanog locus. This allows specific enrichment and purification of the Nanog-interacting regions using streptavidin beads and avoids the less efficient re-circularization and inverse-PCR steps of published 4C methods.
Figure 1. Genome-wide interactions of the Nanog locus in differentiated and pluripotent cells.
(A) Schematic representation of m4C-seq. LM-PCR: Ligation Mediated-PCR, strep-beads: streptavidin-conjugated beads.
(B) Unsupervised clustering and correlation matrix of pluripotent and differentiated cells (3 ESCs, 3 iPSCs and 3 MEFs). Normalized (observed over expected) m4C-seq signals at individual HindIII fragments are clustered, with Spearman correlation (color gradient) and average linkage. Fragments detected in at least 3 out of 9 samples are used.
(C) Venn diagram showing the degree of overlap among the Nanog-interacting HindIII fragments common within each group: ESCs, iPSCs and MEFs.
(D) The upper panels show details of domainogram analysis for broad intra-chromosomal interacting domains in individual samples. Regions around broad interacting domains are shown for a representative ESC sample (ESC1 cell line). The centers of interacting domains are marked in red at the bottom (p-value < 0.0001). The dashed horizontal white line indicates the maximum window size cutoff. The bottom panels show representative 3D DNA FISH in ESCs confirming the interaction of Nanog (green FITC signals) with each of those domains (magenta Alexa 568 signals).
(E) Boxplot for distances between the Nanog locus and the tested domains (n= number of measured nuclei). Intra-chromosomal regions between the positive hits and the bait position were used as negative controls. P-values for Wilcoxon test are reported (see also Figure S2C). Whiskers extend to most extreme values within 1.5 times the inter quartile range (IQR) from the upper or lower quartile.
(F) Differential interactions over large domains (domainogram) for ESCs vs MEFs (upper panel) and iPSC vs MEFs (bottom panel) comparisons in chromosome 6. The green arrow marks Nanog position. Top: interacting domains upregulated in MEF (magenta), Bottom: interacting domains up-regulated in ESC or iPSCs respectively cells (green). In the central part magenta and green marks indicate the regions significantly up-regulated (p-value < 0.001) in MEF or ESC/iPSC, respectively. The dashed horizontal white line indicates the maximum window size cutoff. All replicates for each cell type are taken into account to compute the score for differential interactions.
See also Figure S1, S2, Table S1 and S6
To increase confidence in observed interactions, we used biological replicates, applied multiple filtering and normalization steps, and adjusted for random ligation events and possible technical biases based on a control sample (non-crosslinked genomic DNA, see Experimental Procedures). Technical replicates generated by independent ligation, amplification and sequencing showed high concordance (Spearman correlation coefficient≈0.9) (Figure S1A). We then analyzed three independent biological replicates for ESC lines (R1, V6.5 and KH2-ESC1), MEFs and fibroblast-derived iPSC clones, previously shown to give rise to entirely iPSC-derived mice, thus satisfying the most stringent criteria of pluripotency (Stadtfeld et al., 2010a). The biological replicates of pluripotent cells showed higher variability than the technical replicates as expected, but nevertheless exhibited high correlation (Spearman coefficient≈0.7) (Figures S1A–D). However, MEFs showed notably lower correlation (Spearman coefficient ≈0.3), suggesting that Nanog may have less stable interactions in MEFs, perhaps because the gene is not active.
Unsupervised clustering (Figure 1B) highlighted similarities between ESCs and iPSC, which clustered separately from MEFs. Consistent with this observation, we found extensive overlap (~70%) among the conserved Nanog interactions in ESC and iPSC cells (Table S1), but much less overlap between these pluripotent samples and MEFs (<10% of pluripotent interactions) (Figure 1C). The higher variability in MEF samples resulted in a smaller set of conserved interactions among replicates (Figure 1C and Figure S1C) (Table S1). These results show distinct Nanog interactomes in differentiated and pluripotent cells.
Given that Nanog is located in a gene-rich genomic region containing other pluripotency loci, we first examined a 200kb window around its promoter. We detected several interaction partners, including the Nanog enhancer, Aicda, Apobec1 and Scl2a3 genes (Figure S1E). We observed signal for 11 out of 12 loci that have been previously detected in ESCs by chromosome conformation capture (3C) (Levasseur et al., 2008). We also identified broad interacting domains in more distal regions on chromosome 6, visualized in the form of a “domainogram” (Figure S2A) (Bantignies et al., 2011). Randomly selected interactions in the broad domains were verified by 3D DNA fluorescent in situ hybridization (FISH) (Figure 1D–E, Figure S2B) and by 3C analysis among single HindIII fragments using independent cell preparations (Figure S2D). FISH results were independently confirmed for a subset of nuclei (~250 nuclei for 3 probes in total) at higher resolution, which allowed for more accurate measurement of co-localized signals (Figure S2C).
Broad interaction domains with differential strengths in ESCs and MEFs are shown in Figure 1F. MEF-derived iPSCs and ESCs showed similar differential domainogram patterns when compared to MEFs, suggesting that reprogramming restored the ESC-specific 3D structure along chromosome 6. Furthermore, cis interaction patterns observed in published Hi-C data for ESCs (Dixon et al., 2012) exhibited a higher correlation to those we detected in ESC/iPSC than in MEF (Figure S2E). Together, these data document that Nanog forms a pluripotency-specific interactome with multiple genomic regions along its entire chromosome in both ESCs and iPSCs.
Nanog participates in pluripotency-specific inter-chromosomal associations
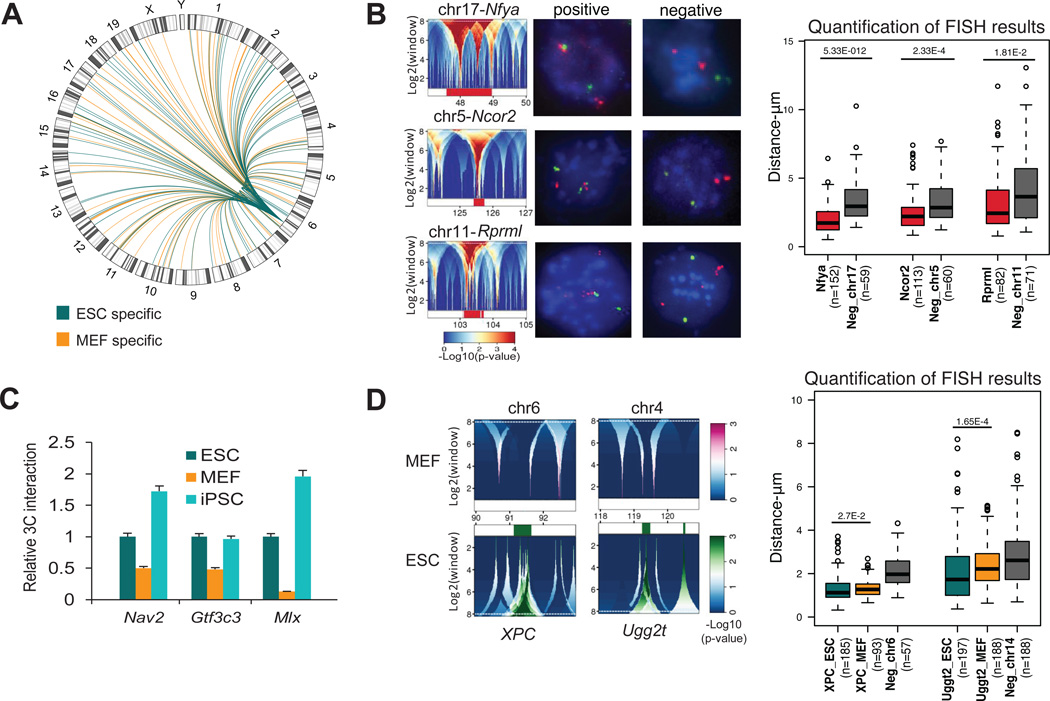
Many of the detected contacts were found to be trans interactions of Nanog with other chromosomes (Figure 2A and Table S1). Although previous studies using conventional 4C-seq protocols did not detect such a high number of trans associations (Simonis et al., 2009; Simonis et al., 2006), our results are consistent with a similar 4C adaption termed “enhanced-4C” (e4C) (Schoenfelder et al., 2010). We believe that m4C-seq and e4C approaches using universal adapters and streptavidin-based purification/enrichment of the bait locus enable greater sensitivity. The high number of observed inter-chromosomal interactions is further supported by the tendency of the Nanog locus to localize on the edge or outside of its chromosome territory (Figure S2F). Moreover, reanalysis of recently published Hi-C data from mouse ESCs (Dixon et al., 2012) showed that over 60% of the Nanog’s trans interactions overlapped significantly with our m4C-seq interactions in ESCs and iPSCs, but not in MEFs (Figure S2G). Selected interacting regions in ESCs, localized on three different chromosomes, were tested by 3D-DNA FISH in ESCs and they showed closer proximity to the Nanog locus compared with non-interacting regions on the respective chromosomes (Figure 2B).
