Abstract
Merkel cell polyomavirus (MCV) causes ~80% of primary and metastatic Merkel cell carcinomas (MCCs). By comparing digital transcriptome subtraction deep-sequencing profiles, we found that transcripts of the cellular survivin oncoprotein [BIRC5a (baculoviral inhibitor of apoptosis repeat-containing 5)] were up-regulated sevenfold in virus-positive compared to virus-negative MCC tumors. Knockdown of MCV large T antigen in MCV-positive MCC cell lines decreased survivin mRNA and protein expression. Exogenously expressed MCV large T antigen increased survivin protein expression in non-MCC primary cells. This required an intact retinoblastoma protein–targeting domain that activated survivin gene transcription as well as expression of other G1-S–phase proteins including E2F1 and cyclin E. Survivin expression is critical to the survival of MCV-positive MCC cells. A small-molecule survivin inhibitor, YM155, potently and selectively initiates irreversible, nonapoptotic, programmed MCV-positive MCC cell death. Of 1360 other chemotherapeutic and pharmacologically active compounds screened in vitro, only bortezomib (Velcade) was found to be similarly potent, but was not selective in killing MCV-positive MCC cells. YM155 halted the growth of MCV-positive MCC xenograft tumors and was nontoxic in mice, whereas bortezomib was not active in vivo and mice displayed serious morbidity. Xenograft tumors resumed growth once YM155 treatment was stopped, suggesting that YM155 may be cytostatic rather than cytotoxic in vivo. Identifying the cellular pathways, such as those involving survivin, that are targeted by tumor viruses can lead to rapid and rational identification of drug candidates for treating virus-induced cancers.
INTRODUCTION
Merkel cell carcinoma (MCC) is an aggressive cutaneous neoplasm treated by surgical excision with or without adjuvant radiation therapy (1, 2). No defined chemotherapeutic regimen has proven effective, and those drug combinations in use are frequently based on MCC’s histological similarity to small-cell lung carcinoma (3, 4). Data from the Surveillance, Epidemiology, and End Results Program (SEER) of the National Cancer Institute (NCI) indicate that MCC incidence has tripled from 0.15 per 100,000 in 1986 to 0.6 per 100,000 in 2006, resulting in ~1500 new cases per year in the United States alone (5–7). MCC is a cancer of elderly (>65 years of age) and immunosuppressed populations that may be triggered by ultraviolet or ionizing radiation exposure (1, 5, 6, 8). Given that these populations are increasing in number, it is likely that MCC incidence will also show an increase.
Merkel cell polyomavirus (MCV) was discovered in 2008 using digital transcriptome subtraction during a directed search for the viral cause of MCC (9). Although seven new human polyomaviruses have been described since 2007 using genomic technologies, MCV is currently the only polyomavirus established to be a cause of human cancer (10–12). Digital transcriptome subtraction is a deep-sequencing method based on generation of high-fidelity (Hi-Fi) sequence data sets from tumor and control sample complementary DNAs (cDNAs) (13). Known human cellular transcript sequences are “subtracted” from the tumor Hi-Fi data set, leaving candidate sequences that might belong to a new viral cDNA. Digital transcriptome subtraction can also be used to quantitate relative cellular gene expression.
MCV, similar to other polyomaviruses, is a double-stranded DNA virus that normally replicates as an episomal viral infection (12, 14). MCV-positive MCC tumors are characterized by two distinctive viral mutations: MCV genome integration into the human Merkel cell genome and a mutation that causes C-terminal truncation of the MCV large tumor (T) antigen. These tumor signature mutations eliminate virus replication but leave the tumor suppressor–targeting domains of the virus intact (9, 15, 16). An additional important risk factor for MCC is loss of host immune surveillance due to immunosuppression caused by AIDS, transplantation, or aging (7, 8), such that virus-positive tumor cells are not cleared by the host immune response (17–19). MCV is a common infection of human skin, and MCV-positive MCC represents an intriguing example where mutations to a commensal viral genome, rather than the host genome, are central to the initiation of an aggressive cancer (20).
As with other polyomaviruses, differentially spliced MCV large T and small T antigen oncoproteins are expressed from the T antigen early locus. MCV large T antigen binds to members of the transcriptional repressor retinoblastoma protein (RB1) family through an N-terminal LXCXE motif that is not affected by MCV tumor-specific mutations (16). For simian virus 40 (SV40) polyomavirus, binding of large T antigen to RB1, with subsequent release of active E2F transcription factors, promotes synthesis of genes required for entry into S phase of the cell cycle (21, 22). Cyclin E and E2F1 are positively regulated by E2F signaling and promote intracellular conditions for active DNA synthesis and centrosome replication (23). SV40 large T antigen also binds to and inhibits the proapoptotic tumor suppressor protein p53 (24, 25), but there is currently no evidence that MCV large T antigen directly targets p53 in tumors because the corresponding region of MCV large T antigen is deleted by tumor-specific mutations. Other prosurvival pathways that might be targeted by MCV, such as the survivin oncoprotein, which is also regulated by RB1 signaling (26, 27), have not been investigated. MCV small T antigen, which is unaffected by tumor-specific mutations, transforms cells and activates cap-dependent translation by targeting the translation regulator 4E-BP1 (28). When small T antigen alone is knocked down, MCV-positive cells undergo cell cycle arrest (28), whereas knockdown of both small T and large T antigens together causes necroptotic cell death (29). Thus, both large T and small T antigen MCV oncoproteins are likely to contribute to the transformed phenotype of MCC.
We show here that discovery of a new cancer virus can be leveraged to rapidly identify rational anticancer therapies. We find that MCV up-regulates survivin oncoprotein transcription in MCC through its RB1-sequestering large T antigen domain and that a survivin inhibitor selectively targets MCV-positive tumor cells. In contrast, a screen of 1360 pharmacologically active compounds failed to identify any candidates with selective activity against MCV-positive MCC, and only a few compounds (bortezomib and topoisomerase I and II inhibitors) show pharmacologically relevant activity against MCC cell lines. YM155, a transcriptional inhibitor of survivin (30, 31), however, is highly potent at inducing the cell death of MCV-positive MCC. It is cytostatic to MCC xenografts and well tolerated in mice. These findings show that basic genome investigations into cancer causation can lead to potential new therapies for often intractable human cancers.
RESULTS
Survivin expression in MCV-positive MCC
To identify pathways perturbed by MCV infection in MCC, we analyzed digital transcriptome subtraction data sets to identify cellular genes differentially regulated between MCV-positive and MCV-negative MCC (9). We compared 400,000 cellular transcripts from MCV-positive and MCV-negative MCC tumors and found 1096 of 11,531 (9.5%) genes elevated more than threefold for the MCV-positive compared to the MCV-negative library.
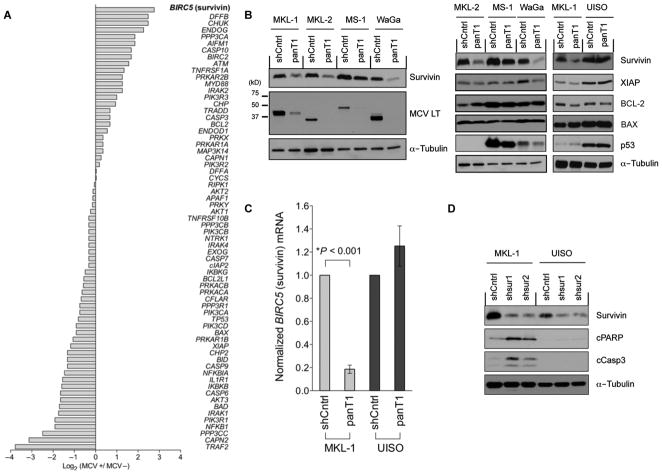
We next identified 64 genes using Gene Ontology (GO) gene definitions (32) directly involved in programmed cell death or cell cycle regulation (Fig. 1A). BIRC5a (baculoviral inhibitor of apoptosis repeat-containing 5) mRNA encoding the survivin oncoprotein was increased sevenfold (P = 2.90 × 10−10) for virus-positive compared to virus-negative MCC tumors (Fig. 1A and table S1). Other genes regulating programmed cell death, including TP53, cIAP2, XIAP, BAX, BCL-2, and caspase 3/6 transcripts, were not differentially expressed (Fig. 1A and table S1). Notably, transcripts from some genes including TRAF2 and NFKB1 were markedly reduced in virus-positive compared to virus-negative MCC tumor libraries.
