Abstract
Compounding evidence supports the important role in pathogenesis that the metabolism of cholesterol by Mycobacterium tuberculosis (M. tuberculosis) plays. Elucidating the pathway by which cholesterol is catabolized is necessary to understand the molecular mechanism by which this pathway contributes to infection. Based on early metabolite identification studies in multiple actinomycetes, it has been proposed that cholesterol side chain metabolism requires one or more acyl-CoA dehydrogenases (ACADs). There are 35 genes annotated as encoding ACADs in the M. tuberculosis genome. Here we characterize a heteromeric ACAD encoded by Rv3544c and Rv3543c, formerly named fadE28 and fadE29, respectively. We now refer to genes Rv3544c and Rv3543c as chsE1 and chsE2 in recognition of their validated activity in cholesterol side chain dehydrogenation. Analytical ultracentrifugation and LC/UV experiments establish that ChsE1-ChsE2 forms an α2β2 heterotetramer, a new architecture for an ACAD. Our bioinformatic analysis and mutagenesis studies reveal that heterotetrameric ChsE1-ChsE2 has only two active sites. E241 in ChsE2 is required for catalysis of dehydrogenation by ChsE1-ChsE2. Steady state kinetic analysis establishes the enzyme is specific for an intact steroid ring system compared to hexahydroindanone substrates with specificity constants (kcat/KM) of 2.5 × 105 ± 0.5 s-1 M-1 vs 9.8 × 102 ± s-1 M-1 respectively, at pH 8.5. The characterization of a unique ACAD quaternary structure involved in sterol metabolism that is encoded by two distinct cistronic ACAD genes opens the way to identification of additional sterol metabolizing ACADs in M. tuberculosis and other actinomycetes through bioinformatic analysis.
Mycobacterium tuberculosis (M. tuberculosis), the causative agent of tuberculosis, is the second most deadly infection in the adult population worldwide. 8.8 million cases were reported in 2010 and 1.4 million deaths attributed to the disease (1). During infection, M. tuberculosis bacilli are phagocytosed by macrophages and the host inflammatory response results in the formation of a granuloma (2). Bacteria can persist within the granuloma for decades.
M. tuberculosis metabolism of cholesterol, an abundant host lipid, is important for persistence of infection in vivo (3, 4). Disruption of an Mce4 ABC transporter mutant results in defective cholesterol transport into the mycobacterium (4, 5). In infected mice, the mce4 mutant's growth is slowed in the chronic phase of infection. fadA5, a cholesterol-regulated gene that encodes a thiolase, is required for growth of M. tuberculosis on cholesterol as a carbon source (6). The number of bacteria in the lungs of mice infected with the fadA5 mutant declined after induction of the cellular immune response demonstrating a persistence phenotype identical to that seen upon mutation of the Mce4 transporter (4). Thiolases catalyze the reverse Claisen condensation step in lipid β-oxidation. Similarly, the M. tuberculosis igr operon, which encodes six proteins, is important for growth on cholesterol in vitro and for mycobacterial survival in vivo and encodes homologs of lipid-modifying enzymes (7, 8).
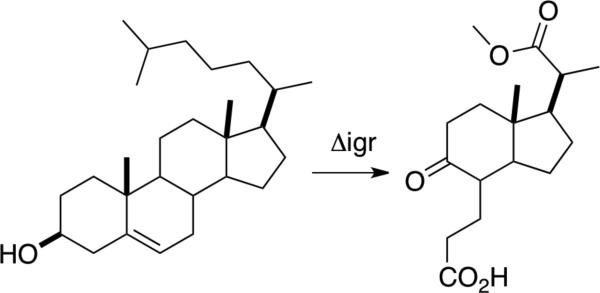
The M. tuberculosis igr operon (Rv3545c-Rv3540c) is required for complete degradation of the cholesterol side chain. Upon disruption of this operon, steroid derived metabolite, methyl 1β-(2’-propanoate)-3aα-H-4α-(3’-propanoic acid)-7aβ-methylhexahydro- 5-indanone accumulates in M. tuberculosis Δigr cultures grown in the presence of cholesterol (Scheme 1) (9). Rv3544c and Rv3543c, annotated as potential acyl-CoA dehydrogenases, are encoded by two adjacent cistronic genes. Heterologously co-expressed N-His6 tagged Rv3544c and tagless Rv3543c co-purify upon immobilized metal affinity chromatography, indicating they form a quaternary complex. We demonstrated that the complex is a functional acyl-CoA dehydrogenase that dehydrogenates steroid-derived substrates analogous to the metabolite isolated from an igr knockout strain (9).
Scheme 1.
An igr disrupted strain of M. tuberculosis accumulates methyl 1β-(2’-propanoate)-3aα-H-4α-(3’-propanoic acid)-7aβ-methylhexahydro- 5-indanone when cultured with cholesterol.
To our knowledge this was the first report of a heteromeric acyl-CoA dehydrogenase (ACAD). All ACADs characterized to date form homotetrameric assemblies comprised of four ~43 kDa monomers, with one exception. The ACAD sub-families specific for very long chain fatty acids (VLCAD and ACAD9) form homodimers from two ~73 kDa monomers (10, 11).
Herein, we present full enzymatic and biophysical characterization of Rv3543c-Rv3544c and demonstrate that Rv3543c-Rv3544c is an α2β2 heterotetramer. Neither Rv3543c nor Rv3544c is functionally competent by itself. Extensive sequence alignments of Rv3543c and Rv3544c with acyl-CoA dehydrogenases of known three-dimensional structure reveal that a canonical active site glutamate required for general base catalysis is conserved in Rv3543c, but not in Rv3544c. Mutagenesis in combination with kinetic assays confirmed that Glu241 of Rv3543c is the base required for catalysis by this enzyme. Furthermore, Rv3543c-Rv3544c has only two conserved FAD binding sites per tetramer. Formation of the Rv3543c-Rv3544c complex is required to constitute functional binding sites and consequently catalytic activity. We propose that this unique heteromeric structure is required for effecting dehydrogenation of steroyl-CoA substrates, and may be a motif that is utilized for polycyclic-CoA esters in general. Therefore, we now refer to the Rv3544c and Rv3543c acyl-CoA dehydrogenase genes as chsE1 and chsE2, respectively, for cholesterol side chain metabolism E, to distinguish them from members of the bacterial fade, fatty acid degradation E acyl Co-A dehydrogenase, gene family.
