Abstract
Polymorphisms in the fat mass and obesity-associated gene (FTO) are associated with human obesity and obesity-prone behaviors, including increased food intake and a preference for energy-dense foods. FTO demethylates N6-methyladenosine, a potential regulatory RNA modification, but the mechanisms by which FTO predisposes humans to obesity remain unclear. In adiposity-matched, normal-weight humans, we showed that subjects homozygous for the FTO “obesity-risk” rs9939609 A allele have dysregulated circulating levels of the orexigenic hormone acyl-ghrelin and attenuated postprandial appetite reduction. Using functional MRI (fMRI) in normal-weight AA and TT humans, we found that the FTO genotype modulates the neural responses to food images in homeostatic and brain reward regions. Furthermore, AA and TT subjects exhibited divergent neural responsiveness to circulating acyl-ghrelin within brain regions that regulate appetite, reward processing, and incentive motivation. In cell models, FTO overexpression reduced ghrelin mRNA N6-methyladenosine methylation, concomitantly increasing ghrelin mRNA and peptide levels. Furthermore, peripheral blood cells from AA human subjects exhibited increased FTO mRNA, reduced ghrelin mRNA N6-methyladenosine methylation, and increased ghrelin mRNA abundance compared with TT subjects. Our findings show that FTO regulates ghrelin, a key mediator of ingestive behavior, and offer insight into how FTO obesity-risk alleles predispose to increased energy intake and obesity in humans.
Introduction
FTO is an AlkB-like 2-oxoglutarate–dependent nucleic acid demethylase of uncertain cellular function (1). Genome-wide association studies (GWAS) have reliably established that SNPs within the first intron of FTO are robustly associated with increased BMI and adiposity across different ages and populations (2–6). Subjects homozygous for the “obesity-risk” A allele of FTO rs9939609 have a 1.7-fold increased risk for obesity compared with subjects homozygous for the low-risk T allele (2). Evidence to date suggests that the association between SNPs in FTO and BMI is predominantly driven by increased energy intake. Subjects homozygous for the obesity-risk A allele of rs9939609 exhibit overall increased ad libitum food-intake (7–9), particularly fat consumption (7, 9–11), and impaired satiety (12, 13). Furthermore, preschool AA children (AA denotes homozygosity for the A obesity-risk allele in the rs9939609 FTO variant) exhibit obesity-prone eating behaviors, including increased food responsiveness and a tendency to eat in response to external cues, prior to the development of an association between FTO rs9939609 and BMI (14). FTO rs9939609 also has per-allele effects on feeding behavior (10) and BMI (2).
Several lines of evidence from rodent studies are consistent with FTO playing a key role in regulating energy homeostasis. First, Fto is highly expressed in brain regions controlling feeding and energy expenditure, such as the hypothalamus (1, 15). Second, hypothalamic Fto expression is modulated by fasting (1, 16–20) and restricted access to food (21), although both upregulation (17, 18) and downregulation (1, 16, 19) have been reported, apparently depending on the severity of caloric restriction. Exposure to a high-fat diet also modulates hypothalamic Fto expression, with downregulation reported with short-term exposure (20) and upregulation with more prolonged exposure (22). Further evidence suggesting that Fto is nutritionally regulated comes from in vitro studies in mouse and human cell lines in which glucose and total amino acid deprivation decreases FTO expression (23, 24). Finally, phenotypic analyses of transgenic mice in which FTO function is either eliminated or enhanced further implicate a role for FTO in regulating energy homeostasis. Mice with constitutive Fto deletion exhibit a tendency toward leanness (25–27), and similarly, mice generated with a dominant missense mutation in the C-terminal of FTO, leading to a partial loss of function, also display a lean phenotype (28). In contrast, mice with global Fto overexpression have increased food intake, body weight, and fat mass (29).
FTO directly demethylates N6-methyladenosine (m6A), a naturally occurring adenosine modification in RNA (30). Two studies combining m6A-specific methylated RNA immunoprecipitation with next-generation sequencing have defined human and mouse m6A RNA methylomes for specific cell types and suggest that m6A methylation of RNA plays a fundamental role in the regulation of gene expression (31, 32). In addition, recent cellular studies have implicated a role for FTO in the coupling of amino acid levels to mammalian target of rapamycin complex 1 (MTORC1) signaling, suggesting that FTO SNPs may influence the ability of cells to sense amino acids (24). However, despite the compelling genetic evidence that SNPs in FTO are associated with increased BMI and the recent advances in our understanding of FTO targets, the mechanisms by which FTO SNPs lead to obesity-prone eating behavior and obesity remain unknown.
Gut hormones are released from the gastrointestinal tract in response to nutrient ingestion and regulate appetite and body weight (33). Circulating levels of the orexigenic hormone, acyl-ghrelin, and the satiety hormone, peptide YY3-36 (PYY3-36), alter appetite, food intake (34–36), and modulate brain activity within both homeostatic and reward-related brain regions (33, 37–39). Thus, we hypothesized that the FTO rs9939609 genotype impacts circulating PYY3-36 and/or acyl-ghrelin levels and the neural responses to food cues, thereby resulting in the reduced satiety (12, 13), increased energy intake (7–9), preference for energy-dense foods (7, 9–11), and increased food-cue responsivity (14) seen with the AA genotype.
To test this hypothesis, we assessed appetite and circulating ghrelin and PYY3-36 levels in AA and TT (TT denotes homozygosity for the T low obesity-risk allele for the rs9939609 FTO variant) adiposity-matched male subjects in response to a standard test meal. We found that AA subjects exhibited attenuated postprandial suppression of both hunger and circulating acyl-ghrelin levels. In parallel, we used blood oxygen level–dependent (BOLD) functional MRI (fMRI) scanning to assess the effect of the FTO genotype on neural responsiveness to food cues and endogenous changes in acyl-ghrelin concentrations. Next, we undertook a series of studies to explore the mechanistic link between FTO and ghrelin. We measured ghrelin levels in Fto KO mice (25) and examined the effect of FTO overexpression on ghrelin mRNA abundance and m6A methylation and total ghrelin and acyl-ghrelin concentrations in both MGN3-1 cells, a ghrelin-producing mouse cell line (40), and in HEK293T human cells. Finally, we analyzed FTO expression, ghrelin mRNA m6A methylation, and ghrelin expression in peripheral blood cells from AA and TT subjects.
Results
Normal-weight subjects homozygous for the obesity-associated FTO rs9939609 A variant exhibit attenuated postprandial suppression of hunger and circulating acyl-ghrelin.
We recruited 359 healthy, normal-weight (BMI = 22.5 ± 0.1 kg/m2) males of mixed European descent, aged 18–35 years (age = 22.9 ± 0.3 years), who underwent FTO rs9939609 genotyping and body composition analysis. Forty-five subjects (13%) were homozygous for the A allele, 149 (41%) were homozygous for the T allele, and 165 (46%) were AT heterozygotes. AA subjects exhibited higher BMI, body fat mass, and visceral fat area (VFA) than TT subjects (P < 0.05, Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI44403DS1), consistent with the reported effects of the A allele. Thus, in order to remove the confounding effects of adiposity from our subsequent studies, all our AA and TT study groups were matched for age, BMI, fat mass, and VFA.
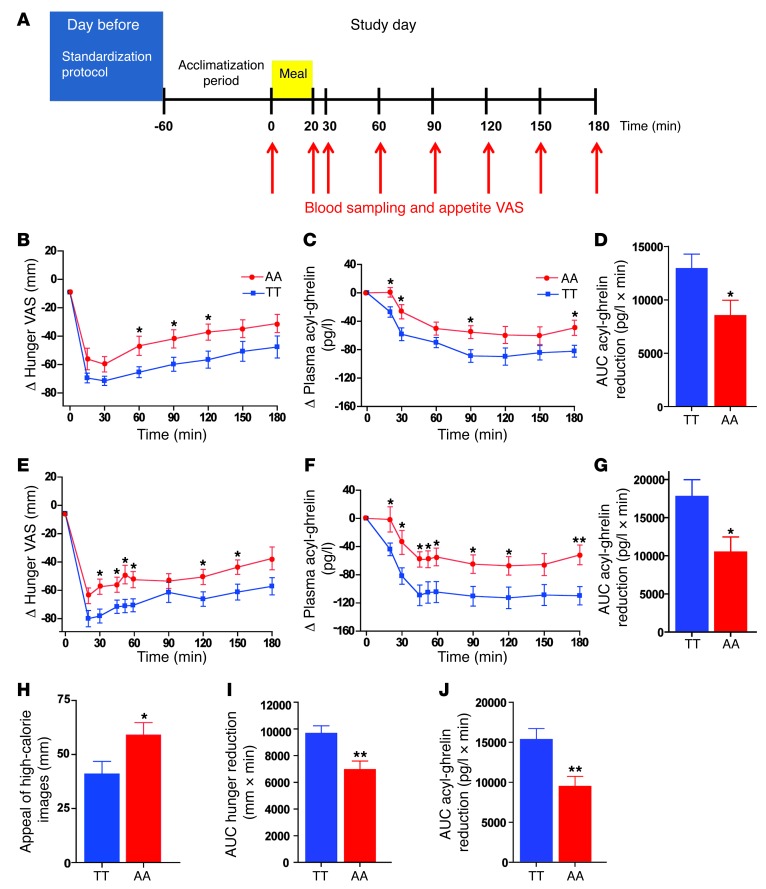
Ten AA and 10 TT overnight-fasted subjects (Supplemental Table 2) underwent a standard test meal study (Supplemental Table 3) using our established protocols (Figure 1A) (41). Subjective appetite was evaluated using visual analog scales (VASs) (39, 41) preprandially, at 20 and 30 minutes following meal ingestion, and every 30 minutes thereafter until 180 minutes postmeal. Simultaneously, blood sampling for the measurement of circulating concentrations of the anorectic hormone, PYY3-36, and the orexigenic hormone, acyl-ghrelin, was undertaken. Fasting appetite VASs were similar in AA and TT groups (fasting hunger [mm]: TT = 70 ± 2, AA = 64 ± 6, P = 0.33; fasting fullness [mm] TT = 11 ± 3, AA = 16 ± 5, P = 0.34). However, postprandially, AA subjects exhibited an attenuated suppression of hunger compared with TT subjects (Figure 1B, effect of genotype on AUC Δ hunger suppression, F = 5.09, P = 0.037). We observed no effect of the rs9939609 FTO genotype on circulating fasted plasma leptin levels (fasted plasma leptin concentration [ng/ml]: TT = 2.1 ± 0.3, AA = 1.9 ± 0.3, P = 0.68); PYY3-36 levels (fasted plasma PYY3-36 concentration [pmol/l]: TT = 29 ± 3, AA = 27 ± 6, P = 0.86; AUC Δ PYY3-36 increase [pmol/l × min]: TT = 3,219 ± 431, AA = 3487 ± 571, P = 0.71); or postprandial changes in fullness scores (AUC Δ fullness [mm × min]: TT = 8,302 ± 972, AA = 7469 ± 781, P = 0.50). While there was a trend toward reduced fasting acyl-ghrelin concentrations in AA compared with TT subjects (P = 0.10) (Supplemental Table 4), the ratio of acyl-ghrelin to total ghrelin was comparable in both genotypes (P = 0.90, Supplemental Table 4). As anticipated, meal ingestion suppressed circulating acyl-ghrelin levels in TT subjects. However, in response to the same caloric load, AA subjects failed to suppress acyl-ghrelin appropriately (effect of genotype on AUC Δ acyl-ghrelin suppression, F = 5.70, P = 0.028) (Supplemental Table 4 and Figure 1, C and D). The ratio of circulating acyl-ghrelin to total ghrelin was also increased postprandially in AA subjects (Supplemental Figure 1 and Supplemental Table 4). These findings suggest that altered postprandial circulating ghrelin might contribute to the altered appetite and food intake seen in AA subjects.