Fig. 1.
Survivin oncoprotein mRNA expression is increased in MCV-positive MCC. (A) Digital transcriptome subtraction comparison of 64 genes involved in programmed cell death and cell cycle regulation showed that survivin (BIRC5) mRNA transcripts (highlighted) were sevenfold higher in an MCV-positive compared with an MCV-negative MCC cDNA library. The relative expression of genes was normalized to total sequence reads for each MCC library (also see table S1). (B) MCV T antigen is required for survivin expression. Lentiviral MCV T antigen exon 1 knockdown (panT1) decreased survivin protein expression (left) but did not alter XIAP, BCL-2, BAX, or p53 protein levels (right) in four MCV-positive MCC cell lines: MKL-1, MKL-2, MS-1, and WaGa. UISO, MCV-negative cell line; shCntrl, scrambled shRNA control lentivirus; LT, large T antigen. (C) MCV T antigen is required for survivin transcription. Survivin mRNA levels were reduced in the MKL-1 but not the UISO cell line after T antigen knockdown, indicating that T antigen activates survivin transcription. Survivin mRNA was measured by qRT-PCR and normalized to β-actin mRNA. The experiments were performed in triplicate and repeated twice (mean ± SEM, two-tailed t test). (D) Survivin expression is required for MCV-positive MCC tumor cell survival. Survivin was targeted for knockdown using two shRNA lentiviral vectors, shsur1 and shsur2, in MKL-1 and UISO cells. MKL-1 cells initiate apoptosis after survivin knockdown, with increased expression of cleaved PARP (cPARP) and caspase 3 (cCasp3), whereas UISO cells are resistant to survivin knockdown-induced apoptosis. α-Tubulin is used as a loading control.
MCV large T antigen induces survivin expression
To determine whether MCV T antigen increases survivin expression in MCC, we used a short hairpin RNA (shRNA) lentivirus (panT1) to perform RNA interference (RNAi) targeting of the MCV T antigen exon 1 sequence that selectively knocks down all MCV T antigen isoforms in MCC cells (29). MCV-positive MCC cells infected with this lentivirus undergo nonapoptotic cell death (necroptosis) when MCV oncoprotein expression is inhibited (29). MCV T antigen reduction correlated with survivin reduction in all MCV-positive MCC cell lines, but not in any of the MCV-negative cell lines (Fig. 1B). The decrease in survivin with T antigen knockdown ranged from modest in MS-1 to near complete in WaGa cell lines (Fig. 1B). This effect is at the level of transcription rather than translation because BIRC5a mRNA is markedly reduced by T antigen knockdown (Fig. 1C). Use of a shRNA lentivirus that selectively targets only the small T antigen isoform (28), however, did not affect survivin expression (fig. S1), indicating that increased survivin transcription was dependent on MCV large T antigen but not small T antigen (knockdown of large T antigen alone is difficult due to the overlapping structure of the T antigen cistron). In contrast to survivin, no consistent changes in protein expression for p53, XIAP, BCL-xL, MCL-1, BAD, BIK, BMF, BIM, PUMA, BCL-2, or BAX proteins (Fig. 1B) (29) or for cleaved poly(adenosine diphosphate–ribose) polymerase (PARP), cleaved caspase 3, or cleaved caspase 9 proteins (fig. S2) were seen after panT1 knockdown in MCV-positive cells. These results are consistent with our digital transcriptome subtraction findings, suggesting that MCV T antigen selectively activates BIRC5a transcription. We next sought to determine the importance of survivin expression to survival of MCV-positive and MCV-negative MCC cells. Although only partial suppression of survivin expression was achieved by shRNA BIRC5a targeting, MCV-positive MKL-1 cells, but not MCV-negative UISO cells, underwent apoptosis (Fig. 1D).
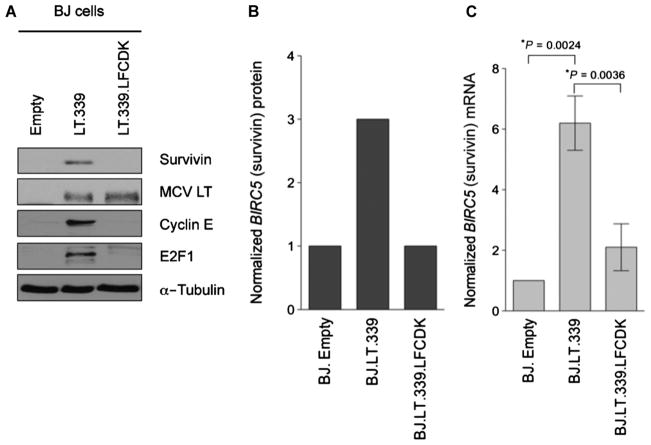
To confirm that large T antigen is the MCV T antigen isoform responsible for survivin activation, we cloned and transduced a tumor-derived large T antigen (LT339) cDNA into nontransformed, primary human BJ fibroblasts. Survivin protein levels increased threefold when large T antigen was expressed compared to the empty vector control (as measured by quantitative LI-COR, an immunoblotting technique that uses antibodies tagged with near-infrared dyes) (Fig. 2, A and B). Also consistent with our knockdown experiments (Fig. 1C), large T antigen directly activates BIRC5a promoter transcription in BJ cells (Fig. 2C). This is mediated by a specific domain (LXCXE) in large T antigen responsible for sequestration of RB1. Both cyclin E and E2F1, required for entry into S phase of the cell cycle (23), are repressed by active RB1 and are used as markers for RB-regulated gene expression. MCV LT339 activates expression of both cyclin E and E2F1 proteins in BJ cells, whereas LT339 having a point mutation that prevents RB1 binding (changing LFCDE to LFCDK) (16) abolishes induction of cyclin E, E2F1, and survivin (Fig. 2A). MCV small T antigen expressed alone in BJ cells did not increase survivin oncoprotein expression (fig. S3). In contrast to primary BJ fibroblasts, we did not find that MCV large T antigen markedly increased survivin expression in cell lines that had already been transformed, including U2OS cells (fig. S4).
Fig. 2.
MCV large T antigen isoform induces survivin oncoprotein expression in human BJ cells by targeting RB. (A) BJ cells were transduced with either empty vector, a tumor-derived large T antigen cDNA (LT.339), or a large T antigen cDNA with an inactive RB binding domain (LT.339LFCDK). Immunoblotting reveals that MCV LT.339 induces survivin expression but LT.339LFCDK does not. A similar pattern is seen for other S-phase cell cycle proteins such as E2F1 and cyclin E that are also transcriptionally repressed by RB. (B) LI-COR quantitative immunoblotting for survivin in (A), normalized to α-tubulin (arbitrary units). (C) Survivin mRNA levels increased in BJ cells expressing LT.339 protein but not in cells expressing the RB1 binding mutant LT.339LFCDK. BJ cells expressing either empty vector, LT.339, or LT.339LFCDK were serum-starved for 48 hours and then harvested for RNA. Survivin mRNA was measured by qRT-PCR and normalized to β-actin mRNA. The experiments were performed three times in duplicate (mean ± SEM, two-tailed t test).