Materials and Methods
Materials, strains, media, and general methods
Ferricenium hexafluorophosphate and ergocalciferol were purchased from Sigma-Aldrich (St. Louis, MO). Coenzyme A was purchased from MP Biomedicals (Solon, Ohio). Isopropyl β-D-1thiogalactopyranoside was from Denville Scientific (Metuchen, NJ). Tryptone, HEPES, TRIS, and ampicillin were purchased from Fisher Scientific (Pittsburgh, PA). L-arabinose, chloramphenicol, and sodium chlorite were purchased from Acros Organics (New Jersey). Tetracycline is from US Biochemical Corp (Cleveland, OH) and kanamycin is from IBI Scientific (Peosta, IA). Yeast extract was purchased from Research Products International Co. (Mount Prospect, IL). iProof DNA polymerase was from Bio-Rad (Hercules, CA). Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, and protein ladder were from New England Biolabs (Beverly, MA). HisTrap FF columns, MonoQ, and Superdex 75 HiLoad 16/60 and 10/300 GL columns were from GE Healthcare Biosciences Corp. (Piscataway, NJ). Oligonucleotides were from IDT Inc. (Coralville, IA). Total genomic DNA of M. tuberculosis H37Rv was obtained from the TB Research Materials Facility at Colorado State University (Fort Collins, CO) (NIAD NO1-AI40091). MALDI mass spectra were acquired on a Bruker Autoflex II TOF/TOF. Big Dye DNA sequencing (Applied Biosystems, Foster City, CA; performed by the Stony Brook University Sequencing Facility) was used to verify the coding sequence of the expression plasmids. BL21(DE3) E. coli was obtained from BioRad and chaperone plasmid pG-KJE8 was obtained from Takara. M. smegmatis mc2155 strain was obtained from E. Dubnau (PHRI). 2 × YT is composed of 16 g tryptone, 10 g yeast extract, and 5 g NaCl per liter. LC/UV/MS analysis was conducted on a Waters UPLC/MS with diode array and SQD detectors. Buffer A: 20 mM Tris-HCl buffer pH 8.0, supplemented with 300 mM NaCl, 1 mM TCEP, and 10 mM imidazole. Buffer B: 50 mM Tris-HCl buffer pH 8.0. Buffer C: 50 mM Tris-HCl buffer pH 8.0, supplemented with 200 mM NaCl, and 1 mM TCEP. Buffer D: 20 mM sodium phosphate buffer pH 7.5, supplemented with 200 mM NaCl and 1 mM TCEP.
Expression plasmid construction
The desired genes were amplified from M. tuberculosis H37Rv total genomic DNA by PCR using forward and reverse primers. The PCR product was digested with the appropriate restriction endonuclease and ligated into a similarly digested vector (Table 1). DNA sequencing of the plasmids confirmed that the sequence was correct and that no mutations were introduced during the cloning procedures. ChsE2 glutamate 241 was mutated to a glutamine in pigr5 using the method of Moore (12). The mutation was confirmed by DNA sequencing.
Table 1.
Plasmids constructed
| Construct name | Plasmid | Gene | Fusion tag | Restriction sites used | Antibiotic marker | Source/Reference |
|---|---|---|---|---|---|---|
| pET20b | amp | Novagen | ||||
| pET28b | kan | Novagen | ||||
| pSD31 | hygro | (13) | ||||
| pG-KJE8 | cam | Takara | ||||
| pChsE1N | pET28b | Rv3544c | N-terminal His6 | NdeI/NotI | kan | This work |
| pChsE2N | pET28b | Rv3543c | N-terminal His6 | NdeI/XhoI | kan | This work |
| pChsE1 | pET20b | Rv3544c | ---- | NdeI /NotI | amp | This work |
| pChsE2Ms | pSD31 | Rv3543c | N-terminal His6 | EcoRv | hygro | This work |
| pigr5 | pET28b | Rv3544c-Rv3540c | N-terminal His6 | NdeI/HindIII | kan | (9) |
| pigr5E241Q | pET28b | Rv3544c-Rv3540c | N-terminal His6 | NdeI/HindIII | kan | This work |
Individual gene expression
pChsE1N or pChsE2N constructs were transformed into BL21(DE3) Escherichia coli, single colonies were selected on LB plates containing 30 μg/mL kanamycin, and cultured in 2 × YT media at 37 °C. Expression was induced at A600 = 0.6–0.8 by the addition of 50 μM to 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG), and cells were grown 20 h at 16 or 25 °C.
Gene expression in M. smegmatis
pChsE2Ms was electroporated into M. smegmatis mc2155 and single colonies were selected on 7H10 plates supplemented with 100 μg/mL ampicillin, 10 μg/mL cycloheximide, and 50 μg/mL hygromicin and grown in 7H9 media supplemented with 0.2% glycerol. After 2 days, expression was induced with 0.2% acetamide and cultures were grown for an additional 24 h.
Gene co-expression
Constructs pChsE2N and pChsE1 were co-transformed into BL21(DE3) E. coli::pG-KJE8 (chaperone plasmid, Takara). Single colonies were selected on LB plates containing the appropriate antibiotics (100 μg/mL ampicillin for pET20b, 30 μg/mL kanamycin for pET28b, and 20 μg/mL chloramphenicol for pG-KJE8) and cultured in 2 × YT media at 37 °C. Chaperone expression was induced upon inoculation with 2 mg/mL L-arabinose and 5 ng/mL tetracycline. ChsE expression was induced at A600 = 0.6–0.8 with the addition of 1 mM IPTG and cells were grown 20 h at 25 ° C. Gene co-expression in cis with construct pigr5 was performed as reported previously (9). Similarly, ChsE2E241Q mutant protein was prepared with construct pigr5E241Q following the expression conditions for pigr5.
Protein purification
ChsE1 or ChsE2 expressing cells were lysed by French press in Buffer A and cellular debris was removed by centrifugation at 125,000 × g for 1 h. Proteins were purified by immobilized metal affinity chromatography using Hisbind resin (Novagen) following the manufacturer's protocol. Soluble ChsE2 was buffer exchanged to Buffer B and further purified by anion exchange chromatography on a MonoQ column (1 mL) equilibrated in Buffer B. Protein was eluted at a flow rate of 0.5 mL/min with a linear gradient from 100% Buffer B to 100% Buffer B supplemented with 1 M NaCl. After injection, the column was washed with five column volumes (CV) of Buffer B, then changed to 80% Buffer B over 20 CV. Next, the gradient was changed from 80% to 25% Buffer B over 10 CV, then to 0% Buffer B over 5 CV. Eluted protein was further purified by size exclusion chromatography on a Superdex 200 HiLoad 16/60 column (GE Healthcare) equilibrated with Buffer C. ChsE1-ChsE2 and ChsE1-ChsE2E241Q were purified as detailed for ChsE2, excluding the anion exchange chromatography step. All protein samples were analyzed by reducing SDS-PAGE and protein band identities were confirmed by in-gel tryptic digestion as reported previously (14). ChsE proteins were stored in 50 mM HEPES buffer pH 8.0 at -80 °C.
Identification and quantification of flavin cofactor
A 15 μM (0.16 mg/mL) solution of ChsE1-ChsE2 in Buffer C was denatured by boiling for 2 min. The sample was chilled on ice and centrifuged to pellet precipitated protein. The supernatant was analyzed by reverse phase C18 liquid chromatography-mass spectrometry in ESI positive mode and compared to flavin adenine dinucleotide standard. The quantity of FAD obtained was determined using the absorbance and extinction coefficient of FAD at 260 nm, ε260 = 37,000 M-1cm-1. The protein pellet was dissolved in 6 M guanidine-HCl and the concentration determined using the calculated extinction coefficients at 280 nm (ε280 (ChsE1)= 35,410 M-1 cm-1; ε280 (ChsE2)=58,900 M-1cm-1; ε280 (ChsE1-ChsE2)=188,620 M-1cm-1).
Biophysical analysis of ChsE proteins
ChsE2, ChsE1-ChsE2, and ChsE1-ChsE2E241Q samples were analyzed by analytical gel-filtration on a Superdex 75 10/300 GL column (GE Healthcare) equilibrated with Buffer D. Samples were eluted isocratically in the same buffer, monitoring at 220 and 280 nm.