Figure 1. Hunger scores, plasma acyl-ghrelin levels, and appeal of hedonic images in TT and AA subjects.
(A) Standard test meal study protocol. On the day prior to the study, subjects consumed the same evening meal at 20:00, then fasted and drank only water. On the study day, a venous cannula was inserted into a left forearm vein, and a period of 1 hour was allowed for acclimatization. At t0, a baseline blood sample and appetite VAS were taken. Subjects were then given a standard test meal to consume within 20 minutes. Blood samples and appetite VAS were taken at t20 and t30 and every 30 minutes thereafter until t180. (B–D) Postprandial responses in 10 TT (blue squares) and 10 AA (red circles) adiposity-matched subjects to a meal at t0. (B) Δ Hunger, (C) Δ plasma acyl-ghrelin concentrations, and (D) AUC Δ acyl-ghrelin in TT (blue bar) and AA (red bar) subjects. (E–G) fMRI fed–study day postprandial responses in 12 TT (blue squares) and 12 AA (red circles) adiposity-matched subjects to a meal at t0. (E) Δ Hunger, (F) Δ plasma acyl-ghrelin concentrations, and (G) AUC Δ acyl-ghrelin suppression in TT (blue bar) and AA (red bar) subjects. (H) Postprandial appeal ratings of hedonic, high-calorie food images from 12 TT (blue bar) and 12 AA (red bar) fMRI study subjects. (I and J) Combined postprandial (I) AUC Δ hunger reduction and (J) AUC Δ acyl-ghrelin suppression in 22 TT (blue bars) and 22 AA (red bars) subjects. Data are presented as the mean ± SEM. *P < 0.05; **P < 0.01.
FTO rs9939609 genotype affects the neural responses to food images.
To identify the neural circuits underlying the altered appetite responses and hedonic food susceptibility of AA compared with TT subjects and to assess the impact of the dysregulated circulating acyl-ghrelin upon such circuits, we examined the neural responses to visual food cues in the fasted and fed states using fMRI. From our database, a new cohort consisting of 12 AA and 12 TT age- and adiposity-matched, right-handed, normal-weight males of mixed European descent (Supplemental Table 5) was studied on 2 separate occasions: once after an overnight fast (fasted study day) and once following our standard test meal (fed study day) in a randomized crossover design (Supplemental Figure 2). On both study days, subjects underwent fMRI scanning to measure their BOLD responses while viewing images of low-calorie foods, high-calorie foods, and non-food items that were pseudorandomly presented in a block design format (Supplemental Figure 3, A and B). Blood was sampled concurrently with the appetite-related VASs for the measurement of plasma acyl-ghrelin (Supplemental Figure 2). An additional VAS question presented at the end of each block of images was used to rate the pleasantness of the images projected and to evaluate individual encoding of the reward value of visual food cues (Supplemental Figure 3B).
In agreement with our previous study, the AA subjects displayed an attenuated postprandial hunger reduction (effect of genotype AUC Δ hunger reduction, F = 6.538, P = 0.018) (Figure 1E and Supplemental Table 6) and reduced postprandial suppression of acyl-ghrelin (effect of genotype on Δ acyl-ghrelin, F = 7.02, P = 0.012) (Figure 1, F and G). Furthermore, AA subjects rated high-calorie food images as significantly more appealing in the postprandial state compared with TT subjects (Figure 1H). The combined results from the 2 meal studies, using linear regression analysis to account for potential differences between studies, revealed a robust effect of the FTO genotype on both postprandial hunger reduction (AUC Δ hunger reduction [mm × min]: TT = 9671 ± 566, AA = 6957 ± 641, P = 0.003) (Figure 1I) and acyl-ghrelin suppression (AUC Δ acyl-ghrelin suppression [pg/l × min]: TT = 15298 ± 1408, AA = 9439 ± 1291, P = 0.002) (Figure 1J).
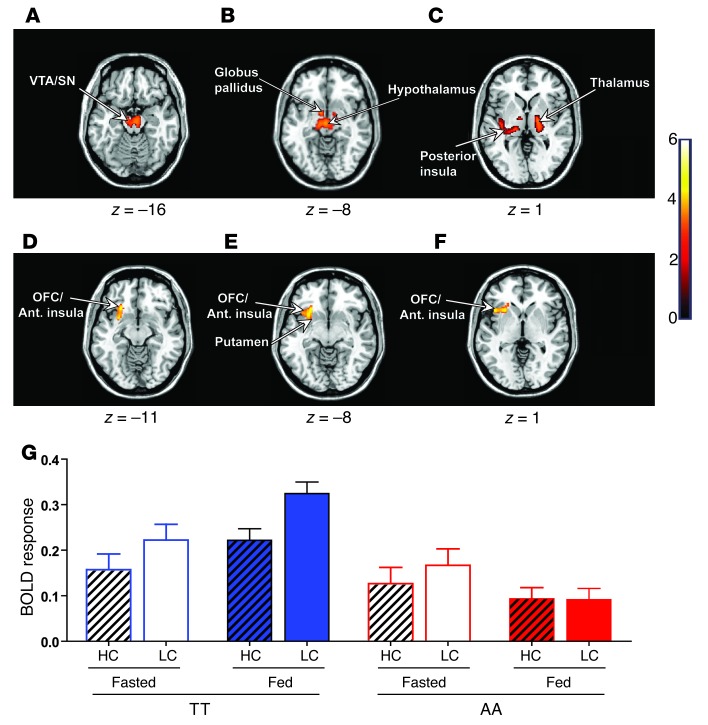
For our fMRI analyses, we examined 2 primary contrasts of interest, one to explore generic (homeostatic) food-related processes (all food images greater than non-food images) and a second to explore hedonic responses (high-calorie food images greater than low-calorie food images). We explored the effects of the FTO genotype on these 2 BOLD contrasts using 2 primary random effects (group level) analyses: (a) separate group comparisons for fed or fasted states, and (b) a full state-by-genotype interaction. We only report the results that survived correction for multiple comparisons. Under fasted conditions, whole-brain analysis revealed that the FTO genotype significantly affected the neural responses to food images (all food greater than non-food) in homeostatic and reward-related brain regions. Compared with TT individuals, AA subjects showed a reduced BOLD response to food-related images within the hypothalamus, left ventral tegmental area/substantia nigra (VTA/SN), left posterior insula, left globus pallidus, left thalamus, and left hippocampus (Figure 2, A–C, and Supplemental Table 7), at a stringent whole-brain corrected threshold. Given that AA subjects preferentially select high-calorie/hedonic foods (7, 9–11), we next compared the BOLD response with high-calorie versus low-calorie food images in the fed versus fasted states. Whole-brain analysis revealed a significant interaction between nutritional state (fed versus fasted), food picture incentive value (high-calorie greater than low-calorie), and genotype (TT versus AA) in a cluster region within the left anterior insula extending into the lateral orbitofrontal cortex/Brodmann area 47 (OFC/BA47) and the left putamen (Figure 2, D–F, and Supplemental Table 8), key brain regions implicated in encoding food reward and controlling goal-directed behavior (39, 42). Post-hoc analysis revealed that the greatest difference in BOLD response to high-calorie and low-calorie images in the TT subjects was seen in the fed compared with the fasted state. Importantly, activity in this region was greatest for low-calorie images. In contrast, AA subjects exhibited the opposite effect, with a reduced difference in BOLD response between high-calorie and low-calorie images in the fed compared with the fasted state (Figure 2E). These findings demonstrate a striking divergence in neural responsivity to food cues within brain regions linked to reward and behavioral control in AA subjects (compared with TT subjects) which, in turn, may underlie their food preferences and obesity predisposition.
Figure 2. Effect of the FTO rs9939609 genotype on BOLD responses to food and non-food images.
