Abstract
Protein quality control and metabolic homeostasis are integral to maintaining cardiac function during stress; however, little is known about if or how these systems interact. Here we demonstrate that C terminus of HSC70-interacting protein (CHIP), a regulator of protein quality control, influences the metabolic response to pressure overload by direct regulation of the catalytic α subunit of AMPK. Induction of cardiac pressure overload in Chip–/– mice resulted in robust hypertrophy and decreased cardiac function and energy generation stemming from a failure to activate AMPK. Mechanistically, CHIP promoted LKB1-mediated phosphorylation of AMPK, increased the specific activity of AMPK, and was necessary and sufficient for stress-dependent activation of AMPK. CHIP-dependent effects on AMPK activity were accompanied by conformational changes specific to the α subunit, both in vitro and in vivo, identifying AMPK as the first physiological substrate for CHIP chaperone activity and establishing a link between cardiac proteolytic and metabolic pathways.
Introduction
Pathological cardiac hypertrophy (subsequently referred to as cardiac hypertrophy) is an adaptive response to increased afterload brought about by hypertension, decreased arterial compliance, and other cardiac stressors (1). During the development of cardiac hypertrophy, numerous cellular processes come into play, including protein synthesis and proteolysis to account for the structural changes that accompany hypertrophy, as well as changes in cardiac metabolism to cope with increased energy demands. Despite the well-known fact that protein turnover and metabolic changes accompany cardiac hypertrophy, surprisingly little is known about how stress-dependent protein quality control mechanisms and metabolic regulation are coordinated, if at all, in the heart.
C terminus of HSC70-interacting protein (CHIP, also known as STUB1) is a 35-kDa cytosolic protein initially identified in a screen for proteins that interact with the mammalian chaperones HSC70, HSP70 (2), and HSP90 (3). CHIP is ubiquitously expressed in mammalian tissues but is expressed at highest levels in the adult heart (2). CHIP plays an important dual role as both a cochaperone and a ubiquitin ligase (2–5). More recently, however, an additional role for CHIP in protein quality control has been suggested. Evidence that CHIP can act as an autonomous chaperone, independent of its association with HSPs, has been reported (6). However, these observations were made with a nonphysiological substrate (luciferase), and, to date, verification of CHIP’s chaperone activity and identification of a physiological substrate have not been made.
Changes in cardiac metabolism, such as oxidative substrate preference, mitochondrial function, and biogenesis, play a key role in the adaptive response to cardiac stress (7). As cardiomyocyte hypertrophy progresses, energy demand increases and cellular ATP levels fall. One of the cellular responses to decreased ATP levels is the activation of AMP-activated kinase (AMPK) (8–10). AMPK is an integral global coordinator of the metabolic stress response through its ability to function as a metabolic sensor. The AMPK holoenzyme consists of 3 subunits, the catalytic α subunit (α1 or α2, the latter being the dominant subunit in the heart; ref. 11) and the β and γ regulatory subunits. AMPK is regulated by AMPK kinase phosphorylation of the α subunit (for example, by the kinase LKB1) and allosterically through the AMP/ATP binding domain found in the γ subunit (11). The α2 subunit in particular plays a cardioprotective role during pressure overload-induced hypertrophy (10) and has recently garnered attention as a therapeutic target for cardiac pathologies (12).
Given recent observations linking defective protein quality control and cardiac dysfunction (13, 14), we used Chip–/– mice to measure the response to acute pressure overload-induced cardiac hypertrophy in order to explore the relationship between protein quality control mechanisms and cardiac function. Surprisingly, we discovered that the cardiac dysfunction associated with pressure overload in Chip–/– mice is likely due to the fact that CHIP mediates metabolic adaptation to increased stress by directly regulating AMPK activity. This observation not only identifies AMPK as the first in vivo chaperone substrate of CHIP but also demonstrates that the processes of stress-dependent protein quality control and regulation of energy metabolism are not only coordinated but are essential for withstanding the increased cardiac demands brought about by hemodynamic stress.
Results
Cardiac pressure overload causes pathological hypertrophy and decreased cardiac function in Chip–/– mice.
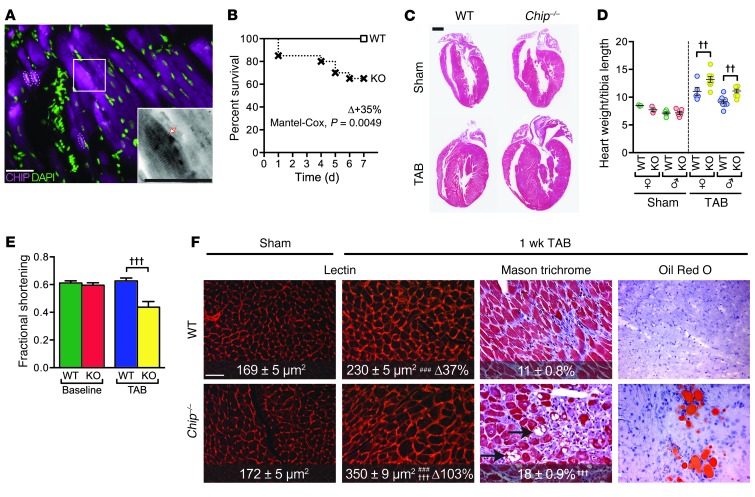
CHIP is expressed in both myocytes and nonmyocyte cells throughout the heart (Supplemental Figure 1, A–D; supplemental material available online with this article; doi: 10.1172/JCI69080DS1). Within myocytes, CHIP expression is localized to the sarcomere as well as nuclear and perinuclear regions (Figure 1A and Supplemental Figure 1, B–D). To examine the consequences of cardiac CHIP deletion during the adaptive response to pressure overload-induced hypertrophy, we subjected wild-type and Chip–/– mice to either a sham surgical procedure or trans-aortic banding (TAB). During the first 2 to 4 weeks of pressure overload, hearts are in a compensatory growth stage, in which the increase in resistance caused by the banding is met by an increase in ventricular mass to normalize wall stress and maintain cardiac output. Surprisingly, 1 week following TAB, Chip–/– mice exhibited 35% mortality (compared with no mortality in wild-type mice; Figure 1B), markedly enhanced ventricular hypertrophy (Figure 1C), and a doubling in heart weight across sexes (65% ± 7% versus 30% ± 7%, respectively; Figure 1D). Despite these adaptive changes, conscious echocardiography revealed that Chip–/– mice had a 26.7% ± 7.6% decrease in fractional shortening relative to that of wild-type mice that maintained normal cardiac function (Figure 1E and Supplemental Table 1). Compared with those from wild-type hearts, histological sections from Chip–/– banded hearts exhibited the hallmark features of a more acute pathological hypertrophic response, including increased myocyte hypertrophy and interstitial fibrosis (Figure 1F and Supplemental Figure 1, E–G). In addition, sections taken from banded Chip–/– hearts also contained extensive lipid accumulation (Figure 1F, right). Given that cardiac lipid accumulation is seen in pressure-overloaded human hearts of individuals with metabolic syndrome (15), and is accompanied by decreases in mitochondrial function and biogenesis (16), we hypothesized that the discordant increase in cardiac mass with a decreased cardiac function seen in Chip–/– mice after only 1 week of banding could be due to the fact that CHIP is required for modifying stress-dependent metabolic responses and/or mitochondrial function in the heart.
Figure 1. Pressure overload-induced cardiac hypertrophy in Chip–/– mice.
