Abstract
The genus Echinacea is used as an herbal medicine to treat a variety of ailments. To better understand its potential chemical variation, 40 Echinacea accessions encompassing broad geographical and morphological diversity were evaluated under controlled conditions. Metabolites of roots from these accessions were analyzed by HPLC-photo diode array (HPLC-PDA), GC-MS, and multivariate statistical methods. In total, 43 lipophilic metabolites, including 24 unknown compounds, were detected. Weighted principal component analysis (WPCA) and clustering analysis of the levels of these metabolites across Echinacea accessions, based on Canberra distances, allowed us to test two alternative taxonomic treatments of the genus, with the further goal of facilitating accession identification. A widely used system developed by McGregor based primarily on morphological features was more congruent with the dendrogram generated from the lipophilic metabolite data than the system more recently developed by Binns et al. Our data support the hypothesis that Echinacea pallida is a diverse allopolyploid, incorporating the genomes of Echinacea simulata and another taxon, possibly Echinacea sanguinea. Finally, most recognized taxa of Echinacea can be identified by their distinct lipophilic metabolite fingerprints.
Keywords: Echinacea (Asteraceae), alkamides, alkylamides, HPLC, GC-MS, PCA
Introduction
Echinacea extracts, particularly from roots, have historically been used by Native Americans and, more recently, by Western cultures as herbal remedies to treat ailments ranging from snake bites to pain, burns, cough, sore throats, and toothache [1], [2], [3]. Echinacea products are promoted for immune system enhancement and are among the best-selling herbal preparations in the United States [4]. Many unusual phytochemicals have been found in Echinacea, some with reports of bioactivity in animal cells or animals, including alkamides and ketones, caffeic acid derivatives, glycoproteins, and unusual polysaccharides (as reviewed by Bauer [5]). Despite its popularity as an herbal dietary supplement, and many pertinent pharmacological and clinical studies, little is known regarding the specific compounds primarily responsible for observed bioactivity or whether they are consistently efficacious in humans [6].
Although morphologically distinct Echinacea groups exhibit differences in phytochemical composition [5] and bioactivity [7], most studies have focused on three major medicinal species: Echinacea angustifolia DC., E. pallida (Nutt.) Nutt., and E. purpurea (L.) Moench. The most comprehensive study to date was conducted by Binns et al. [8], who reported on phytochemical variation in Echinacea from roots and capitula of wild and cultivated populations representing all nine Echinacea species recognized by McGregor [9]. However, in that report, most alkamides and ketones were identified by UV spectra and relative retention times compared with one major pair of alkamide standards. Baum et al. [10], in a recent review of the status of Echinacea systematics and phytochemistry, indicated that Echinacea taxa are readily distinguishable on the basis of HPLC profiles and that HPLC profiles for lipophilic compounds contain more information than those based on caffeic acid derivatives. This is noteworthy because of the existence of two alternative taxonomic treatments for Echinacea. The older, developed by McGregor [9], was based on field observations, common garden studies, and cytological and anatomical analyses. McGregor's classification has been widely used by botanists and herbalists [11] and serves as the basis for the recent Flora North America treatment [12]. Binns et al. [13] proposed a revision, based on morphometric data and phytochemical data from greenhouse-grown and wild plants, using canonical discriminant and cladistic analyses. This revision recognizes all but one of McGregor's taxa, the most significant changes being a reduction in the number of species, an increase in the number of varieties, and, in particular, the incorporation of 5 morphologically diverse clades characterized as species by McGregor [9] into a single species. This Binns et al. [13] revision is controversial in the botanical community [11].
The current DNA-based molecular marker evidence is not yet refined sufficiently to generate accession-level systematics [14], [15]. Even an extensive DNA-sequencing study using these identical accessions and based on multiple loci has thus far been unable to completely evaluate systematic relationships among these accessions [16].
Here we have taken a targeted, metabolite-profiling approach to investigate the accumulation of putatively bioactive alkamides and ketones in 40 geographically and morphologically diverse Echinacea populations, which already had been well characterized morphologically and as to origin. We used as standards authentic alkamides and ketones that were purchased or synthesized by our group [17], [18], [19], as well as structural information obtained by a combination of HPLC-PDA and GC-MS, for more comprehensive compound identification. Because we lacked reference standards for many of the metabolites reported in this study (43 in all), we used relative instead of absolute metabolite concentrations to compare overall lipophilic-metabolite profiles across the 40 accessions. This approach, coupled with weighted principal component analysis (WPCA) and clustering analysis based on Canberra distances [20], provides an opportunity to test these two taxonomic classifications. Furthermore, these metabolic profiles may help standardize Echinacea products, characterize plant material of unknown provenance, and identify genetic sources to select for increased production of desired compounds.