(A–F) Axial slices with superimposed group functional activity. (A–C) Brain regions where the TT and AA genotypes exhibited significantly different BOLD responses while viewing food images (all food greater than non-food) in the fasted state, with TT subjects showing a greater BOLD response than AA subjects. (D–F) Brain regions where a marked interaction between the FTO genotype (TT versus AA) and nutritional state (fed versus fasted) was found in BOLD responses to high-incentive- versus low-incentive-value food images (high-calorie greater than low-calorie). Left side of each panel is the left side of the brain. z is the MNI space z coordinate of the axial slice. T color scale reflects the T value of the functional activity. Results are presented at a threshold of P < 0.05, FWE corrected on the basis of cluster extent. Ant. insula, anterior insula. (G) Mean response (β coefficients) for the left anterior insula/OFC (BA47) cluster displaying genotype-by-nutritional state interaction for the high-calorie versus low-calorie contrast. Graph shows cluster mean parameter estimates ± SEM for high-calorie (hatched bars) and low-calorie food images (bars without hatching) in the fasted (open bars) and fed (solid bars) states in TT (blue bars) and AA (red bars) subjects. HC, high-calorie; LC, low-calorie.
Circulating acyl-ghrelin levels differentially modulate the neural responses to food cues in AA versus TT subjects.
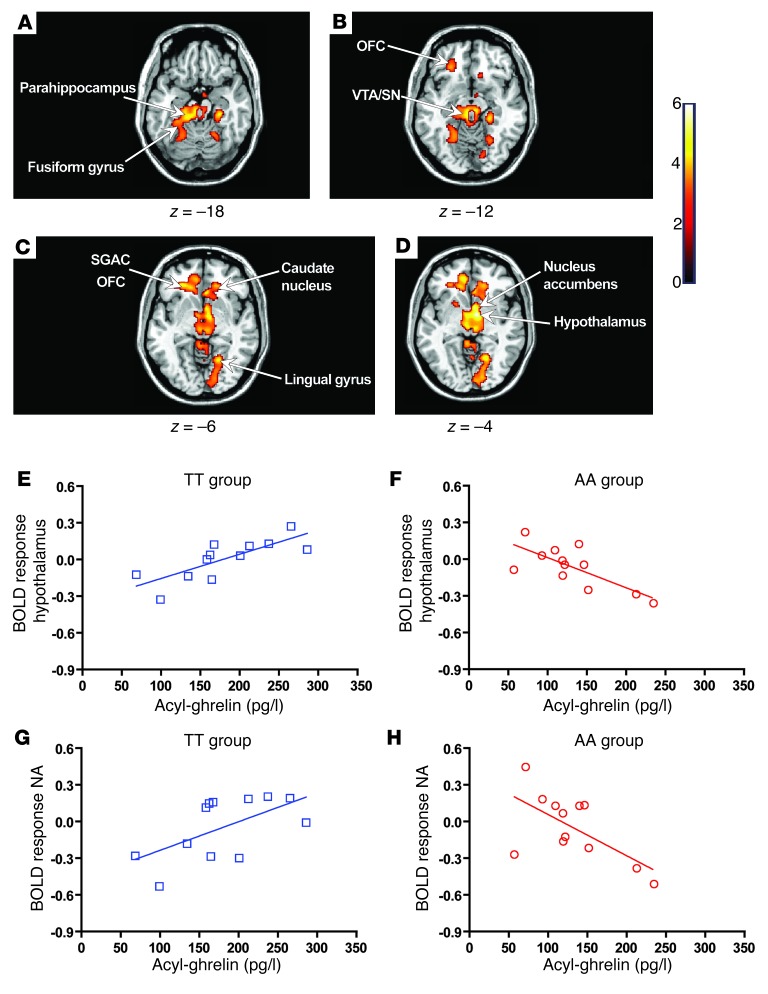
Using whole-brain analysis, we examined the effect of the FTO rs9939609 genotype on the relationship between circulating acyl-ghrelin levels and neural responses to visual food cues. In the fasted state, the magnitude of the neural response to hedonic food images (high-calorie greater than low-calorie) in AA and TT subjects was differentially modulated by fasted circulating acyl-ghrelin concentrations within a distributed network of subcortical and cortical appetitive regions. These included the nucleus accumbens, hypothalamus, thalamus, OFC, parahippocampus, and cingulate (Figure 3, A–D, and Supplemental Table 9). For all of these brain regions, post-hoc analysis showed that AA and TT subjects exhibited divergent responses to the modulatory effects of circulating acyl-ghrelin. In TT subjects, there was a positive relationship between circulating acyl-ghrelin and BOLD response (Figure 3, E and G), whereas in AA subjects, a negative relationship was found (Figure 3, F and H). These findings suggest that the FTO genotype alters CNS responsivity to acyl-ghrelin in the fasted state.
Figure 3. Modulatory effects of acyl-ghrelin on BOLD responses in the fasted state.
(A–D) Axial slices with superimposed group functional activity. Brain regions where TT and AA subjects significantly differed in their relationship between BOLD response to high-incentive- versus low-incentive-value food images (high-calorie greater than low-calorie) and circulating acyl-ghrelin in the fasted state. The left side of each panel is the left side of the brain. z is the MNI space z coordinate of the axial slice. T color scale reflects the T value of the functional activity. Results are presented at a threshold of P < 0.05, FWE corrected on the basis of cluster extent. (E–H) Regression plots between the BOLD response to high-calorie versus low-calorie food images and circulating acyl-ghrelin in the TT (blue, open squares) and AA (red, open circles) groups in the fasted state. A positive relationship (E and G) was found in the TT group, whereas a negative regression coefficient (F and H) was seen in the AA group. (E and F) The plotted β coefficients were extracted from the cluster peak within the hypothalamus (MNI space x, y, and z coordinates for the peak cluster voxel [8, –4, –4]). (G and H) Plotted β coefficients were extracted from the cluster peak within the nucleus accumbens (NA) (MNI space x, y, and z coordinates for the peak cluster voxel [6, –6, –4]).
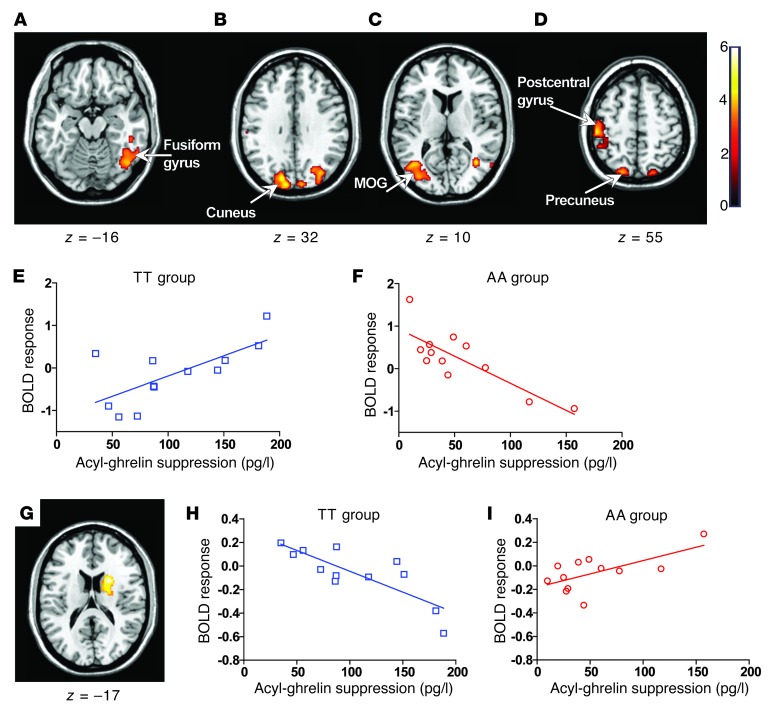
Next, we undertook whole-brain analysis to investigate whether the FTO rs9939609 genotype altered the relationship between postprandial acyl-ghrelin suppression and BOLD response either to all food images (all food greater than non-food) or to hedonic food images (high-calorie greater than low-calorie). We found that neural responses to all food images within the occipitotemporal portion of the fusiform gyrus (BA37) (43), the postcentral gyrus, and the cuneus, a brain region involved in visual attention and encoding the incentive value of visual cues (44), were differentially sensitive to postprandial acyl-ghrelin suppression in the AA and TT subjects (Figure 4, A–D, and Supplemental Table 10). This effect was driven by a positive relationship between the BOLD response within these regions and meal-induced acyl-ghrelin suppression in the TT subjects, while the AA subjects displayed a negative relationship (Figure 4, E and F). In response to high-calorie images, the AA and TT subjects exhibited a significantly divergent response within the right caudate nucleus (z = 3.91 [22, 8, 18]) (Figure 4G), a brain region that controls incentive motivation and that has been shown to exhibit an increased BOLD response to hedonic food images in obese compared with normal-weight subjects (45). In AA subjects, the BOLD response within the caudate was positively related with postprandial acyl-ghrelin suppression, whereas in TT subjects, a negative relationship was observed (Figure 4, H and I). Together, these findings demonstrate altered neural responses to circulating acyl-ghrelin in subjects homozygous for the FTO rs9939609 risk allele.
Figure 4. Modulatory effects of acyl-ghrelin on BOLD responses in the fed state.
(A–D and G) Axial slices with superimposed group activity. MOG, middle occipital gyrus. (A–D) Brain regions where the TT and AA groups significantly differed in their relationship between food-related (all food greater than non-food) BOLD response and postprandial circulating acyl-ghrelin suppression (t0–t54). P < 0.05, FWE corrected. The left side of each panel is the left side of the brain. z is the MNI space z coordinate of the axial slice. T color scale reflects the T score of the interaction. (E and F) Regression plots between food-related BOLD response and circulating postprandial acyl-ghrelin suppression (t0–t54) in TT (blue, open squares) and AA (red, open circles) subjects. Positive regression and negative coefficients (β) were found in the TT (E) and AA (F) groups, respectively. Plotted coefficients were extracted from the cluster peak within the left cuneus (MNI space x, y, and z coordinates for the peak cluster voxel cluster [–12, –92, 28]). (G) Right caudate nucleus where the TT and AA groups exhibited a divergent relationship between BOLD response to hedonic food images and postprandial circulating acyl-ghrelin suppression (t0–t54). (H and I) Regression plots between BOLD response to hedonic food images and circulating postprandial ghrelin suppression in TT (blue, open squares) and AA (red, open circles) subjects. Negative and positive βs were found in the TT (H) and AA (I) groups, respectively. Plotted coefficients were extracted from the cluster peak within the right caudate nucleus (MNI space x, y, and z coordinates for the peak cluster voxel [22, 8, 18].