(A) Indirect immunofluorescence images of CHIP expression (magenta) and nuclei (green) in mouse heart. The arrow indicates sarcomere localization; dashed areas indicate nuclear localization (white). Scale bar: 50 μm. (B) Survival curves after TAB surgery in wild-type or Chip–/– mice (n = 12 and 20, respectively). (C) Hematoxylin and eosin–stained whole heart sections from the indicated conditions. Scale bar: 1 mm. (D) Heart weight normalized to tibia length (mg/mm; n = 3 and 6 male and female wild-type sham mice, respectively; n = 5 and 8 male and female wild-type TAB mice, respectively; n = 3 and 5 male and female Chip–/– sham mice, respectively; n = 8 male and female Chip–/– TAB mice, respectively; P < 0.01 across all sexes and genotypes, sham vs. TAB; ††P < 0.01 sex-matched wild-type vs. Chip–/– mice). (E) Fractional shortening (%) after sham or TAB surgery (n = 6 and 7 for sham and 8 and 6 for TAB in wild-type and Chip–/– mice, respectively; †††P < 0.001 vs. all other groups). (F) Representative histological staining of whole heart sections from wild-type and Chip–/– mice. Lectin images represent 25 areas from 4 mice per condition. In Masson trichrome images, numbers represent percentage interstitial fibrosis (n = 13). ###P < 0.001 TAB vs. sham; †††P < 0.001 wild-type vs. Chip–/–. Arrows indicate intercellular and intracellular regions void of tissue. Scale bar: 100 μm.
Chip–/– mice have decreased mitochondrial content compared with wild-type mice following TAB.
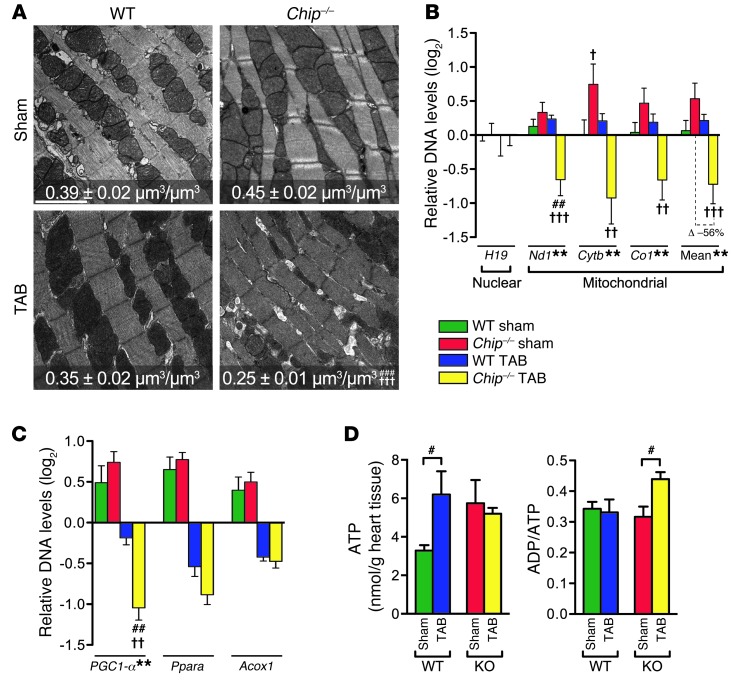
To determine whether the decrease in cardiac function in banded Chip–/– mice is related to a decrease in energy production due to abnormal mitochondrial form or function, we used electron microscopy to examine mitochondrial structure and volume density after pressure overload in both wild-type and Chip–/– hearts. Tissue sections from banded Chip–/– hearts exhibited a 44% and 29% decrease in mitochondrial density compared with Chip–/– sham surgery hearts and banded wild-type hearts, respectively, with no observable changes in overall morphology (such as disputed cristae) that have been observed in models of contractile dysfunction (ref. 17, Figure 2A, and Supplemental Figure 2A). In addition, qPCR analysis of the relative abundance of mitochondrial DNA normalized to a nuclear DNA target revealed that the genomic levels of mitochondrial genes were decreased in Chip–/– (but not wild-type) hearts after banding (56% decrease; Figure 2B), confirming the phenotype seen in the electron micrographs and suggesting that CHIP is critical for maintaining mitochondrial content during cardiac stress.
Figure 2. Pressure overload-induced changes in mitochondrial biogenesis and cardiac gene expression in Chip–/– mice.
(A) Electron micrographs and mitochondrial volume density (represented by the mean ± SEM, n > 5 mice represented per condition) in heart sections. Scale bar: 2 μm. (B) Log2 of mean-centered DNA levels measured via qPCR of nuclear (H19) or mitochondrial (Nd1, Cytb, Co1, and the mitochondrial mean) DNA (represented by the mean ± SEM from 6 biological replicates per condition). The surgery-dependent percentage change (Δ) in mean mitochondrial biogenesis (TAB vs. sham) in wild-type vs. Chip–/– mice is significant at P = 0.004. (C) Log2 of mean-centered mRNA levels in mouse hearts measured via qPCR (represented by the mean ± SEM from 4 biological replicates); changes in Ppara and Acox1 are significant at the level of surgery (P < 0.0001), with no significant difference between wild-type and Chip–/– mice at either sham or TAB conditions. (D) Total ATP levels and ADP/ATP ratios in mouse hearts measured via HPLC (represented by the mean ± SEM from 4 biological replicates per condition). **P < 0.01 surgery-genotype interaction via 2-way ANOVA; post-hoc test, #P < 0.05, ##P < 0.01, and ###P < 0.001 sham vs. TAB surgery; †P < 0.05, ††P < 0.01, and †††P < 0.001 Chip–/– vs. wild-type genotypes.
Pressure overload in Chip–/– hearts causes decreased expression of gene clusters related to energy metabolism.
Given the numerous pathways involved in maintaining mitochondrial function, biogenesis, and energy metabolic homeostasis, we used microarray analysis to analyze global changes in gene expression in wild-type and Chip–/– mice following TAB (expanded reporting of the results obtained from this analysis is included in the Supplemental Methods, Supplemental Table 2, and Supplemental Figure 2, B–D). Not surprisingly, microarray analysis revealed differential expression of numerous genes involved in myocyte function (Supplemental Figure 2C, right), including multiple metabolic gene sets, such as oxidative phosphorylation (OxPhos), muscle contraction, and mitochondrion, all of which were downregulated in banded Chip–/– mice (Supplemental Figure 2C, middle). The OxPhos category was the most highly enriched functional group between banded Chip–/– and wild-type hearts, comprising 9 differentially expressed genes encoding for subunits of multiple OxPhos complexes (confirmed via qPCR; Supplemental Figure 2, E–G, and Supplemental Tables 3 and 4). One of these differentially expressed genes was the transcription factor PGC1-α (also known as Ppargc1a), which was the fourth most downregulated gene comparing banded Chip–/– and wild-type hearts (Supplemental Table 2; 2.0-fold change, P = 0.00018, confirmed via qPCR; Figure 2C). PGC1-α is a primary regulator of the OxPhos subunit gene transcription as well as a direct regulator of mitochondrial biogenesis. As such, the decrease in cardiac PGC1-α expression in banded Chip–/– mice was consistent with a decrease in cardiac energy-generating capacity in these mice. To rule out other possible causes for the failure of banded Chip–/– mice to generate cardiac energy, we examined the expression of Ppara, the gene encoding the nuclear receptor PPARα, which is primarily responsible for mediating the switch between energy substrates that commonly occurs during cardiac stress (18, 19). However, the expression pattern of Ppara and its canonical target gene Acox1 did not differ between banded wild-type or Chip–/– hearts (Figure 2C), suggesting a more global defect in energy generation, rather than a defect in the ability to switch from one energy substrate to another, is the primary metabolic defect in banded Chip–/– hearts. Taken together, the stress-dependent decrease in PGC1-α expression in the banded Chip–/– hearts, along with the decreased mitochondrial content (Figure 2, A and B) and transcription of nuclear-encoded components of the OxPhos pathways (Supplemental Figure 2G), suggest a pivotal role for CHIP in regulating cardiac ATP generation and the metabolic stress response at a global level.