Materials and Methods
Plant materials
We selected 40 well-characterized Echinacea accessions (Table 1) representing a broad geographic and morphological sampling of the germplasm conserved by the U.S. National Plant Germplasm System, USDA-ARS North Central Regional Plant Introduction Station, Ames, Iowa [21]. Initially we looked at roots of two ages of plants: 6-month-old and 3-year-old. We found that the relative levels of metabolites vary, but the same identified alkamides, ketones, and unknown metabolites are present at both ages (Fig. 1S, Supporting Information); therefore, we focused on 6-month-old plants because we are able to grow them under well-controlled conditions. Characterization data for a broad range of (> 40) morphological traits are available at Germplasm Resources Information Network database (http://www.ars-grin.gov/cgi-bin/npgs/html/desc_form.pl?221). Accessions were keyed to species (or subspecies) during initial regeneration on the basis of McGregor [9], and we converted McGregor identifications to the treatment of Binns et al. [13] via Table 2 in [10]. Growth conditions and sampling methods are available in the Supporting Information.
Table 1.
Accession information for Echinacea evaluated in this study
| Taxon (sensu McGregor) [9] | Taxon (sensu Binns et al.) [13] | Accession number | Location of source population |
|---|---|---|---|
| E. angustifoliaa | E. pallida var. angustifolia | PI631267 | Murray County, OK |
| E. angustifolia var. angustifolia | E. pallida var. angustifolia | PI631272 | Comanche County, OK |
| E. angustifolia var. angustifolia | E. pallida var. angustifolia | PI631285 | Lyon County, IA |
| E. angustifolia var. angustifolia | E. pallida var. angustifolia | PI631318 | Rooks County, KS |
| E. angustifolia var. strigosa | E. pallida var. angustifolia | PI631266 | Murray County, OK |
| E. angustifolia var. strigosa | E. pallida var. angustifolia | PI631320 | Pontotoc County, OK |
| E. atrorubens | E. atrorubens var. atrorubens | PI631255 | Douglas County, KS |
| E. atrorubens | E. atrorubens var. atrorubens | PI631260 | Bryan County, OK |
| E. atrorubens | E. atrorubens var. atrorubens | PI631262 | Murray County, OK |
| E. atrorubens | E. atrorubens var. atrorubens | PI631299 | Osage County, KS |
| E. laevigata | E. laevigata | PI631310 | Oconee County, SC |
| E. laevigata | E. laevigata | PI631312 | Oconee County, SC |
| E. laevigata | E. laevigata | PI631314 | Granville County, NC |
| E. laevigata | E. laevigata | PI631316 | Franklin County, VA |
| E. pallida | E. pallida var. pallida | PI631275 | Osage County, OK |
| E. pallida | E. pallida var. pallida | PI631290 | Sac County, IA |
| E. pallida | E. pallida var. pallida | PI631293 | Stone County, AR |
| E. pallida | E. pallida var. pallida | PI631296 | Taney County, MO |
| E. pallida | E. pallida var. pallida | PI631315 | Granville County, NC |
| E. paradoxa var. neglecta | E. atrorubens var. neglecta | PI631263 | Murray County, OK |
| E. paradoxa var. neglecta | E. atrorubens var. neglecta | PI631264 | Murray County, OK |
| E. paradoxa var. neglecta | E. atrorubens var. neglecta | PI631265 | Murray County, OK |
| E. paradoxa var. paradoxa | E. atrorubens var. paradoxa | PI631292 | Stone County, AR |
| E. paradoxa var. paradoxa | E. atrorubens var. paradoxa | PI631301 | Camden County, MO |
| E. paradoxa var. paradoxa | E. atrorubens var. paradoxa | PI631321 | Camden County, MO |
| E. purpurea | E. purpurea | PI631307 | Franklin County, MO |
| E. purpurea | E. purpurea | PI631313 | Madison County, NC |
| E. purpurea | E. purpurea | PI633669 | Caldwell Parish, LA |
| E. sanguinea | E. pallida var. sanguinea | PI631257 | Vernon Parish, LA |
| E. sanguinea | E. pallida var. sanguinea | PI631258 | Vernon Parish, LA |
| E. sanguinea | E. pallida var. sanguinea | PI633672 | Bienville Parish, LA |
| E. simulata | E. pallida var. simulata | PI631249 | Grayson County, KY |
| E. simulata | E. pallida var. simulata | PI631304 | Franklin County, MO |
| E. simulata | E. pallida var. simulata | PI631308 | Rutherford County, TN |
| E. tennesseensis | E. pallida var. tennesseensis | PI631250 | Wilson County, TN |
| E. tennesseensis | E. pallida var. tennesseensis | PI631324 | Wilson County, TN |
| E. tennesseensis | E. pallida var. tennesseensis | PI631325 | Wilson County, TN |
| E. tennesseensis | E. pallida var. tennesseensis | PI631326 | Wilson County, TN |
| Putative hybrid population involving E. paradoxa var. paradoxa and E. pallida | n/ab | PI631294 | Stone County, AR |
| Putative hybrid population involving E. paradoxa var. paradoxa and E. simulata | n/ab | PI631306 | Franklin County, MO |
Extraction, HPLC, and GC-MS analysis
Plant extraction, HPLC, and GC-MS analyses were performed as in [17], [18], and [22].