Figure 2. Detection and validation of inter-chromosomal associations of the Nanog locus in pluripotent and differentiated cells.
(A) Circos plot for differential inter-chromosomal interactions in ESCs (green) compared to MEFs (orange) as detected from broad domains analysis using domainograms (Figure 1F) in each chromosome.
(B) Three inter-chromosomal Nanog-interacting domains confirmed by 3D-DNA FISH in ES cells. The domainograms refer to the ESC1 line and are representative of other ESCs. Representative 3D-DNA FISH photos show the Nanog alleles (Green FITC signals) interacting with each of those domains (left) or their corresponding negative controls (right) (Magenta Alexa 568 signals). Boxplots report for 3D-DNA FISH results (n=number of nuclei; p=Wilcoxon test p-value) (whiskers like Figure 1E). Negative controls were selected in regions within 2Mb of the targets.
(C) 3C-PCR confirmation of selected differential inter-chromosomal interactions of the Nanog locus in ESCs and iPSCs vs MEF. For each primer pair the PCR signal was calculated relative to the corresponding signal in ESCs (Relative 3C Interaction) after normalization with the PCR signal of primers designed at the bait locus (see Table S6). Error bars indicate standard deviations (n=3 technical replicates). All 3C-PCR products were isolated and analyzed by Sanger sequencing.
(D) Domainograms details for differential interactions around XPC and Ugg2t, which were found to interact with Nanog preferentially in ESCs. Upper (magenta) and bottom panels (green) refer to interaction enrichment in MEFs and pluripotent cells, respectively. 3D-DNA FISH results for the two regions are shown in the boxplot similarly to panel B (whiskers like Figure 1E).
The distribution of broad differential intra- and inter-chromosomal interaction domains in pluripotent (ESCs) vs. differentiated (MEFs) cells is visualized in Figure 2A. In addition, differential interactions selected at single fragment level are reported in Table S2 and shown in Figure S1B. We confirmed several of the differential interactions between MEFs and ESCs either by 3C (Figure 2C) or 3D-DNA FISH (Figure 2D) using independent cell preparations. Collectively, these results show that Nanog forms a complex genomic interaction network with multiple chromosomes that differs between pluripotent and differentiated cells, and is restored in iPSCs.
Nanog-interacting loci are enriched for open chromatin features as well as binding sites for pluripotency factors and cohesin/Mediator
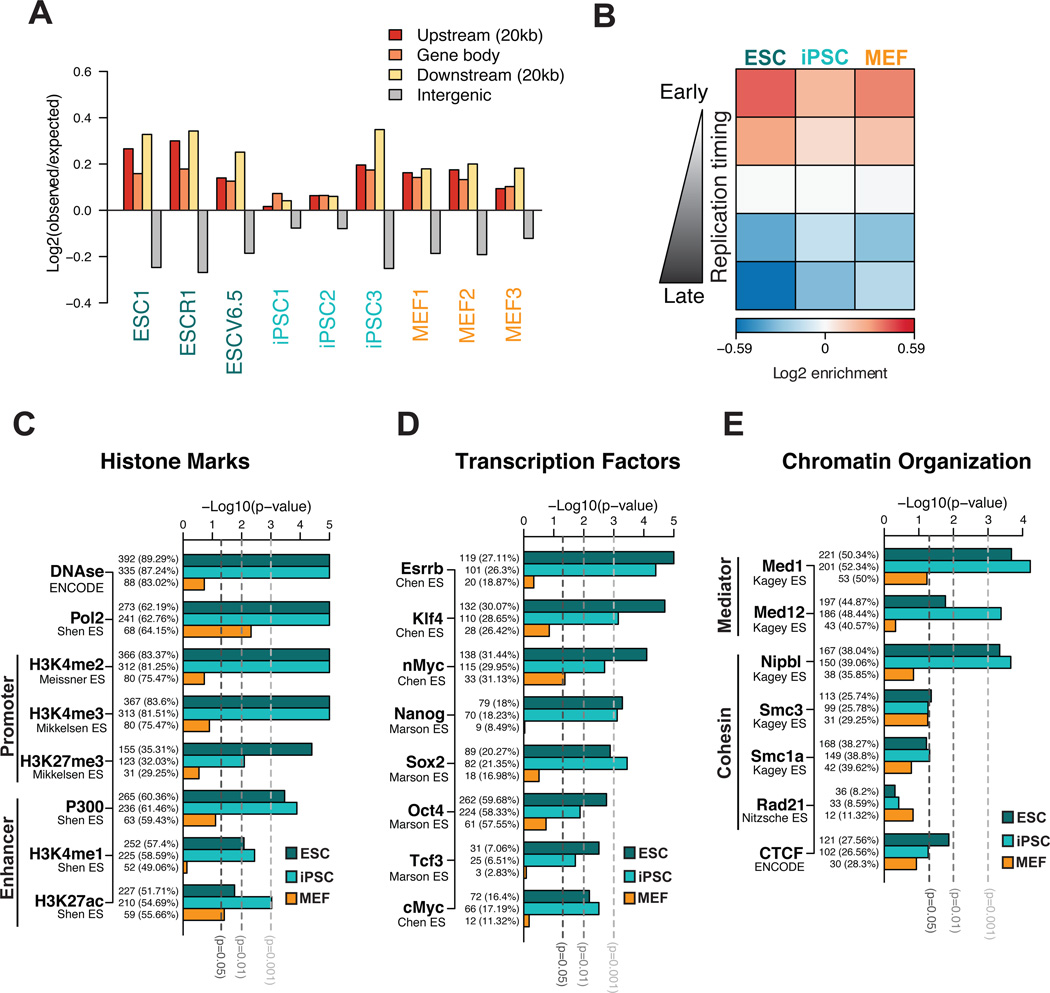
To determine whether Nanog-interacting loci share common genomic features, we compared our results with published data (Table S3). We first noticed consistent enrichment for gene bodies and surrounding regulatory regions among interactions in both ESCs/iPSCs and MEFs (Figure 3A) as well as for early-replicating domains, which typically exhibit an open chromatin structure (Figure 3B). The latter correlation is consistent with the fact that Nanog replicates early in both cell types despite its transcriptional silencing in MEFs (Hiratani et al., 2010; Hiratani et al., 2008).