To test the theory that CHIP is involved in ATP generation, adenine nucleotide levels (ATP and ADP) were compared in hearts isolated from Chip–/– and wild-type mice after 1 week of either sham or TAB surgery. This early time point following banding was chosen due to the quick decline in the survival of Chip–/– mice following the induction of pressure overload (Figure 1B). Following 1 week of banding, total ATP levels in extracts isolated from wild-type mice increased 1.8 fold (Figure 2D and Supplemental Table 5), consistent with recent metabolic studies demonstrating energetic adaptation to maintain cardiac power during the initial compensation to pressure overload (20, 21). In contrast, total ATP levels failed to increase in extracts isolated from Chip–/– banded hearts, supporting the theory that CHIP may be involved in ATP generation during cardiac pressure overload. Despite the lack of increase in ATP levels, banded Chip–/– hearts did contain elevated ratios of total ADP/ATP, a phenotype consistent with either accelerated ATP consumption or disruption of ATP synthesis (22). In metabolically healthy systems, a rise in the ADP/ATP ratio (amplified further by the AMP/ATP ratio through the myokinase reaction) is a physiological cue to restore ATP homeostasis through the activation of the energy-sensing kinase AMPK (22). The activation and increased activity of AMPK promotes multiple ATP-generating pathways, including PGC1-α–dependent mitochondrial biogenesis pathways, while at the same time inhibiting energy-consuming processes, such as protein synthesis. Given the elevated ratios of ADP/ATP in banded Chip–/– hearts, without a concomitant increase in ATP levels, we hypothesized that a failure of activation of AMPK may be responsible for the inability of Chip–/– hearts to maintain energy homeostasis in response to pressure overload.
Deficient AMPK activation, signaling, and activity during pressure overload in Chip–/– hearts.
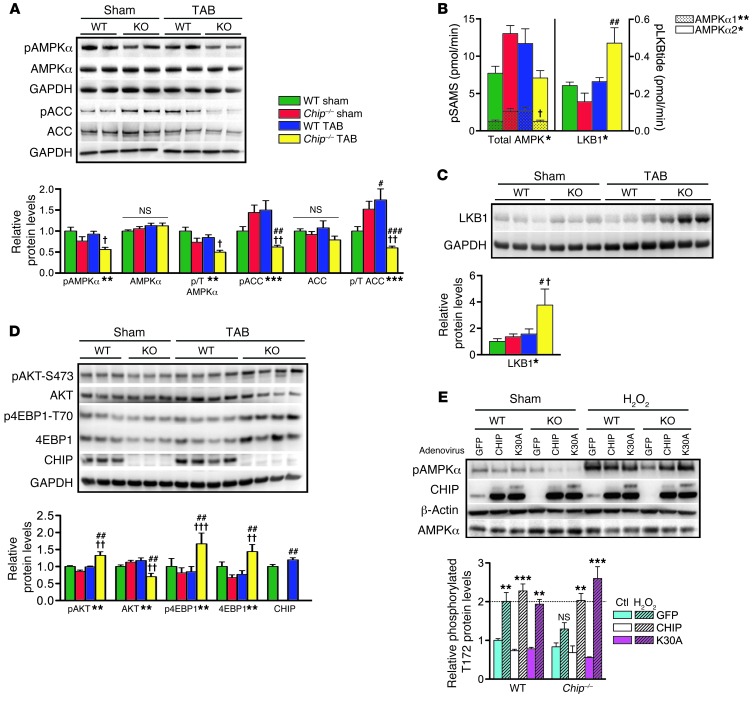
As a first step in determining whether there is a defect in the AMPK signaling pathway in banded Chip–/– hearts, the activation and activity of the catalytic subunits of AMPK were measured after 1 week following sham or TAB surgery in wild-type and Chip–/– hearts (Figure 3A). The primary readout of AMPK activation, phosphorylation of AMPKα-T172, was substantially decreased in banded Chip–/– hearts, as was phosphorylated ACC (a well-characterized in vivo AMPK substrate), indicating an inhibition of AMPK activity. To confirm that AMPK activity was indeed inhibited in Chip–/– hearts, we measured the rate of phosphorylation of a synthetic AMPK substrate (SAMS peptide) in immunoisolated AMPKα1 and AMPKα2 complexes. Consistent with the decrease in ACC phosphorylation (Figure 3A), SAMS phosphorylation was also decreased in Chip–/– banded hearts (Figure 3B), confirming an inhibition of AMPK activity. Taken together, these data demonstrate that, in Chip–/– banded hearts, AMPK activation (illustrated by phosphorylated AMPKα-T172) and activity (illustrated by phosphorylated ACC and SAMS) are significantly inhibited, offering a plausible explanation for why Chip–/– hearts fail to compensate and maintain energy homeostasis in response to pressure overload.
Figure 3. AMPK pathway and activity during pressure overload.
(A) Representative immunoblots and densitometry analysis of AMPKα, pAMPKα-T172, pACC-S79, and ACC after sham or TAB surgery in wild-type or Chip–/– (KO) mouse hearts. (B) AMPKα1, AMPKα2, total AMPK, and LKB1 activity determined by phosphorylation of SAMS (pSAMS) or LKB1 (pLKBtide) peptides in immunoprecipitates from heart lysates (represented by the mean ± SEM from 3–6 biological replicates per condition). (C and D) Representative immunoblots and densitometry analysis of (C) LKB1 and (D) pAKT-S473, AKT, p4EBP1-T70, 4EBP1, and CHIP protein after sham or TAB surgery. Relative cardiac protein levels from A, C, and D normalized to GAPDH are expressed as mean ± SEM from 3 to 6 biological replicates per condition. *P < 0.05, **P < 0.01, ***P < 0.001 surgery-genotype interaction via 2-way ANOVA; post-hoc test, #P < 0.05, ##P < 0.01, and ###P < 0.001 sham vs. TAB surgery; †P < 0.05, ††P < 0.01, and †††P < 0.001 Chip–/– vs. wild-type genotypes. (E) Representative immunoblots and densitometry analysis of protein extracts from wild-type or Chip–/– MEFs treated with adenoviruses as labeled followed by exposure to H2O2. Relative protein levels of phosphorylated AMPKα-T172 normalized to β-actin are expressed as mean ± SEM from 3 independent experiments. **P < 0.01, ***P < 0.001 via Dunnett’s comparison to wild-type GFP control condition and control-treated cells for each genotype/adenovirus pairing. The dashed line indicates the amount of phosphorylation in wild-type cells in response to H2O2.
LKB1 is the primary kinase involved in the energy status–dependent phosphorylation of AMPKα-T172 in the heart (23). To determine whether decreases in LKB1 activity or expression contribute to the lack of AMPK activation and activity in banded Chip–/– hearts, we measured the rate of phosphorylation of a synthetic LKB1 substrate as well as steady-state LKB1 protein expression levels. Surprisingly, both LKB1 activity (Figure 3B) and expression (Figure 3C) were increased 2.0 and 2.8 fold, respectively, in Chip–/– banded hearts, suggesting that the deficit in AMPK activation in these hearts is not due to decreased LKB1 activity or expression.
Collectively, these phenotypes in the Chip–/– banded hearts are consistent with a loss in AMPK function in response to a need for increased cardiac power and ATP (Figure 3, A–C). A recent genetic model determined a critical role for AMPKα2 (the predominant AMPK catalytic subunit in the heart) in response to pressure overload in which levels of pAKT-S273 and p4EBP1-T70, key regulators of protein translation usually inhibited by AMPK signaling, were increased in response to banding in AMPKα2–/– hearts (24). AKT and 4EBP1 are not substrates of AMPK but instead regulate global protein translation and are responsive to the activity levels of AMPK (10, 24, 25). Consistent with our hypothesis of reduced AMPK function in banded Chip–/– hearts, we observed a nearly identical increase in the activation of these pathways (Figure 3D) to those found in the banded AMPKα2–/– hearts. Notably, CHIP expression itself was increased approximately 30% in wild-type mouse hearts after 1 week of TAB (Figure 3D), a trend that continued through longer periods of pressure overload (Supplemental Figure 3A) and correlated with increased AMPK activation and activity (Supplemental Figure 3, A and B). An additional known target of AMPK, estrogen-related receptor α (ERRα) expression (26), was also decreased in banded Chip–/– hearts (Supplemental Figure 3C). ERRα regulates mitochondrial biogenesis independently of PGC1-α (26, 27), and our observation of decreased ERRα and PGC1-α expression is consistent with reduced AMPK function and decreased mitochondrial biogenesis in banded Chip–/– hearts. These results indicate that, despite allosteric cues (increased ADP/ATP ratio) and increased expression of LKB1, AMPK activation and activity are significantly diminished in banded Chip–/– hearts, suggesting that either proteins regulated by CHIP or CHIP itself plays a key role in AMPKα activation and activity toward downstream targets during cardiac pressure overload.