Survivin as a target for MCC chemotherapy
Given the apparent importance of MCV-induced survivin expression to MCC cell survival, we examined YM155, an imidazolium small-molecule inhibitor of the survivin promoter that is currently undergoing phase 2 trials for prostate cancer (30, 31, 33). YM155 is both highly active and selective for inhibiting MCV-positive MCC cell growth in vitro as measured by CellTiter-Glo assays [median effective concentration (EC50), 1.34 to 12.2 nM] (Fig. 3A). MCV-negative MCC cell line growth is also inhibited by YM155, but this occurs at concentrations one to two orders of magnitude higher than for MCV-positive MCC cells. YM155 treatment for 48 hours at 10 to 100 nM preferentially killed MCV-positive MKL-1 cells compared to MCV-negative UISO cells as measured by trypan blue staining (Fig. 3B). Survivin protein was reduced in MKL-1 cells after YM155 treatment, consistent with YM155’s proposed mechanism of action of inhibiting the BIRC5a promoter (Fig. 3C).
Fig. 3.
The survivin promoter inhibitor YM155 inhibits MCV-positive MCC cell line growth. (A) Dose-dependent growth curves at 48 hours for YM155-treated cell lines. MCV-negative MCC13, BJ, and BJhTERT cells showed relative resistance to YM155 treatment, whereas all MCV-positive cell lines (MKL-1, MKL-2, MS-1, and WaGa) were sensitive to YM155. MCV-negative UISO and MCC26 cell lines had intermediate sensitivity to YM155. (B) Trypan blue vital dye exclusion assay showed dose-dependent cell killing at 48 hours for MKL-1 cells (blue bars), whereas UISO cells (red bars) are relatively less sensitive and BJ cells (green bars) are resistant to YM155. (C) Dose-dependent decrease in MKL-1 cell survivin protein expression after 12 hours of YM155 treatment.
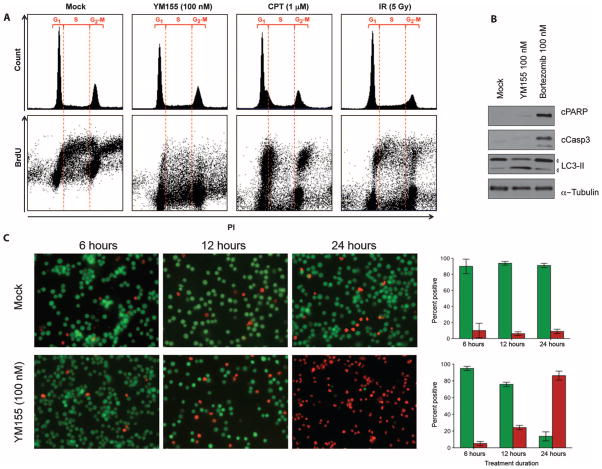
We next examined mechanisms of cell killing by YM155 in MCV-positive MCC cells (Fig. 4). Survivin plays a role in mitotic progression, and loss of survivin can lead to cell death through mitotic catastrophe in some tumor cells (34, 35). MCV-positive MCCs, however, are slowly cycling cells (doubling time of 3 days) (29) and do not undergo G1 arrest or G2-M pileup as would occur with mitotic checkpoint activation in rapidly cycling cells (Fig. 4A, top panels). Instead, bromodeoxyuridine (BrdU) incorporation into DNA reveals profound early inhibition of DNA synthesis by YM155. Figure 4A (bottom panel, left) shows the normal inverted U pattern for BrdU incorporation during S-phase DNA synthesis in MKL-1 cells. When MKL-1 cells are treated with the topoisomerase I inhibitor camptothecin or with 5-Gy γ-irradiation (Fig. 4A, bottom panel, right), S-phase BrdU is reduced and BrdU-positive cells accumulate in G1 and G2-M as expected for cell cycle checkpoint activation. YM155 treatment, in contrast, markedly inhibits all BrdU incorporation, consistent with disruption of DNA synthesis but not with mitotic catastrophe.
Fig. 4.
Cell death phenotype of YM155-treated MCV-positive MCC. (A) Cell cycle analysis reveals that YM155-treated cells do not undergo mitotic catastrophe. MKL-1 cells were treated with DMSO, YM155 (100 nM), camptothecin (CPT, 1 μM), or 5-Gy γ-irradiation (IR). They were stained for propidium iodide (PI) and BrdU and harvested 12 hours later. Upper panel shows cell cycle profiles, in which no evidence of G2-M pileup is seen (a marker for mitotic catastrophe) after YM155 treatment. Bottom panel shows the corresponding propidium iodide versus BrdU uptake for these same cells showing new DNA synthesis for cells in G1, S, and G2-M. Mock-treated cells have an inverted U-shaped curve showing BrdU incorporation during S-phase DNA synthesis. Both camptothecin and γ-irradiation primarily reduce S-phase BrdU incorporation, consistent with checkpoint activation. In contrast, YM155 reduces all early DNA synthesis as measured by BrdU incorporation. (B) YM155 induces nonapoptotic cell death associated with autophagy in MKL-1 cells. MKL-1 cells were treated with DMSO, YM155 (100 nM), or bortezomib (100 nM) and immunoblotted for cleaved PARP (cPARP), cleaved caspase 3 (cCasp3), LC3-II, and α-tubulin. In contrast to YM155, the proteasome inhibitor bortezomib activates MKL-1 cell apoptosis (also see fig. S2). (C) YM155 treatment initiates programmed cell death within 12 to 24 hours after treatment. MKL-1 cells were costained with CFDA (green, live) and propidium iodide (red, dead). Column graphs (right panel) represent mean and range of percent CFDA-positive (green) and percent propidium iodide–positive cells (red).
YM155 treatment also results in early commitment of MCV-positive MCC cells to nonapoptotic programmed cell death. No evidence of apoptosis was present, as measured by caspase 3 and PARP cleavage (Fig. 4B), when MKL-1 cells were treated with 100 nM YM155 for 48 hours. However, YM155 did initiate LC3-II accumulation, a marker for cell autophagy (36). This is not due to complete loss of apoptosis pathway signaling in MKL-1 cells because treatment with the proteasome inhibitor bortezomib (Velcade) activated PARP and caspase cleavage (Fig. 4B and fig. S5). Commitment to YM155-dependent cell death occurs relatively quickly and irreversibly: When MKL-1 cells were treated with 100 nM YM155 for 3, 6, or 12 hours, followed by washout with complete cell culture medium, only 51, 3, and 1.8% of cells, respectively, remained viable at 48 hours, as measured by trypan blue dye exclusion. This was confirmed using a cell viability assay with propidium iodide (dead) and carboxyfluorescein di-acetate (CFDA, alive) costaining (Fig. 4C). Twelve to 24 hours of 100 nM YM155 treatment causes MKL-1 cells to lose membrane integrity, such that they become positive for propidium iodide and negative for CFDA staining.
To search for other MCC chemotherapeutics, we performed a two-stage cytotoxicity library screen on 1360 pharmacologically active compounds (see table S2 and fig. S6, A and B), including 1280 drugs from the Library of Pharmacologically Active Compounds (LOPAC) (Sigma-Aldrich), 89 drugs from the NCI’s Approved Oncology Drug Set II (19 compounds in common with LOPAC1280), 6 compounds targeting SV40 large T antigen adenosine triphosphatase (ATPase) activity (37, 38), and 4 compounds targeting MDM2, a negative regulator of p53 (39, 40). These compounds were screened at 10−5 M for >90% inhibition of MKL-1 cell growth in a CellTiter-Glo assay (Promega). Notably, mammalian target of rapamycin (mTOR) inhibitors (everolimus, rapamycin, etc.), antiviral compounds (ribavirin, acyclovir), and MDM2 inhibitors active in other viral cancers (for example, nutlin-3) (41, 42) were not active in our screen. This is consistent with previous findings that MCV loses replication activity (16) and activates cap-dependent translation downstream of mTOR (28) in MCC tumor cells.