Compound identification and relative abundance
In addition to 19 known alkamides and ketones, another 24 unknown lipophilic metabolites were detected and grouped according to their retention times and UV spectra (Table 1S, Supporting Information). Methods for determining the relative abundance of metabolites are provided in Supporting Information.
Statistical analysis
Multivariate analyses by WPCA and hierarchical clustering analysis were performed in R software, version 2.2.1 (http://www.r-project.org/). Detailed information is provided in the Supporting Information.
Supporting information
Detailed methods and additional data are available as Supporting Information.
Results and Discussion
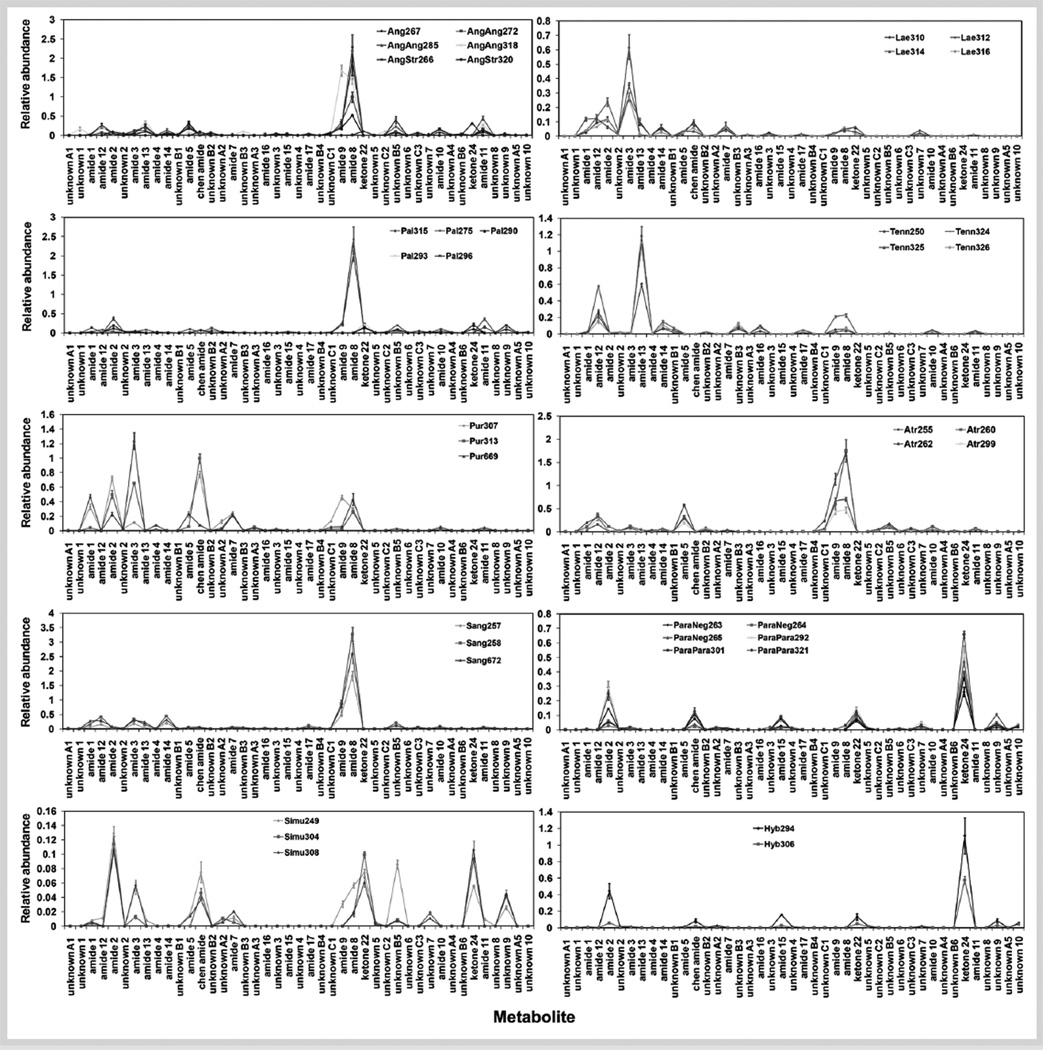
Most Echinacea extractions use dry materials and rigorous methods that last 1–24 hours (e. g., soxhlet extraction [23], ultrasonic extraction [24], and shaking [8]). To minimize possible degradation of unstable metabolites during extraction, we used a quick extraction method by powderizing a small amount of fresh tissue with liquid N2 and extracting at low temperature [17], [18]. By using authentic synthesized standards, combined with GC-MS and HPLC-PDA, we evaluated 40 accessions (Fig. 1) and detected 43 UV-absorbing lipophilic metabolites. Of these, 19 metabolites were identified, including 16 alkamides and 2 ketones among those reported so far by the pioneering studies of Bauer and colleagues [5] and another recently reported alkamide, herein referred to as “Chen alkamide” [25] (for structures, see Fig. 2S, Supporting Information). In addition, we detected 24 unknown lipophilic metabolites, some of which (e. g., unknown B5 and unknown 9) are relatively abundant. Five unknowns (unknowns A1–A5) have UV spectra similar to the 2,4-diene alkamides (1, 2, 3, 4, 7, 10, and 11). Six unknowns (B1–B6) have UV spectra similar to the monoene alkamides (12, 13, 14, 16, and 17) (Table 1S, Supporting Information). Interestingly, 10 unknown metabolites have atypical UV spectra. Most of the unknowns (3–10) are highly lipophilic and thus elute at later times. Identification of these unknowns is currently being conducted by HPLC-tandem mass spectrometry (LC-MS/MS), semi-preparative HPLC, and NMR.