CHIP is required for AMPK activation in response to stress.
Our previous data suggest that, whereas CHIP is not required for AMPK activation under basal conditions, CHIP is essential for optimal activation of AMPK in response to pressure overload. However, it is not known whether the reliance on CHIP expression is specific to stress caused by pressure overload or whether it occurs also in other metabolic stress models involving AMPK. To test this, we cultured mouse embryonic fibroblasts (MEFs) isolated from either wild-type or Chip–/– mice, induced acute oxidative stress and ATP depletion via H2O2 exposure, and measured the AMPK response. As previously shown (28), AMPK activation and activity increased substantially in wild-type MEFs in response to H2O2 exposure, as measured by phosphorylation of AMPKα-T172 and SAMS phosphorylation (Figure 3E and Supplemental Figure 3D). In contrast, AMPK activation and activity were markedly attenuated in Chip–/– MEFs; however, this attenuation in both AMPK activation and activity could be reversed by the reintroduction of CHIP (Figure 3E and Supplemental Figure 3D), confirming that CHIP is essential for proper stress-dependent activation of AMPK. Interestingly, AMPK activation was also rescued by treatment of Chip–/– MEFs with the mutant construct CHIP-K30A (Figure 3E) that is unable to bind with HSP70/90 (29), indicating that the ability of CHIP to activate AMPK is not reliant on CHIP’s direct interaction with HSPs and is therefore unlikely to be due to the cochaperone function of CHIP. The ability of CHIP to activate AMPK was also tested in mice using the AMPK agonist AICAR (which is metabolized to ZMP, an AMP mimetic). In these experiments, as in the cell culture experiments, AICAR activation of AMPK was found to be dependent on the presence of CHIP (Supplemental Figure 3E). These data demonstrate that CHIP is required for AMPK activation in response to a variety of stressors. However, whether or not CHIP interacts directly with AMPK, or through mediation via another protein, remains unknown.
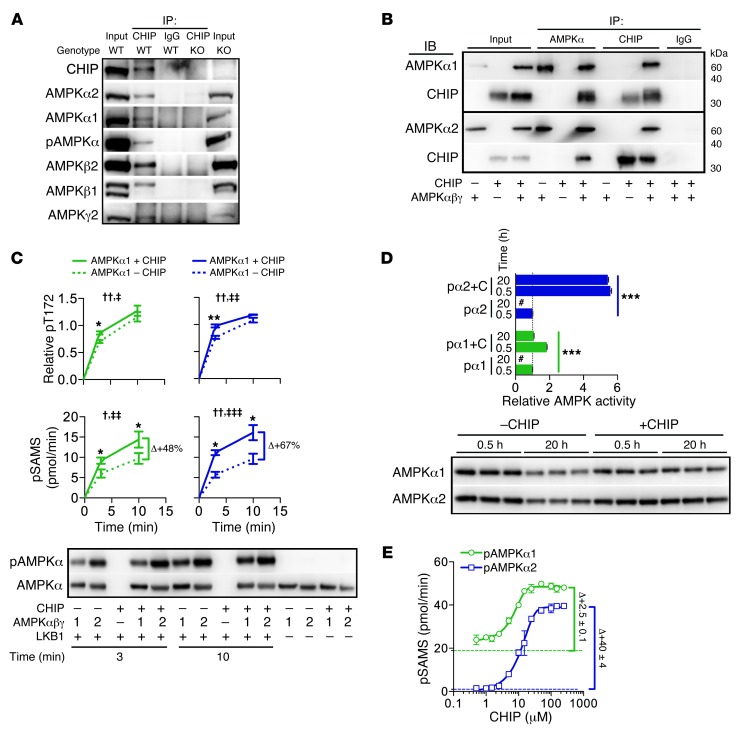
CHIP forms a complex with AMPK in vivo and directly interacts with AMPK in vitro.
To determine whether CHIP interacts with AMPK, we performed coimmunoprecipitation assays on purified heart extracts from wild-type mice. Both α1 and α2 AMPK catalytic subunits (including the active, phosphorylated form, AMPKα-T172) as well as the β1, β2, and γ2 regulatory subunits coeluted with CHIP immunoprecipitation (Figure 4A and Supplemental Figure 4, A and B), confirming that CHIP forms a complex with AMPK holoenzymes in the heart. Quantitative analysis of the cardiac proteome demonstrated that CHIP protein exists in over a 3:1 molar ratio to AMPKα2 (30), demonstrating that abundance of CHIP and AMPK protein in the heart are sufficient for CHIP to make substantial contributions to AMPK function (Supplemental Table 6). The evidence of a CHIP-AMPK complex suggested that CHIP functions directly with AMPK to increase both its activation and activity. The two known molecular functions of CHIP are as a ubiquitin ligase, to mediate either the polyubiquitination of chaperone-bound proteins for eventual degradation by the proteasome, or as a cochaperone, to assist heat shock chaperones in the refolding of damaged or unfolded proteins. Given the positive effect of CHIP on AMPK function, as well as the ability of the K30A mutant to rescue the Chip–/– phenotype in response to stress, CHIP did not appear to be functioning in either of these capacities with respect to regulating AMPK activity or activation. A newly proposed function of CHIP is as an autonomous chaperone, acting independently of other molecular chaperones to recognize and bind misfolded proteins. However, to date, this role for CHIP has only been demonstrated on luciferase, a nonphysiological substrate ligand (6). Therefore, to test whether CHIP acts as an autonomous chaperone to AMPK, we established an in vitro system whereby the interaction between CHIP and AMPK could be studied without the influence of extraneous molecules and in the absence of the HSP cochaperone and ubiquitin ligase functions of CHIP. Evidence of direct interaction between recombinant CHIP and AMPK in solution was demonstrated through coimmunoprecipitation of a CHIP-AMPK complex with antibodies that recognize either AMPK or CHIP (Figure 4B). The CHIP-AMPK complex formed independent of other cellular cofactors and consisted of the trimeric AMPK holoenzyme (irrespective of α-T172 phosphorylation status) and a CHIP dimer, the well-defined in vivo functional oligomeric state of CHIP (ref. 6 and Supplemental Figure 4, C–F).
Figure 4. Analysis of the CHIP-AMPK interaction.
(A) Immunoblot analysis from CHIP or control (IgG) immunoprecipitations in wild-type and Chip–/– heart lysates. (B) Immunoprecipitations of reactions containing recombinant CHIP, AMPKα1β1γ1, AMPKα2β1γ1, or combined proteins, subsequently immunoblotted with the indicated antibodies. (C) Densitometry of AMPKα-T172 phosphorylation (pT172) and AMPK activity measured by SAMS phosphorylation (pSAMS) in reactions containing combinations of proteins as indicated. Relative pT172 was normalized to total AMPKα. Data are represented by the mean ± SEM from 3 independent experiments (2-way ANOVA, †P < 0.05, ††P < 0.01,comparing time; ‡P < 0.05, ‡‡P < 0.01, and ‡‡‡P < 0.0001, comparing the presence of CHIP; t test, *P < 0.05 and **P < 0.01, comparing the presence of CHIP per time point). (D) Activated pAMPKα1β1γ1 (pα1, green) and pAMPKα2β1γ1 (pα2, blue) activity in the absence or presence of CHIP (+C) measured by SAMS phosphorylation (represented by mean ± SEM from 3 independent experiments) (ANOVA post-hoc test, ***P < 0.001 comparing all pairs of conditions). Values were normalized to control conditions (0.5 hours in the absence of CHIP); “#” indicates activity measurements not significantly above background. The samples were also immunoblotted for total AMPKα1 or AMPKα2 from the indicated conditions. (E) AMPK activity measured by pSAMS (represented by mean ± SEM from 3 biological replicates) using reactions containing either activated pAMPKα1β1γ1 (green) or pAMPKα2β1γ1 (blue) mixed with increasing amounts of recombinant CHIP. Dashed green and blue lines represent baseline activities of pα1β1γ1 and pα2β1γ1, respectively.