Molecular weights of ChsE2 and ChsE1-ChsE2 were determined using analytical ultracentrifugation sedimentation equilibrium with a Beckman Optima XL-A centrifuge. ChsE2 (8.5, 3.4, and 1.7 μM) in 5 mM sodium phosphate buffer pH 7.5 and ChsE1-ChsE2 (10.6 μM, 5.3 μM, and 2.6 μM) in buffer B, were centrifuged at speeds of 20k, 25k, and 30k at 20 °C. Scans were acquired after 18 and 20 h of centrifugation at each speed monitoring at 280 nm. The protein partial-specific volume of 0.7336 for ChsE2 and 0.7350 for ChsE1-ChsE2 and the solvent density 0.9994 for ChsE2 and 1.0079 for ChsE1-ChsE2 were calculated using SEDNTERP (University of New Hampshire, Durham NH). Data were fit globally to the ideal, single-species model using Heteroanalysis (University of Connecticut, Storrs CT) to determine the molecular weight.
Protein complex stoichiometry of ChsE1-ChsE2 was confirmed by LC/UV/MS (Waters UPLC/diode array/SQD). ChsE1-ChsE2 were separated on a Waters XBridge BEH300 C4 3.5 μm column (2.1 × 100 mm) at 40 °C with a linear gradient from 95% A to 95% B over 15 min, where A is 5% isopropanol/0.1% trifluoroacetic acid and B is 99.9% isopropanol/0.1% trifluoroacetic acid. MS spectra were collected in ESI positive ion mode with a cone voltage of 40 V, a capillary voltage of 4.5 kV, and source temperature of 150 °C. MS spectra were deconvolved using ESIprot 1.0 (15) and peaks in the UV 280 nm chromatograms were integrated using R. The integrated peak areas of each protein were divided by the corresponding molar extinction coefficient for the protein to yield the molar concentrations. Protein stoichiometry was determined from the ratio of the molar concentrations.
Bioinformatic analysis
ACAD protein sequences were obtained from UniProt and sequence alignments were conducted with ClustalW2 (EMBL-EBI, European Bioinformatics Institute, Cambridge, U.K.) using the default parameters (16). FAD binding and CoA binding in published ACAD PDB structures was investigated with PoseView (Center of Bioinformatics, University of Hamburg) (17). A ChsE1-ChsE2 ligand binding model was created using MCAD homodimer, PDB 1EGC, as a template. The model is based on a ChsE1-ChsE2 heterodimer assembly and highlights conserved residues determined from the ACAD sequence alignment with ChsE1 and ChsE2.
Dehydrogenase assay
CoA thioesters, 3-oxo-4-pregnene-20-carboxyl-CoA (1) and 1β- (2’-propanoyl-CoA)-3aα-H-7aβ-methylhexahydro-4-indanone (2), were prepared and purified as reported previously (9, 18). The identities and purity of CoA thioesters was assessed by LC/UV/MS.
Dehydrogenase activity was tested with artificial electron acceptor ferricenium hexafluorophosphate as reported previously (19). Product formation was monitored spectroscopically at 300 nm and 25 °C. Initial velocities were determined for the first 100 s of reaction. Enzyme activity was optimized for pH using the three component buffer system reported by Ellis and Morrison with 50 μM of acyl-CoA 1 (20). Subsequent assays were conducted in 100 mM TAPS pH 8.5 buffer. The optimal ionic strength was investigated over NaCl concentrations of 0 to 1.0 M. Protein stability and substrate 1 solubility were tested at concentrations of 100 nM and 50 μM respectively with and without 0.2 M NaCl by dynamic light scattering on a 90 Plus partical size analyzer from Brookhaven Instruments Corporation. Data was acquired in triplicate at 25 °C.
Steady state kinetic analysis was conducted in 100 mM TAPS buffer pH 8.5 with CoA thioesters 1 and 2. The rates of product formation were fit to the Michaelis-Menten equation (1) to determine the KM for substrates 1 and 2 and kcat for enoyl-CoA product formation.
| (1) |
where v is initial velocity, Vmax is the maximum velocity, S is the varied substrate, and KM is the Michaelis-Menten constant,
NMR analysis of assay product
The product of ChsE1-ChsE2 assay with substrate 3-oxo-4-pregnene-20-carboxyl-CoA was purified on a 3 cc sep-pak C18 cartridge (Waters) equilibrated in 5% MeOH/95% 10 mM ammonium acetate pH 8.0. Following loading of the sample the cartridge was washed with 6 mL of the equilibration buffer followed by 6 mL of 25% MeOH/75% 10 mM ammonium acetate pH 8.0. Product was eluted in 45% MeOH/55% 10 mM ammonium acetate pH 8.0. Dried compound was dissolved in D2O for NMR analysis on a Bruker 700 MHz NMR. 1H spectra of substrate 1 and purified product were acquired using excitation sculpting for water suppression (21).
Results
ChsE2 is isolated as a monomeric apoenzyme when heterologously expressed in either E. coli or M. smegmatis
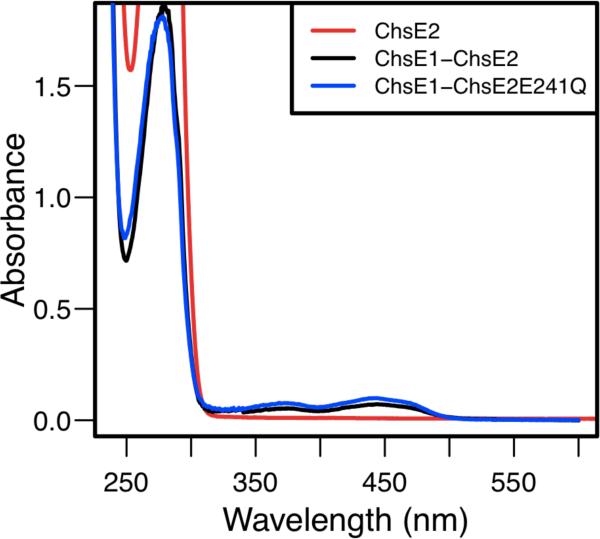
Initially, chsE1 and chsE2 were expressed individually in E. coli as N-terminal His6 tagged proteins. ChsE1 was found to be insoluble and was localized in inclusion bodies. ChsE2 was isolated and purified by immobilized metal affinity, anion exchange, and size exclusion chromatographies (Figure S1). UV-visible spectroscopy revealed the protein was purified as the apoprotein, without the flavin cofactor necessary for catalysis (Figure 1). Attempts to reconstitute with FAD were not successful.
Figure 1. UV-visible spectra of purified ChsE proteins.
Purified ChsE2 (50 μM), ChsE1-ChsE2 (15 μM), and ChsE1-ChsE2E241Q (15 μM). Relative spectral maxima at 370 nm and 446 nm for ChsE1-ChsE2 and ChsE1-ChsE2E241Q are characteristic of oxidized flavin. No flavin absorbances were observed for ChsE2 alone.