Fto KO mice exhibit altered circulating ghrelin levels.
To begin exploring the mechanisms by which FTO might regulate ghrelin function, we undertook studies in mice lacking Fto and their WT littermates. After a 16-hour overnight fast, total ghrelin concentrations were higher in Fto KO mice (total ghrelin [ng/ml]: WT = 5.4 ± 0.4, Fto KO = 9.6 ± 1.6, P = 0.05) and acyl-ghrelin levels were nonsignificantly decreased (acyl-ghrelin [ng/ml]: WT = 2.8 ± 0.6, Fto KO = 1.4 ± 0.5, P = 0.16), with a reduced acyl-ghrelin/total ghrelin ratio (acyl-ghrelin: total ghrelin: WT = 0.50 ± 0.11, Fto KO = 0.14 ± 0.07, P = 0.02). In the fed state, Fto KO mice exhibited increased total ghrelin (total ghrelin [ng/ml]: WT = 3.4 ± 0.3, Fto KO = 4.7 ± 0.5, P = 0.03), but similar acyl-ghrelin levels (acyl-ghrelin [ng/ml]: WT = 1.2 ± 0.2, Fto KO = 0.9 ± 0.2), resulting in a decreased acyl-ghrelin/total ghrelin ratio (acyl-ghrelin/total ghrelin: WT = 0.34 ± 0.03, Fto KO = 0.20 ± 0.04, P = 0.02). These studies show that loss of FTO function in vivo in the mouse leads to an opposite effect on circulating ghrelin function to that seen in human AA subjects, and together with the obesity phenotype reported in mice overexpressing Fto (20), suggest that FTO obesity-risk alleles might lead to a gain of function. Therefore, we next studied the effect of upregulating FTO expression in cellular models to define the molecular link between FTO and the altered ghrelin levels seen in vivo in humans.
FTO overexpression in MGN3-1 cells and HEK293T cells regulates ghrelin levels.
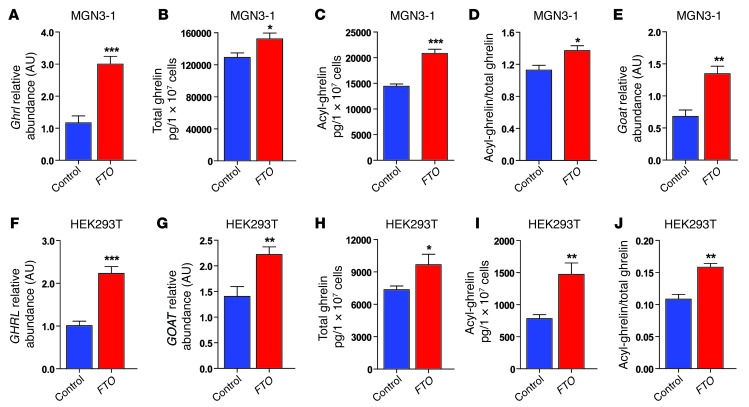
Initially we undertook studies in MGN3-1 cells, a mouse cell line that produces and processes ghrelin (40), and in HEK293T cells, which endogenously express ghrelin. Overexpression of FTO in MGN3-1 cells increased Ghrl mRNA abundance (Figure 5A) and increased cell lysate total ghrelin and acyl-ghrelin concentrations (Figure 5, B and C), leading to an increased acyl-ghrelin/total ghrelin ratio (Figure 5D). Given that acyl-ghrelin is generated from ghrelin through acylation by ghrelin-O-acyltransferase (GOAT) (46), we also measured Goat (Mboat4) mRNA and found that FTO overexpression increased Goat mRNA abundance (Figure 5E). Similarly, FTO overexpression in HEK293T cells resulted in increased GHRL and GOAT mRNA levels (Figure 5, F and G), increased cell lysate total ghrelin and acyl-ghrelin concentrations, and an increased acyl-ghrelin/total ghrelin ratio (Figure 5, H–J). Together, these findings suggest that FTO can directly regulate ghrelin production in a cell-autonomous manner.
Figure 5. Effect of FTO overexpression on ghrelin mRNA abundance, total ghrelin, acyl-ghrelin, and GOAT mRNA abundance in MGN3-1 and HEK293T cells.
(A–E) Effect of FTO overexpression (red bars) compared with the control vector (blue bars) in MGN3-1 cells on (A) Ghrl mRNA abundance, (B) total ghrelin, (C) acyl-ghrelin, (D) acyl-ghrelin/total ghrelin, and (E) Goat mRNA abundance. (F–J) Effect of FTO overexpression (red bars) compared with control vector (blue bars) in HEK293T cells. (F) GHRL mRNA abundance, (G) GOAT mRNA abundance, (H) total ghrelin, (I) acyl-ghrelin, and (J) acyl-ghrelin/total ghrelin. Data are presented as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
FTO overexpression reduces ghrelin pre-mRNA m6A-specific methylation.
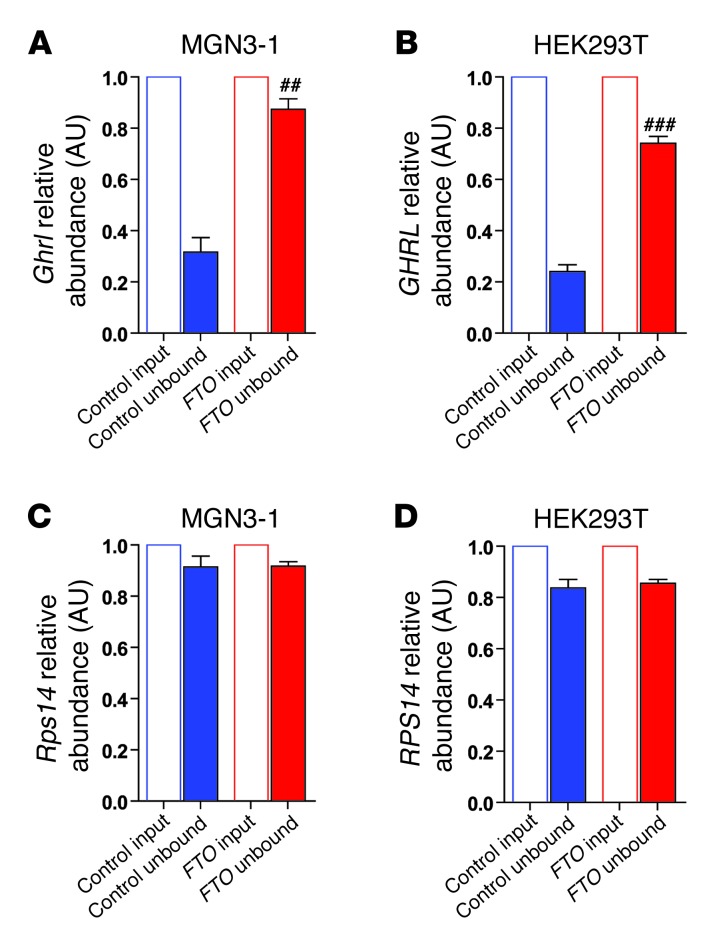
FTO directly demethylates N6-methyladenosine (m6A), a naturally occurring adenosine modification in RNA (30) thought to regulate gene expression (31, 32). In light of our findings that FTO overexpression increased ghrelin mRNA levels in MGN3-1 and HEK293T cells, we next examined whether the ghrelin pre-mRNA m6A methylation status was altered by FTO overexpression by conducting m6A-specific methylated RNA immunodepletion studies (32) on total cellular mRNA from MGN3-1 and HEK293T cells. These studies demonstrate that FTO overexpression specifically reduced m6A methylation of ghrelin mRNA in both cell lines (Figure 6, A and B), but not that of RPS14 (Figure 6, C and D), a non–m6A methylated mRNA (32).
Figure 6. Effect of FTO overexpression on immunodepletion of m6A-methylation of ghrelin mRNA and Rps14 mRNA from total cellular mRNA in (A and C) MGN3-1 cells and (B and D) HEK293T cells.
mRNA abundance was determined by qPCR and calculated relative to the input sample, with transcripts normalized to the amount of Gusb mRNA (a non-m6A–containing mRNA). n = 4 per study group, and results are representative of 2 independent experiments. Data are the mean ± SEM. Differences between normalized control unbound and normalized FTO unbound; ##P < 0.01; ###P < 0.001.
Peripheral blood cells from rs9939609 AA subjects have an increased abundance of FTO and GHRL mRNA.