CHIP directly increases AMPK activation and functions as an AMPK agonist independent of extraneous factors.
Chaperones function though transient interactions with substrates until a particular conformation is achieved (31). Activation of AMPK by upstream kinases, dephosphorylation by phosphatases, and activity of AMPK are regulated allosterically primarily through conformational changes throughout the AMPK molecule. Consequently, we next tested CHIP’s influence on the multiple mechanisms that can influence AMPK phosphorylation and activity using our in vitro model. In the reaction without CHIP, LKB1 robustly phosphorylated and activated AMPK holoenzymes in vitro. However, when CHIP was present, phosphorylation of AMPK holoenzymes occurred more rapidly (<5 minutes) prior to saturation (>10 minutes) (Figure 4C). Similar CHIP-dependent effects were measured with LKB1-mediated phosphorylation of monomeric AMPKα2 (Supplemental Figure 4G). Surprisingly, in the presence of CHIP, AMPK holoenzyme-specific activity determined by SAMS phosphorylation was increased even when AMPKα-T172 phosphorylation levels were identical to those in the treatment conditions in which CHIP was absent (Figure 4D), suggesting that CHIP can enhance AMPK activity independent of α-T172 phosphorylation.
Next, we tested the ability of CHIP to protect activated AMPK from PP2Cα-mediated dephosphorylation at α-T172 in the presence and absence of AMP binding to the regulatory γ subunit of AMPK. The presence of CHIP did not blunt the dephosphorylation of AMPK holoenzymes or change the AMP-dependent attenuation of dephosphorylation (Supplemental Figure 4H), suggesting CHIP does not affect AMPKγ subunit function. In the absence of PP2Cα, CHIP once again increased AMPK activity (Supplemental Figure 4H, bottom) without any change in the phosphorylation levels of α-T172 (Supplemental Figure 4H, top). The CHIP-dependent increase in AMPK activity independent of α-T172 phosphorylation is consistent with our similar observation in the LKB1 phosphorylation assays (Figure 4C), again suggesting CHIP-dependent effects on the kinase activity of AMPK consistent with our chaperone model.
An additional mechanism of chaperone function is to help maintain substrate stability and solubility. Recombinant proteins vary in their stability at physiological temperatures. Activated AMPK maintains 100% stability for up to 2 hours at 37°C but loses all measurable activity after 20 hours at 37°C (Figure 4D). Remarkably, coincubation of CHIP with the activated AMPK holoenzyme resulted in the complete retention of AMPK activity after 20 hours. A similar affect of CHIP on protein stability was also observed with monomeric AMPKα2 (Supplemental Figure 4G), demonstrating that CHIP can affect AMPK stability in vitro and elicit effects specifically toward the catalytic (α) subunit of AMPK.
In all of our in vitro experiments measuring AMPK activity (Figure 4, C and D, and Supplemental Figure 4, H and I), reactions containing both activated AMPK and CHIP consistently resulted in higher SAMS phosphorylation compared with reactions containing activated AMPK alone, consistent with CHIP functioning as an AMPK agonist independent of α-T172 phosphorylation. To directly test the agonist function of CHIP, and to rule out any nonspecific artifacts in our experiments, we confirmed that CHIP exhibited no kinase activity itself toward either AMPKα-T172 (Figure 4C) or SAMS peptide (data not shown) and that the presence of CHIP did not result in nonspecific binding of radiolabeled phosphate (Supplemental Figure 4I). However, CHIP did directly increase activated AMPK kinase activity in a dose-dependent manner that resulted in a 2.5- and 40-fold increase in α1- and α2-containing AMPK complexes, respectively (Figure 4E), including significant increases in activity at submicromolar concentrations of CHIP (Supplemental Figure 4J). This is the first such report of an endogenous AMPK agonist that directly affects the catalytic subunit independent of phosphorylation that, unlike other AMPK agonists, does not act as a low-energy AMP mimetic. Taken together, these data ascribe a new function to CHIP as being an AMPK agonist that directly augments the ability of AMPK to regulate energy metabolism in the heart during periods of stress, a finding that has immense therapeutic value.
CHIP alters AMPK tertiary structure.
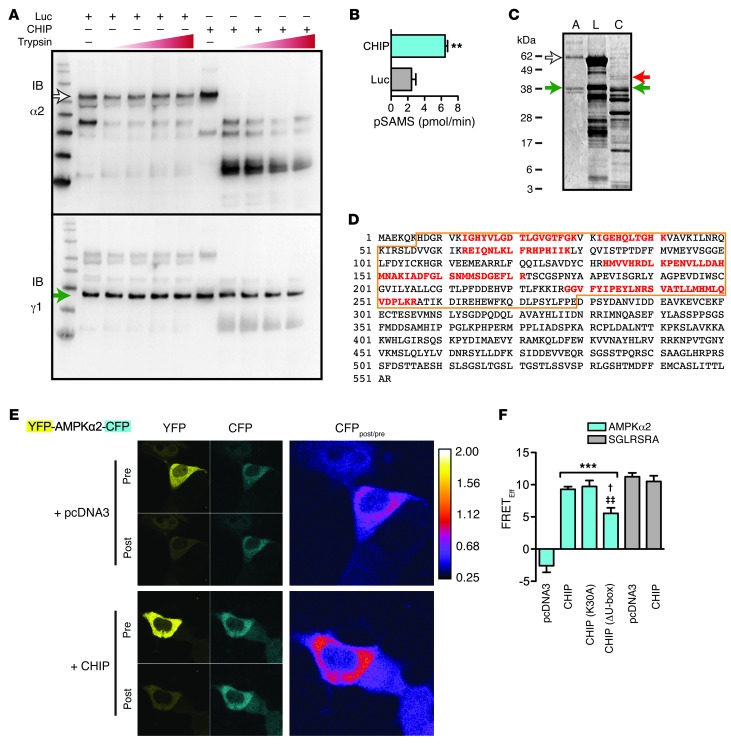
Given that AMPK activation and activity are augmented through conformational changes, such as the conformation rearrangement of the autoinhibitory domain (AID), the α-hook, and nucleotide binding to the γ subunit, we hypothesized that CHIP’s ability to increase AMPK activation and activity may involve a change to the tertiary structure of AMPK. To determine whether CHIP is able to induce conformational changes to AMPK, we used both in vitro and in vivo systems. For the in vitro studies, we used limited proteolysis analysis, which is predicated on the fact that the accessibility of specific protein residues recognized by proteases is dependent on the native 3-dimensional form of the protein. Therefore, incubating intact folded proteins with low amounts of proteases and examining the resultant fragments can detect structural changes through changes in the protease site susceptibility. Using recombinant proteins in conditions identical to those used to measure functional CHIP-dependent functional changes in AMPK (Figure 4, C and D) and limited proteolysis analysis (32), we determined that the proteolytic fingerprint (trypsin) of the α subunit of the activated AMPK holoenzyme (pAMPKα2β1γ1) was markedly changed in the presence of CHIP (Figure 5A, right) compared with that in the presence of luciferase (Figure 5A). CHIP’s effect appeared to be localized to the catalytic subunit (consistent with the potent effect of CHIP on increasing the specific activity of AMPK; Figure 5B), given that there was no difference in the γ subunit fingerprint, which was completely intact when the pAMPKα2 holoenzyme was incubated with either CHIP or luciferase (Figure 5A, bottom). We confirmed CHIP-dependent conformational changes in the proteolytic fingerprints using proteinase K in place of trypsin (Supplemental Figure 5A). In an effort to better localize the specific regions within the α subunit subject to CHIP-induced conformational changes, we resolved the proteolytic fragments in identical conditions via SDS-PAGE and visualized them with a protein stain to identify intact portions of AMPK that were present only in reactions containing CHIP. As expected, in reactions containing a molar excess of either luciferase or CHIP relative to AMPK, the predominant proteolytic fragments identified via sequencing were luciferase or CHIP (Figure 5C, lanes 2 and 3). However, we also identified a unique fragment in the reactions containing CHIP (Figure 5C, red arrow, and Supplemental Table 7) that corresponded to the intact protein kinase domain of AMPKα2 (NCBI GenInfo Identifier: 110590560), with peptide coverage that spans residues 13–256 (Figure 5D, red). Identical to the immunoblot data for the γ subunit, we identified the intact full-length γ1 band in the same reactions via mass spectroscopy sequencing (Figure 5C, green arrow, and Supplemental Table 7). Given the size and content of the intact α subunit fragment (approximately 42 kDa) it is likely that CHIP affects C-terminal regions of AMPKα, which include the α-hook regulatory domain or components of the β subunit interaction domain. Taken together, these data are consistent with CHIP altering the tertiary structure of the AMPK holoenzyme.