Figure 3. Nanog- interacting regions are enriched for open chromatin features and pluripotency factor binding in pluripotent cells.
(A) Distribution of the Nanog-interacting loci detected at single fragment level in each sample. Log ratios of observed over expected fragments in different genomic regions show a consistent overrepresentation of interactions in genes and surrounding regions (20kb upstream or downstream).
(B) Association of the Nanog-interacting regions with replication timing (RT). Genomic segments were divided into 5 groups (from early to late) based on their replication timing data in each cell type (Hiratani et al., 2010). The median association of interacting fragments (observed over expected log ratio) across biological replicates is plotted as a heatmap.
(C) Association of conserved Nanog interactions within each cell type (ESCs, iPSCs or MEFs) with active or repressive chromatin features. Conserved Nanog interactions were identified by gene-level analysis; ChIP peaks in ESCs were linked to genes when overlapping with a −5Kb/+1Kb window at transcript start. The barplots show the significance of association between Nanog-interacting genes and genes enriched for a given mark, tested independently for each cell type. The number and the percentage of interacting genes with a given chromatin mark are reported for each bar.
(D) and (E) show similar analyses of association to genes bound by pluripotency transcription factors in ESCs and genes bound by components of cohesin and Mediator complexes and CTCF in ESCs, respectively.
We next examined chromatin features of pluripotent cells including histone marks (Table S3) and DNase I hypersensitivity among Nanog-interacting genes using data from the Encyclopedia of DNA Elements (ENCODE) project (2011). Nanog-interacting genes in pluripotent cells were enriched for the activating histone marks H3K4me3 and H3K4me2 and enhancer marks (H3K27ac, H3K4me1 and p300) as well as for DNase I-hypersensitive sites characterizing open chromatin areas (Figure 3C and Figure S3A). A weak correlation was also detected for the repressive H3K27me3 mark and for bivalent promoters (p-value<0.05 in ESC and iPSC). However, we were unable to detect significant and consistent enrichment for binding sites of the Polycomb complex, which deposits H3K27me3 (Figure 3C, Figure S3A). Thus, Nanog interacts mostly with active genes and regulatory elements in pluripotent cells.
To gain mechanistic insights into how the identified interactions are established, we searched for enrichment of pluripotency transcription factor binding sites among the Nanog-interacting loci using published chromatin immunoprecipitation-sequencing (ChIP-seq) datasets (Table S3). Indeed, target sites for Esrrb, Klf4, c-Myc and Sox2 were among the most consistently and significantly enriched sequences, whereas enrichment of Nanog and Oct4 targets varied across datasets (Figure 3D and Figure S3B). We also found a pluripotency-specific association with binding of additional factors of the pluripotency network (Chen et al., 2008) including Tcf3, Tcfcp2l1, Nr5a2 and Zfx (Figure 3D and Figure S3B). Together, these data show that genes interacting with Nanog in ESC/iPSC are strongly enriched for binding of essential pluripotency factors. It remains to be elucidated whether this result reflects that coregulated genes are spatially connected or that some of these factors might be actively involved in chromatin looping.
We also examined occupancy of cohesin, Mediator and CTCF molecules, proteins reported to mediate long-range interactions, among the ESC-specific contacts (Table S3). We found a significant association of Nanog interactions in pluripotent cells with binding of the Mediator (Med1, Med12) and cohesin (Smc1a, Nipbl and Smc3) complexes and a less consistent correlation with CTCF depending on the dataset (Figure 3E and Figure S3C). Collectively, these results suggest that key pluripotency transcription factors might collaborate with molecules known to mediate promoter-enhancer looping and general chromatin organization to establish the observed pluripotency-specific Nanog interactome.
Nanog interactions are dependent on Mediator and cohesin subunits in ESCs
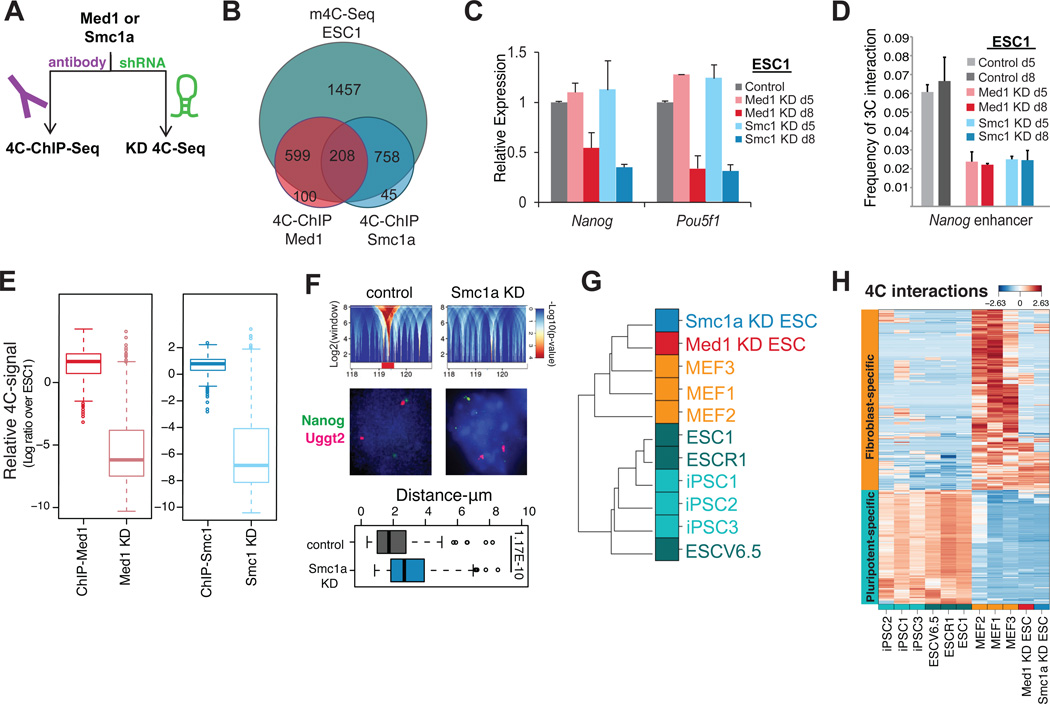
Given their role in promoter-enhancer looping and their enrichment among the Nanog-interacting regions in pluripotent cells, we next asked how many of those regions were indeed bound by the Mediator and cohesin complexes in ESCs. To this end, we performed “4C-ChIP-seq” (Figure 4A), where ChIP for the Med1 and Smc1 proteins is carried out before sequencing of the Nanog-centered m4C libraries (Figure S4A and Experimental Procedures). Loci bound by Med1, Smc1 or both accounted for about 40% of all ESC-specific interactions (Figure 4B) (Table S4). These data reinforce the results of our association analysis with published data and show that a large portion of the ESC-specific Nanog interactions involve the Mediator and cohesin complex.