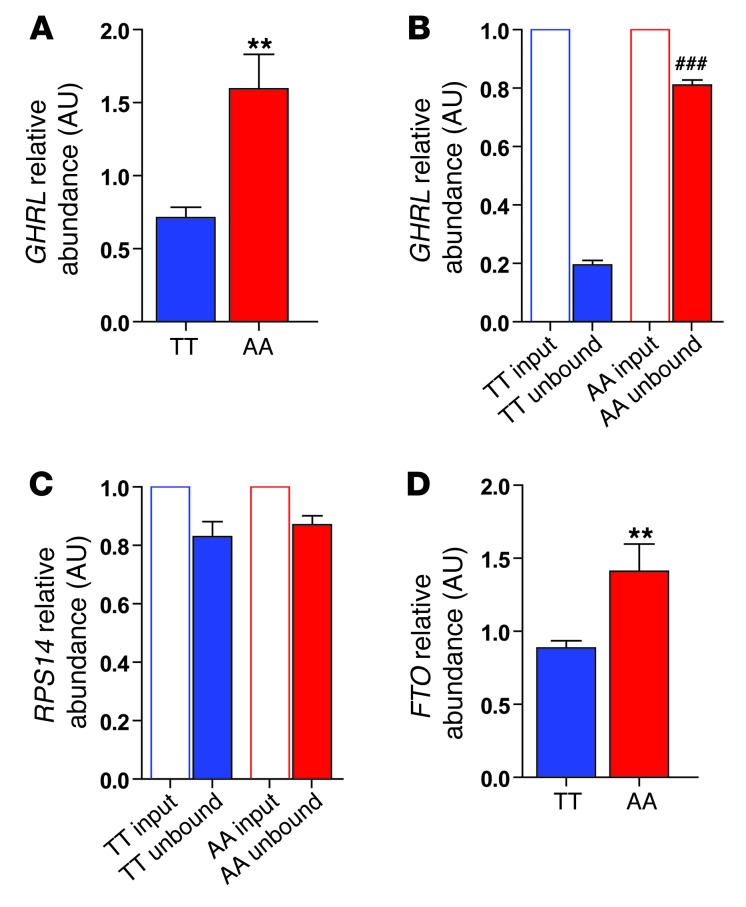
Human peripheral blood cells express FTO (47) and GHRL (48). Thus, we recruited a third cohort of overnight-fasted, normal-weight, adiposity-matched AA and TT subjects (Supplemental Table 11) in order to assess the impact of the rs9939609 A allele on FTO and GHRL mRNA levels in peripheral blood cells. In addition, we undertook m6A-specific methylated RNA immunodepletion studies of peripheral blood cells from these subjects. We found a 2.5-fold increase in ghrelin mRNA abundance in AA compared with TT subjects (Figure 7A). Furthermore, immunodepletion studies revealed that AA subjects had reduced m6A methylation of ghrelin mRNA (Figure 7, B and C). Finally, we assessed the effect of the rs9939609 A allele on FTO mRNA levels, and found that AA subjects had increased FTO mRNA abundance compared with TT subjects (Figure 7D). Together, these results suggest that the A obesity–risk rs9939609 allele is associated with increased FTO expression, reducing m6A ghrelin mRNA demethylation and leading to altered ghrelin production.
Figure 7. Effect of the rs9939609 FTO genotype on GHRL mRNA abundance, immunodepletion of m6A-methylated GHRL mRNA, and RPS14 mRNA in peripheral blood cells.
(A) GHRL mRNA abundance in peripheral blood cells from TT (blue bars) and AA (red bars) study subjects. Effect of the FTO genotype on immunodepletion of m6A-methylated (B) GHRL mRNA and (C) RPS14 mRNA abundance from total cellular mRNA in TT and AA study subjects. mRNA abundance was determined by qPCR and calculated relative to the input sample, with transcripts normalized to the amount of GUSB mRNA (a non-m6A–containing mRNA). (D) FTO mRNA abundance in peripheral blood cells. Data are presented as the mean ± SEM; n = 10 per genotype. **P < 0.01. Difference between normalized TT unbound and normalized AA unbound; ###P < 0.001.
Discussion
In the current studies performed in normal-weight, adiposity-matched individuals with either the FTO rs9939609 TT or the obesity-risk AA genotype, we found that AA subjects exhibited attenuated postprandial suppression of both hunger and circulating acyl-ghrelin levels. Our fMRI studies showed that in both fasted and fed states, in response to viewing food cues, AA subjects exhibited different neural activity in homeostatic and brain reward regions compared with TT subjects. Moreover, neural responses in brain regions known to regulate appetite, reward, and incentive motivation were differentially sensitive to circulating acyl-ghrelin concentrations in AA and TT subjects. In mechanistic studies, we found that Fto KO mice (25) had a reduced acyl-ghrelin/total ghrelin ratio compared with WT mice, the opposite ghrelin pattern to that observed in our AA subjects. FTO overexpression in cell lines increased ghrelin mRNA abundance, while specifically reducing its m6A methylation, and led to increased total ghrelin and acyl-ghrelin concentrations and an increased abundance of Goat mRNA. Finally, we found that peripheral blood cells from AA human subjects exhibited increased FTO expression, reduced m6A ghrelin mRNA methylation, and increased ghrelin expression compared with TT subjects. Our results identify a direct link between FTO and ghrelin, and together with our fMRI findings, offer what we believe to be the first insights into the mechanisms underlying the obesity-prone eating behavior of AA rs9939609 subjects.
A key question arising from our studies is whether perturbation of the ghrelin system offers a biologically plausible explanation for the obesity-prone feeding behavior and subsequent increased adiposity observed in subjects homozygous for the A obesity–risk FTO rs9939609 variant. Ghrelin, the only known orexigenic peptide hormone, regulates both homeostatic and reward-related feeding (37, 38, 49–51). Posttranslational addition of an acyl group to serine-3 by GOAT, resulting in the generation of acyl-ghrelin, is essential for the appetite-stimulatory and reward-related effects of ghrelin, which are mediated via GHSR (52, 53). The effects of ghrelin on appetite and food intake are well established; acyl-ghrelin stimulates hunger and increases food intake (34, 49), and chronic administration increases adiposity (54). Although some studies have shown little or no effect of genetic or pharmacological blockade of ghrelin signaling on feeding or body weight (55, 56), other studies suggest that intact ghrelin signaling is required for normal eating behavior and body-weight responses, especially to rewarding high-fat diets (38, 51, 53, 57, 58). Not only does acyl-ghrelin increase the intake of freely available food, but it also shifts food preference toward diets rich in fat, enhances operant responding for food rewards, and induces conditioned place preference for food rewards (38, 51). More recently, ghrelin has been shown to play a key role in food cue–potentiated feeding (59), and exogenous administration of acyl-ghrelin to humans increases neural responses to food cues in several regions implicated in hedonic feeding (37). These known actions of acyl-ghrelin are strikingly similar to the reported feeding phenotype of FTO rs9939609 AA subjects: altered appetite, increased food consumption, preference for high-fat foods, enhanced food responsivity, and increased food cue–potentiated feeding, suggesting that altered ghrelin could contribute to the feeding phenotype observed in these subjects.
While our findings of altered ghrelin function in FTO rs9939609 AA subjects provide a parsimonious explanation for the obesity-risk phenotype seen in these subjects, a number of other mechanisms could also be at work. First, the human and mouse m6A RNA methylomes consist of several thousand mRNA species, including genes that have been implicated in the regulation of energy homeostasis in a range of tissues (31, 32). Studies on such mechanisms are at an early stage in terms of validation, but these findings suggest that FTO might have pleiotropic and complex effects on the regulation of metabolism at the cellular and organismal level. Indeed, it has recently been shown in cell-based studies that FTO plays a role in amino acid sensing, perhaps providing a link between amino acid levels and mTORC1 signaling (24), as cells lacking FTO display decreased activation of the mTORC1 pathway. How this finding impacts whole-body energy homeostasis or relates to the demethylase function of FTO remains to be defined, but a link between amino acid sensing and the CNS regulation of energy balance has been established in mouse models (60, 61). Second, in addition to mRNA, FTO demethylates other RNA modifications and other RNA species with regulatory roles (62, 63). These include the 3-methyluracil modifications that are more prevalent in transfer RNAs (tRNAs) and rRNAs, and indeed, altered tRNA function may provide the link between the FTO and amino acid sensing described above. m6A methylation has also been mapped to many long noncoding RNA species that have yet to be implicated in the central regulation of energy homeostasis, but have been shown to be involved in pancreatic β cell function (64). Together, these observations suggest the existence of additional molecular mechanisms linking FTO function to metabolism. It will be important in future studies to define, for example, the m6A RNA methylome in subjects carrying FTO obesity–risk alleles in order to determine which pathways to study in cell-, mouse-, and human-based studies.
Our fMRI studies provide what we believe to be additional novel insights into the neurobiological mechanisms by which FTO obesity-risk alleles lead to obesity-prone eating behavior and also provide further evidence of a contributory role for ghrelin. In the fasted state, we observed a significant effect of the FTO rs9939609 genotype on the BOLD response to food images within the hypothalamus, mid-brain dopaminergic reward areas (left VTA/SN), limbic areas (left globus pallidus and left thalamus), and within brain regions that are implicated in monitoring bodily state and cognitive integration. Postprandially, AA subjects rated high-calorie food images as more appealing than did TT subjects, suggesting that the FTO rs9939609 genotype impacts the mechanisms or neural circuits that normally modulate motivational changes to hedonic food cues. In support of this notion, a comparison of BOLD responses to hedonic food cues in the fed state versus the fasted state revealed differences between AA and TT subjects within the left anterior insula extending into the lateral OFC, brain regions responsible for integrating internal and external determinants of eating behavior and encoding food reward, and also within the left putamen, a region implicated in regulating goal-directed behavior (65). Taken together, these results show that the FTO rs9939609 genotype markedly impacts neural responsivity to food cues in brain regions controlling energy homeostasis and reward and incentive motivation. Furthermore, when we examined the impact of ghrelin, we found that in the fasted state, in response to viewing hedonic food cues, TT and AA subjects differed in their BOLD responses to circulating endogenous ghrelin within key ghrelin-sensitive dopaminergic mesolimbic reward regions, limbic and paralimbic regions, visual processing areas, and brain regions involved in executive decision making. In TT subjects, for all of these regions, acyl-ghrelin concentrations positively covaried with an increased BOLD response, i.e. the greater the fasting acyl-ghrelin levels, the greater the BOLD response. In contrast, in AA subjects, fasting acyl-ghrelin concentrations negatively covaried with the BOLD response to hedonic food images within these regions. Our findings in TT subjects are in agreement with a recent study by Kroemer and colleagues (66), who found that fasting ghrelin concentrations covaried with the BOLD response elicited by viewing palatable food images within the hypothalamus, VTA/SN, thalamus, cingulate, and fusiform gyrus. Postprandially, AA and TT subjects exhibited a differential relationship between postprandial acyl-ghrelin suppression and BOLD responses to food-related images within brain regions involved in visual attention and in encoding visual cue incentive value (44) and integrative/executive decision making. In addition, divergent acyl-ghrelin suppression and BOLD responses to hedonic food cues were observed within the right caudate nucleus, a region implicated in controlling incentive motivation, anticipatory food reward, and food craving (45). Collectively, these results show that the FTO genotype alters CNS responsivity to circulating ghrelin levels. Importantly, these neural responsivity changes to food cues are present in adiposity-matched, normal-weight groups prior to the development of increased BMI and thus potentially play a causative role in mediating the link between FTO-risk alleles and obesity predisposition. Studies aimed at understanding the cellular and molecular basis for this altered neural sensitivity will be an important future undertaking.