Figure 5. CHIP-dependent effects on AMPK tertiary structure.
(A) Limited proteolysis analysis of pAMPKα2β1γ1 in the presence of either luciferase (Luc) or CHIP. (B) Specific activity of AMPK in the presence of luciferase or CHIP prior to protease treatment (represented by mean ± SEM; Student’s t test, **P < 0.01; n = 3). (C) Nondigested pAMPKα2β1γ1 (A) or fragments of limited proteolysis reactions containing pAMPKα2β1γ1 in the presence of either luciferase (L) or CHIP (C) visualized with SYPRO-Ruby gel stain. (A and C) The white arrows indicate intact AMPKα2; the green arrows indicate intact γ1 subunit; and the red arrow indicates a fragment of AMPKα2 determined via protein sequencing that was detected only reactions containing CHIP. (D) Sequence of human AMPKα2 protein. The outlined area indicates the kinase domain, with the sequences identified by mass spectroscopy (red). (E) Representative live cell micrographs from before and after photobleaching conditions for both YFP and CFP channels and the resulting image ratio (CFPpost/pre) for cells cotransfected with a control vector (top) or CHIP expression vector (bottom). (F) Quantification of the apparent FRET efficiencies of AMPKα2 protein (cyan) or the positive control FRET protein (gray) coexpressed with the indicated vectors on the x axis (n = 45 cells from 3 independent transfections; ***P < 0.001 compared with pcDNA3 conditions; †P < 0.05 and ‡‡P < 0.01 compared with CHIP and CHIP(K30A), respectively, via ANOVA).
To determine whether CHIP also influences AMPKα folding or structure in vivo, we created a tricistronic YFP-AMPKα2-CFP vector (33, 34) to use with acceptor photobleaching, a form of fluorescent resonance energy transfer (FRET) that detects changes in 3-dimensional space between 2 fluorophores. We initially confirmed that the tricistronic vector incorporated into intact holoenzyme molecules, responded to agonist, and was functionally active (Supplemental Figure 5, B–E). Measurable FRET efficiency (E) of YFP-AMPKα2-CFP was only detected with coexpression of CHIP, both wild-type CHIP (E = 9.3%, range 2.0%–17.3%) and mutants that abolish either the HSP-interacting motif or ubiquitin ligase domain of CHIP (E = 9.7% and 5.6%, respectively), demonstrating that CHIP expression results in conformational changes to AMPKα that force the distance between the 2 fluorophores to a distance less than 10 nm (the Förster radius) or reorientation of their relative diploes (35). The effect of CHIP on FRET was specific to the AMPKα2 insert, as CHIP had no effect on a positive FRET control YFP-CFP vector. This in vitro and in vivo data demonstrating CHIP-dependent conformational changes to AMPKα offers a plausible mechanism by which CHIP is able to directly influence AMPK activation. The sum total of the data presented here supports a direct role for CHIP in regulating cardiac metabolism and function via specific chaperoning activity on AMPK.
Discussion
Along with CHIP’s well-documented role as a both a ubiquitin ligase and cochaperone (29), an additional role for CHIP in protein quality control has been suggested due to the autonomous chaperone activity of CHIP that is independent of its associations with HSPs (6). However, before now, no known physiological substrate for CHIP’s chaperone activity has been identified. The data presented here demonstrate that CHIP acts as a molecular chaperone to AMPK, inducing conformational changes to the α catalytic subunit both in vitro and in vivo (Figure 5, A, C, E, and F, and Supplemental Figure 5A), thereby regulating not only AMPK stability (Figure 5D) and activation (Figure 3, A, E, and C; Supplemental Figure 3E; and Supplemental Figure 4G), but also directly increasing the rate of activated AMPK’s ability to phosphorylate its substrate (Figure 4E and Supplemental Figure 4J). The influence of CHIP expression in mediating AMPK activation and activity appears to be essential for maintaining cardiac function in the acute response to pressure overload, thereby suggesting a direct link between cardiac protein quality control mechanisms and regulators of cardiac energy metabolism. Uniquely, these molecular actions are independent of CHIP’s interactions with the HSP family of chaperones (Figure 3E and Supplemental Figure 3C). In addition, the steady-state expression level of a well-characterized CHIP ubiquitin ligase substrate, serum- and glucocorticoid-regulated kinase SGK (36), was not increased in Chip–/– mice during pressure overload (Supplemental Figure 3C). Although this does not preclude other CHIP ubiquitin ligase substrates from contributing to the pressure overload phenotype, this observation is consistent with the fact that ubiquitin ligase activity is not required for the direct effect of CHIP on AMPK both in vitro and in cell-based models (Supplemental Figure 4G and Figure 5F). The ability of CHIP to interact with and regulate α1- and α2-containing holoenzymes, in addition to other CHIP substrates, likely contributes to the more rapid onset and severe cardiac hypertrophy in response to pressure overload in Chip–/– mice (Figure 1) compared with the inactivation of only the AMPKα2 gene (10). It also bares noting that the overall effect of CHIP on the pressure overload phenotype reported here may in fact be underestimated, given the 35% mortality in Chip–/– mice.
Interestingly, Chip–/– mice at baseline (or under sham surgery conditions) are essentially healthy in regards to cardiac function, suggesting that CHIP is dispensable for baseline regulation of AMPK. This also suggests that there are adaptive physiological mechanisms capable of compensating for the loss of CHIP expression. This adaptation may account for the enhanced cardiac metabolic phenotype observed in Chip–/– mice at baseline that includes increased mitochondrial content (Figure 2B) and trends toward increased mitochondrial volume (Figure 2A), ATP content (Figure 2D), and AMPK activity (pACC in Figure 3A and pSAMS in Figure 3B); these changes also likely contribute to the enhanced diastolic function measured in sham-operated Chip–/– mice (13% increase; Supplemental Table 1). As seen in both Chip–/– mice and several AMPK knockout and transgenic mouse lines, neither CHIP nor AMPK are essential to cardiac development or normal growth and function. However, the importance of both molecules becomes vastly apparent under stressful conditions, with CHIP and AMPK activating protein quality control and energy homeostatic pathways, respectively.
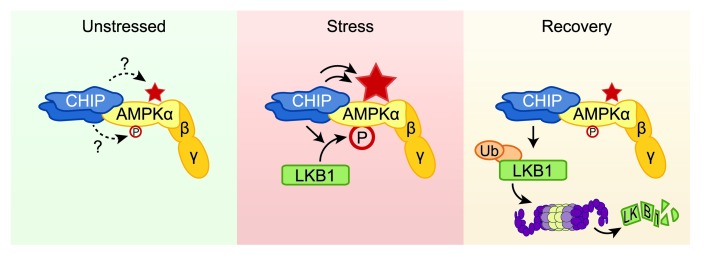
The CHIP and AMPK pathways have previously not been thought to work in a coordinated manner; however, in cell culture models, CHIP does promote the polyubiquitination of LKB1, resulting in decreased LKB1 protein levels (37). Likewise, transient silencing of CHIP expression leads to an increase in LKB1 protein expression (37), suggesting another possible intersection between the CHIP and AMPK pathways. Following TAB surgery in Chip–/– mice, we observed a stress-dependent 4-fold increase in LKB1 protein levels (Figure 3C) and a 2-fold increase in LKB1 activity (Figure 3B) following induction of pressure overload, consistent with a role for CHIP in regulating LKB1 protein expression. However, in these Chip–/– hearts with increased LKB1 expression and activity, AMPK was still not activated (Figure 3, A and B), supportive of CHIP functioning in a dual role in regulating AMPK activity under stressful conditions. That is, whereas CHIP may in fact be a bona fide E3 ligase for LKB1 under stress conditions, it also enhanced LKB1-mediated phosphorylation (Figure 4C) and was necessary for stress-induced activation of AMPK (Figure 3E). This places CHIP in a unique regulatory role, one in which it is capable of fine-tuning the stress-mediated activation of AMPK by promoting its activation, enhancing its activity, and attenuating the signal through the degradation of LKB1 (Figure 6). This is similar to the role CHIP plays in mediating the heat shock stress response both at the level of initiation and also in levels returning to baseline (38). Furthermore, the observation of modest increases in CHIP expression in both pressure overload (Figure 3D) and AICAR-stimulated mice (Supplemental Figure 3E) may be the result of a feedback loop necessary for regulating stress-induced pathways that can contribute to the regulation of AMPK activity.