Figure 4. Mediator and cohesin coordinate Nanog’s genomic interactions in pluripotent cells.
(A) Two-pronged strategy to test the role of candidate proteins in the Nanog interactome in ESCs.
(B) Venn diagram depicting the overlap of Nanog-interacting HindIII fragments detected by m4C-ChIP-seq for either Med1 or Smc1a compared to m4C-seq in ESC line ESC1.
(C) RT-PCR analysis for pluripotency genes Nanog and Pou5f1 in ESCs treated with shRNAs against Med1 or Smc1 for 5 (d5) or 8 days (d8). Error bars indicate standard deviation (n=3 technical replicates). m4C-seq analysis was performed on day 5, before downregulation of Nanog or Pou5f1 and apparent differentiation of cells.
(D) 3C-PCR quantifying the interaction frequency between the Nanog promoter and enhancer in control ESCs and in ESCs harvested 5 (d5) or 8 days (d8) after knocking down Med1 or Smc1a. For each primer pair the PCR signal was normalized to the PCR signal of primers designed at the bait locus (see Table S6). Error bars indicate standard deviations (n=3 technical replicates).
(E) Boxplot reporting the relative change in 4C-seq normalized signal of the 4C-ChIP selected fragments compared to ESC1 (log2 ratio) (whiskers like Figure 1E).
(F) Top: Domanograms details showing the interaction of Nanog with Uggt2 locus in control ESC1 and its disruption in Smc1a KD ESC1. Middle: Representative DNA FISH photos for Nanog (FITC signal) and Uggt2 (Magenta signal) in control or Smc1a knockdown ESCs. Bottom: Boxplot for distances between the Nanog and Ugg2t as measured by DNA FISH (whiskers like Figure 1E). The difference is significant (Wilcoxon test).
(G) Unsupervised clustering of samples is performed as in Figure 1B with the addition of the ESC samples for Med1 or Smc1a knock down (KD).
(H) Heatmap showing the relative change in m4C-seq signal for the set of 4C fragments selected as differential interactions between ESCs and MEFs, clearly showing that in Med1 or Smc1a knock down sample the pluripotency specific interactions have been lost. Rows refer to individual HindIII fragments and columns are different 4C-seq samples. Color refers to standardized values across samples (z-score) for log transformed normalized 4C read counts.
To test whether the Mediator complex is required for Nanog interactions, we performed m4C-seq in ESCs transduced with lentiviral vectors expressing short hairpins against Smc1 or Med1 (Figure 4A and Figure S4B and Table S6). Chromatin was isolated 5 days after viral transduction when protein levels were substantially reduced (Figure S4B) but before the onset of differentiation, as assessed by their undifferentiated morphology (Figure S4C) and the ESC-like mRNA and protein levels of several pluripotency factors (Figure 4C, Figure S4D and S4E). Importantly, Nanog’s promoter-enhancer interaction was already disrupted at day 5 of Med1 or Smc1a knockdown (KD) (Figure 4D), although Nanog transcription was still detectable by RT-PCR (Figure 4C) and by the presence of PolII Phospho-Ser2 on the Nanog promoter (Figure S4F). Med1 and Smc1a- mediated Nanog interactions were severely reduced or completely abrogated in the day 5 KD 4C-seq samples (Figure 4E). Loss of chromatin contacts was confirmed by DNA FISH for one of the interacting candidate loci (Figure 4F). RNA-seq analysis of Med1 and Smc1a KD ESCs confirmed downregulation of pluripotency-related genes and up-regulation of differentiation-related genes by day 8, while the changes were less evident on day 5 (Figure S4G). The altered transcriptional profiles of our KD cells at day 8 resembled those of previously published ESCs infected with shRNAs against Med12 (another Mediator subunit) or Smc1a (Kagey et al., 2010) (Figure S4H). The faster kinetics of differentiation upon Med12 and Smc1a KD reported in that study likely resulted from a more efficient depletion with a different vector system. Remarkably, the m4C-seq profiles of KD ESCs indicated that the majority of the ESC-specific interactions were lost (Figure 4H and S4I), while many of the MEF-specific interactions were established, presumably in a Med1/Smc1a-independent manner (Figure 4G, H). Thus, Smc1a and Med1 depletion led to rearrangement of chromatin from a pluripotent- to a differentiation-specific state, even though cells still showed phenotypic and transcriptional features of the pluripotent state.
The Nanog interactome undergoes dramatic changes during somatic cell reprogramming
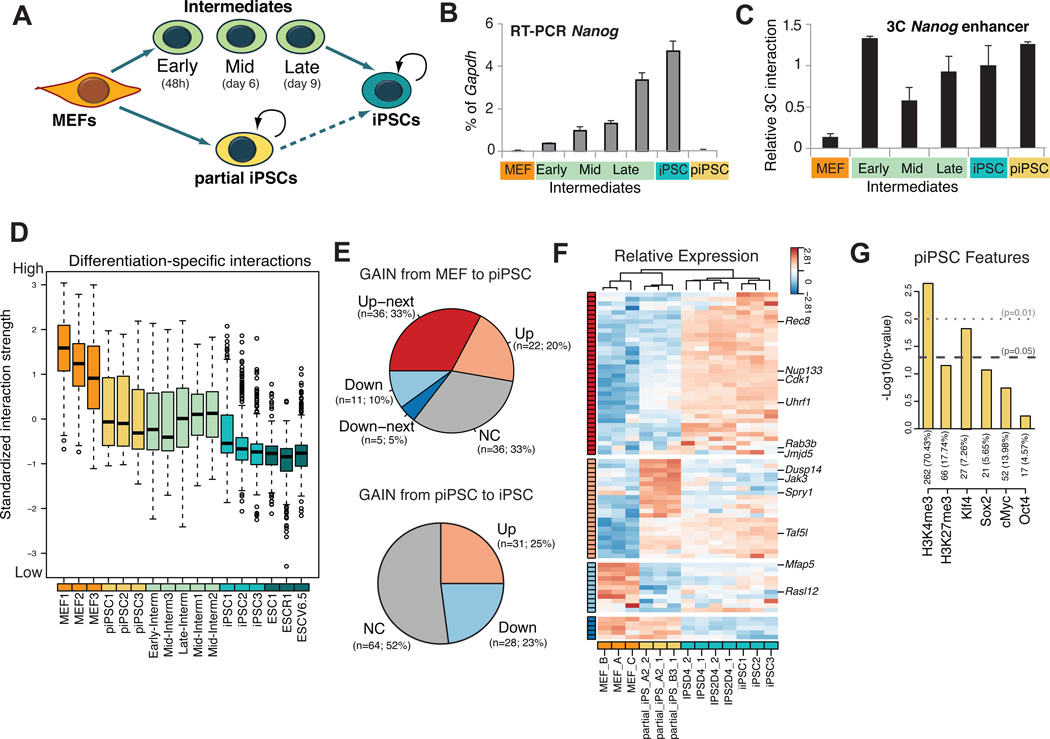
As iPSCs have reset the Nanog interactome from a somatic to a pluripotent state, we asked when chromatin rearrangements occur during reprogramming and how these relate to gene expression changes. Specifically, we compared the kinetics of chromatin looping with gene expression using partially reprogrammed iPSC lines (piPSCs) and sorted SSEA1+ intermediates at different stages of reprogramming (Figure 5A). Importantly, both piPSCs and SSEA1+ intermediates have exited the somatic state and are poised to forming iPSCs under different conditions, consistent with previous observations (Figures S5A and S5B) (Sridharan et al., 2009; Stadtfeld et al., 2008). In further agreement with those previous reports, we found that Nanog is not yet expressed in piPSCs, whereas it is gradually upregulated during mid-to-late stages of reprogramming (Figure 5B). Surprisingly, 3C analysis revealed that looping between the Nanog enhancer and promoter was established in both piPSC and in SSEA1+ intermediates before detectable transcriptional activation of Nanog (Figure 5C). We extended this analysis by performing 3C analysis in piPSCs for Oct4, Phc1 and Lefty1, which form promoter-enhancer loops in ESCs (Figure S5C) (Kagey et al., 2010). While Phc1 already exhibited looping and expression in piPSCs, Oct4 had neither initiated looping nor activated expression. In contrast, Lefty1 had initiated looping, but not yet expression. These results support the conclusion that the looping at the examined pluripotency-associated genes precedes, but is not sufficient for, transcriptional activation in the context of cellular reprogramming.