Eighteen (1.3%) of these 1360 drugs met our initial screening criterion for anti-MCC activity and were selected for secondary dose-dependent screening on MCV-positive and MCV-negative cell lines (Table 1). Only the proteasome inhibitor bortezomib (43) was active in vitro at low doses (EC50, 1.3 to 13.2 nM; figs. S5 and S6C). This activity, however, was not selective for MCV-positive cells (fig. S5). Other agents, particularly topoisomerase I and II inhibitors, also inhibited MCC cell growth but were generally far less potent or had variable activity among different MCC cell lines (Table 1 and fig. S6, A and B).
Table 1.
EC50 (μM) concentrations for MCC cell lines.
| Drugs | MKL-1 | MKL-2 | MS-1 | WaGa | UISO |
|---|---|---|---|---|---|
| Proteasome inhibitor | |||||
| Bortezomib | 0.013 | 0.005 | 0.002 | 0.001 | 0.003 |
| Topoisomerase inhibitors | |||||
| Ellipticine | 3.2 | 6.5 | 6.9 | 3.0 | 1.1 |
| Amsacrine hydrochloride | 0.11 | 3.0 | 7.5 | 0.25 | 2.6 |
| Teniposide | 0.010 | 2.8 | 3.0 | 0.026 | 7.3 |
| Valrubicin | 0.23 | 2.8 | 9.9 | 0.32 | 4.0 |
| Mitoxantrone | 0.006 | 1.3 | 1.6 | 0.014 | 0.97 |
| Daunorubicin | 0.015 | 0.086 | 0.15 | 0.018 | 1.4 |
| Doxorubicin | 0.21 | 0.022 | 0.37 | 0.022 | 0.25 |
| Topotecan | 0.028 | 0.43 | 0.62 | 0.015 | 0.17 |
| Others | |||||
| Iodoacetamide | 0.29 | 0.30 | 0.64 | 0.37 | 2.5 |
| Sanguinarine chloride | 5.3 | 8.4 | 4.9 | 2.5 | 6.5 |
| NSC95397 | 1.2 | 1.5 | 1.8 | 0.73 | 2.8 |
| Chelerythrine chloride | 0.60 | 0.65 | 0.52 | 0.70 | 3.7 |
| Calmidazolium chloride | 2.2 | 1.7 | 2.1 | 2.1 | 2.0 |
| Tetraethylthiuram disulfide | 0.49 | 0.19 | 6.3 | 1.1 | 13 |
| Bay 11-7085 | 1.4 | 1.7 | 2.7 | 1.2 | 4.5 |
| Quinacrine dihydrochloride | 5.1 | 4.6 | 4.9 | 5.2 | 7.1 |
Effect of YM155 on human MCC xenografts in mice
We developed an MKL-1 xenograft mouse model to test the in vivo efficacy of YM155 and bortezomib on human MCC tumor cells transplanted into nonobese diabetic–severe combined immunodeficient γ (NOD-SCIDγ) (NSG) mice. Subcutaneous injection of MKL-1 cells generates tumors that are positive for MCV large T antigen and the MCC diagnostic marker CK20 (Fig. 5A) and progress to endpoint (2-cm tumor diameter) within 2 to 4 weeks after tumors are first detected. We treated these mice with bortezomib, YM155, or saline for 3 weeks once tumors became palpable. Bortezomib was administered at levels effective on multiple myeloma xenografts (44) (1 mg/kg, twice weekly subcutaneous injections). This did not significantly affect MCC tumor progression or volume compared to treatment with saline alone (P = 0.53, log-rank test). Bortezomib administration at this level was associated with mouse lethargy and weight loss, requiring temporary use of heat blankets and hydrogel to prevent death of animals during initial treatment.
Fig. 5.

YM155 inhibits growth of human MKL-1 MCC xenografts in NOD-SCIDγ mice. (A) MKL-1 xenograft tumors were stained with hematoxylin and eosin (H&E), and for MCV large T antigen and cytokeratin 20 (CK20) expression (magnification, ×40). (B) MKL-1 xenograft survival curves after drug treatment. Mice were subcutaneously injected with 20 million MKL-1 cells and assigned to a 3-week drug treatment after tumors became palpable. No significant difference was found between saline and bortezomib treatment. Tumor progression was significantly delayed by YM155, with none of the YM155-treated mice dying during treatment (up to day 19) compared to 23 of 31 (74%) saline-treated and 14 of 21 (67%) bortezomib-treated mice (P < 0.0001, log-rank test). Tumor progression recurred for all YM155-treated mice once treatment was stopped. (C) Piecewise linear hierarchical Bayesian model for tumor volumes in treated mice (59) (see the Supplementary Materials for details). Colored lines show estimated central population tumor volumes, with shaded regions representing 95% credible intervals. Actual tumor volumes (gray lines) for each mouse are shown for comparison. YM155 treatment retards tumor growth compared to saline or bortezomib treatment. (D) Table showing day of termination and tumor volumes for MS-1 and UISO xenograft mice treated with YM155 or saline.
In contrast to bortezomib, YM155 [2 mg/kg, subcutaneous, five times weekly (30, 45)] markedly delayed MKL-1 xenograft growth and significantly prolonged survival compared to either saline or bortezomib (P < 0.0001, log-rank test; Fig. 5, B and C). This YM155 treatment regimen was chosen because it is the standard dosing for mouse xenograft experiments based on pharmacokinetic and treatment response studies of Nakahara et al. on six different human cancer xenografts (prostate, bladder, melanoma, breast, and lung cancer) in mice and >100 human tumor cell lines (45, 46). Whereas 66.7 to 74.2% of the bortezomib or saline-treated mice reached the euthanasia endpoint during the 3-week treatment period, none of the YM155-treated mice reached this endpoint during treatment. Partial tumor regression occurred among some YM155-treated mice, but all tumors resumed growth once YM155 was stopped, indicating that a single 3-week treatment course was insufficient for tumor eradication (Fig. 5B). For most of the mice, MKL-1 tumor volumes were unchanged or showed delayed growth during YM155 treatment, suggesting that this drug may be cytostatic rather than cytocidal for MKL-1 xenografts (Fig. 5C and appendix S1). YM155 was well tolerated, and no adverse effects or acute toxicities were noted. In smaller cohorts of mice bearing MS-1 (MCV-positive) and UISO (MCV-negative) tumor cell xenografts, final tumor volumes for YM155-treated mice were 43 to 55% (median) of saline-treated control mice at the end of the 3-week treatment period (Fig. 5D).
DISCUSSION
The discovery of a virus as the molecular cause for most MCCs has led to the description of a rationally targeted therapeutic in less than 4 years. MCV was discovered in 2008, followed by identification of the viral oncogenes 1 year later. This enabled us to apply a rational drug screen, resulting in the identification of a survivin inhibitor that may have activity against MCC. Our library screen of 1360 drugs, including the entire NCI Approved Oncology Drug Set II, identified only one drug (bortezomib) that was highly active in vitro, and confirmed that MCC is very chemoresistant. Bortezomib, however, was not active in vivo against MCC xenografts in mice. Digital transcriptome subtraction, a quantitative cDNA deep sequencing method, not only identified MCV as a new human polyomavirus in MCC but also helped to uncover cell signaling pathways that are potential targets for MCV-positive MCC treatment.