Fig. 1.
Relative abundance of lipophilic metabolites in roots from 6-month-old plants of 40 accessions of Echinacea. Error bars indicate standard deviations of means of triplicate experiments.
Our observation of 43 lipophilic metabolites (Table 1S, Supporting Information) can be contrasted with the findings of Binns et al. [8], who distinguished 15 unique alkamides, 2 pairs of alkamides (alkamide 8/9 and alkamide 5/15), and 3 ketones. We detected the presence of all but 4 of these, 3 of the alkamides and 1 of the ketones. Our ability to distinguish more than twice the compounds from these samples is likely attributable to two factors: rapid extraction under low temperature, minimizing possible degradation, and a more sensitive HPLC separation method with extended retention times.
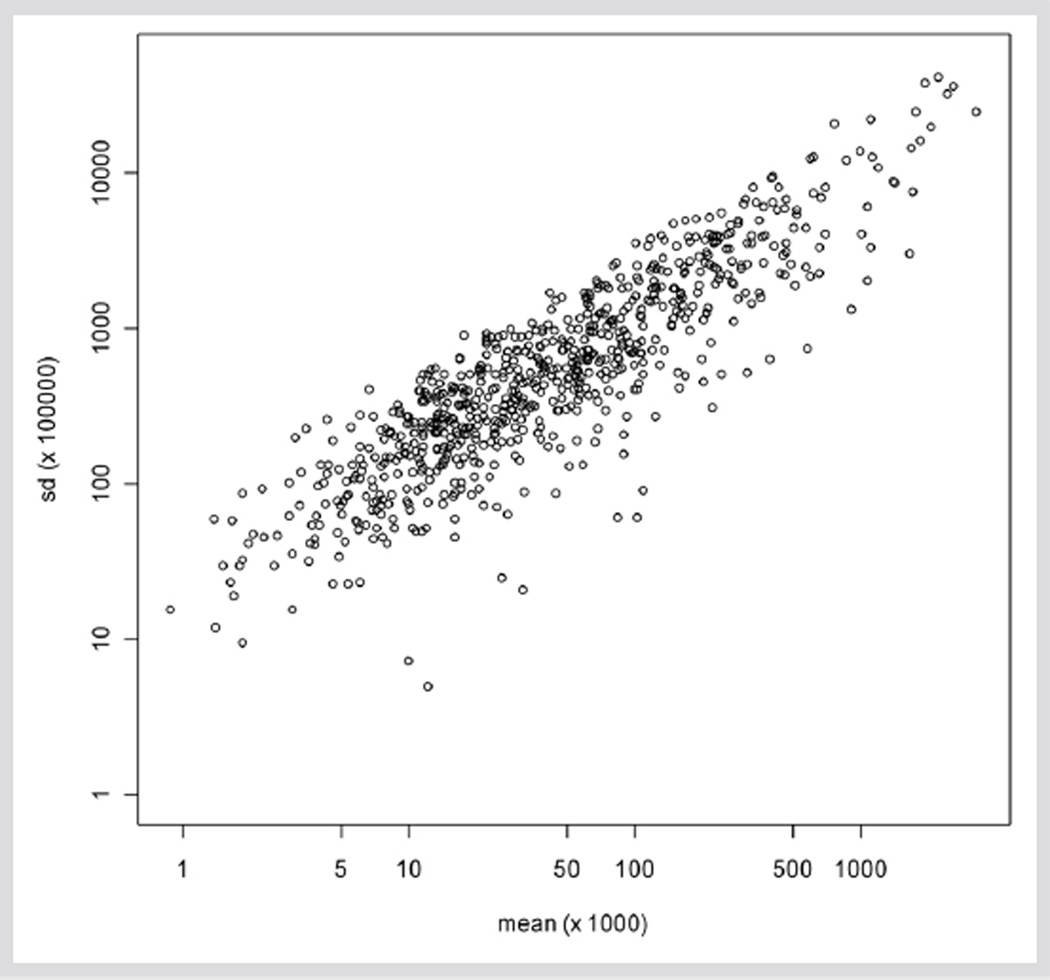
To elucidate how Echinacea accessions are related in terms of their overall metabolite profiles, two multivariate statistical approaches were used: WPCA and clustering analysis. Traditional PCA assumes that all observations of a particular metabolite have the same variance, although variances may differ between metabolites. However, for our dataset, standard deviations among biological replicates increase with metabolite abundance, i. e., the abundant compounds are more variable (Fig. 2). We used WPCA to account for this pattern. Each element of the data is given a corresponding “weight”, proportional to the inverse of the variance. Thus, smaller peaks with smaller errors are given a larger weight, placing more emphasis on these less abundant compounds.