Figure 5. Dynamic change of Nanog interactome during cellular reprogramming into iPSCs.
(A) Isolation and study of reprogramming intermediates and partially reprogrammed cells (piPSCs).
(B) RT-PCR analysis for Nanog mRNA in each cell type. The Nanog expression is normalized over Gapdh (% of Gapdh). The error bars indicate standard deviation (n=3 technical replicates).
(C) 3C analysis of relative interaction frequency between the Nanog promoter and enhancer during reprogramming and in the partial iPSCs. The PCR signal is relative to ESCs (Relative 3C Interaction) after normalization with bait locus primers (see Table S6). Error bars are standard deviation (n=3 technical replicates).
(D) Boxplot for standardized interaction strength for differentiation-specific fragments (whiskers like Figure 1E). The fragments were selected as differential fragments up-regulated in MEFs vs. ESCs. Five groups of samples are shown: ESCs, iPSCs, SSEA1+ intermediates, partially reprogrammed iPSCs (piPSC) and MEFs. SSEA1-intermediates and piPSCs show an intermediate interaction strength between stronger MEFs and weaker ESCs/iPSCs. For each fragment, the log transformed normalized 4C read counts are standardized by subtracting the mean value across all of the samples, then dividing over standard deviation (z- score) (see also Figure S5D).
(E) Pie charts showing the number of genes, which have established (gain) interactions with Nanog during the transition from MEFs to piPSCs (upper panel) or from piPSC to iPSC (lower panel). Genes are grouped based on the change of expression detected by microarray data (FDR 0.05, fold change 1.3) (Sridharan and Hochedlinger datasets – Table S3 and Figure S5G). Up/Down are up-/down-regulated in the transition from MEFs to piPSCs (upper panel) or from piSPC to iPSC (lower panel); Up-/Down-next (ONLY for the upper panel) are Up-/Down-regulated in the next stage, i.e. the transition from piPSCs to iPSCs (see also panel F); NC is for genes without statistically significant change in expression. The number of genes and percentage over the total are indicated. We found significant enrichment in the “Up-next” group (one tail Fisher test p 0.001). Gene level interactions detected in all piPSC replicates and in none of the MEFs were used. Alternative selection of differentially or piPSC specific (transient) interacting genes supported the same conclusions.
(F) Heatmap showing in expression of Nanog-interacting genes gained in the MEF to piPSC transition as in (E). Rows are genes and columns are microarray samples (Table S3). Expression pattern groups were defined as in (E) and marked accordingly with the side color bar. Some genes showed significant up-regulation in both the MEFs to piPSCs and in the piPSCs to iPSCs transitions. In this case they were assigned to the “Up-next” group as well. The statistically significant enrichment in “Up-next” pattern is confirmed even if these genes are assigned to the “Up” group. The heatmap shows standardized gene expression levels across samples (z-score).
(G) Association of conserved Nanog-interacting genes in piPSCs with H3K4me3, H3K27me3 and pluripotency TFs binding in the same cell type. Number and percentage of interacting genes with ChIP enrichment is reported for each bar. Similar analysis criteria as in Figure 3.
At a genome-wide scale, m4C-seq analysis of piPSCs and SSEA1+ intermediates showed that both cell populations had lost a large fraction of the MEF-specific interactions and had gained a small number of ESC-specific interactions (Figure 5D, S5D, S5E). Unexpectedly, we also observed a number of reprogramming-specific interactions detectable neither in MEFs nor in iPSCs (Table S5). Transient interactions were variable among SSEA1+ samples from independent reprogramming experiments, probably reflecting the heterogeneity of the SSEA1+ population (see single-cell RT-PCR of Figure S5F and (Polo et al., 2012)). We therefore focused on piPSCs, which are of clonal origin and hence more homogeneous. Notably, these transient interactions in piPSCs (Table S5) were preferentially associated with pluripotency-rather than differentiation- related genes (p-value=0.014). Thus, forced expression of reprogramming factors readily extinguished fibroblast-specific interactions and induced a large number of transient chromatin interactions enriched for pluripotency-associated genes.
We next correlated the reorganization of Nanog’s interactome during reprogramming with transcriptional changes of associated genes. Notably, more than 50% of genes that established interactions with Nanog during the transition of MEFs into piPSCs became transcriptionally upregulated in piPSCs (“Up”), or at the subsequent (iPSC) stage (“Up-next”) (Figures 5E, 5F and Figure S5G). These results extend, at a genome-wide level, our previous observations that the gain of Nanog-centered chromatin contacts during early reprogramming coincides with or precedes transcriptional changes of genes. Unexpectedly, the interactions gained during the piPSC-to-iPSC transition showed a weaker correlation with transcriptional changes, suggesting lesser impact of Nanog interactions on gene expression during the late stages of reprogramming. We conclude that Nanog’s chromatin associations during early stages of reprogramming mostly involve genes that are either immediately upregulated or poised for activation in iPSCs.
To investigate which molecules might mediate Nanog’s interactions during reprogramming, we compared m4C-seq results on piPSCs with published ChIP-chip data of reprogramming factors and histone modifications in the same cell type (Sridharan et al., 2009). This analysis revealed a positive correlation with the active histone mark H3K4me3 and a significant association of Nanog’s interacting loci with Klf4 binding, further supporting its possible role in regulating long-range chromatin interactions (Figure 5G). Thus, forced expression of Oct4, Sox2, Klf4 and c-Myc induces reorganization of chromatin architecture and facilitates interactions of the Nanog locus with other Klf4 target genes, as well as with open chromatin domains.