Survivin, a member of the inhibitor of apoptosis protein family (47), contributes to chemoresistance of melanoma (48) and is overexpressed in many cancers including MCC (49). Its expression increases during nonneoplastic JC polyomavirus infection (47, 50, 51) and through an E2F-regulated mechanism in response to SV40 large T antigen expression (26, 27). Our MCV T antigen knockdown and expression experiments confirm survivin to be activated in response to sequestration of RB family transcription repressors by MCV large T antigen. Survivin has pleiotropic activities in both preventing apoptosis and activating cell cycle entry (50). These findings are consistent with RB or other pocket proteins repressing survivin expression in primary Merkel cells (26), which is relieved when MCV large T antigen is expressed. MCV large T antigen expression did not increase survivin levels in transformed cells (U2OS cells) that already have dysregulated RB signaling. MCV activation of survivin through RB targeting may typically serve to promote virus replication but is aberrantly activated during MCV tumorigenesis (10).
YM155 is highly cytotoxic in vitro to MCV-positive MCC cells, causing these cells to undergo necroptotic cell death. Unexpectedly, YM155 leads to rapid loss of new DNA synthesis and breakdown of membrane integrity in MCC cells but not apoptosis. In other forms of cancer, survivin knockdown causes mitotic catastrophe (35); however, we did not find this to significantly contribute to MCC tumor cell death. There was no distinct G1- or G2-M–phase accumulation of cells during YM155 treatment, and irreversible commitment to cell death occurs too quickly for most MCC cells to have an opportunity to transit the cell cycle. YM155 does increase expression of the autophagy marker LC3-II, but this may reflect a consequence rather than a cause of cell death (52). Intriguingly, a similar cell death phenotype occurs when T antigens are knocked down in MCV-positive MCC cells (29), whereas targeted knockdown of survivin alone by RNAi causes apoptosis rather than necroptosis. We therefore cannot exclude the possibility that YM155 targets other molecules in addition to survivin (fig. S7), or that differing levels of survivin inhibition result in different forms of programmed cell death.
There are several important caveats to our current study. Although YM155 was highly active in vitro, it was only cytostatic—not cytotoxic—in most mouse xenografts during short-term treatment. Once YM155 treatment was stopped, tumors reemerged in all of the treated mice. Either prolonged YM155 treatment or combined use with other drugs (for example, bortezomib and topoisomerase inhibitors) may more effectively control MCC. YM155 was relatively nontoxic in our study, making it potentially more suitable for prolonged or combined therapy. Practical limitations prevented us from extensively measuring YM155 activity on a variety of different MCC cell line xenografts so we cannot exclude the possibility of drug resistance among other MCCs. But it is encouraging that YM155 reduced tumor masses in two additional MCC xenografts (MS-1 and UISO), including one from an MCV-negative tumor. Finally, MCV small T antigen is also expressed in MCC cells and increases cap-dependent protein translation (28), which could contribute to YM155 resistance by transcription-independent induction of survivin protein (53).
Digital transcriptome subtraction (13) is a useful method to discover (9) or exclude (13) viruses present in human cancers. Here, we show that digital transcriptome subtraction is also useful in measuring cellular gene expression by demonstrating that BIRC5 transcription is activated in MCV-positive MCC. If carefully performed and rigorously analyzed, digital transcriptome subtraction data yield useful information even when no cancer viruses are discovered (13). Before discovery of MCV, few clues were available about the molecular causes for MCC (4). Now that MCV has been shown to be central to the formation of most MCCs, rational targeting of survivin and other cellular pathways perturbed by MCV may lead to discovery of additional treatments that may be more effective and less toxic than current therapies.
MATERIALS AND METHODS
Cell culture
Seven MCC cell lines [MKL-1, MKL-2, MS-1, UISO, MCC13, MCC26, and WaGa (gift of J. Becker) (29, 54, 55)], NCI-H69 small-cell lung cancer cell line [American Type Culture Collection (ATCC)], 293 human embryonic kidney cells (ATCC), U2OS osteosarcoma cell line (gift of O. Gjoerup), BJhTERT immortalized foreskin fibroblast cell line (gift of O. Gjoerup), and BJ primary foreskin fibroblasts (ATCC) were used to screen and evaluate the small molecules examined in this study (29, 54, 55). The Merkel cell lines and NCI-H69 were grown in RPMI 1640 supplemented with 10% fetal calf serum, penicillin, and streptomycin at 37°C in humidified air containing 5% CO2. The remaining cell lines were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum.
Compounds
A total of 1360 compounds were used in the screening survey and are listed in table S1: 1280 compounds from LOPAC1280 library (Sigma-Aldrich, accessed through University of Pittsburgh Drug Discovery Institute), 89 compounds from NCI’s Approved Oncology Drug Set II [from the NCI/Developmental Therapeutics Program (DTP) Open Chemical Repository, http://dtp.cancer.gov], 6 SV40 large T antigen ATPase inhibitors [MAL2-11B, MAL3-101, MAL2-51, DMT3084, bithionol, and hexachlorophene (37, 38)], and 4 compounds that target and inhibit p53 and MDM2 binding [nutlin-3, YH264A, Y2H265A, and KK_NW_16A (39, 40)]. Nineteen compounds that were in common between the LOPAC1280 and the NCI Approved Oncology Drug Set II library were screened twice with comparable results. NCI Approved Oncology Drug Set II, SV40 large T antigen ATPase inhibitors, and MDM2 inhibitors were reconstituted as recommended by the supplier. YM155 (4,9-dihydro-1-(2-methoxyethyl)-2-methyl-4,9-dioxo-3-(2-pyrazinylmethyl)-1H-naphth[2,3-d]imidazolium bromide) was purchased from Active Biochemicals Co. Ltd. Reconstituted compounds were diluted in cell culture medium to obtain a 100× stock concentration before addition to cells. Doxorubicin (positive control) was obtained from Sigma-Aldrich, and dimethyl sulfoxide (DMSO) (negative control) was obtained from Fisher Bioreagents.
For EC50 measurements, dose-response curves were established for 17 drugs initially identified by library screening, and then each compound was obtained individually from specific suppliers. Iodoacetamide, sanguinarine chloride, NSC95397, chelerythrine chloride, calmidazolium chloride, tetraethylthiuram disulfide, Bay 11-7085, quinacrine dihydrochloride, ellipticine, amsacrine hydrochloride, and nutlin-3 were purchased from Sigma-Aldrich. Mitoxantrone, daunorubicin HCl, valrubicin, topotecan HCl, teniposide, and bortezomib were provided by the NCI/DTP Open Chemical Repository (http://dtp.cancer.gov).
MKL-1 cytotoxicity screen
MKL-1 cells were seeded at a density of 6000 cells in 50 μl of medium per well (120 cells/μl) in opaque polypropylene 384-well microplates (Greiner Bio-One). Cells were incubated at 37°C in humidified air containing 5% CO2 for 24 hours. Thereafter, 25 μl of medium containing 3× drug per well was added to the plates, which were incubated for an additional 48 hours. Cell viability was measured with CellTiter-Glo (Promega) following the manufacturer’s instructions. The validity of CellTiter-Glo results in measuring cell viability was confirmed by trypan blue exclusion staining and WST-1 assays (Roche) in pilot studies.
The LOPAC1280 library was screened at a final concentration of 10 μM for each compound, and the NCI library was screened at a final concentration of 1 μM for each compound. MAP-C (Titertek Instruments Inc.) and Janus MDT (PerkinElmer Inc.) were used for automated resuspension and the addition of LOPAC library drugs to assay plates. NCI library compounds were added to wells by manual pipetting.
CellTiter-Glo assays were performed in duplicate with 384-well plates, each containing 24 wells with 1% DMSO (negative control) and 32 wells with 200 μM doxorubicin (positive control). Screening results were evaluated on the basis of percentage cell survival normalized to the DMSO control (100%). Positive candidates were identified with a cutoff value of <10% cell survival. The average Z factor was 0.61 (range, 0.34 to 0.74) for the LOPAC library screen and 0.82 for the NCI library screen (range, 0.75 to 0.91).