Fig. 2.
Relationship between the standard deviation (sd) and mean across all combinations of accessions and metabolites. Both sd and mean are plotted on log scales.
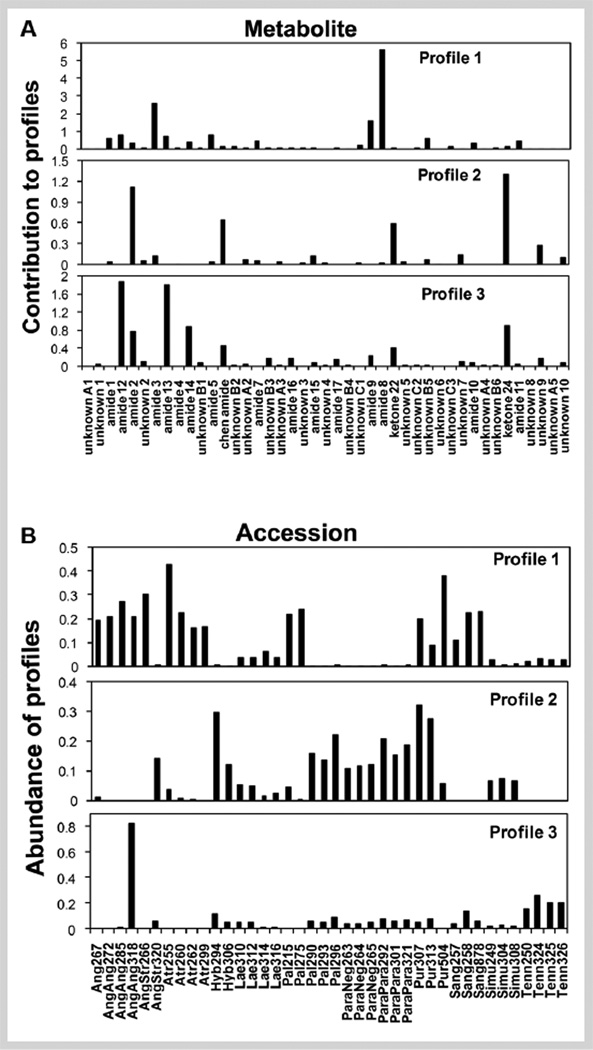
WPCA reveals large quantitative and qualitative differences in lipophilic metabolites among Echinacea populations (Fig. 3). Each composite metabolite profile indicates metabolites that tend to be present together or absent together (Fig. 3A). The accession profiles indicate the relative abundance of corresponding metabolite profiles for each accession (Fig. 3B). Profile 1 (WPC1) focuses on the most abundant compounds, primarily amides 8 and 9, with smaller amounts of amides 3, 5, 11, and 12. Profile 1 is most abundant in accessions identified on the basis of McGregor [9] as E. angustifolia, E. atrorubens (Nutt.) Nutt., E. purpurea, E. pallida, and E. sanguinea Nutt. Profile 2 (WPC2) contains predominantly amides 2 and 3, the Chen alkamide, ketones 22, and 24, and unknown 8. Profile 2 is most abundant in E. purpurea as well as in many other species, except E. angustifolia, E. atrorubens, and E. tennesseensis (Beadle) Small. Profile 3 (WPC3) is almost exclusively composed of amides 12, 13, and 14 and is most abundant in E. angustifolia and E. tennesseensis. The three-profile solution explains 94.8% of the variance of the whole dataset.
Fig. 3.
Weighted principal component analysis (WPCA) of lipophilic metabolite profiles in roots from 6-month-old plants of 40 accessions of Echinacea, illustrating the differences among Echinacea accessions. (A) The relative importance of each metabolite to each of the three WPCs. (B) The relative abundance of each component in different accessions.