Despite the need for further mechanistic studies, our current findings may have immediate implications for both the investigation of the neurobiological basis of human obesity and for its treatment. First, our studies highlight the utility of combining detailed physiological phenotyping with a parallel assessment of neural function in genotyped subjects before the development of obesity in order to gain insights into the underlying biology of risk alleles. Second, our findings argue for a genotype-tailored therapeutic approach, targeting the hormonal and neurobiological abnormalities that underlie the development of obesity. For example, AA subjects may gain the greatest benefits from preventative and treatment strategies aimed at targeting the ghrelin system and modulating reward responsiveness.
In summary, our studies offer what we believe to be the first insights into the physiological and neurobiological abnormalities that may underlie the effects of the FTO variant rs9939609 on obesity risk in humans, delineating a mechanism that stems from perturbed epitranscriptomic regulation of ghrelin mRNA abundance through to altered neural response to ghrelin in key brain reward and appetite regulatory regions.
Methods
Subjects.
A total of 359 normal-weight (BMI range, 18–25 kg/m2) males of mixed European descent, aged 18–35 years, were recruited. Exclusion criteria were medical or psychiatric illness, medication use, substance abuse, smoking, food allergies, and weight fluctuation greater than 3 kg in the 3 months prior to enrollment. Adiposity affects circulating gut hormone levels (67) and neural responses to food cues (68). Thus, to avoid these confounding factors, our study groups were matched for BMI, a surrogate marker of body adiposity (69). However, BMI alone does not distinguish between muscle and fat mass and has been criticized as being an inaccurate assessment of body fatness (70), especially in young, fit adults (71). Thus, in light of previous studies reporting that total fat mass (67) and visceral fat (72) impact circulating ghrelin levels, we also matched our AA and TT groups for total fat mass and visceral fat. Three separate cohorts of age- and adiposity-matched AA and TT subjects were recruited for additional studies: 10 AA and 10 TT subjects were recruited for the initial test meal study (Supplemental Table 1), 12 AA and 12 TT subjects participated in the fMRI study (Supplemental Table 5), and a third cohort of matched 10 AA and 10 TT subjects were recruited for the peripheral blood cell study (Supplemental Table 11). Additional exclusion criteria for the fMRI study were: left-handedness, claustrophobia, presence of a hearing aid or ferromagnetic material, food preferences/dislikes inconsistent with our food images, and irregular eating habits. Our established protocols for subject standardization and acclimatization/habituation to the study environment were used (41). Subjects recruited to the fMRI study participated in a simulated scan in a mock-MRI scanner before the first scanning session.
Anthropometric phenotyping.
Bioelectric impedance (InBody 720 Analyzer; Biospace) was used for body composition analysis. Differences between genotypes were compared using 1-way ANOVA followed by Student’s and Neuman-Keuls multiple comparison tests.
FTO rs9939609 genotyping.
DNA was extracted from whole blood using the QIAamp DNA Blood Midi-Kit (QIAGEN), and FTO rs9939609 allelic discrimination was performed with an ABI7900HT detector (Applied Biosystems) set at an automatic allele-calling quality value of 0.95 using a TaqMan probe (assay ID:c_30030620_10; Applied Biosystems).
Appetite VAS.
Validated appetite VASs were used to obtain subjective appetite ratings (39). Each VAS was 100 mm long, with text expressing extreme positive and extreme negative ratings anchored at each end.
Standard test meal study.
Ten AA and 10 TT subjects were given a standard test meal (1,840 kcal) (Supplemental Table 3) at t = 0 minutes (referred to hereafter as t0; all times herein are in minutes) to consume within 20 minutes. Blood from a forearm cannula and appetite VAS were taken preprandially and at t20, t30, and every 30 minutes thereafter until t180 postmeal (Supplemental Figure 1A).
Imaging study protocol.
BOLD fMRI (3.0 Tesla, HDX Excite II scanner; GE Healthcare) responses were collected from 12 AA and 12 TT age- and adiposity-matched subjects (Supplemental Table 5) in both fasted and fed states in a counterbalanced, crossover manner (Supplemental Figure 2). During fMRI scanning, subjects viewed high-calorie and low-calorie food images and non-food images presented in a pseudorandomized block design format (see fMRI task in Supplemental Figure 3). Blood for acyl-ghrelin measurement and appetite ratings were obtained concurrently at set time points during fMRI and outside the scanner. On the fed study day at t0, subjects were given a standard test meal (1,840 kcal, Supplemental Table 3) to consume within 20 minutes. Blood and appetite VASs were taken at t0, t20, and t30. At t35, subjects underwent a conventional T2-weighted scan, a fluid-attenuated inversion recovery (FLAIR) scan, and an fMRI (task). Task initiation coincided with t45 postprandium, and blood and appetite VASs were taken at t45, t49.5, and t54 and subsequently out of the scanner at t90, t120, t150, and t180 (Supplemental Figure 2). On the fasted study day, participants underwent a high-resolution echo-planar imaging (EPI) scan and a volumetric 3D coronal spoiled gradient recalled echo (SPGR) scan, followed by fMRI (task). Time was recorded at zero upon task initiation. Appetite VASs and blood were taken at t0, t4.5, and t9.
Imaging data acquisition.
All scanning was performed between 09:00 and 11:30. One hundred eighty-two volumes were collected (in-plane matrix size of 64 × 64 and a field of view [FOV] of 211 mm), with whole-brain acquisition for each functional time point; (repetition time [TR]: TR = 3 s, echo time [TE]: TE =25 ms, flip angle = 90°, 43 slices per volume collected in an interleaved manner, slice thickness = 3 mm, between-slice gap = 0.3 mm parallel to the anterior commissure to posterior commissure [AC-PC] line). The higher-resolution EPI scan had a slice thickness of 3.3 mm, composed of 43 slices, a matrix of 128 × 128, and a FOV of 240 mm (TE = 30 ms, flip angle = 90°). The 3D SPGR had a matrix size of 256 × 256 × 196 voxels, with an isotropic resolution of 1.1 mm (TE/TR and inversion time = 2.812/6.616/450 ms, excitation flip angle = 20°).
Visual stimuli and fMRI task.
The stimuli consisted of 192 color pictures taken with a digital camera then scanned and standardized for size, resolution, luminance consistency, and complexity. The food images were subdivided into low-calorie and high-calorie categories, each consisting of 64 unique images. Low-calorie food images included low-fat items (steamed vegetables, fruit, and boiled fish and chicken). High-calorie foods were sweet and savory high-calorie items (cakes, waffles, ice cream, burger meals, etc.). Control stimuli consisted of 64 non-food–related household and office items. The images were prevalidated on a manual 9-point Likert scale (73) by 30 independent, healthy, normal-weight males matched for age and educational background with the fMRI participants. During imaging, the first 4 volumes were discarded (discarded dummy acquisitions [DDAs]) to allow for equilibration of the longitudinal magnetization. During the task, visual stimuli were presented in a block design format (Supplemental Figure 3). Each epoch began with 24 seconds of blank screen, with the central fixation cross (rest period) followed by a block of low-calorie food images, a block of high-calorie food images, and a block of non-food items pseudorandomly presented. This sequence was presented 4 times during the task with a pseudorandomized order of conditions. Within each 24-second block of food images or non-food images, 8 individual images were presented for 3 seconds each. At the end of each block, a single VAS question, “How pleasant were the images?” was projected. This question was included in the design to ensure that the subjects remained attentive and to control for intersubject variability in terms of image valence. Appetite VASs were presented before the first cycle of blocks, between the second and third cycle, and at the end of the task run (i.e., after the fourth cycle) (Supplemental Figure 4). Participants responded to the VAS interrogations by moving a joystick placed in their right hand. Images were projected through the radiofrequency (RF) window onto a screen at the bottom of the scanner bed using a Sanyo PLC-XV40 projector and viewed via a head coil–mounted twin mirror system. The visual display FOV was 24° × 18°. The total duration of the task was 9:06 minutes. When out of the scanner, participants completed the VAS on a laptop using identical in-house software to that used for the VAS during the task.
fMRI analysis.
The Statistical Parametric Mapping suite, version 8 (SPM8; Wellcome Trust Centre for Neuroimaging) was used for image processing and data analysis. Preprocessing procedures included correction for volume-to-volume head movements, coregistration to a high-resolution gradient echo structural image, normalization to Montreal Neurological Institute (MNI) spacing using the SPM EPI template, and smoothing with an 8-mm full width at half maximum (FWHM) Gaussian smoothing kernel. During first-level analysis, statistical contrasts were generated for regional BOLD responses to food (high-calorie and low-calorie images combined) versus non-food images (all food greater than non-food) and high-calorie versus low-calorie images (high-calorie greater than low-calorie). These contrasts were then entered into 2 separate random effects (i.e., second level) flexible factorial ANOVAs to assess the interaction of genotype and nutritional state. The primary contrasts of interest were the main effects of genotype, nutritional state, and their interaction. For all whole-brain analyses, we took a stringent statistical approach and only report clusters that survived full family-wise error (FWE) correction for multiple comparisons (i.e., whole-brain correction) on the basis of cluster extent (P < 0.05). This involves selecting a cluster-forming voxel threshold (P < 0.001, uncorrected here) and using the Gaussian random field theory (74) to calculate the minimum cluster size (i.e., number of contiguous voxels) that survives correction for multiple comparisons (P < 0.05, FWE corrected). Finally, the map was adjusted to remove clusters that were too small to survive correction.