Figure 6. Proposed physiological model for the CHIP-dependent effects on the regulation of AMPK function.
In unstressed conditions, CHIP forms a complex that contains the AMPK holoenzyme; however, the consequence of this interaction is not understood, as CHIP does not appear to directly affect either baseline AMPK activation status, depicted by phosphorylation at α-T172 (P), or baseline AMPK activity (indicated by the star). In cell stress conditions, such as oxidative stress and cardiac pressure overload, or in the presence of AMPK agonists, CHIP is necessary for LKB1-mediated activation of AMPK, as observed by increased phosphorylation of the α (catalytic) subunit, which leads to an increase in AMPK activity. In addition to increasing LKB1-mediated phosphorylation of AMPK, in cell-free systems, CHIP robustly increases AMPK activity independent of phosphorylation, acting as a chaperone of AMPK, amplifying the positive effect of CHIP on AMPK activity. CHIP may also play a key role in the recovery phase in returning the cell to metabolic homeostasis. CHIP mediates polyubiquitination of LKB1, leading to degradation of LKB1 via a proteasome-dependent mechanism that may contribute to restoring AMPK activation status to baseline levels.
Previous studies of AMPK provide clues to the structure-function relationship between CHIP and AMPKα. Both α1 and α2 contain a stretch of residues that comprise an AID that is sufficient for a conformational-dependent inhibition of kinase activity (39, 40) as well as the α-hook domain that propagates conformational changes across the AMPK subunits (41). Given the regulatory role of both the AID and α-hook on allosteric regulation of AMPK function, and conversely the ability of CHIP to directly mediate increases in AMPK substrate turnover (Figure 4E), CHIP chaperone activity potentially alters either of these domains, as demonstrated by our limited proteolysis analysis (Figure 5, A, C, and D, and Supplemental Figure 5A). The chaperone activity of CHIP appears to directly affect the catalytic α subunit compared with other well-characterized AMPK agonists that are either indirect effectors or low-energy mimetics (such as ZMP) that target the regulatory γ subunit; therefore, the AMPK agonist function of CHIP described here is unique to any known biological or synthetic effector molecule of AMPK.
Further characterization of our CHIP chaperone model will be needed to elucidate the exact interaction between AMPK and CHIP and the physiological repercussions of AMPK conformational changes. Chaperones function through a constant transition of substrate-bound and -unbound states until a stable or altered conformation of the substrate is reached. This likely explains how increasing CHIP concentrations in excess of a 2:1 molar ratio can have such a potent effect on AMPK activity by increasing the likelihood of interaction (Figure 4E and Supplemental Figure 4J). The continued increase in AMPK activity when CHIP is in molar excess is distinct from CHIP’s cochaperone activity of augmenting HSC70 activity. Interestingly, we found no further increase in the ability of CHIP to alter HSC70 activity whether CHIP is used in molar excess or molar equivalency (2). The results from our chaperone model also predict that changes in local concentrations of CHIP in vivo, either through increases in expression or through localization, would elicit potent effects on AMPK activation and activity. It is interesting to note that AMPKα2-containing complexes preferentially localize to the nucleus under stress conditions (42), much like the redistribution of CHIP during stress (43), suggesting that CHIP may also chaperone the rather uncharacterized nuclear functions of AMPK (42). Although this study focuses primarily on the role of CHIP during acute periods of stress, CHIP-dependent regulation of AMPK may also play a broader role in metabolic homeostasis during aging. We previously characterized an accelerated aging phenotype in Chip–/– mice (44). In isolated skeletal and cardiac muscle, we observed decreased levels of AMPKα-T172 phosphorylation in aging Chip–/– mice relative to that in age-matched controls (our unpublished observations). Recent studies highlight the role for AMPK as a key regulator in aging through regulation of PGC1-α and mitochondrial biogenesis (45) as well as a protective role for AMPKα2 in age-induced defects in heart function (17). Taken together, these studies suggest that the role of CHIP on aging may extend to AMPK-dependent pathways. Furthermore, our results, taken in context of previous studies, offer the possibility that regulating the mechanisms that trigger AMPK and CHIP interactions may prove to be an efficacious strategy to combat both acute and chronic metabolic diseases.
Methods
Further details are provided in the Supplemental Methods.
Global gene expression microarrays.
Complete, MIAME-compliant data sets are archived with the Gene Expression Omnibus of the National Center for Biotechnology Information and are accessible through the GEO series accession no. GSE24209.
AMPK phosphorylation and activity assays.
Protein complexes were isolated by immunoprecipitation using AMPKα1, AMPKα2, pan-AMPKα, or LKB1 antibodies. Kinase activity was determined by measuring the amount of SAMS or LKB1 peptide phosphorylation as described previously (46). AMPK phosphorylation by LKB1/STRADα/MO25α was measured as described previously (47) in the presence of recombinant CHIP where indicated. AMPK activity using purified proteins was assayed using the SAMS peptide as described previously (46).
Limited proteolysis analysis of AMPK holoenzyme.
AMPKα2β1γ1 was incubated alone or in the presence of luciferase or CHIP for 30 minutes and then digested with increasing amounts of trypsin or proteinase K for 1 hour and analyzed by immunoblotting as described previously (48).
Twisted FRET plasmid and assay.
The full-length cDNA encoding rat AMPKα2 from plasmid 15991 (Addgene; ref. 49) was PCR subcloned into pCR 2.1-TOPO. The purified XhoI/EcoRI restriction fragment was ligated into EYPF-ECFP (provided by B.K. Berdiev; ref. 33), resulting in the EYFP-AMPKα2-ECFP vector. Transient transfection of COS7 cells was used with the twisted FRET (double-tagged ECFP-EYFP) method as previously described (33, 34) to quantify changes in AMPKα2 protein conformation in live cells. Apparent FRET efficiency (E) was calculated using emission spectra before and after photobleaching (Dpre and Dpost, respectively) using the following equation: E = (Dpost – Dpre)/Dpost corrected for bleaching efficiency as described previously (50).
Statistics.
Data are presented as mean ± SEM. Significance was evaluated using either 2-way or 1-way ANOVA, with Newman-Keuls multiple comparison or Bonferroni post-hoc tests where appropriate. Additional statistical tests used are indicated in the figure legends. GraphPad Prism was used for all calculations. P < 0.05 was considered statistically significant.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee of The University of North Carolina at Chapel Hill.
Supplementary Material
Acknowledgments
We would like to thank Michael Chua and the Michael Hooker Microscopy Facility for the expert help in FRET imaging, Ashutosh Tripathy and the UNC Macromolecular Interactions Facility, Michael Miley and the UNC Protein Expression and Purification Core Facility, Hongyu Ren for outstanding technical support, and Andrea Portbury and Monte S. Willis for the critical review of this manuscript. This work was supported in part by NIH grants R01-GM61728 and R37-HL65619 (to C. Patterson) and by the Fondation Leducq (to C. Patterson).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Citation for this article: J Clin Invest. 2013;123(8):3588–3599. doi:10.1172/JCI69080.