Reprogramming factors and Mediator cooperate during the establishment of Nanog-centered interactions
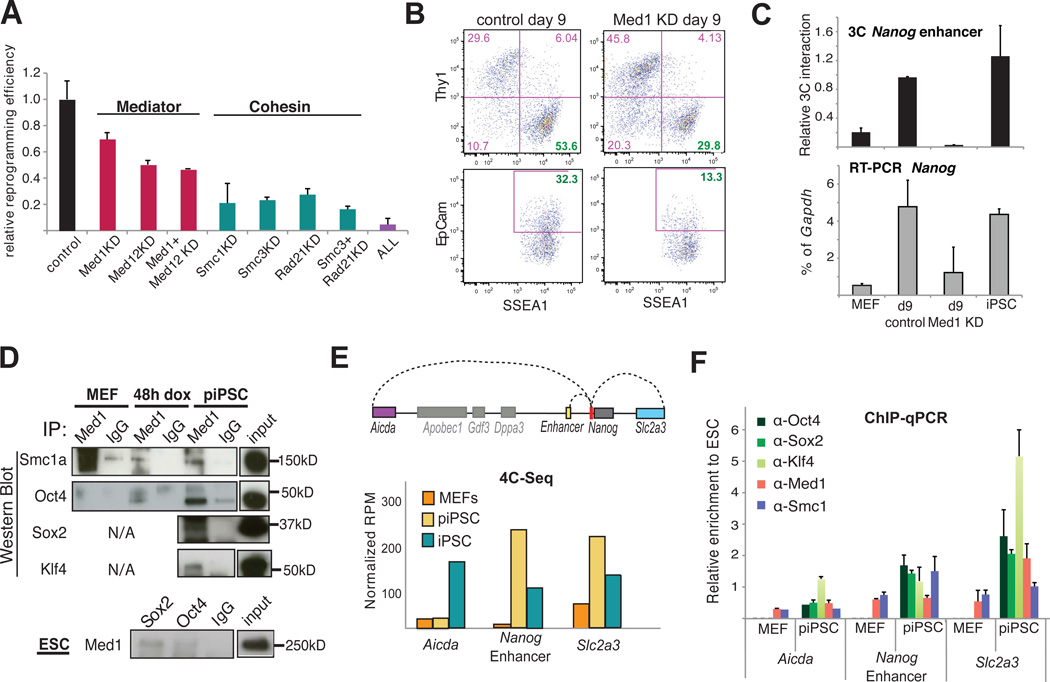
To investigate whether Mediator and cohesin are involved in the acquisition of pluripotency, we assayed the potential to generate iPSCs from reprogrammable MEFs when subunits of Mediator (Med1, Med12) and/or cohesin (Smc1a, Smc3, Rad21) were depleted (Figure S6A). Indeed, knockdown of Mediator and/or cohesin components significantly decreased reprogramming efficiencies (Figure 6A).
Figure 6. Role of Mediator and cohesin on the reprogramming of MEFs to iPSCs.
(A) Graph comparing the reprogramming efficiency of tetO-OKSM MEFs after infection with empty vector (control) or or shRNA vectors (KD) against individual subunits of Mediator (Med1, Med12) or cohesin (Smc1a, Smc3, Rad21) complexes or combinations of them. The efficiency was calculated as a ratio of AP-positive colonies to the starting number of cells, expressed as a fraction of the analogous ratio for the control MEFs. Error-bars indicate standard deviation (n=3 biological replicates)
(B) FACS plots of SSEA1 positive or EpCam positive cells on day 9 of reprogramming starting with either wild type (left) or Med1-knocked down (KD, right) reprogrammable MEFs. SSEA1 and EpCam are picked as early or late, respectively, surface markers of pluripotency.
(C) RT-PCR (bottom) for Nanog expression and 3C assay (top) for Nanog enhancer-promoter interaction in MEFs, iPSCs and reprogramming Intermediates of control or Med1 knockdown MEFs (Med1 KD) on day 9. 3C PCR signal was calculated relative to ESCs (Relative 3C Interaction) after normalization with bait locus primers (Table S6). Error bars are standard deviation (n=2 technical replicates). RT-PCR Nanog signal was normalized to Gapdh levels and the error bars indicate standard deviation (n=4 replicates).
(D) Med1 protein immunoprecipitation (upper panels) in reprogrammable MEFs before (MEF) and after doxycyclin induction (MEF 48hr) and in partial iPSC (piPSC). In the bottom panel, the interaction of Med1 with Oct4, Sox2 and Nanog was also confirmed in ESCs, this time using antibodies for the reprogramming factors for the pull down.
(E) Schematic representation of the genomic regions found to interact in cis with the Nanog promoter (red) in a pluripotent specific way (top). Barplot of m4C-seq signal for each of the indicated regions in MEFs, partial iPSC (piPSC) and ESCs. The signal is expressed in reads per million (RPM) and represents the average value of 3 biological replicates.
(F) Chromatin Immunoprecipitation (ChIP) experiments of the reprogramming factors Oct4, Sox2 and Klf4 as well as Med1 and Smc1a on the indicated genomic regions in MEFs and partial iPSCs (piPSC). All the ChIP-qPCR signals are first normalized to the input and then, expressed relative to the corresponding signal in ESCs (See also Figure S6). Error bars indicate standard deviation.
Fewer iPSC colonies upon knockdown of Mediator and cohesin components could result either from deficient reprogramming or from immediate differentiation of newly formed iPSCs. To distinguish between these possibilities, we analyzed early (SSEA1) and late (EpCam) markers of pluripotency at intermediate stages of reprogramming (Polo et al., 2012). We focused on Med1 KD cells since Med1 is expressed most differentially between somatic and pluripotent cells (Figure S6B) (Kagey et al., 2010; Polo et al., 2012). Figure 6B shows that Med1 KD MEFs gave rise to fewer SSEA1+ and EpCam+ reprogramming intermediates at day 9 of reprogramming factor overexpression. 3C analysis at this time point showed that Nanog promoter-enhancer looping was not efficiently established in the absence of Med1, concordant with decreased transcription (Figure 6C). Together, these data suggest that Med1 is important for acquiring pluripotency-specific chromatin loops and gene expression in addition to its established role in the maintenance of pluripotency.
We hypothesized that Med1 might cooperate with reprogramming factors to reorganize 3D chromatin architecture and to control gene expression during iPSC formation. Co-immunoprecipitation experiments in piPSCs showed association of Med1 with the reprogramming factors Oct4, Sox2 and Klf4 in (Figure 6D), as well as with Med12 and Smc1 (Figure S6C), which have been previously reported to interact with Med1 in ESCs (Borggrefe and Yue, 2011; Kagey et al., 2010). Importantly, these protein-protein interactions were detected as early as 48h after expression of the reprogramming factors, suggesting an early function. Med1 interactions with Oct4 and Sox2 were also confirmed in ESCs (Figure S6C). These results indicate that Mediator components and pluripotency factors form a multiprotein complex throughout cellular reprogramming and in pluripotent cells.
Lastly, we asked how reprogramming factors and Mediator/cohesin might collaborate to form chromatin loops during reprogramming. We investigated the binding of these proteins to three regions (Aicda, Nanog enhancer and Slc2a3) found to interact with the Nanog promoter in pluripotent cells based on m4C-seq data (Figure 6E). This analysis showed that Klf4, Oct4, Sox2, Med1 and Smc1 were bound to all three loci in pluripotent cells (Figure S6D). Similarly, the loci that had already established chromatin loops with the Nanog promoter (Nanog enhancer and Slc2a3) in piPSC lines were occupied by all tested factors (Figure 6F). In contrast, Aicda, which interacted with Nanog promoter only in established iPSCs but not yet in piPSCs, was bound solely by Klf4 in piPSC. This result suggests that a minimum set of pluripotency proteins may be required by cohesin and Mediator to bridge distal chromatin elements.