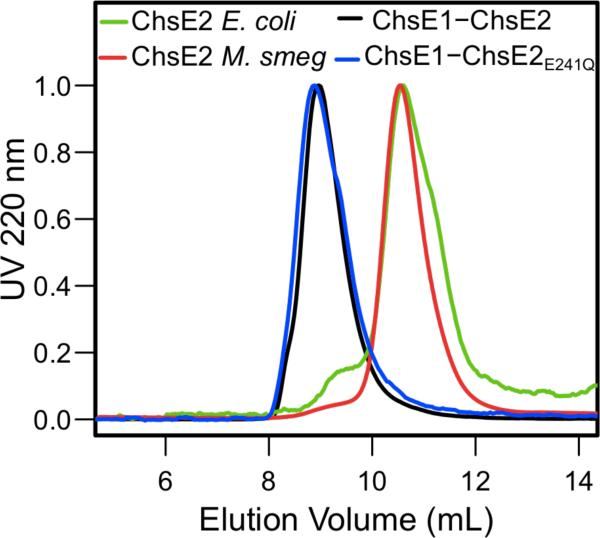
ChsE2 was analyzed by analytical size exclusion chromatography (Figure 2) and analytical ultracentrifugation (AUC) sedimentation equilibrium experiments (Figure 3A). The predicted monomer molecular weight with a His6 tag is 44.9 kDa. The AUC data fit an ideal model with a molecular weight of 42.0 ± 0.5 kDa, where the reported error is the measure of the fit and indicates the chosen model is satisfactory. Thus, under the conditions tested, ChsE2 is a monomer. In crystal structures of ACADs, one molecule of FAD binds non-covalently per monomer and the adenosine lies at the interface of the two monomers. Therefore, two monomer chains comprise the FAD binding pocket, and ChsE2 may not bind FAD due to its monomeric state.
Figure 2. Analytical gel filtration of ChsE2 and ChsE1-ChsE2 complexes.
ChsE2 expressed in E. coli and M. smegmatis and ChsE1-ChsE2 and ChsE1-ChsE2E241Q expressed in E. coli were analyzed by analytical gel filtration on a Superdex 75 column under identical conditions. Signal at 220 nm was normalized to 1.0. The ChsE1-ChsE2 protein formed a stable complex with a higher molecular weight than ChsE2 expressed alone in E. coli or M. smegmatis.
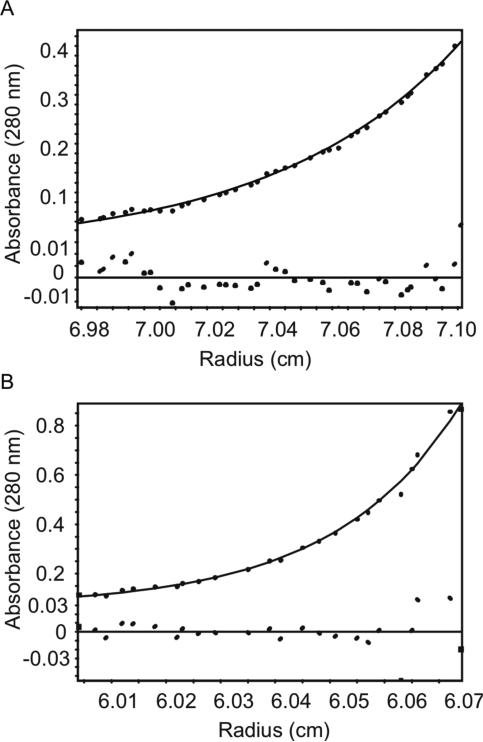
Figure 3. Analytical ultracentrifugation sedimentation equilibrium data for ChsE proteins.
(A) ChsE2 and (B) ChsE1-ChsE2 were analyzed at three concentrations ranging from 1 μM to 11 μM at centrifugation speeds of 20k, 25k, and 30k at 20 °C. A representative fit for each sample is shown. The solid line shows the fit of the data to the ideal species model and the residuals of the fit are graphed below. The global fit for each protein provided molecular weights of 42.0 ± 0.5 kDa and 156 ± 1 kDa for ChsE2 and ChsE1-ChsE2, respectively.
Next, chsE2 was expressed in M. smegmatis, a nonpathogenic, faster-growing relative of M. tuberculosis, as a N-terminal His6 tagged protein. The protein was isolated by immobilized metal affinity chromatography (IMAC) and further purified by anion exchange and size exclusion chromatographies. Analysis by analytical size exclusion chromatography indicated that M. smegmatis expressed ChsE2 was also monomeric (Figure 2). Analysis of ChsE2 expressed in E. coli and in M. smegmatis demonstrates that the protein is not in the predicted quaternary structure of ACADs, lacks FAD, and therefore is inactive.
ChsE1-ChsE2 forms an obligate α2β2 tetramer with noncovalently bound FAD
Isolation of monomeric ChsE2 apoprotein and insoluble ChsE1 indicated they do not form native structures as individual proteins. Previously, we had reasoned that these proteins might form a complex since they are encoded in a single operon. Co-expression would allow assembly of ChsE1 and ChsE2 that would otherwise be insoluble or inactive when expressed individually. Indeed, when chsE1 and chsE2 were expressed in a cistronic construct on a single plasmid, pigr5, a complex of ChsE1 and ChsE2 was isolated in a pull-down assay (9).
Here, we further investigated expression conditions and constructs. We found that the ChsE1-ChsE2 complex was obtained upon expressing chsE1 as a tagless protein and chsE2 as a N-terminal His6 tagged protein from separate plasmids (in trans) in E. coli with coexpression of chaperones. We used E. coli harboring plasmid pG-KJE8, which encodes for 5 folding chaperones, dnaK, dnaJ, grpE, groES, and groEL. The ChsE1-ChsE2 protein complex was isolated regardless of which protein, ChsE1 or ChsE2, contained a N terminal His6 tag. The identity of ChsE1 and ChsE2 in the complex was confirmed by SDS-PAGE (Figure S1) and in-gel tryptic digest followed by MALDI-TOF-MS analysis.
Under both in cis and in trans expression conditions, the isolated complex was yellow in appearance and had a characteristic UV-visible flavin spectrum, with absorbance maxima at 370 nm and 446 nm (Figure 1). Expression in cis however yielded higher A446/A280 ratios, indicative of higher FAD occupancy. Kinetic studies were conducted with ChsE1-ChsE2 expressed with construct pigr5.
The flavin cofactor was released from ChsE1-ChsE2 by heat denaturation of the protein and was analyzed by LC/UV/MS. A protonated molecular ion at m/z = 786 was observed (Figure S2), confirming the identity of the cofactor as FAD. In addition, the retention time and absorbance spectrum were identical to those of a FAD standard. Therefore, ChsE1-ChsE2 is isolated as a holoprotein with FAD cofactor.
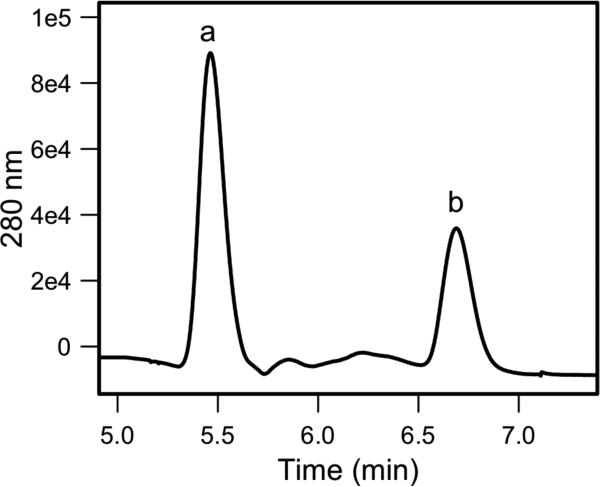
Analysis of the complex by analytical size exclusion chromatography confirmed that a stable quaternary complex is formed (Figure 2). Further analysis by AUC sedimentation equilibrium experiments established that ChsE1-ChsE2 exists as a tetramer with a molecular weight of 156 ± 1 kDa (Figure 3B). The predicted molecular weights of N tagged-ChsE1 and ChsE2 are 37.6 kDa and 42.7 kDa respectively. To determine precisely the stoichiometry of the complex, the assembly was analyzed by LC/UV/MS (Figure 4). The two observed peaks were identified as ChsE1 and ChsE2 by ESI+ MS. The absorbance peaks at 280 nm for ChsE1 and ChsE2 were integrated and using the extinction coefficient for each protein the relative molar stoichiometry of ChsE1 to ChsE2 was calculated to be 1 to 1. This ratio indicates that ChsE1-ChsE2 forms an α2β2 complex.