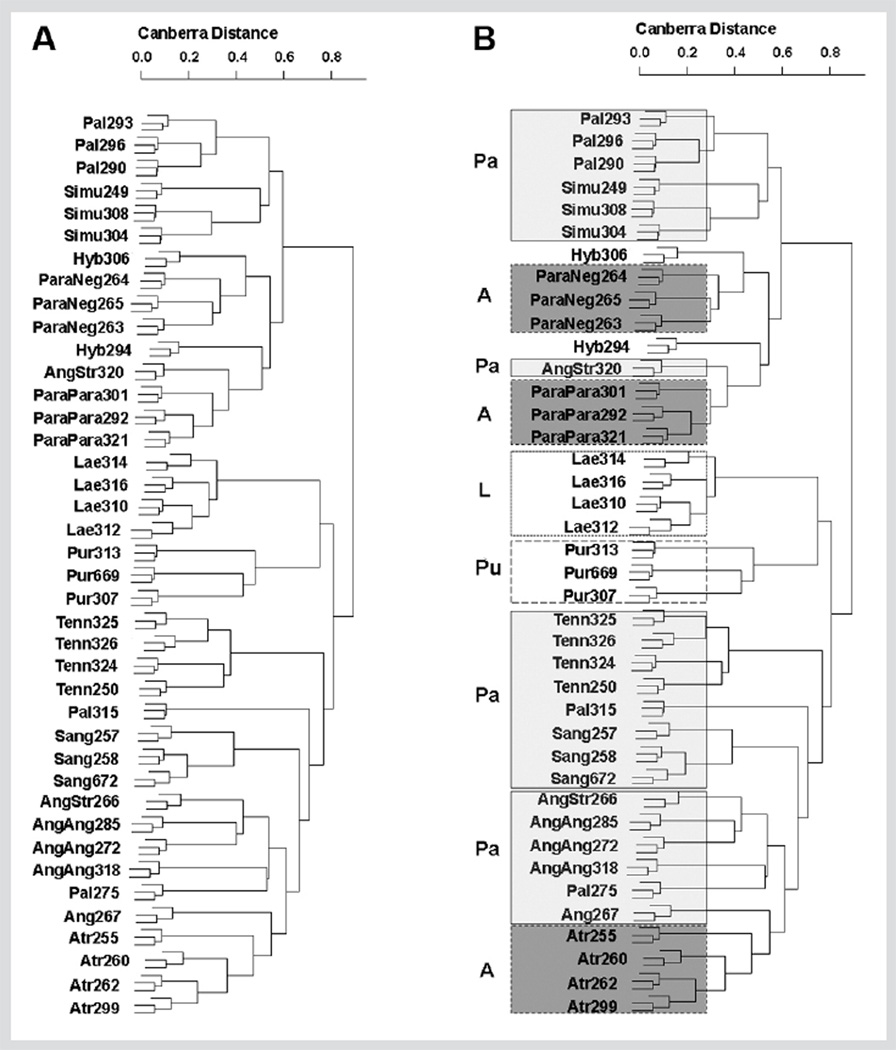
The hierarchical dendrogram constructed from Canberra distances [20] for all pairs of 40 accessions (3 plants per accession), based on average linkage (Fig. 4), displays phenetic relationships among accessions, labeled on the basis of McGregor [9] (Fig. 4A) and coded for the four species recognized by Binns et al. [13] (Fig. 4B). The primary area of agreement between the two taxonomic schemes is the recognition of a distinct species pair, E. purpurea and E. laevigata (C. L. Boynton & Beadle) S. F. Blake, which share a distinct stem anatomy, leaf shape, and phyllary structure, clustered in adjacent groups in Fig. 4.
Fig. 4.
Hierarchical dendrogram constructed from profiles generated from 43 lipophilic metabolites, using Canberra distance, illustrating the distances between 40 Echinacea accessions (3 plants per accession). Panel A is marked only with abbreviations of the accessions following McGregor [9] and the final three digits of the accession number. Panel B also includes blocking corresponding to the four species recognized by Binns et al. [13], with A = E. atrorubens, L=E. laevigata, Pa=E. pallida, and Pu = E. purpurea.
The major difference between the two treatments centers on the remaining taxa, which McGregor [9] treated as seven species with two additional varieties and Binns et al. [13] treated as only two species with six additional varieties. The dendrogram presented in Fig. 4 lends little support for the circumscription of the two, diverse species as proposed by Binns et al. [13], for two primary reasons. First, the observed degree of differentiation that distinguishes E. laevigata from E. purpurea in Fig. 4, recognized by Binns et al. [13] as a clear distinction at the subgeneric level, would support the recognition of three additional subgenera, which we do not feel is warranted based on all other relevant data. In addition, the two inclusive species recognized by Binns et al. [13] as E. pallida and E. atrorubens cluster in an intercalated fashion within the dendrogram, above and below the cluster containing E. laevigata and E. purpurea. Thus, lipophilic metabolic profiles do not support the broad species combinations proposed by Binns et al. [13].
In contrast, there is a better correspondence to the species and at least one of the varieties recognized by McGregor [9] (Fig. 4). Of the nine species recognized by McGregor [9], accessions from six cluster together into a single branch in the tree: E. laevigata, E. purpurea, E. tennesseensis, E. sanguinea, E. atrorubens, and E. angustifolia. The three that do not are E. pallida, E. simulata McGregor, and E. paradoxa (Norton) Britton.
