Abstract
Sickle cell hemoglobin conveys resistance to malaria. In this issue of Cell Host & Microbe, LaMonte et al. (2012) demonstrate a surprising mechanism for this innate immunity. A microRNA enriched in sickle red blood cells is translocated into the parasite, incorporated covalently into P. falciparum mRNAs and inhibits parasite growth.
Haldane was the first to suggest that individuals with the red blood cell disorder β-thalassemia are more frequent in malaria endemic regions because the mutation confers resistance to the parasite (Haldane, 1949). Several other red blood cell disorders are associated with malaria resistance, including G6PD deficiency, pyruvate kinase deficiency, and α-thalassemia. Most strikingly the variant of the β-globin gene that causes sickle cell disease, HbS, protects against clinical malaria caused by infection with the most virulent species, Plasmodium falciparum (Hill et al., 1991).
Our current understanding of the molecular mechanism for this protection rests on several observations. First, under conditions of low O2 tension, both homozygous HbSS sickle cells and heterozygous HbAS sickle cell trait cells do not support growth of P. falciparum, in contrast to wild-type HbAA red blood cells (Pasvol et al., 1978). This was attributed to the enhanced propensity of HbSS to polymerize, creating an environment not conducive to parasite growth. A second mechanism posits that HbSS erythrocytes have an enhanced ability to elicit immunity, resulting in a more rapid clearance of parasitized erythrocytes (Ayi et al., 2004). It has also been suggested that presentation of the major virulence protein, PfEMP1, on the surface of parasite-infected erythrocytes is impaired in HbSS and HbCC red blood cells (Fairhurst et al., 2005). Recently, a molecular mechanism for this altered presentation has been suggested to result from an aberrant actin remodeling in HbSS and HbCC red blood cells (Cyrklaff et al., 2011).
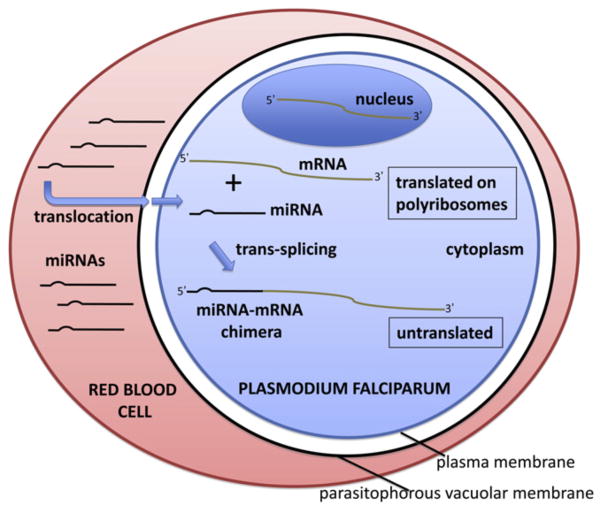
LaMonte et al. (2012) now add a further layer of complexity to the interactions between the host erythrocyte and the P. falciparum parasite (Figure 1). They find that at least two human microRNAs (miRNAs), miR-451 and miR-233, are more abundant in both sickle cells and in the epidemiologically relevant sickle cell trait cells compared to normal red blood cells. In tightly controlled experiments, they show that miR-451 is translocated into the parasite, where it is incorporated covalently into P. falciparum mRNAs, leading to an inhibition of mRNA translation and a modest but significant reduction in parasite growth. When overexpressed in normal erythrocytes, miR-451 and miR-223 reduced parasite growth; conversely, blocking the erythrocyte to parasite translocation of miR-451 and miR-223, using 2′-O-methyl anti-sense oligonucleotides, diminished malaria resistance in both sickle cells and sickle cell trait cells.
Figure 1. Model for the Inhibition of Plasmodium falciparum Growth by Red Blood Cell miRNAs.
The P. falciparum parasite invades and grows within the host erythrocyte. Red blood cell miRNAs, enriched in sickle cells, translocate to the parasite cytoplasm, crossing over both the parasitophorous vacuolar membrane and the parasite plasma membrane. Within the parasite cytoplasm, the miRNAs are trans-spliced onto the 5′ ends of specific parasite mRNA transcripts, forming miRNA-mRNA chimeras. These chimeras are unable to be loaded onto poly-ribosomes or translated, resulting in the inhibition of parasite growth.
miRNAs are a recently discovered class of small noncoding eukaryotic RNAs 18–24 nucleotides long that downregulate target genes at a posttranscriptional level. The majority of miRNA genes are transcribed by RNA polymerase II into long primary (pri) miRNA transcripts, processed by the nuclear nuclease Drosha into ~60 bp hairpins termed precursor (pre) miRNAs, and further cleaved in the cytosol by the Dicer nuclease into mature miRNAs. Mature miRNAs are then incorporated into the multiprotein RNA-induced silencing complex (RISC), exerting posttranscriptional repression of target mRNAs, either by inducing mRNA degradation or blocking mRNA translation.