Figure 4. Reverse phase LC/UV chromatogram of ChsE1-ChsE2.
Peak a and peak b were identified as ChsE2 and ChsE1, respectively, by deconvolution of multiple charged states in the corresponding ESI+ MS spectra. The absorbance peaks were integrated and relative concentrations were determined from the calculated extinction coefficients of ChsE1 and ChsE2, ε280(ChsE1)= 35,410 M-1 cm-1; ε280 (ChsE2)=58,900 M-1cm-1.
Tetrameric ChsE1-ChsE2 has two cofactor binding sites
The FAD stoichiometry in the protein complex was determined spectrophotometrically after protein denaturation. The protein concentration was determined for unfolded protein using the calculated extinction coefficient of ChsE1 and ChsE2 at 280 nm. The concentration of FAD was calculated from the extinction coefficient at 260 nm after removal of precipitated ChsE1-ChsE2. The FAD to protein molar ratio was 1.4 ± 0.1 FAD molecules per α2β2 ChsE1-ChsE2 tetramer. This result suggests that ChsE1-ChsE2 has two FAD bindings sites in contrast to typical ACAD tetramers that have four FAD binding sites.
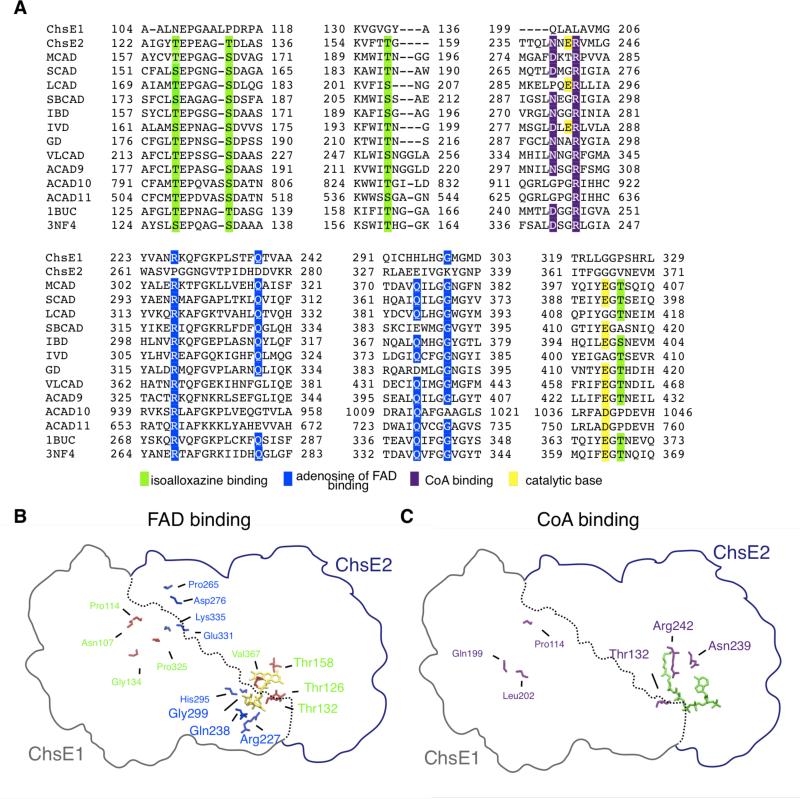
To corroborate the FAD stoichiometry in heterotetrameric ChsE1-ChsE2, we analyzed the protein sequences of ChsE1 and ChsE2 for FAD binding residues that are conserved in other ACADs. First, we examined ligand protein interactions between FAD and several ACADs in reported crystal structures (PDB code shown in parentheses), including human SCAD (2VIG), MCAD (1EGC), SBCAD (2JIF), IBD (1RX0), IVD (1IVH), GD (1SIQ), VLCAD (3B96), and ACAD11 (2WBI) and bacterial ACADS from Megsphaera elsdenii (1BUC) and Mycobacterium thermoresistibile (3NF4). In each structure, a single FAD cofactor-binding site is comprised of two chains from the ACAD tetramer or dimer, in the case of VLCAD and ACAD9. Each chain interacts with adenosine in one binding pocket and with riboflavin in a second binding pocket. The polar side chains of Thr161A, Ser167A, Thr193A, and Thr403A form hydrogen bonds with the isoalloxazine and ribityl diphosphate moieties (numbering from MCAD). These Ser and Thr residues are conserved within the ACAD family (Figure 5A).
Figure 5. Bioinformatic analysis of ChsE1 and ChsE2.
(A) Sequence alignment of ChsE1 and ChsE2 against human and bacterial ACADS; MCAD (P11310), SCAD (P16219), LCAD (P28330), SBCAD (P45954), iBD (Q9UKU7), IVD (P26440), GD (Q92947), VLCAD (P49748), ACAD9 (Q9H845), ACAD10 (Q6JQN1), ACAD11 (Q709F0), 1BUC (SCAD Megasphaera elsdenii, Q06319), 3NF4 (Mycobacterium thermoresistibile, G7CDN2). Residues in green bind the isoalloxazine and ribityl diphosphate moieties of FAD, residues in blue bind adenosine of FAD, residues in purple bind CoA, and residues in yellow are the catalytic base. (B) Homology model of FAD binding residues and (C) octanoyl-CoA binding residues in ChsE1-ChsE2 based on human MCAD homodimer (PDB 1EGC). FAD is displayed in yellow and octanoyl-CoA is displayed in green. FAD and CoA cofactors are shown adjacent to conserved binding residues.
Analysis of the ChsE2 and ChsE1 sequence alignments with these ACADs revealed that Thr161A, Ser167A, and Thr193A are conserved in ChsE2 as residues Thr126, Thr132, and Thr158, respectively. Thr403A is not conserved in ChsE2 (Figure 5A). In contrast, Thr161A, Ser167A, Thr193A, and Thr403A are not conserved in ChsE1.
Residues involved in forming hydrogen bonds with adenosine of FAD, Arg306A’, Gln317A’, Gln374A’, and Gly378A’, are from the second chain that contributes to the FAD cofactor binding sites in ACADs and are highly conserved. Sequence analysis of ChsE1 revealed that Arg306A’, Gln317A’, Gln374A’and Gly378A’ align with residues Arg227, Gln238, His295, and Gly299 (Figure 5A). However, Arg306A’, Gln317A’, Gln374A’, and Gly378A’ are not conserved in ChsE2.
In all ACAD crystal structures solved to date, two FAD cofactors bind at each dimer interface. In contrast, only one set of FAD binding site residues is conserved in the homology model of the ChsE1-ChsE2 interface. The lack of conserved riboflavin binding residues in ChsE1 and adenosine binding residues in ChsE2 indicates that one FAD cofactor binds at the ChsE1-ChsE2 heterodimer interface (Figure 5B), and that two heterodimeric units comprise the heterotetramer. This homology model is consistent with the experimentally determined FAD stoichiometry for ChsE1-ChsE2. Isolation of monomeric apo-ChsE2 suggests it does not self-associate to form a homodimer and further supports our model.