References
- 1.Katz AM. Heart failure: a hemodynamic disorder complicated by maladaptive proliferative responses. J Cell Mol Med. 2003;7(1):1–10. doi: 10.1111/j.1582-4934.2003.tb00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connell P, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 5.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 6.Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282(31):22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- 7.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115(3):547–555. doi: 10.1172/JCI24405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingwall JS, Weiss RG. Is the failing heart energy starved? On using chemical energy to support cardiac function. Circ Res. 2004;95(2):135–145. doi: 10.1161/01.RES.0000137170.41939.d9. [DOI] [PubMed] [Google Scholar]
- 9.Tian R, Musi N, D’Agostino J, Hirshman MF, Goodyear LJ. Increased adenosine monophosphate-activated protein kinase activity in rat hearts with pressure-overload hypertrophy. Circulation. 2001;104(14):1664–1669. doi: 10.1161/hc4001.097183. [DOI] [PubMed] [Google Scholar]
- 10.Zhang P, et al. AMP activated protein kinase-alpha2 deficiency exacerbates pressure-overload-induced left ventricular hypertrophy and dysfunction in mice. Hypertension. 2008;52(5):918–924. doi: 10.1161/HYPERTENSIONAHA.108.114702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim AS, Miller EJ, Young LH. AMP-activated protein kinase: a core signalling pathway in the heart. Acta Physiol (Oxf). 2009;196(1):37–53. doi: 10.1111/j.1748-1716.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 12.Wong AK, Howie J, Petrie JR, Lang CC. AMP-activated protein kinase pathway: a potential therapeutic target in cardiometabolic disease. Clin Sci (Lond). 2009;116(8):607–620. doi: 10.1042/CS20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattison JS, Robbins J. Protein misfolding and cardiac disease: establishing cause and effect. Autophagy. 2008;4(6):821–823. doi: 10.4161/auto.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanbe A, et al. Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2005;102(38):13592–13597. doi: 10.1073/pnas.0503324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marfella R, et al. Myocardial lipid accumulation in patients with pressure-overloaded heart and metabolic syndrome. J Lipid Res. 2009;50(11):2314–2323. doi: 10.1194/jlr.P900032-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren J, Pulakat L, Whaley-Connell A, Sowers JR. Mitochondrial biogenesis in the metabolic syndrome and cardiovascular disease. J Mol Med. 2010;88(10):993–1001. doi: 10.1007/s00109-010-0663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turdi S, et al. AMP-activated protein kinase deficiency exacerbates aging-induced myocardial contractile dysfunction. Aging Cell. 2010;9(4):592–606. doi: 10.1111/j.1474-9726.2010.00586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107(7):825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schisler JC, Willis MS, Patterson C. You spin me round: MaFBx/Atrogin-1 feeds forward on FOXO transcription factors (like a record). Cell Cycle. 2008;7(4):440–443. doi: 10.4161/cc.7.4.5451. [DOI] [PubMed] [Google Scholar]
- 20.Doenst T, et al. Decreased rates of substrate oxidation ex vivo predict the onset of heart failure and contractile dysfunction in rats with pressure overload. Cardiovasc Res. 2010;86(3):461–470. doi: 10.1093/cvr/cvp414. [DOI] [PubMed] [Google Scholar]
- 21.Abel ED, Doenst T. Mitochondrial adaptations to physiological vs. pathological cardiac hypertrophy. Cardiovasc Res. 2011;90(2):234–242. doi: 10.1093/cvr/cvr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto K, et al. Deficiency of LKB1 in heart prevents ischemia-mediated activation of AMPKalpha2 but not AMPKalpha1. Am J Physiol Endocrinol Metab. 2006;290(5):E780–E788. doi: 10.1152/ajpendo.00443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarrinpashneh E, et al. AMPKalpha2 counteracts the development of cardiac hypertrophy induced by isoproterenol. Biochem Biophys Res Commun. 2008;376(4):677–681. doi: 10.1016/j.bbrc.2008.09.057. [DOI] [PubMed] [Google Scholar]
- 25.Chan AY, Soltys CL, Young ME, Proud CG, Dyck JR. Activation of AMP-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J Biol Chem. 2004;279(31):32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 26.Hu X, et al. AMP activated protein kinase-alpha2 regulates expression of estrogen-related receptor-alpha, a metabolic transcription factor related to heart failure development. Hypertension. 2011;58(4):696–703. doi: 10.1161/HYPERTENSIONAHA.111.174128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karamanlidis G, Nascimben L, Couper GS, Shekar PS, del Monte F, Tian R. Defective DNA replication impairs mitochondrial biogenesis in human failing hearts. Circ Res. 2010;106(9):1541–1548. doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi SL, et al. The regulation of AMP-activated protein kinase by H(2)O(2). Biochem Biophys Res Commun. 2001;287(1):92–97. doi: 10.1006/bbrc.2001.5544. [DOI] [PubMed] [Google Scholar]
- 29.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:CALBTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aye TT, et al. Proteome-wide protein concentrations in the human heart. Mol Biosyst. 2010;6(10):1917–1927. doi: 10.1039/c004495d. [DOI] [PubMed] [Google Scholar]
- 31.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2(2):R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 33.Berdiev BK, et al. Molecular proximity of cystic fibrosis transmembrane conductance regulator and epithelial sodium channel assessed by fluorescence resonance energy transfer. J Biol Chem. 2007;282(50):36481–36488. doi: 10.1074/jbc.M708089200. [DOI] [PubMed] [Google Scholar]
- 34.Xu H, Zhang Z, Li M, Zhang R. MDM2 promotes proteasomal degradation of p21Waf1 via a conformation change. J Biol Chem. 2010;285(24):18407–18414. doi: 10.1074/jbc.M109.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol. 2003;160(5):629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belova L, Sharma S, Brickley DR, Nicolarsen JR, Patterson C, Conzen SD. Ubiquitin-proteasome degradation of serum- and glucocorticoid-regulated kinase-1 (SGK-1) is mediated by the chaperone-dependent E3 ligase CHIP. Biochem J. 2006;400(2):235–244. doi: 10.1042/BJ20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaude H, et al. Molecular chaperone complexes with antagonizing activities regulate stability and activity of the tumor suppressor LKB1. Oncogene. 2012;31(12):1582–1591. doi: 10.1038/onc.2011.342. [DOI] [PubMed] [Google Scholar]
- 38.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459(7250):1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 40.Pang T, et al. Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits. J Biol Chem. 2007;282(1):495–506. doi: 10.1074/jbc.M605790200. [DOI] [PubMed] [Google Scholar]
- 41.Xiao B, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472(7342):230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oakhill JS, Scott JW, Kemp BE. Structure and function of AMP-activated protein kinase. Acta Physiol (Oxf). 2009;196(1):3–14. doi: 10.1111/j.1748-1716.2009.01977.x. [DOI] [PubMed] [Google Scholar]
- 43.Dai Q, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22(20):5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min JN, Whaley RA, Sharpless NE, Lockyer P, Portbury AL, Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol Cell Biol. 2008;28(12):4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Lluch G, Irusta PM, Navas P, de Cabo R. Mitochondrial biogenesis and healthy aging. Exp Gerontol. 2008;43(9):813–819. doi: 10.1016/j.exger.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keyse SM. Stress Response: Methods And Protocols. Totowa, New Jersey, USA: Humana Press; 2000. [Google Scholar]
- 47.Jaleel M, et al. The ubiquitin-associated domain of AMPK-related kinases regulates conformation and LKB1-mediated phosphorylation and activation. Biochem J. 2006;394(pt 3):545–555. doi: 10.1042/BJ20051844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Z, Cyr DM. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273(10):5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 49.Mu J, Brozinick JT, Valladares O, Bucan M, Birnbaum MJ. A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol Cell. 2001;7(5):1085–1094. doi: 10.1016/S1097-2765(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 50.Trinkle-Mulcahy L, Chusainow J, Lam YW, Swift S, Lamond A. Visualization of intracellular PP1 targeting through transiently and stably expressed fluorescent protein fusions. Methods Mol Biol. 2007;365:133–154. doi: 10.1385/1-59745-267-X:133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.