Why any miRNAs are present in mature erythrocytes is a mystery, since there is no ongoing protein synthesis. Red blood cell miRNAs like miR-451 and miR-223 are generated during erythroblast proliferation and differentiation, prior to enucleation. miR-451 is induced during red blood cell development and is important for formation of erythrocytes (Dore et al., 2008), but it is unclear why it is retained in mature erythrocytes or why its level is higher in sickle than normal red blood cells. Experimentally, miRNAs can be introduced into mature erythrocytes, so exogenous miR-223 or miR-451 could possibly be used therapeutically in treating malaria.
How miRNAs are incorporated into intracellular malaria parasites is also unknown. Plasmodium parasites are contained within a parasitophorous vacuolar membrane, and thus the miRNA would have to cross from the red blood cell cytosol through both this membrane and the parasite’s plasma membrane. Direct visualization of mir-451 in the parasitophorous vacuolar membrane will need to be reconciled with its activity within the parasite. There is a precedent for this, however, as one can use preloaded erythrocytes to introduce plasmid DNA into the parasite (Deitsch et al., 2001), although the efficiency of this process is very low at ~1 in 106 cells. It will be of great interest to elucidate the mechanism of miRNA translocation, as this may form the basis for more efficient genetic manipulation of this Plasmodium species.
Plasmodium parasites do not contain Dicer or any Argonaute homologs that comprise the RISC complex. RNA interference, including conventional functions of miRNAs, is not functional in these organisms. Thus, it was a huge surprise that the translocated miRNAs became covalently linked to certain Plasmodium mRNAs, forming chimeric RNAs. Indeed, several transcripts in a Plasmodium EST database contain miR-451 at their 5′ ends, and this linkage was subsequently confirmed by both real-time PCR and northern blots on parasite material. Covalent linkage requires a trans-splicing event, but in unicellular eukaryotes this has only been observed in kinetoplastid organisms, where a leader sequence is spliced onto the 5′ end of all RNA transcripts during processing and maturation (Sutton and Boothroyd, 1986). In P. falciparum, miRNA tagging occurs to a minority of transcripts, including genes such as PKA-R, PEAMT, and the 28S and 18S rRNAs. We do not know what determines the specific enrichment of certain miRNAs or the incorporation of these miRNAs to specifc parasite mRNAs.
This study raises many other questions. Has increased miRNA-451 or miR-223 expression in red blood cells been selected for as a malaria resistance factor, similar to the selection for other red blood cell disorders? Why does a reduction in PKA-R result in reduced parasite proliferation and increased conversion to sexual stage parasites? What, if any, are the functional consequences of the other miRNAs differentially regulated between HbSS and HbAA? Is there conservation of this mechanism in protection against other Plasmodium species? What is the relative contribution of miRNA-based inhibition of parasite growth in HbSS erythrocytes compared to other postulated mechanisms of protection, particularly under low oxygen tension when inhibition of parasite growth is most marked (Pasvol et al., 1978)?
Clearly, Lamonte and colleagues have revealed a startling and unique mechanism of cross species trans-splicing in P. falciparum-infected erythrocytes where the effector molecule of parasitic inhibition is the miRNA itself.
References
- Ayi K, Turrini F, Piga A, Arese P. Blood. 2004;104:3364–3371. doi: 10.1182/blood-2003-11-3820. [DOI] [PubMed] [Google Scholar]
- Cyrklaff M, Sanchez CP, Kilian N, Bisseye C, Simpore J, Frischknecht F, Lanzer M. Science. 2011;334:1283–1286. doi: 10.1126/science.1213775. [DOI] [PubMed] [Google Scholar]
- Deitsch K, Driskill C, Wellems T. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst RM, Baruch DI, Brittain NJ, Ostera GR, Wallach JS, Hoang HL, Hayton K, Guindo A, Makobongo MO, Schwartz OM, et al. Nature. 2005;435:1117–1121. doi: 10.1038/nature03631. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Proc VIII Int Cong Genet Hereditas. 1949;35:267–273. [Google Scholar]
- Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, Chapman L, Telen MJ, Ohler U, Nicchitta CV, Haystead T, Chi J-T. Cell Host Microbe. 2012;12:187–199. doi: 10.1016/j.chom.2012.06.007. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasvol G, Weatherall DJ, Wilson RJ. Nature. 1978;274:701–703. doi: 10.1038/274701a0. [DOI] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]