Tetrameric ChsE1-ChsE2 has two active sites
The FAD binding stoichiometry and binding residue conservation in tetrameric ChsE1-ChsE2 suggest that the complex has two active sites. ACADs require a catalytic base for activity, and this base is highly conserved in the ACAD protein family (Figure 5A). The catalytic base is most often a glutamate and is located at one of two positions in helix G or helices J/K (22). Glutamate at either of these two positions orients in the active site in a catalytically competent configuration in the three-dimensional protein structures of ACADs.
In the ChsE2 sequence alignment with representative ACAD family members, Glu241 aligns with the glutamate of IVD and LCAD in helix G (Figure 5A). There is not a conserved glutamate in helices J/K alignment with ChsE2. Notably, ChsE1 does not have a glutamate or aspartate that aligns with either helix G or helices J/K. The conserved glutamates in helix G and helices J/K align with glycine residues in ChsE1. The lack of a potential active site base, in addition to the lack of riboflavin binding residues in ChsE1 suggests that ChsE1-ChsE2 has only two active sites, in contrast to traditional homotetrameric ACADs, which have four active sites. On the basis of these alignments, we hypothesized that ChsE2 Glu241 is the catalytic base responsible for deprotonation during catalysis.
Analysis of the CoA binding site in the three-dimensional structures of ACADs identified conserved CoA hydrogen-bonding residues Ser142A, Arg256A, and Asp253A (numbering for human MCAD). The side chain of Arg256 forms a hydrogen bond network with the pantetheine group of CoA and is highly conserved in the ACAD family. Arg256 is conserved in ChsE2 as Arg242. In ChsE1, Leu202 aligns with Arg256. Asp253 in MCAD forms a hydrogen bond with the adenine moiety of CoA. In GD and IBD, however, this residue is an Asn and analysis of the crystallographic structure shows the back-bone carbonyl of Asn forms a hydrogen bond with adenine. Asn is conserved in ChsE2 as residue Asn239 and this residue is Val204 in ChsE1. The lack of CoA binding residues in ChsE1 supports our hypothesis that ChsE1 is not an active homtetrameric ACAD.
ChsE1-ChsE2 forms 3-oxo-pregna-4,17-dien-20-carboxyl-CoA(3)
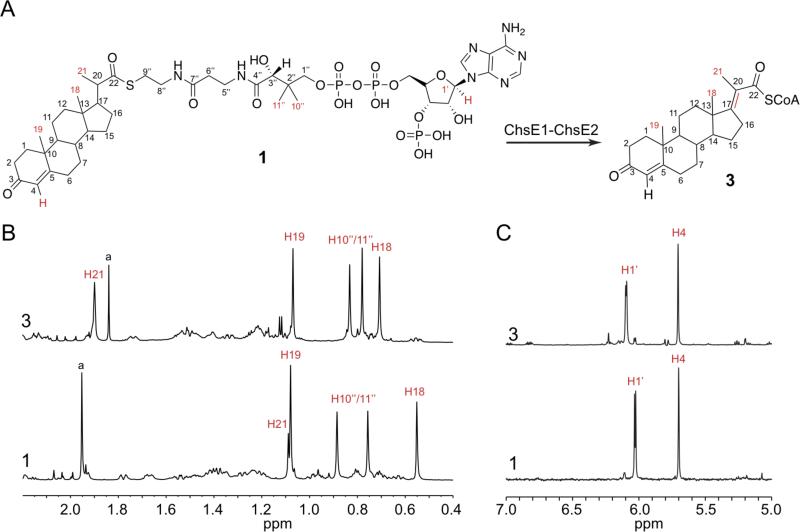
1H spectra of substrate 1 and the product 3 were acquired in D2O (Figure 6). Comparison of substrate and product 1H spectra highlights the disappearance of the C21 methyl hydrogen coupling to H21 (δ1.09, d). The C21 methyl becomes a singlet at 1.84 ppm (Figure 6B). If C20-C21 dehydrogenation had occurred, two new alkene protons would have replaced the C21 methyl in the product spectrum. However, a new proton resonance in the alkene region was not observed (Figure 6C). We conclude that ChsE1-ChsE2 forms the thermodynamically favored tetrasubstituted alkene product 3.
Figure 6. Characterization of dehydrogenated product by ChsE1-ChsE2.
1H spectra (700 MHz) of substrate 1 and ChsE1-ChsE2 assay product 3 illustrating the changes in the methyl (B) and alkene (C) regions. The alkene stereochemistry is represented as (E), but was not determined.
Steady state kinetics of ChsE1-ChsE2
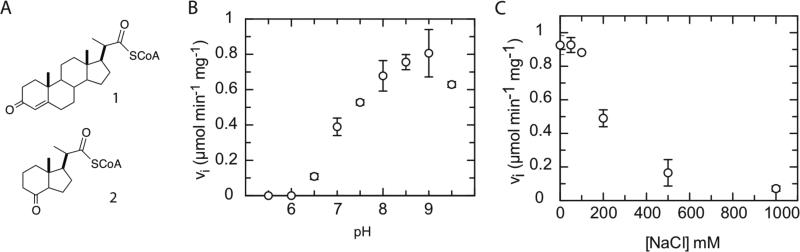
Previously we had demonstrated that the pregnenecarboxyl CoA ester (1) and indanone CoA ester (2) are substrates of ChsE1-ChsE2 (Figure 7A). Here, we determine the pH and ionic strength dependence of ChsE1-ChsE2. Initial velocity data were collected for ChsE1-ChsE2 with substrate 1 from pH 5.5 to pH 9.5 in 0.5 pH unit steps. No activity was observed at pH 6.0 or lower and the highest activity was observed at pH's greater than 8.0 (Figure 7B). Further experiments were conducted at pH 8.5 in TAPS buffer.
Figure 7. Optimization of dehydrogenase activity of ChsE1-ChsE2.
(A) 3-oxo-4-pregnene-20-carboxyl-CoA 1 and 1β- (2’-propanoyl-CoA)-3aα-H-7aβ-methylhexahydro-4-indanone 2, substrates of ChsE1-ChsE2. (B) pH and (C) ionic strength optimization of dehydrogenase activity. Assays were conducted with 50 μM of substrate 1 and 250 μM ferricenium hexafluorophosphate at 25 °C. Assays in C were conducted in 100 mM TAPS buffer pH 8.5.
ChsE1-ChsE2 was then assayed as a function of NaCl concentration to determine the optimal ionic strength (Figure 6C). Initial velocities were determined over NaCl concentrations of 0 to 1.0 M with 50 μM substrate 1. Increasing the ionic strength by addition of NaCl diminished activity and no rate enhancement was observed even at low concentrations of NaCl. We tested the protein stability and substrate solubility at working concentrations using dynamic light scattering at 0 and 0.2 M NaCl. The activity of ChsE1-ChsE2 was reduced by 50% in 0.2 M NaCl. However, neither protein aggregation nor substrate precipitation were observed. The decrease in enzyme activity is not due to protein denaturation or substrate aggregation.
Next, the ionic strength was increased with halide salts KCl, MgCl2, CaCl2, NaI, and NaBr, as well as sodium acetate (Figure S3). The specific activity was reduced in the presence of all the tested salts. Further analysis would be required to determine the cause of rate reduction.