Dose-response studies
Compounds that met the selection criteria <10% cell survival were purchased or obtained in bulk from NCI/National Institutes of Health (NIH) DTP. Serial drug dilutions from 10−4 to 10−9 M were used on MCC and non-MCC cell lines. Cells were seeded into 384-well plates at 6000 cells in 50 μl of medium per well. After 24 hours, 25 μl of 3× drugs was added at increasing concentration to each well. Cell viability was then measured with CellTiter-Glo (Promega) kit following the manufacturer’s instructions as described previously. Each drug concentration was tested in triplicate for each cell line, and experiments were repeated twice. EC50 doses for the drugs were calculated with a four-parameter logistic equation (GraphPad Prism).
Trypan blue dye exclusion assay
Cells were equally seeded and treated with YM155 for 48 hours. To quantitate cell death, we treated the cells with Accutase (Millipore), collected them, resuspended them in phosphate-buffered saline (PBS), mixed them with equal volume of trypan blue (Lonza, 0.4%), and counted them with a hemocytometer under the microscope. Counting was performed three times, in triplicate.
Expression and shRNA lentivirus construction
To express codon-optimized full-length MCV large T antigen, we synthesized the gene (DNA2.0) from the MCV-HF strain large T antigen sequence template (56) (GenBank ID: JF813003) and cloned it into the lentiviral pLVX EF puro vector (28). Truncated tumor LT339 (representing the MCV339 strain, amino acids 1 to 455) and LT339. LFCDK were cloned by site-directed mutagenesis from the codon-optimized full-length large T antigen into pSMPUW-hygro vector (Cell Biolabs Inc.) (28). MCV small T codon-optimized was also cloned into the lentivirus vector. Cells were infected with lentiviruses in the presence of polybrene (1 to 4 μg/ml) for 24 hours, followed by medium change. Stable selection with either puromycin (1 μg/ml) or hygromycin (200 μg/ml) was initiated 48 hours after infection.
shRNA for MCV T antigen knockdown was generated and used as previously described (29, 57), and we renamed shT1 in Houben et al. to panT1 in this study. To knock down survivin gene expression, we cloned the shRNA sequence [shsur1, 5′-ccggCCGCATCTCTACATTCAAGAACTCGAGTTCTTGAATGTAGAGATGCGGtttttg-3′; shsur2, 5′-ccggCCTTTCTGTCAAGAAGCAGTTCTCGAGAACTGCTTCTTGACAGAAAGGtttttg-3′ (lower-cased nucleotides indicate linker sequences used for cloning)] into a pLKO.1puro lentiviral vector. shCntrl is a nontargeting shRNA negative control (29). Cells were infected with lentiviruses, washed after 24 hours, and then harvested for immunoblotting 6 days after infection.
Immunoblotting
Cells were lysed in buffer [radioimmunoprecipitation assay or 10 mM tris-HCl (pH 8.0), 0.6% SDS] containing protease inhibitor cocktail (Roche). Lysates were electrophoresed in 10% SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane (Amersham), and reacted with specific antibodies CM2B4 (1:5000 dilution) (15), CM8E6 (1:500 dilution), cleaved PARP, cleaved caspase 3, survivin, XIAP, p53, BCL-2, BAX (1:1000 dilution, Cell Signaling Technology), E2F1, cyclin E (1:1000 dilution, Santa Cruz Biotechnology), LC3 (1:1000 dilution, Novus Biologicals), or α-tubulin (1:5000 dilution, Sigma) overnight at 4°C, followed by anti-mouse (1:5000 dilution, Amersham) or anti-rabbit immunoglobulin G (IgG)–horseradish peroxidase conjugates (1:3000 dilution, Cell Signaling Technology) for 1 hour at room temperature. Peroxidase activity was detected with Western Lightning Plus ECL reagent (PerkinElmer). For quantitative immunoblotting, Odyssey Infrared Imaging System (LI-COR) was used with IRDye 800–conjugated secondary antibodies (1:5000 dilution, Rockland Immunochemicals).
Quantitative reverse transcription–polymerase chain reaction
RNA was extracted from cell lysates with TRIzol reagent (Invitrogen), and cDNA was synthesized with SuperScript III First-Strand Synthesis (Invitrogen). Quantitative real-time polymerase chain reaction (PCR) for survivin was performed on cDNA with the SYBR Green method (based on the manufacturer’s protocol, Applied Biosystems). Primers used were 5′-CTGCCTGGCAGCCCTTT-3′ (forward) and 5′-CCTCCAAGAAGGGCCAGTTC-3′ (reverse) for survivin (30) and 5′-CACTGGCTCGTGTGACAAGG-3′ and 5′-CAGACCTACTGTGCGCCTACTTAA-3′ for β-actin. The relative change in expression was calculated with the Pffal method (58). Experiments for MCC cell lines were repeated six times (two biological repeats done in triplicate). Experiments for BJ cell lines were repeated six times (three biological repeats done in duplicate). Mean and SEM were calculated and plotted as column graphs for comparison.
Cell cycle analysis
MKL-1 cells were treated with Accutase (Millipore) to break clumps and then resuspended in fresh medium containing drug and treated for 12 hours. BrdU (10 μM concentration) was added 3 hours before harvesting. Cells were then harvested and fixed in chilled 70% ethanol overnight. The cells were then washed, resuspended in 200 μl of 2 M HCl/Triton X (1%), and incubated for 30 min at room temperature. Cells were centrifuged at 2000 rpm for 10 min and neutralized in 200 μl of 0.1 M sodium tetraborate (pH 8.5). Cells were then washed, suspended in 20 μl of PBS containing 0.5% Tween 20, 1% donkey serum, and 2 μl of anti-BrdU antibody (1:10 dilution, BD Biosciences), and incubated overnight at 4°C. Cells were washed and incubated with secondary anti-mouse IgG Alexa Fluor 488 (1:1000) for 1 hour at room temperature. Cells were washed, suspended in PBS containing ribonuclease A (100 μg/ml), propidium iodide (50 μg/ml), and 0.05% Triton X, incubated for 30 min at 37°C in the dark, and then analyzed with an Accuri C6 flow cytometer.
Cell death evaluation by CFDA and propidium iodide staining
After harvesting, cells were resuspended in 2 ml of PBS containing propidium iodide (4 μg/ml) (Sigma) and 10 μl of CFDA (Invitrogen) at room temperature for 10 min. Cells were then rinsed in 1× PBS and examined under the microscope. Quantitation was performed with ImageJ software.
Mouse xenograft studies
Compounds
For in vivo experiments, clinical-grade bortezomib (Velcade) was purchased from the University of Pittsburgh Cancer Institute (UPCI) Pharmacy and YM155 was purchased from Active Biochemicals Ltd. Compounds were dissolved in sterile 0.9% saline solution for administration to animals.
Animals
Six-week-old female triple immunodeficient NOD-SCIDγ mice (Jackson Laboratory) were maintained in a specific pathogen-free environment at the Hillman Cancer Center Mouse Facility, University of Pittsburgh. All animal studies were conducted according to protocols approved by the Animal Ethics Committee of the University of Pittsburgh (Institutional Animal Care and Use Committee Protocol 1102226).
Xenograft drug treatments
MCC cells were checked for viability >90% by trypan blue staining, resuspended in PBS (2 × 107 cells in 100 μl), and inoculated subcutaneously into the right flank of mice. Once tumors were palpable (2 to 4 weeks after injection), mice were assigned sequentially into receiving either bortezomib, YM155, or saline treatment arms.