DISCUSSION
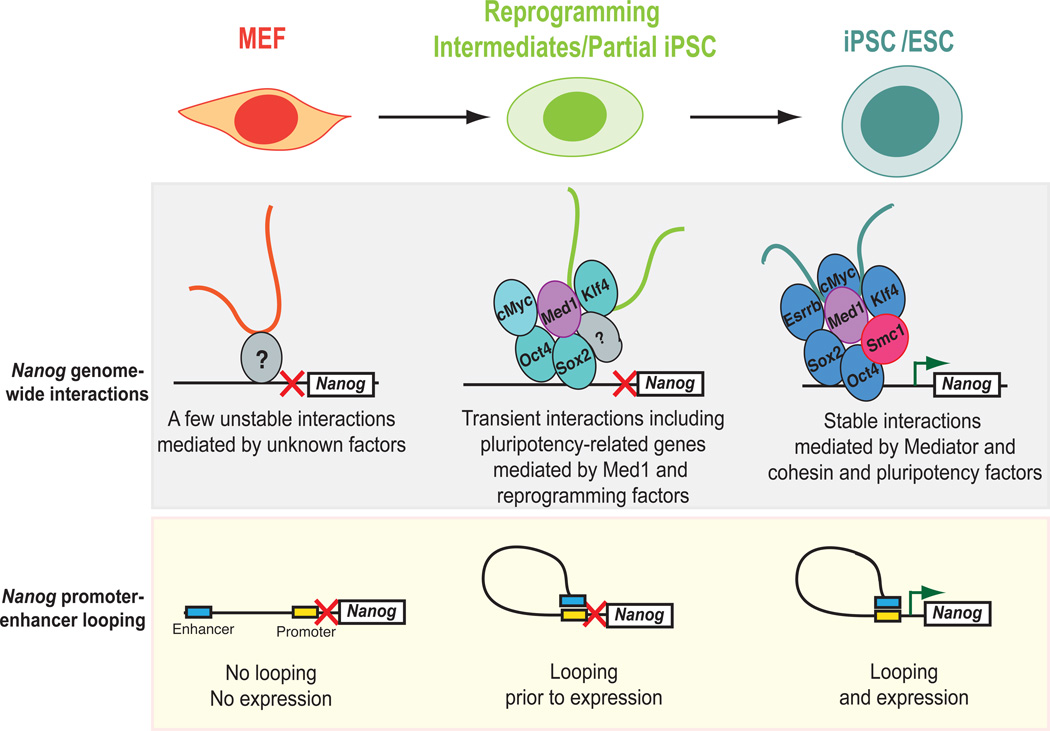
Herein, we provide genetic, biochemical and bioinformatic evidence that Nanog engages in a pluripotency-specific genome-wide chromatin network that resolves into a somatic-specific pattern upon differentiation and is reset in iPSCs (Figure 7). This is the first genome-wide interaction map of a key mouse pluripotency gene at high resolution. Our results extend previous genome-scale transcription factor occupancy and protein interaction studies for pluripotency factors (Chen et al., 2008; Kim et al., 2008) and reveal an unexpectedly complex genomic interactome in pluripotent cells.
Figure 7. Model depicting dynamics of Nanog interactions during differentiation and cellular reprogramming.
The Nanog locus engages in genome-wide chromatin interactions in MEFs (“MEF-specific interactome”) that are highly variable, possibly because the Nanog gene is inactive in differentiated cells. During reprogramming, the complexity of interactions increases, presumably by the cooperative action of the overexpressed reprogramming factors and “bridging” factors, including Mediator components (Med1). The majority of the interactions gained in the partial iPSC lead to upregulation of the involved genes immediately or in iPSCs. Once cells reach the pluripotent state, different and more stable interactions are established. These pluripotency-specific interactions are mainly maintained by cohesin and Mediator complexes as well as the key pluripotency factors. Upon normal differentiation or depletion of either Med1 or Smc1a, the Nanog-interactome is rearranged into the less organized differentiated state.
We document Nanog promoter interactions with individual loci as well as broader domains on the same and on different chromosomes. These interactions were stable and conserved among different pluripotent cell lines, whereas they were less consistent in MEFs (Figure 7). This finding indicates that pluripotency loci might engage in less stable and/or more random interactions in cell types where the bait locus is inactive. Alternatively, it may reflect the heterogeneity of fibroblast populations, which were used as a proxy for differentiated cells. Of note, almost half of the conserved interactions found in MEF samples were also detected in pluripotent cells, indicating a cell-type independent network of presumably structural interactions.
A positive correlation between Nanog-centered interactions and active chromatin marks specifically in pluripotent cells is in accordance with previous studies showing that active genes tend to colocalize in the genome (Gao et al., 2013; Kalhor et al., 2012; Simonis et al., 2006). Notably, binding sites for the key pluripotency factors Oct4, Sox2, Nanog, Esrrb, c-Myc and Klf4 were also enriched among the Nanog-interacting genes in pluripotent cells (Figure 7), suggesting that these proteins might be involved in bringing co-regulated pluripotency-associated genes into physical proximity for subsequent transcriptional activation during the induction and maintenance of pluripotency. Indeed, previous studies documented roles for Oct4 in the maintenance of cis DNA loops around Nanog (Levasseur et al, 2008), for c-Myc in the spatial organization of ribosomal RNA genes in other cell types (Shiue et al., 2009) and for Klf1 in long-range interactions of erythroid genes during blood cell development (Schoenfelder et al., 2010). It is worth mentioning here that forced expression of either of c-Myc, Nanog, Esrrb or Klf4 proteins relieves ESCs from LIF-dependent growth (Festuccia et al., 2012; Jiang et al., 2008; Marks et al., 2012; Smith and Dalton, 2010; Smith et al., 2010), suggesting that the observed interaction network and its constituents may also be functionally connected.
We provide evidence that members of the Mediator/cohesin families are responsible for about 40% of the observed interactions in ESCs. Their depletion from ESCs resulted in a rearrangement of chromatin from a pluripotent to a differentiated state before the transcriptional and phenotypic onset of differentiation. Similarly, their reduction during cellular reprogramming impaired iPSC colony formation, suggesting an additional role in establishing pluripotency. Our observation that Med1 physically associated with the overexpressed Oct4, Sox2 and Klf4 factors during reprogramming and with the corresponding endogenous proteins in established ESCs supports this interpretation and extends previous results on direct interactions of cohesin and Mediator subunits with Oct4 and Nanog in ESCs (Costa et al., 2013; Nitzsche et al., 2011; Tutter et al., 2009; van den Berg et al., 2010). Our results suggest that Mediator and cohesin components, in collaboration with pluripotency transcription factors, play a critical role in establishing and maintaining a broader 3D chromatin network centered around Nanog and possibly other pluripotency loci (Figure 7). We cannot exclude that Mediator and cohesin influence iPSC formation and ESC maintenance by additional mechanisms such as cell cycle, cell signaling (Rocha et al., 2010), mesenchymal-to-epithelial transition (Huang et al., 2012) and/or transcriptional regulation (Malik and Roeder, 2010; Wood et al., 2010).