For E. pallida, three of five accessions cluster together (Fig. 4), but accessions PI 631315 and PI 631275 do not. Interestingly, the three E. pallida accessions that are clustered together are adjacent to E. simulata, which McGregor [9] considered to be very close to E. pallida and a likely progenitor of this tetraploid species. He hypothesized that the other species involved in the parentage of E. pallida was E. sanguinea; the two “atypical” accessions are located on our dendrogram closer to E. sanguinea than to E. simulata. Support for a close biochemical relationship between E. pallida and E. sanguineawas reported recently by Senchina et al. [26], who conducted a phenetic analysis of the immunomodulatory characteristics of seven Echinacea taxa. Binns et al. [13] treated both E. simulata and E. sanguinea as varieties of E. pallida.
For E. paradoxa, the two varieties recognized by McGregor [9] form clean clusters adjacent to each other (Fig. 4). Taken as a single group, the E. paradoxa “cluster” also includes two putative hybrid accessions, both of which were collected from populations where E. paradoxa was sympatric with other taxa (Table 1), and E. angustifolia var. strigosa McGregor PI 631320. Five of six accessions of E. angustifolia cluster together (Fig. 4), the outlier being E. angustifolia var. strigosa PI 631320. We speculate that E. angustifolia var. strigosa occupies some “hybrid middle ground” between E. paradoxa var. neglecta McGregor and E. angustifolia var. angustifolia (supported geographically) and/or that E. angustifolia var. strigosa is not well differentiated based on lipophilic compounds. Variety strigosa has been recognized as problematic by other researchers as well. McGregor [9] considered it to be of hybrid origin, as did Binns et al. [13], and the conversion table presented by Baum et al. [10] does not recognize it as a distinct taxon nor does Flora North America [12].
In general, the dendrogram generated on the basis of Canberra distances for lipophilic metabolite profiles among our 40 accessions supports the taxonomic treatment presented by McGregor [9], with the possible exception of E. angustifolia var. strigosa. The metabolic profiles also indicate that there are diverse chemotypes of E. pallida, consistent with its proposed allopolyploid origin.
Although we sampled a broad geographic distribution of accessions representing each taxon, these accessions generally clustered consistently with taxa as identified by morphological, anatomical, and cytological characteristics used by McGregor [9] rather than by geographic or environmental variables gleaned from their provenance data. Thus, our analyses imply that the distribution and types of alkamide and ketone metabolites in Echinacea do not evolve in a convergent manner in response to particular geo/environmental conditions.
Finally, the relative concentrations of the 43 lipophilic compounds appear to be distinctive enough by taxon to allow us to develop “typical” profiles for Echinacea fingerprinting, which could be validated by evaluating additional populations. Thus, this research expands the basis for the evaluation, standardization, and identification of plant material of unknown provenance for cultivated Echinacea and for commercial Echinacea products. In addition, these data contribute to the identification of genetic resources for the production of specific alkamides and ketones. In the course of this study, we found more than 20 unidentified metabolites, some of which may be alkamides or ketones. Bioactivity-guided fractionation together with compound identification will elucidate these as yet unidentified metabolites.
Supplementary Material
Acknowledgements
This publication was made possible by grant number 9 P50 AT004155-06 from the National Center for Complementary and Alternative Medicine (NCCAM) and was supported by the Hatch Act and State of Iowa funds to the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, Project No.1018. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCCAM or NIH. We thank Dr. Ann Perera and Dr. Jonathan Wendel for suggestions.
Footnotes
Supporting information available online at http://www.thieme-connect.de/ejournals/toc/plantamedica
References
- 1.Greger H. Alkamides: Structural relationships, distribution and biological activity [Asteraceae, Heliantheae, Anthemideae] Planta Med. 1984;50:366–375. doi: 10.1055/s-2007-969741. [DOI] [PubMed] [Google Scholar]
- 2.Foster S. Echinacea: Nature's immune enhancer. Rochester: Healing Arts Press; 1991. pp. 20–23. [Google Scholar]
- 3.Jackson JB. Toothache medicine: A customary use of pale purple cone-flower (Echinacea pallida (Nutt.) Nutt.) among the Yuchi in eastern Oklahoma, USA. Econ Bot. 2006;60:386–388. [Google Scholar]
- 4.Yu H, Kaarlas M. Popularity, diversity, and quality of Echinacea. In: Miller SC, editor. Echinacea: The genus Echinacea. Boca Raton: CRC Press; 2004. pp. 127–149. [Google Scholar]