All treatments were delivered for 3 consecutive weeks. Bortezomib treatment was delivered subcutaneously twice weekly at 1 mg/kg per mouse. To avoid previously observed side effects, we gave mice hydrogel (ClearH2O) and kept them at 30°C (using a heating blanket to heat up half of the cage) during bortezomib treatment. YM155 (2 mg/kg) was given intraperitoneally on 5 consecutive days, followed by a 2-day treatment-free interval. The control group received saline alone (21 mice on the same dosing schedule as bortezomib and 10 mice on the same dosing schedule as YM155). Day 19 was the last day of drug delivery for both schedules and hence the end of treatment. Caliper measurements of the longest perpendicular tumor diameters were performed every other day, and tumor volumes were calculated with the following formula: (width)2 × (length/2). Animals were killed when tumors reached 2 cm in any dimension, >20% weight loss, or when they became moribund. Survival was defined as the time from the first day of treatment until death/killing.
Statistical analysis
Two-tailed paired Student’s t test was used to analyze statistical differences in quantitative reverse transcription–PCR (qRT-PCR) results. Mouse survival curves were estimated with the Kaplan-Meier product-limit method and were compared with the log-rank test (GraphPad Software). A piecewise linear hierarchical Bayesian model (59) was used to characterize differences in tumor volumes and growth between treatments.
Immunohistochemistry
Immunohistochemical staining of mouse tumor tissues was performed as previously described (15).
Supplementary Material
Fig. S1. MCV small T antigen knockdown does not affect survivin expression.
Fig. S2. T antigen knockdown does not affect caspase and PARP cleavage.
Fig. S3. MCV small T antigen isoform does not induce survivin expression in BJ cells.
Fig. S4. MCV large T antigen does not affect survivin oncoprotein expression in U2OS cells.
Fig. S5. Bortezomib induces apoptotic cell death in MCC tumor cells.
Fig. S6. Dose-response curves for drug screen candidates
Fig. S7. Survivin mRNA in xenograft mice.
Table S1. Transcripts in MCC libraries.
Table S2. Compound library screen results (data set).
Appendix S1. Bayesian hierarchical changepoint analysis of the effect of YM155 and bortezomib on tumor xenograft volume.
Acknowledgments
We thank C. Bakkenist for help with irradiation experiments and cell cycle profiles; D. Altieri for survivin reagents; J. Becker for WaGa cells and the pIH survivin construct; O. Gjoerup for U2OS and BJhTERT cells; X. Liu for help with plasmid construction; S. Scudiere for help with immunostaining; J. T. Newsome, K. L. Leschak, and M. L. Lambert for assistance with mouse protocols, handling, and care; and J. Kirkwood, F. Jenkins, and S. Moschos for valuable discussions. NCI/DTP Open Chemical Repository provided drugs tested from the NIH Oncology Drug Set II. We thank F. Rzewski and E. Ziporyn for help with the manuscript.
Funding: This work was supported by American Cancer Society Research Professorships to Y.C. and P.S.M. and by NIH grants CA78039 to A.V., and CA136363 and CA120726 to P.S.M. and Y.C. Preliminary studies were supported in part by a grant from the Al Copeland Foundation. This project used UPCI Shared Resources supported in part by award NIH P30CA047904.
Footnotes
Author contributions: H.F. performed the digital transcriptome subtraction analysis. R.A. and M.S. performed the T antigen and survivin knockdown experiments and immunoblots as well as cell cycle analysis. T.T. and R.A. created BJ stable cell lines and tested for survivin, cyclin E, E2F1, and MCV T antigen expression. R.A., Y.T., and L.L.V. performed the drug screen. R.A. tested individual drugs and performed CellTiter-Glo studies. A.G. and R.A. performed mouse xenograft experiments and qRT-PCR studies. D.N. performed xenograft tumor volume analysis. A.V., J.L.B., and A.D. provided compounds and supervision on drug screening. Y.C., P.S.M., and R.A. interpreted the data and wrote the manuscript.
Competing interests: Y.C. and P.S.M. have submitted a patent application U.S. 12/808,042 entitled “Methods to diagnose and immunize against the virus causing human Merkel cell carcinoma.” This manuscript is the basis of a pending patent application and, if issued, Y.C. and P.S.M. may be entitled to receive royalties under University of Pittsburgh policy.
REFERENCES AND NOTES
- 1.Allen PJ, Bowne WB, Jaques DP, Brennan MF, Busam K, Coit DG. Merkel cell carcinoma: Prognosis and treatment of patients from a single institution. J Clin Oncol. 2005;23:2300–2309. doi: 10.1200/JCO.2005.02.329. [DOI] [PubMed] [Google Scholar]
- 2.Veness M, Foote M, Gebski V, Poulsen M. The role of radiotherapy alone in patients with Merkel cell carcinoma: Reporting the Australian experience of 43 patients. Int J Radiat Oncol Biol Phys. 2010;78:703–709. doi: 10.1016/j.ijrobp.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Eng TY, Boersma MG, Fuller CD, Goytia V, Jones WE, III, Joyner M, Nguyen DD. A comprehensive review of the treatment of Merkel cell carcinoma. Am J Clin Oncol. 2007;30:624–636. doi: 10.1097/COC.0b013e318142c882. [DOI] [PubMed] [Google Scholar]
- 4.Lemos B, Nghiem P. Merkel cell carcinoma: More deaths but still no pathway to blame. J Invest Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- 5.Hodgson NC. Merkel cell carcinoma: Changing incidence trends. J Surg Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 6.Agelli M, Clegg LX. Epidemiology of primary Merkel cell carcinoma in the United States. J Am Acad Dermatol. 2003;49:832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- 7.Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: A population based study. J Cutan Pathol. 2010;37:20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 8.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Peñas PF, Nghiem P. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: The AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gjoerup O, Chang Y. Update on human polyomaviruses and cancer. Adv Cancer Res. 2010;106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- 11.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyoma-virus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, Chang Y, Moore PS. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg Infect Dis. 2011;17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci USA. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: Implications for immunotherapy of a polyomavirus-associated cancer. Curr Oncol Rep. 2011;13:488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer JG, Afanasiev OK, McClurkan C, Paulson K, Nagase K, Jing L, Marshak JO, Dong L, Carter J, Lai I, Farrar E, Byrd D, Galloway D, Yee C, Koelle DM, Nghiem P. Merkel cell polyomavirus-specific CD8+ and CD4+ T-cell responses identified in Merkel cell carcinomas and blood. Clin Cancer Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, Schrama D, Simonson WT, Lemos BD, Byrd DR, Koelle DM, Galloway DA, Leonard JH, Madeleine MM, Argenyi ZB, Disis ML, Becker JC, Cleary MA, Nghiem P. Transcriptome-wide studies of Merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol. 2011;29:1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y, Moore PS. Merkel cell carcinoma: A virus-induced human cancer. Annu Rev Pathol. 2012;7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 22.Ludlow JW, Shon J, Pipas JM, Livingston DM, DeCaprio JA. The retinoblastoma susceptibility gene product undergoes cell cycle-dependent dephosphorylation and binding to and release from SV40 large T. Cell. 1990;60:387–396. doi: 10.1016/0092-8674(90)90590-b. [DOI] [PubMed] [Google Scholar]