Lastly, we document that the reprogramming of somatic cells into iPSCs resets Nanog’s chromatin interactome. We show that fibroblasts rapidly lose MEF-specific interactions upon overexpression of Oct4, Sox2, Klf4 and c-Myc, while they gradually establish pluripotency-specific interactions. This is in accordance with the transcriptional shutdown of the somatic program prior to the activation of the pluripotency program as described recently (Polo et al., 2012; Soufi et al., 2012; Stadtfeld et al., 2008). Unexpectedly, we detected a number of transient, reprogramming-specific contacts, which involved many pluripotencyrelated genes (Figure 7). These genes might be physically brought together with Nanog by forced reprogramming factor expression for coordinated gene activation. The observed protein-protein interactions of Oct4, Sox2 and Klf4 with Med1 in piPSCs support a model whereby reprogramming factors and associated bridging factors act synergistically to orchestrate chromatin rearrangements during reprogramming (Figure 7). However, we cannot rule out the possibility that these interactions might be the consequence of global chromatin changes or aberrant binding of the overexpressed transcription factors during reprogramming (Soufi et al., 2012).
Collectively, our data provide a comprehensive analysis of genomic interactions of a key pluripotency gene and their relation with transcription, epigenetic marks and pluripotency factor binding. Our findings further suggest an important and possibly causative role for chromatin structure in controlling transcriptional patterns and eventually determining cell identity in the context of pluripotency, differentiation and cellular reprogramming. Identifying the interactomes for other pluripotency loci should allow researchers to construct an integrative view of 3D chromatin architecture in pluripotent cells in the future.
EXPERIMENTAL PROCEDURES
Cell Culture and Reprogramming
ESCs, MEF-derived iPSCs (Stadtfeld et al., 2010a) and partial iPSCs (Maherali et al., 2007) were cultured as described before. MEFs were isolated from “reprogrammable” mouse (Stadtfeld et al., 2010b) and reprogrammed in presence of 1ug/ml doxycycline and 50ug/ml ascorbic acid.
shRNA virus production and infection
The shRNA lentiviruses for Med1 and Smc1a were designed according to previous study (Kagey et al., 2010) and cloned into a different vector (Addgene-pSicoR-GFP). The virus production, transduction and reprogramming of infected MEFs are described in Supplemental Experimental Procedures. All the shRNA sequences used for this study are shown in Table S6.
RNA-seq library preparation
The RNA-seq library construction is described in the Supplemental Experimental Procedures.
Protein Co-immunoprecipation (IP)
The antibodies used for this study were: Med1 (Bethyl Laboratories), Smc1 (Bethyl Laboratories), Oct4 (Santa cruz for Western and R&D for IP), Sox2 (R&D), Klf4 (R&D), Nanog (Bethyl Laboratories), actin-HPRT (abcam), Med12 (Bethyla Laboratories), Smc3 (abcam), Rad21 (Santa-Cruz). The exact process is described in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation
The Chromatin Immunoprecipitation was performed as described previously (Stadtfeld et al., 2012) The antibodies used were: Oct4 (R&D), Sox2 (R&D), Klf4 (R&D), Med1 (Bethyl Laboratories), Smc1 (Bethyl Laboratories), IgG (abcam), PolII phospho-Ser2 (abcam). The primers used for the qPCR analysis are listed in Table S6.
3D-DNA FISH and image analysis
3D-DNA FISH analysis was performed as described previously (Xu et al., 2006). The protocol and the BAC clones used for this study are listed in the Supplementary Experimental Procedures.
Modified 4C-seq (m4C-seq), m4C-ChIP-seq and 3C analyses
4C and 3C were performed as has been previously described (Schoenfelder et al., 2010) with some modifications described in detail in Supplementary Experimental Procedures. For m4C-ChIP-seq an immunoprecipitation step with anti-Med1 and anti-Smc1 (Bethyl) antibodies was included. The primers used for these assays are listed in Table S6.
Bioinformatics analyses of m4C-seq and associations to public datasets
Data Availability
All sequencing data are available in SRA (accession number: SRA051554).
Supplementary Material
HIGHLIGHTS.
The Nanog promoter forms a pluripotency-specific genome-wide chromatin interactome
Nanog-interactions in ESCs are enriched for active marks and pluripotency factors
Mediator and cohesin are essential for the maintenance of the Nanog interactome
Many Nanog interactions form before increased gene expression during reprogramming
ACKNOWLEDGEMENTS
We acknowledge Peter Fraser, Frank Grosveld, Jane Skok and Job Dekker for comments and suggestions. We thank Mariann Miscinai and Triantiafyllos Paparountas for discussing bioinformatics analysis; Eda Yildirim and Berni Peyer for advice about FISH experiments; all members of the Hochedlinger and Park groups for suggestions. E.A. was supported by Jane Coffin Childs postdoctoral fellowship. P.J.P. was supported by Sloan Research Fellowship and NIH (RC2HL102815), P.V.K was supported by NIH (K25AG037596) and K.H. was supported by NIH (DP2OD003266 and R01HD058013).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information contains 6 Figures and Supplemental Experimental Procedures and can be found with this article online.
REFERENCES
- A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega V, Wong E, Orlov Y, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1123. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013 doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe M, Silva S, Pinter S, Xu N, Lee J. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–160. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim Y, Lee C, Shendure J, Fields S, Blau C, Noble W. A three-dimensional model of the yeast genome. Nature. 2010;465:363–370. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JD, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell stem cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wei Z, An W, Wang K, Lu W. The interactomes of POU5F1 and SOX2 enhancers in human embryonic stem cells. Sci Rep. 2013;3:1588. doi: 10.1038/srep01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell stem cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, Lyou Y, Townes T, Sch√°beler D, Gilbert D. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS biology. 2008;6 doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C, Prahallad A, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur D, Wang J, Dorschner M, Stamatoyannopoulos J, Orkin S. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes & development. 2008;22:575–655. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum N, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie B, Sabo P, Dorschner M, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, NY) 2009;326:289–382. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein B, Jaenisch R, Lander E, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–104. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, Janssen-Megens E, Hubner N, Schulz H, de Vries I, Ding L, Huebner N, Mann M, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PloS one. 2011;6 doi: 10.1371/journal.pone.0019470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman S, Solovei I, Brugman W, Gr√§f S, Flicek P, Kerkhoven R, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Molecular cell. 2010;38:603–616. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Micsinai M, Kim JR, Hewitt SL, Souza PP, Trimarchi T, Strino F, Parisi F, Kluger Y, Skok JA. Close proximity to Igh is a contributing factor to AID-mediated translocations. Mol Cell. 2012;47:873–885. doi: 10.1016/j.molcel.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137:2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope N, Horton A, Andrews S, Kurukuti S, Mitchell J, Umlauf D, Dimitrova D, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nature genetics. 2010;42:53–114. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shiue CN, Berkson RG, Wright AP. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene. 2009;28:1833–1842. doi: 10.1038/onc.2009.21. [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Homminga I, Galjaard R-J, Rijkers E-J, Grosveld F, Meijerink JP, de Laat W. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nature methods. 2009;6:837–879. doi: 10.1038/nmeth.1391. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nature genetics. 2006;38:1348–1402. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5:947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell stem cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010a;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010b;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell stem cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E, Kadam S. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284:3709–3718. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- van den Berg D, Snoek T, Mullin N, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot R. An Oct4-centered protein interaction network in embryonic stem cells. Cell stem cell. 2010;6:369–450. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tsai C-L, Lee J. Transient homologous chromosome pairing marks the onset of X inactivation. Science (New York, NY) 2006;311:1149–1201. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available in SRA (accession number: SRA051554).