Steady state kinetic constants KM and kcat were determined for CoA thioesters 1 and 2 at the optimized assay conditions of 100 mM TAPS pH 8.5 at 25 °C with 250 μM ferricenium hexafluorophosphate (Table 2). Assays were initiated by the addition of ChsE1-ChsE2 and initial velocities were determined over substrate concentrations of 0 to 200 μM, following the reduction of electron acceptor ferricenium hexafluorophosphate at 300 nm. Controls without enzyme or without substrate showed negligible decreases in absorbance at 300 nm. The calculated specificity constants (kcat/KM) indicate that the 4-ring steroid 1 is the preferred substrate of ChsE1-ChSE2 relative to the 2-ring hexahydroindanone substrate 2.
Table 2.
Michaelis-Menten kinetic constants for ChsE1-ChsE2.1
| Substrate | KM (μM) | kcat (min-1) | kcat/KM (s-1 M-1) |
|---|---|---|---|
| 1 | 5.3 ± 0.9 | 78 ± 1 | 2.5 × 105 ± 0.5 |
| 2 | 86 ± 7 | 5.1 ± 0.2 | 9.8 × 102 ± 0.8 |
ChsE1-ChsE2 was assayed with CoA thioesters 1 and 2 in 100 mM TAPS buffer pH 8.5 with 250 μM ferricenium hexafluorophosphate at 25 °C. Three independent replicates were performed. The errors are the standard deviation of the fit.
Glutamate 241 of ChsE2 is required for dehydrogenase activity of ChsE1-ChsE2
In order to experimentally demonstrate that ChsE2 Glu241 is the active site base of ChsE1-ChsE2, glutamate 241 of ChsE2 was mutated to a glutamine in pigr5. Purified mutant protein showed characteristic flavin absorbances by UV-visible spectroscopy (Figure 1). ChsE1-ChsE2E241Q was analyzed by SDS-PAGE (Figure S1) and analytical size exclusion chromatography (Figure 2), under identical conditions as ChsE1-ChsE2. The elution profiles of the mutant and wild type protein were identical indicating that ChsE1-ChsE2E241Q forms an α2β2 heterotetramer like ChsE1-ChsE2. The activity of ChsE1-ChsE2E241Q was tested under the optimized steady state conditions at pH 8.5 in TAPS buffer with substrate 1. No detectable turnover was observed at concentrations of 5 μM ChsE1-ChsE2E241Q and 200 μM substrate 1.
Discussion
Sterol metabolism by actinomycetes has been extensively studied. However, little is known about the genes required for the β–oxidation of the sterol side chain. Partially metabolized cholesterol side chain intermediates in Nocardia established that cholesterol side-chain metabolism proceeds through C24 and C22 intermediates, presumably via two rounds of conventional fatty acid β-oxidation (23-25). The mechanism of the final loss of propionate has not been established. We previously showed through metabolite characterization in M. tuberculosis igr knockout strain cultured with cholesterol that the igr operon is required for complete metabolism of the cholesterol side chain (9). The enzymes encoded by the igr operon are likely responsible for the final loss of propionate resulting in full side chain metabolism. Here we characterize a cholesterol side chain degrading ACAD, ChsE1-ChsE2, encoded by the M. tuberculosis igr operon.
ACADs have been identified in plants (26), animals, bacteria, nematodes (27), and fungi (28). The family of enzymes has been extensively studied in eukaryotes (29). There are eleven eukaryotic classes of ACADs that vary in their substrate specificities. Seven classes are involved in fatty acid metabolism and the remaining four classes are involved in leucine, isoleucine, valine, lysine, and tryptophan metabolism. Prokaryotic ACADs, also known as Fatty acid degrading E (FadE) enzymes, characterized to date have less distinct substrate specificities than eukaryotic ACADs and have not been categorized into distinct classes (30, 31). However, eukaryotic and bacterial ACAD sequences are highly homologous and typically align well with the eukaryotic ACADs (32).
Our biophysical analysis demonstrates that ChsE1-ChsE2 forms an α2β2 heterotetramer, a unique quartenary structure for an ACAD. Typical ACADs form homotetramers with the exception of the very long chain classes of ACADs that form homodimers and have an additional C terminal 180 residues that bind to the mitochondrial membrane (10, 33).
Bioinformatics analysis and mutagenesis results demonstrate that heteromeric ChsE1-ChsE2 has two FAD binding sites and two active sites, unlike traditional tetrameric ACADs with four active sites, one per subunit. Formation of the ChsE1-ChsE2 complex is necessary for FAD binding. chsE2 expressed in the absence of chsE1 yields monomeric ChsE2 without FAD bound. Bioinformatics analysis show that neither ChsE1 nor ChsE2 have a complete FAD binding site and both chains are required for FAD binding. This analysis further supports the necessity of heteromeric complex formation for forming active protein. The ChsE1-ChsE2 complex from M. tuberculosis represents a third quaternary architecture for ACADs, in which a heterotetrameric ACAD enzyme has two active sites.
The evolution of the ACAD family is highly dynamic and members have evolved to accommodate a variety of substrates by increasing the binding pocket size (32, 34). Highly conserved residues, like the catalytic glutamate, are usually evolutionarily impervious to mutation. It is likely chsE1, chsE2 arose from a gene duplication event, allowing for the evolutionary loss of the chsE1 active site. For a homotetrameric or homodimeric ACAD loss of an active site would result in loss of function. ChsE1 and ChsE2 share only 7% amino acid identity while at the DNA level they share 47% identity. We propose that the complex has evolved to increase the binding pocket size in order to accommodate polycyclic steroid substrates. Further structural analysis is underway to corroborate this hypothesis. Moreover, the distinct differences in protein architecture between M. tuberculosis ChsE1-ChsE2 and human ACADs suggests that specific inhibition of the M. tuberculosis enzyme may be possible.
It is well established that chsE1 and chsE2 are regulated by cholesterol and involved in the cholesterol metabolism of M. tuberculosis. We have previously shown that this enzyme complex is able to bind and turn over steroid substrates. In this work, our experimentally determined specificity constants for 3-oxo-4-pregnene-20-carboxyl-CoA (1) and 1β- (2’-propanoyl-CoA)-3aα-H-7aβ-methylhexahydro-4-indanone (2) demonstrate that ChsE1-ChsE2 strongly prefers the ring nucleus intact species to the hexahydroindinone species, in vitro. This suggests the side chain of cholesterol is fully metabolized prior to aromatization and degradation of the ring system.
Assignment of biochemical function to genes is complicated by the large number of annotated fatty acid oxidation genes in M. tuberculosis (35). It is impossible to assign enzymes to specific biochemical steps in sterol metabolism from sequence data alone. We propose that the heterotetrameric architecture is important for binding sterols and possibly other polycyclic substrates. Our biochemical characterization and bioinformatic analysis of a heteromeric sterol oxidizing ACAD provide information that will be applied to further proper annotation of the M. tuberculosis genome.
Supplementary Material
Acknowledgements
We would like to thank Francis Picart (NMR Coordinator, Stony Brook University) for his assistance in NMR acquisition.
Funding: *This work was supported by the NIH (R21AI092455, R01HL53306, S10RR021008, N.S.S.), NSF-BIO1039771 (NMR) and a DOE-GAANN fellowship (S.T.T.).