- 23.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linzer DI, Levine AJ. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhu JY, Abate M, Rice PW, Cole CN. The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J Virol. 1991;65:6872–6880. doi: 10.1128/jvi.65.12.6872-6880.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA. Aberrant regulation of survivin by the RB/E2F family of proteins. J Biol Chem. 2004;279:40511–40520. doi: 10.1074/jbc.M404496200. [DOI] [PubMed] [Google Scholar]
- 27.Raj D, Liu T, Samadashwily G, Li F, Grossman D. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis. 2008;29:194–201. doi: 10.1093/carcin/bgm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakahara T, Takeuchi M, Kinoyama I, Minematsu T, Shirasuna K, Matsuhisa A, Kita A, Tominaga F, Yamanaka K, Kudoh M, Sasamata M. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 31.Tolcher AW, Quinn DI, Ferrari A, Ahmann F, Giaccone G, Drake T, Keating A, de Bono JS. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol. 2012;23:968–973. doi: 10.1093/annonc/mdr353. [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwasa T, Okamoto I, Takezawa K, Yamanaka K, Nakahara T, Kita A, Koutoku H, Sasamata M, Hatashita E, Yamada Y, Kuwata K, Fukuoka M, Nakagawa K. Marked anti-tumour activity of the combination of YM155, a novel survivin suppressant, and platinum-based drugs. Br J Cancer. 2010;103:36–42. doi: 10.1038/sj.bjc.6605713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu SP, Jiang XH, Lin MC, Cui JT, Yang Y, Lum CT, Zou B, Zhu YB, Jiang SH, Wong WM, Chan AO, Yuen MF, Lam SK, Kung HF, Wong BC. Suppression of survivin expression inhibits in vivo tumorigenicity and angiogenesis in gastric cancer. Cancer Res. 2003;63:7724–7732. [PubMed] [Google Scholar]
- 35.Zhang R, Ma L, Zheng M, Ren J, Wang T, Meng Y, Zhao J, Jia L, Yao L, Han H, Li K, Yang A. Survivin knockdown by short hairpin RNA abrogates the growth of human hepatocellular carcinoma xenografts in nude mice. Cancer Gene Ther. 2010;17:275–288. doi: 10.1038/cgt.2009.68. [DOI] [PubMed] [Google Scholar]
- 36.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 37.Wright CM, Seguin SP, Fewell SW, Zhang H, Ishwad C, Vats A, Lingwood CA, Wipf P, Fanning E, Pipas JM, Brodsky JL. Inhibition of simian virus 40 replication by targeting the molecular chaperone function and ATPase activity of T antigen. Virus Res. 2009;141:71–80. doi: 10.1016/j.virusres.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seguin SP, Evans CW, Nebane-Akah M, McKellip S, Ananthan S, Tower NA, Sosa M, Rasmussen L, White EL, Maki BE, Matharu DS, Golden JE, Aubé J, Brodsky JL, Noah JW. High-throughput screening identifies a bisphenol inhibitor of SV40 large T antigen ATPase activity. J Biomol Screen. 2012;17:194–203. doi: 10.1177/1087057111421630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czarna A, Beck B, Srivastava S, Popowicz GM, Wolf S, Huang Y, Bista M, Holak TA, Dömling A. Robust generation of lead compounds for protein–protein interactions by computational and MCR chemistry: p53/Hdm2 antagonists. Angew Chem Int Ed Engl. 2010;49:5352–5356. doi: 10.1002/anie.201001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popowicz GM, Czarna A, Wolf S, Wang K, Wang W, Dömling A, Holak TA. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle. 2010;9:1104–1111. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 41.Sarek G, Kurki S, Enbäck J, Iotzova G, Haas J, Laakkonen P, Laiho M, Ojala PM. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J Clin Invest. 2007;117:1019–1028. doi: 10.1172/JCI30945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petre CE, Sin SH, Dittmer DP. Functional p53 signaling in Kaposi’s sarcoma-associated herpesvirus lymphomas: Implications for therapy. J Virol. 2007;81:1912–1922. doi: 10.1128/JVI.01757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Einsele H. Bortezomib. Recent Results Cancer Res. 2010;184:173–187. doi: 10.1007/978-3-642-01222-8_12. [DOI] [PubMed] [Google Scholar]
- 44.LeBlanc R, Catley LP, Hideshima T, Lentzsch S, Mitsiades CS, Mitsiades N, Neuberg D, Goloubeva O, Pien CS, Adams J, Gupta D, Richardson PG, Munshi NC, Anderson KC. Proteasome inhibitor PS-341 inhibits human myeloma cell growth in vivo and prolongs survival in a murine model. Cancer Res. 2002;62:4996–5000. [PubMed] [Google Scholar]
- 45.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Kinoyama I, Matsuhisa A, Kudou M, Sasamata M. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Sci. 2011;102:614–621. doi: 10.1111/j.1349-7006.2010.01834.x. [DOI] [PubMed] [Google Scholar]
- 46.Nakahara T, Takeuchi M, Kinoyama I, Minematsu T, Shirasuna K, Matsuhisa A, Kita A, Tominaga F, Yamanaka K, Kudoh M, Sasamata M. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 47.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi H, Morton DL, Elashoff D, Hoon DS. Survivin expression by metastatic melanoma predicts poor disease outcome in patients receiving adjuvant polyvalent vaccine. Int J Cancer. 2005;117:1032–1038. doi: 10.1002/ijc.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, McNiff JM. Nuclear expression of survivin portends a poor prognosis in Merkel cell carcinoma. Mod Pathol. 2008;21:764–769. doi: 10.1038/modpathol.2008.61. [DOI] [PubMed] [Google Scholar]
- 50.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 51.Piña-Oviedo S, Urbanska K, Radhakrishnan S, Sweet T, Reiss K, Khalili K, Del Valle L. Effects of JC virus infection on anti-apoptotic protein survivin in progressive multifocal leukoencephalopathy. Am J Pathol. 2007;170:1291–1304. doi: 10.2353/ajpath.2007.060689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kroemer G, Levine B. Autophagic cell death: The story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santhanam AN, Bindewald E, Rajasekhar VK, Larsson O, Sonenberg N, Colburn NH, Shapiro BA. Role of 3′UTRs in the translation of mRNAs regulated by oncogenic eIF4E—A computational inference. PloS One. 2009;4:e4868. doi: 10.1371/journal.pone.0004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronan SG, Green AD, Shilkaitis A, Huang TS, Das Gupta TK. Merkel cell carcinoma: In vitro and in vivo characteristics of a new cell line. J Am Acad Dermatol. 1993;29:715–722. doi: 10.1016/0190-9622(93)70236-m. [DOI] [PubMed] [Google Scholar]
- 55.Rosen ST, Gould VE, Salwen HR, Herst CV, Le Beau MM, Lee I, Bauer K, Marder RJ, Andersen R, Kies MS, Moll R, Franke WW, Radosevich JA. Establishment and characterization of a neuroendocrine skin carcinoma cell line. Lab Invest. 1987;56:302–312. [PubMed] [Google Scholar]
- 56.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houben R, Schrama D, Alb M, Pföhler C, Trefzer U, Ugurel S, Becker JC. Comparable expression and phosphorylation of the retinoblastoma protein in Merkel cell polyoma virus positive and negative Merkel cell carcinoma. Int J Cancer. 2010;126:796–798. doi: 10.1002/ijc.24790. [DOI] [PubMed] [Google Scholar]
- 58.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao L, Morgan MA, Parsels LA, Maybaum J, Lawrence TS, Normolle D. Bayesian hierarchical changepoint methods in modeling the tumor growth profiles in xenograft experiments. Clin Cancer Res. 2011;17:1057–1064. doi: 10.1158/1078-0432.CCR-10-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. MCV small T antigen knockdown does not affect survivin expression.
Fig. S2. T antigen knockdown does not affect caspase and PARP cleavage.
Fig. S3. MCV small T antigen isoform does not induce survivin expression in BJ cells.
Fig. S4. MCV large T antigen does not affect survivin oncoprotein expression in U2OS cells.
Fig. S5. Bortezomib induces apoptotic cell death in MCC tumor cells.
Fig. S6. Dose-response curves for drug screen candidates
Fig. S7. Survivin mRNA in xenograft mice.
Table S1. Transcripts in MCC libraries.
Table S2. Compound library screen results (data set).
Appendix S1. Bayesian hierarchical changepoint analysis of the effect of YM155 and bortezomib on tumor xenograft volume.