Whole-brain random effects multiple regression analyses were undertaken to explore the relationship between BOLD response to food images (all food greater than non-food) in the control (TT) group, with circulating acyl-ghrelin on the fasted study day, and between BOLD response and postprandial acyl-ghrelin suppression on the fed study day. Subsequently, whole-brain multiple regression analyses were implemented to test for genotype by covariate interaction, looking for regions of significant difference between the 2 groups in their relationships between BOLD response and acyl-ghrelin. The results of these ghrelin-BOLD regression analyses were fully corrected for FWE across the whole brain on the basis of spatial extent (P < 0.05, FWE corrected), with a cluster-forming voxel threshold of P < 0.005. Where we had a priori hypotheses regarding the effects of postprandial ghrelin suppression on brain activation, we corrected for multiple comparisons within independently derived regions of interests (ROIs), i.e., small-volume correction (SVC). We used spheres (radius of 10 mm) around peak coordinates drawn from Batterham et al. (39) (OFC; MNI [–46, 27, –10] and VTA; MNI [–2, –20, –10]) and anatomical ROIs for the hypothalamus and striatum (caudate nucleus and putamen) constructed within the WFU PickAtlas software package (Wake Forest University). Given its small size, the hypothalamus mask was dilated (×1) to accommodate between-subject anatomical variability. In all ROI analyses, the threshold for significance was defined as P < 0.05 following correction for multiple comparisons within a mask formed from the combination of all these ROIs.
Mouse studies.
Fto KO mice (25) and their 20-week-old WT littermates were sacrificed after a 16-hour overnight fast or in the ad libitum fed state (n = 6–9 per group). Blood was collected and processed as previously described (41). Mouse total and acyl-ghrelin plasma levels were determined by ELISA (Millipore). Differences between genotypes were analyzed using an unpaired 2-tailed Student’s t test.
Human hormone assays.
Blood was collected as previously described (39, 41). Plasma leptin was measured by ELISA and PYY3-36 by RIA. In the meal study, acyl-ghrelin and total plasma ghrelin were assayed by ELISA. In the fMRI study, acyl-ghrelin study was assayed by RIA. ELISA and RIA kits were purchased from Millipore.
Human peripheral blood cell isolation.
After a 14-hour overnight fast, venepuncture was performed and blood was collected into PAXgene tubes (QIAGEN) according to the manufacturer’s instructions and then frozen at –70°C until RNA extraction was performed.
Cell culture and FTO overexpression.
HEK293T cells were cultured in DMEM, and MGN3-1 cells were cultured in DMEM using published protocols (40). FTO overexpression experiments were carried out by transfecting HEK293T or MGN3-1 cells with a synthesized FTO cDNA cloned into pIRES2-EGFP or with empty vector. Lipofectamine 2000 (Invitrogen) transfection was performed according to the manufacturer’s instructions, and cells were cultured for 48 hours prior to RNA or protein extraction. In MGN3-1 cells, ghrelin cell lysate studies were also performed using previously described protocols (40), incubating cells with 50 μM octanoate (Sigma-Aldrich) in serum-free medium for 24 hours prior to cell harvesting. Studies of HEK293T cells were performed in a similar manner.
RNA isolation, processing, and m6A immunoprecipitation.
Total RNA was isolated from HEK293T and MGN3-1 cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Total RNA was extracted from whole blood using the PAXgene Blood RNA system (QIAGEN) following the manufacturer’s instructions. Total RNA was subjected to Ribominus (Invitrogen) treatment to deplete rRNA and fragmented using Ambion RNA Fragmentation Reagent (Invitrogen) according to the manufacturer’s instructions. m6A immunoprecipitation (IP) was performed with rabbit polyclonal anti-m6A antibody (Synaptic Systems) using an adaptation of recently described protocols (31, 32). In brief, anti-m6A antibody was coupled to sheep anti-rabbit Dynabeads (Invitrogen), and fragmented denatured RNA was immunoprecipitated in 300 μl of IP buffer (10 mM Na phosphate, 0.05% Triton-X, 140 mM NaCl) with 300 μl of washed antibody-coupled beads for 2 hours at 4°C. Beads were then washed in 5× 500 μl of 140 mM IP buffer, retaining unbound RNA fractions at each stage. RNA was then eluted from the beads by incubation in 300 μl of elution buffer (5 mM Tris-HCl, 1 mM EDTA, and 0.05% SDS with 4.2 μl proteinase K [20 mg/ml]) for 1.5 hours at 50°C. RNA was isolated from input and unbound fractions using phenol/chloroform extraction and ethanol precipitation and then resuspended in H2O.
Real-time quantitative PCR analysis.
Abundance of ghrelin and FTO mRNAs and immunodepletion of m6A- and non-m6A–containing mRNAs from cellular total mRNA samples following IP were measured using quantitative PCR (qPCR). cDNA was generated from input and unbound RNA samples using a SuperScript VILO cDNA Synthesis Kit (Invitrogen) according to the manufacturer’s instructions. qPCR analysis was performed using the TaqMan RT system (Applied Biosystems) with proprietary sequence FAM/TAMRA primers on an Applied Biosystems Fast Real-Time PCR 7900HT thermocycler. The immunodepletion of methylated ghrelin mRNA and a non-m6A–containing mRNA, Rps14 (in MGN3-1 or HEK293T cells with and without FTO transfection or in peripheral blood cells from AA and TT subjects), was determined by qPCR and calculated relative to the input sample with transcripts normalized to the amount of Gusb mRNA (a non-m6A–containing mRNA). The Applied Biosystems primers used were: Ghrl Mm1190295_m1; Gusb Mm01197698_m1; Mboat4 Mm01200389_m1; Rps14 Mm00849906_g1; GHRL Hs03654068_m1; MBOAT4 Hs01074954_s1; GUSB Hs00939627_m1; FTO Hs01057145_m1; and RPS14 Hs00852033_g1.
Statistics.
Data are presented as the mean ± SEM and were analyzed with GraphPad Prism 6 (GraphPad) and the Statistical Package for the Social Sciences (SPSS) version 21.0. Differences between 2 groups were analyzed using a 2-tailed Student’s t test. Where several groups were compared, the differences were analyzed by 1-way ANOVA with a post-hoc Newman-Keuls multiple comparison test. General linear model for repeated-measure analyses, using a between-subject factor (genotype) and a within-subject factor (minutes), were performed to assess plasma hormone levels and appetite scores in the meal and fMRI studies. Where significant differences were present between genotypes, an independent Student’s t test was performed for each time point. The integrated AUC was calculated using the trapezoid method, and the difference between 2 groups was analyzed using a 2-tailed Student’s t test. For all analyses, P < 0.05 was considered statistically significant.
Study approval.
Approval for the human studies was obtained from the University College London and King’s College London Ethics Committees, and all subjects gave written informed consent. Care of all animals was carried out within institutional animal care committee guidelines, and all animal experiments were approved by the local government (Landesamt für Natur, Umwelt und Verbraucherschutz, North Rhine-Westphalia, Germany).
Supplementary Material
Acknowledgments
We are indebted to the subjects who participated in our studies. We also thank J. Dalton and C. Andrew for their technical assistance with the imaging paradigm development, J. Jones and A. Pucci for assistance with human studies, and S. Blanco, P. Hajkova, K. Meyer, and S. Jaffrey for advice regarding the m6A IP studies. This work was supported by Rosetrees Trust, University College London Hospital (UCLH) Charities, and University College London/UCLH Comprehensive Biomedical Research Centre, which received a proportion of its funding from the Department of Health’s NIHR Biomedical Research Centres. A.I. Choudhury and D.J. Withers are funded by the Medical Research Council (MC-A654-5QB40), and E. Karra is a Wellcome Trust Clinical Research Fellow. The authors wish to acknowledge the support of the King’s Centre of Excellence in Medical Engineering, funded by the Wellcome Trust and the EPSRC (WT 088641/Z/09/Z), for the imaging elements of this research project.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(8):3539–3551. doi:10.1172/JCI44403.