Abbreviations
- M. tuberculosis
Mycobacterium tuberculosis
- ACAD
acyl-CoA dehydrogenase
- igr
intracellular growth
- IPTG
isopropyl β-D-1thiogalactopyranoside
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- TRIS
tris(hydroxymethyl)aminomethane
- IMAC
immobilized metal ion affinity chromatography
- TAPS
N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid
- TCEP
tris(2-carboxyethyl)phosphine
- FAD
flavin adenine dinucleotide
- M. smegmatis
Mycobacterium smegmatis
- AUC
analytical ultracentrifugation
- CoA
coenzyme A
Footnotes
Supporting Information Available
Supplementary figures including SDS-PAGE of purified proteins (Figure S1), LC/UV/MS analysis of isolated flavin from ChsE1-ChsE2 (Figure S2), and the salt dependence of dehydrogenase activity (Figure S3) can be found in the supporting information. These materials are available free of charge online at http://pubs.acs.org.
References
- 1.WHO Global Tuberculosis Control 2011. 2011.
- 2.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. The actinobacterial mce4 locus encodes a steroid transporter. J Biol Chem. 2008;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesbitt N, Yang X, Fontán P, Kolesnikova I, Smith I, Sampson NS, Dubnau E. A thiolase of M. tuberculosis is required for virulence and for production of androstenedione and androstadienedione from cholesterol. Infect Immun. 2010;78:275–282. doi: 10.1128/IAI.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang JC, Miner MD, Pandey AK, Gill WP, Harik NS, Sassetti CM, Sherman DR. igr Genes and Mycobacterium tuberculosis cholesterol metabolism. J Bacteriol. 2009;191:5232–5239. doi: 10.1128/JB.00452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang JC, Harik NS, Liao RP, Sherman DR. Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. J Infect Dis. 2007;196:788–795. doi: 10.1086/520089. [DOI] [PubMed] [Google Scholar]
- 9.Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J Biol Chem. 2011;286:43668–43678. doi: 10.1074/jbc.M111.313643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, Cao X. Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem Biophys Res Commun. 2002;297:1033–1042. doi: 10.1016/s0006-291x(02)02336-7. [DOI] [PubMed] [Google Scholar]
- 11.Souri M, Aoyama T, Hoganson G, Hashimoto T. Very-long-chain acyl-CoA dehydrogenase subunit assembles to the dimer form on mitochondrial inner membrane. FEBS Lett. 1998;426:187–190. doi: 10.1016/s0014-5793(98)00343-3. [DOI] [PubMed] [Google Scholar]
- 12.Moore S. 'Round-the-horn site-directed mutagenesis. Wikiomics. 2012 [Google Scholar]
- 13.Daugelat S, Kowall J, Mattow J, Bumann D, Winter R, Hurwitz R, Kaufmann SH. The RD1 proteins of Mycobacterium tuberculosis: expression in Mycobacterium smegmatis and biochemical characterization. Microbes Infect. 2003;5:1082–1095. doi: 10.1016/s1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 14.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 15.Winkler R. ESIprot: a universal tool for charge state determination and molecular weight calculation of proteins from electrospray ionization mass spectrometry data. Rapid Commun Mass Spectrom. 2010;24:285–294. doi: 10.1002/rcm.4384. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stierand K, Maass PC, Rarey M. Molecular complexes at a glance: automated generation of two-dimensional complex diagrams. Bioinformatics. 2006;22:1710–1716. doi: 10.1093/bioinformatics/btl150. [DOI] [PubMed] [Google Scholar]
- 18.Capyk JK, Casabon I, Gruninger R, Strynadka NC, Eltis LD. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J Biol Chem. 2011;286:40717–40724. doi: 10.1074/jbc.M111.289975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehman TC, Thorpe C. Alternate electron acceptors for medium-chain acyl-CoA dehydrogenase: use of ferricenium salts. Biochemistry. 1990;29:10594–10602. doi: 10.1021/bi00499a004. [DOI] [PubMed] [Google Scholar]
- 20.Ellis KJ, Morrison JF. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 1982;87:405–426. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 21.Hwang TL, Shaka AJ. Water suppression that works - excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson Ser A. 1995;112:275–279. [Google Scholar]
- 22.Djordjevic S, Dong Y, Paschke R, Frerman FE, Strauss AW, Kim JJ. Identification of the catalytic base in long chain acyl-CoA dehydrogenase. Biochemistry. 1994;33:4258–4264. doi: 10.1021/bi00180a021. [DOI] [PubMed] [Google Scholar]
- 23.Sih CJ, Tai HH, Tsong YY. The mechanism of microbial conversion of cholesterol into 17-keto steroids. J Am Chem Soc. 1967;89:1957–1958. doi: 10.1021/ja00984a039. [DOI] [PubMed] [Google Scholar]
- 24.Sih CJ, Tai HH, Tsong YY, Lee SS, Coombe RG. Mechanisms of steroid oxidation by microorganisms. XIV. Pathway of cholesterol side-chain degradation. Biochemistry. 1968;7:808–818. doi: 10.1021/bi00842a039. [DOI] [PubMed] [Google Scholar]
- 25.Sih CJ, Wang KC, Tai HH. Mechanisms of steroid oxidation by microorganisms. XIII. C22 acid intermediates in degradation of cholesterol side chain. Biochemistry. 1968;7:796–807. doi: 10.1021/bi00842a038. [DOI] [PubMed] [Google Scholar]
- 26.Bode K, Hooks MA, Couee II. Identification, separation, and characterization of acyl-coenzyme A dehydrogenases involved in mitochondrial beta-oxidation in higher plants. Plant Physiol. 1999;119:1305–1314. doi: 10.1104/pp.119.4.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komuniecki R, Fekete S, Thissen-Parra J. Purification and characterization of the 2-methyl branched-chain Acyl-CoA dehydrogenase, an enzyme involved in NADH-dependent enoyl-CoA reduction in anaerobic mitochondria of the nematode, Ascaris suum. J Biol Chem. 1985;260:4770–4777. [PubMed] [Google Scholar]
- 28.Kionka C, Kunau WH. Inducible beta-oxidation pathway in Neurospora crassa. J Bacteriol. 1985;161:153–157. doi: 10.1128/jb.161.1.153-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunau WH, Dommes V, Schulz H. beta-oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res. 1995;34:267–342. doi: 10.1016/0163-7827(95)00011-9. [DOI] [PubMed] [Google Scholar]
- 30.Campbell JW, Cronan JE., Jr. The enigmatic Escherichia coli fadE gene is yafH. J Bacteriol. 2002;184:3759–3764. doi: 10.1128/JB.184.13.3759-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen YQ, Lang BF, Burger G. Diversity and dispersal of a ubiquitous protein family: acyl-CoA dehydrogenases. Nucleic Acids Res. 2009;37:5619–5631. doi: 10.1093/nar/gkp566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swigonova Z, Mohsen AW, Vockley J. Acyl-CoA dehydrogenases: Dynamic history of protein family evolution. J Mol Evol. 2009;69:176–193. doi: 10.1007/s00239-009-9263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAndrew RP, Wang Y, Mohsen AW, He M, Vockley J, Kim JJ. Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase. J Biol Chem. 2008;283:9435–9443. doi: 10.1074/jbc.M709135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiltunen JK, Qin Y. beta-oxidation - strategies for the metabolism of a wide variety of acyl-CoA esters. Biochim Biophys Acta. 2000;1484:117–128. doi: 10.1016/s1388-1981(00)00013-5. [DOI] [PubMed] [Google Scholar]
- 35.Cole ST, Brosch R, Parkhil J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.