- 5.Bauer R. Echinacea: Biological effects and active principles. In: Lawson LD, Bauer R, editors. Phytomedicines of Europe: chemistry and biological activity; American Chemical Society Symposium Series 691. ACS; Washington, DC. 1998. pp. 140–157. [Google Scholar]
- 6.Linde K, Barrett B, Wolkart K, Bauer R, Melchart D. Echinacea for preventing and treating the common cold. Cochrane Database Syst Rev. 2006;25:CD000530. doi: 10.1002/14651858.CD000530.pub2. [DOI] [PubMed] [Google Scholar]
- 7.Barnes J, Anderson LA, Gibbons S, Phillipson JD. Echinaceas species (Echinacea angustifolia (DC.) Hell., Echinacea pallida (Nutt.) Nutt., Echinacea purpurea (L.) Moench): A review of their chemistry, pharmacology and clinical properties. J Pharm Pharmacol. 2005;57:929–954. doi: 10.1211/0022357056127. [DOI] [PubMed] [Google Scholar]
- 8.Binns SE, Livesey JF, Arnason JT, Baum BR. Phytochemical variation in Echinacea Moench (Helianthese: Asteraceae) from roots and flower-heads of wild and cultivated populations. J Agric Food Chem. 2002;50:3673–3687. doi: 10.1021/jf011439t. [DOI] [PubMed] [Google Scholar]
- 9.McGregor RL. The taxonomy of the genus Echinacea (Compositae) Univ Kansas Sci Bull. 1968;68:113–142. [Google Scholar]
- 10.Baum BR, Binns SE, Arnason JT. Integrating recent knowledge about the genus Echinacea: Morphology, molecular systematics, phytochemistry. Herbal Gram. 2006;72:32–46. [Google Scholar]
- 11.Blumenthal M, Urbatsch LE. Echinacea taxonomy – is the re-classification of the genus warranted? Herbal Gram. 2006;72:30–31. 80. [Google Scholar]
- 12.Urbatsch LE, Neubig KM, Cox PB. Echinacea Moench, Methodus. Flora of North America. eFloras.org. 2006;21:43, 64–65, 88. [Google Scholar]
- 13.Binns SE, Baum BR, Arnason JT. A taxonomic revision of the genus Echinacea (Heliantheae; Asteraceae) Syst Bot. 2002;27:610–632. [Google Scholar]
- 14.Mechanda SM, Baum BR, Johnson DA, Arnason JT. Analysis of diversity of natural populations and commercial lines of Echinacea using AFLP. Can J Bot. 2004;82:461–484. [Google Scholar]
- 15.Kim DH, Heber D, Still DW. Genetic diversity of Echinacea species based on amplified fragment length polymorphism markers. Genome. 2004;47:102–111. doi: 10.1139/g03-086. [DOI] [PubMed] [Google Scholar]
- 16.Flagel L, Rapp RA, Grover CE, Widrlechner MP, Hawkins J, Grafenberg JL, et al. Phylogenetic, morphological, and chemotaxonomic incongru-ence in the North American endemic genus Echinacea Moench. Am J Bot. 2008;95:756–765. doi: 10.3732/ajb.0800049. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Bae J, Kraus G, Wurtele ES. Diacetylenic isobutylamides of Echinacea: synthesis and natural distribution. Phytochemistry. 2004;65:2477–2484. doi: 10.1016/j.phytochem.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Kraus GA, Bae J, Wu L, Wurtele ES. Synthesis and natural distribution of anti-inflammatory alkamides from Echinacea. Molecules. 2006;11:758–767. doi: 10.3390/11100758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus GA, Bae J, Wu L, Wurtele ES. The synthesis and natural distribution of the major ketone constituents in Echinacea pallida. Molecules. 2007;12:406–414. doi: 10.3390/12030406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Digby PG, Kempton RA. Multivariate analysis of ecological communities. New York: Chapman and Hall/Methuen; 1987. [Google Scholar]
- 21.Widrlechner MP, McKeown KA. Assembling and characterizing a comprehensive Echinacea germplasm collection. In: Janick J, Whipkey A, editors. Trends and new crops and new uses. Alexandria: ASHS Press; 2002. pp. 506–508. [Google Scholar]
- 22.Senchina DS, Wu L, Flinn GN, Konopka DE, McCoy JA, Widrlechner MP, et al. Year-and-a-half old, dried Echinacea spp. roots retain cytokine-modulating capabilities in an in vitro human older adult model of influenza vaccination. Planta Med. 2006;72:1207–1215. doi: 10.1055/s-2006-947254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer R, Remiger P. TLC and HPLC analysis of alkamides inEchinacea drugs. Planta Med. 1989;55:367–371. doi: 10.1055/s-2006-962030. [DOI] [PubMed] [Google Scholar]
- 24.Molgaard P, Johnsen S, Christensen P, Cornett C. HPLC method validated for the simultaneous analysis of cichoric acid and alkamides in Echinacea purpurea plants and products. J Agric Food Chem. 2003;51:6922–6933. doi: 10.1021/jf026158f. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Fu T, Tao T, Yang J, Chang Y, Wang M, et al. Macrophage activating effects of new alkamides from the roots of Echinacea species. J Nat Prod. 2005;68:773–776. doi: 10.1021/np040245f. [DOI] [PubMed] [Google Scholar]
- 26.Senchina DS, Flagel LE, Wendel JF, Kohut ML. Phenetic comparison of seven Echinacea species based on immunomodulatory characteristics. Econ Bot. 2006;60:205–211. doi: 10.1663/0013-0001(2006)60[205:pcoses]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.