References
- 1.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 4.Hardy R, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19(3):545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55(4):981–995. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauria F, et al. Prospective analysis of the association of a common variant of FTO (rs9939609) with adiposity in children: results of the IDEFICS study. PLoS One. 2012;7(11):e48876. doi: 10.1371/journal.pone.0048876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 8.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring). 2008;16(8):1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 9.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond). 2009;33(1):42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 10.Timpson NJ, et al. The fat mass- and obesity-associated locus and dietary intake in children. Am J Clin Nutr. 2008;88(4):971–978. doi: 10.1093/ajcn/88.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanofsky-Kraff M, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr. 2009;90(6):1483–1488. doi: 10.3945/ajcn.2009.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wardle J, Carnell S, Haworth CM, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 13.den Hoed M, Westerterp-Plantenga MS, Bouwman FG, Mariman EC, Westerterp KR. Postprandial responses in hunger and satiety are associated with the rs9939609 single nucleotide polymorphism in FTO. Am J Clin Nutr. 2009;90(5):1426–1432. doi: 10.3945/ajcn.2009.28053. [DOI] [PubMed] [Google Scholar]
- 14.Velders FP, et al. FTO at rs9939609, food responsiveness, emotional control and symptoms of ADHD in preschool children. PLoS One. 2012;7(11):e49131. doi: 10.1371/journal.pone.0049131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McTaggart JS, Lee S, Iberl M, Church C, Cox RD, Ashcroft FM. FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS One. 2011;6(11):e27968. doi: 10.1371/journal.pone.0027968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stratigopoulos G, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294(4):R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredriksson R, et al. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149(5):2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- 18.Olszewski PK, et al. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci. 2009;10:129. doi: 10.1186/1471-2202-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, et al. Involvement of leptin receptor long isoform (LepRb)-STAT3 signaling pathway in brain fat mass- and obesity-associated (FTO) downregulation during energy restriction. Mol Med. 2011;17(5–6):523–532. doi: 10.2119/molmed.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez-Aguilar R, Kim DH, Woods SC, Seeley RJ. Expression of new loci associated with obesity in diet-induced obese rats: from genetics to physiology. Obesity (Silver Spring). 2012;20(2):306–312. doi: 10.1038/oby.2011.236. [DOI] [PubMed] [Google Scholar]
- 21.Boender AJ, van Rozen AJ, Adan RA. Nutritional state affects the expression of the obesity-associated genes Etv5, Faim2, Fto, and Negr1. Obesity (Silver Spring). 2012;20(12):2420–2425. doi: 10.1038/oby.2012.128. [DOI] [PubMed] [Google Scholar]
- 22.Tung YC, et al. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS One. 2010;5(1):e8771. doi: 10.1371/journal.pone.0008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung MK, Gulati P, O’Rahilly S, Yeo GS. FTO expression is regulated by availability of essential amino acids. Int J Obes (Lond). 2013;37(5):744–747. doi: 10.1038/ijo.2012.77. [DOI] [PubMed] [Google Scholar]
- 24.Gulati P, et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc Natl Acad Sci U S A. 2013;110(7):2557–2562. doi: 10.1073/pnas.1222796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer J, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 26.Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5(11):e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurray F, et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2013;9(1):e1003166. doi: 10.1371/journal.pgen.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Church C, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5(8):e1000599. doi: 10.1371/journal.pgen.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Church C, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42(12):1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 32.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummings DE, Overduin J. Gastrointestinal regulation of food intake. J Clin Invest. 2007;117(1):13–23. doi: 10.1172/JCI30227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wren AM, et al. Ghrelin enhances appetite and increases food intake in humans. J Clin Endocrinol Metab. 2001;86(12):5992. doi: 10.1210/jc.86.12.5992. [DOI] [PubMed] [Google Scholar]
- 35.Batterham RL, et al. Gut hormone PYY(3-36) physiologically inhibits food intake. Nature. 2002;418(6898):650–654. doi: 10.1038/nature00887. [DOI] [PubMed] [Google Scholar]
- 36.Batterham RL, et al. Inhibition of food intake in obese subjects by peptide YY3-36. N Engl J Med. 2003;349(10):941–948. doi: 10.1056/NEJMoa030204. [DOI] [PubMed] [Google Scholar]
- 37.Malik S, McGlone F, Bedrossian D, Dagher A. Ghrelin modulates brain activity in areas that control appetitive behavior. Cell Metab. 2008;7(5):400–409. doi: 10.1016/j.cmet.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Perello M, et al. Ghrelin increases the rewarding value of high-fat diet in an orexin-dependent manner. Biol Psychiatry. 2010;67(9):880–886. doi: 10.1016/j.biopsych.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batterham RL, et al. PYY modulation of cortical and hypothalamic brain areas predicts feeding behaviour in humans. Nature. 2007;450(7166):106–109. doi: 10.1038/nature06212. [DOI] [PubMed] [Google Scholar]
- 40.Iwakura H, et al. Establishment of a novel ghrelin-producing cell line. Endocrinology. 2010;151(6):2940–2945. doi: 10.1210/en.2010-0090. [DOI] [PubMed] [Google Scholar]
- 41.Chandarana K, et al. Subject standardization, acclimatization, and sample processing affect gut hormone levels and appetite in humans. Gastroenterology. 2009;136(7):2115–2126. doi: 10.1053/j.gastro.2009.02.047. [DOI] [PubMed] [Google Scholar]
- 42.Balleine BW, O’Doherty JP. Human and rodent homologies in action control: corticostriatal determinants of goal-directed and habitual action. Neuropsychopharmacology. 2010;35(1):48–69. doi: 10.1038/npp.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kroemer NB, et al. Hum Brain Mapp. (Still) longing for food: Insulin reactivity modulates response to food pictures [published online ahead of print March 28, 2012]. doi: 10.1002/hbm.22071 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Litt A, Plassmann H, Shiv B, Rangel A. Dissociating valuation and saliency signals during decision-making. Cereb Cortex. 2011;21(1):95–102. doi: 10.1093/cercor/bhq065. [DOI] [PubMed] [Google Scholar]
- 45.Nummenmaa L, et al. Dorsal striatum and its limbic connectivity mediate abnormal anticipatory reward processing in obesity. PLoS One. 2012;7(2):e31089. doi: 10.1371/journal.pone.0031089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132(3):387–396. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Lappalainen T, et al. Gene expression of FTO in human subcutaneous adipose tissue, peripheral blood mononuclear cells and adipocyte cell line. J Nutrigenet Nutrigenomics. 2010;3(1):37–45. doi: 10.1159/000320732. [DOI] [PubMed] [Google Scholar]
- 48.Mager U, et al. Expression of ghrelin gene in peripheral blood mononuclear cells and plasma ghrelin concentrations in patients with metabolic syndrome. Eur J Endocrinol. 2008;158(4):499–510. doi: 10.1530/EJE-07-0862. [DOI] [PubMed] [Google Scholar]
- 49.Tschop M, Smiley DL, Heiman ML. Ghrelin induces adiposity in rodents. Nature. 2000;407(6806):908–913. doi: 10.1038/35038090. [DOI] [PubMed] [Google Scholar]
- 50.Abizaid A, et al. Ghrelin modulates the activity and synaptic input organization of midbrain dopamine neurons while promoting appetite. J Clin Invest. 2006;116(12):3229–3239. doi: 10.1172/JCI29867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perello M, Zigman JM. The role of ghrelin in reward-based eating. Biol Psychiatry. 2012;72(5):347–353. doi: 10.1016/j.biopsych.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402(6762):656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 53.Davis JF, et al. GOAT induced ghrelin acylation regulates hedonic feeding. Horm Behav. 2012;62(5):598–604. doi: 10.1016/j.yhbeh.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wren AM, et al. Ghrelin causes hyperphagia and obesity in rats. Diabetes. 2001;50(11):2540–2547. doi: 10.2337/diabetes.50.11.2540. [DOI] [PubMed] [Google Scholar]
- 55.Sun Y, Butte NF, Garcia JM, Smith RG. Characterization of adult ghrelin and ghrelin receptor knockout mice under positive and negative energy balance. Endocrinology. 2008;149(2):843–850. doi: 10.1210/en.2007-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albarran-Zeckler RG, Sun Y, Smith RG. Physiological roles revealed by ghrelin and ghrelin receptor deficient mice. Peptides. 2011;32(11):2229–2235. doi: 10.1016/j.peptides.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zigman JM, et al. Mice lacking ghrelin receptors resist the development of diet-induced obesity. J Clin Invest. 2005;115(12):3564–3572. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wortley KE, et al. Absence of ghrelin protects against early-onset obesity. J Clin Invest. 2005;115(12):3573–3578. doi: 10.1172/JCI26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker AK, Ibia IE, Zigman JM. Disruption of cue-potentiated feeding in mice with blocked ghrelin signaling. Physiol Behav. 2012;108:34–43. doi: 10.1016/j.physbeh.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cota D, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312(5775):927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 61.Cota D, Matter EK, Woods SC, Seeley RJ. The role of hypothalamic mammalian target of rapamycin complex 1 signaling in diet-induced obesity. J Neurosci. 2008;28(28):7202–7208. doi: 10.1523/JNEUROSCI.1389-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38(4):204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moran I, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shin AC, Zheng H, Berthoud HR. An expanded view of energy homeostasis: neural integration of metabolic, cognitive, and emotional drives to eat. Physiol Behav. 2009;97(5):572–580. doi: 10.1016/j.physbeh.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kroemer NB, et al. Addict Biol. Fasting levels of ghrelin covary with the brain response to food pictures [published online ahead of print September 13, 2012]. doi: 10.1111/j.1369-1600.2012.00489.x . [DOI] [PubMed] [Google Scholar]
- 67.Fagerberg B, Hulten LM, Hulthe J. Plasma ghrelin, body fat, insulin resistance, and smoking in clinically healthy men: the atherosclerosis and insulin resistance study. Metabolism. 2003;52(11):1460–1463. doi: 10.1016/S0026-0495(03)00274-9. [DOI] [PubMed] [Google Scholar]
- 68.Batterink L, Yokum S, Stice E. Body mass correlates inversely with inhibitory control in response to food among adolescent girls: an fMRI study. NeuroImage. 2010;52(4):1696–1703. doi: 10.1016/j.neuroimage.2010.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallagher D, Heymsfield SB, Heo M, Jebb SA, Murgatroyd PR, Sakamoto Y. Healthy percentage body fat ranges: an approach for developing guidelines based on body mass index. Am J Clin Nutr. 2000;72(3):694–701. doi: 10.1093/ajcn/72.3.694. [DOI] [PubMed] [Google Scholar]
- 70.Ramel A, et al. Relationship between BMI and body fatness in three European countries. Eur J Clin Nutr. 2013;67(3):254–258. doi: 10.1038/ejcn.2013.6. [DOI] [PubMed] [Google Scholar]
- 71.Ode JJ, Pivarnik JM, Reeves MJ, Knous JL. Body mass index as a predictor of percent fat in college athletes and nonathletes. Med Sci Sports Exerc. 2007;39(3):403–409. doi: 10.1249/01.mss.0000247008.19127.3e. [DOI] [PubMed] [Google Scholar]
- 72.Sondergaard E, Gormsen LC, Nellemann B, Vestergaard ET, Christiansen JS, Nielsen S. Visceral fat mass is a strong predictor of circulating ghrelin levels in premenopausal women. Eur J Endocrinol. 2009;160(3):375–379. doi: 10.1530/EJE-08-0735. [DOI] [PubMed] [Google Scholar]
- 73.Stoeckel LE, Weller RE, Cook EW, 3rd, Twieg DB, Knowlton RC, Cox JE. Widespread reward-system activation in obese women in response to pictures of high-calorie foods. NeuroImage. 2008;41(2):636–647. doi: 10.1016/j.neuroimage.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 74.Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited--again. NeuroImage. 1995;2(3):173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.