Abstract
Current techniques used in stem cell research only crudely mimic the physiological complexity of the stem cell niches. Recent advances in the field of micro- and nanoengineering have brought an array of in vitro cell culture models enabling development of novel, highly precise and standardized tools that capture physiological details in a single platform, with greater control, consistency, and throughput. In this review, we describe the micro- and nanotechnology-driven modern toolkit for stem cell biologists to design novel experiments in more physiological microenvironments with increased precision and standardization, while cautioning them against potential challenges that the modern technologies may present.
Extending the conventions
The world experienced and sensed by living cells can be utterly complex, consisting of various soluble and insoluble biochemical cues, including those secreted by adjacent and remote cells; mechanical inputs, including the local rigidity and topography of the extracellular matrix (ECM); other biophysical and biochemical components, including oxygen tension, and pH, influencing various cellular phenotypes [1–3]. Moreover, most of these interactions are frequently present in a combinatorial fashion, and dynamically change both in space and time. A cell, in other words, divorced from its microenvironmental context, can be an altogether different cell.
Often enough, in vitro (or more appropriately, in culture) experiments with stem cells are conducted without providing the context in which these cells might reside in vivo. Some of this could be attributed to the tendency to simplify a complex world, which scientists and humans, more generally, are not immune to, but perhaps more to the unavailability of platforms that can mimic the physiological cell microenvironments. Furthermore, in a state ominous for making clinical and biological stem cell research and application, the mainstream methodologies used to perturb the cell microenvironment suffer from the lack of reproducibility, expense associated with the use of cells and reagents, imprecise spatial and temporal control, low throughput, and scalability [4–7].
That, however, is changing rapidly; bringing to the stem cell biologists an array of engineered tools that can be integrated with the standardized experimental techniques for cell biology and allow precise control of biochemical, mechanical, and physical perturbations [4, 5]. Many of these techniques have been standardized for laboratory use, and many new techniques are in development. Though there are existing limitations that prohibit performing many conventional assays on these platforms, which biologists would be wise to be cautioned against, many new techniques allow conducting new kinds of experiments that might otherwise be close to impossible using the traditional cell biology techniques. For the stem cell biologists these technologies allow maintaining stem cells in environment more closely mimicking their natural microenvironment, perform experiments difficult or impossible to conduct using conventional technologies, and attain much enhanced temporal and spatial control of perturbation with greater precision and throughput. Attention to these methodologies is important, as in spite of the many advantages offered and wide availability, we believe that these tools remain underutilized in basic science explorative research.
In their own world
We are used to experiments conducted on our own scale – on the scale we can see and touch what we are dealing with, the scale of liter-sized beakers and centimeter-sized plates. Physics at the micro-scale in which cells reside, differs significantly from the one observed in the macro-world, in which conventional experiments are conducted [6]. For example, an epithelial stem cell residing in its niche is subjected to various microenvironmental maintenance and perturbation cues. The cell, like other stem cell types, can detect and respond to gradients of extracellular cues, both soluble and surface-bound [8, 9], responds to juxtracrine cell-cell communication signals from adjacent cells, and is supported by and responds to signaling by the extracellular matrix architecture and nanotopographical features. Cells also interact with extracellular matrix (ECM) molecules present in the niche either via signaling domains in ECM proteins like fibronectin, or with growth factors that are presented to the cells by being tethered to the ECM. In addition, the cell also is influenced by other factors like hypoxia, which together influence its fate, proliferation, migration and other phenotypes [1, 10, 11].
Since the natural cell environment is tightly controlled on this very small scale, arguably a lot can be gained from being able to hone the technology for cell manipulation on their rather than our scales. Most of the above perturbations are difficult to present to stem cells, even singly, using conventional laboratory techniques in a controlled manner.
Fortunately, with the advent of soft lithography techniques, many of which were pioneered by semi-conductor industry, it is now increasingly possible to control cell microenvironment on the micro- and nano-scopic levels. In particular, microfluidics, a technology involving nanoliters of fluids perfused through micro-scale devices of various complexities, has emerged as an important tool to control shear, and delivery of soluble factors to cells on a microscale, complement existing cell biology techniques. Within microfluidics, fluidic flow is laminar, i.e., it occurs in a non-turbulent and predictable fashion allowing application of highly developed electrical circuit theory to design precise networks for the delivery of a single drug or a combination of drugs at desired spatial locations, correct time points, and accurate concentrations [6]. Many simple ready-to-use microfluidic tools are available commercially, and therefore lithography need not be performed in the lab. As discoveries on the role of various growth factors, cytokines and other soluble cues that are presented to the stem cells in their niche have accumulated; there is a greater need to quantitatively analyze the spatial, temporal, and absolute dose response of these cues in influencing various cellular phenotypes. Modern technology like micropatterning or protein microarray can be harnessed to screen many biomaterials and ECM combinations for cell culture, as has been done by Anderson et al. using high throughput protein micropatterning to investigate stem cell responses to more than 1000 biomaterials that were used in tissue engineering disciplines [12], or by Yang et al. by a high throughput optimization of stem cell microenvironment [13].
Probing with a sharper eye
The advantages of microfluidic technology extend far beyond the increase in experimental precision. One can now perform experiments that would be hard, or nearly impossible, to do otherwise. For example, stem cells home to site of injury in response to chemotactic cues, while continuously being influenced by the mechanical and physical microenvironment present in their path [14–16]. The conventional assays that probe cell responses to spatially-graded signals are extremely limited in both experimental control and the ability to yield interpretable data. For instance, while it is hard to examine many (say dozens) cells, control both the average concentration and the gradient value at the same time in the micropipette experiments, it is also impossible to arbitrarily vary the gradient shapes. Boyden chambers do not permit careful analysis of cell migration trajectories or details of spatiotemporal dynamics. Microfluidic devices, on the other hand, can generate temporally controlled highly precise spatial gradients of soluble and insoluble factors [17–19]. For example, Wnt signaling, an important signaling pathway involved in stem cell renewal was studied using gradients generated by microfluidics (Fig. 2c). Recent advances in lab on a chip technology also allow formation of precise gradients of biophysical factors such as substratum rigidity [20], shear stress [21], and oxygen tension [22]. Since these devices can be easily incorporated with high resolution live fluorescence microscopy, they do not require any addition of expensive equipments to operate, and existing imaging modalities can be easily applied to acquire the experimental data. These platforms allow conducting stem cell based experiments in the soluble, physical, and mechanical environment matching the in vivo microenvironments, and allow precise dosage control of cytokines, growth factors, ECM, shear stress, and even hypoxia both temporally and spatially.
Figure 2.
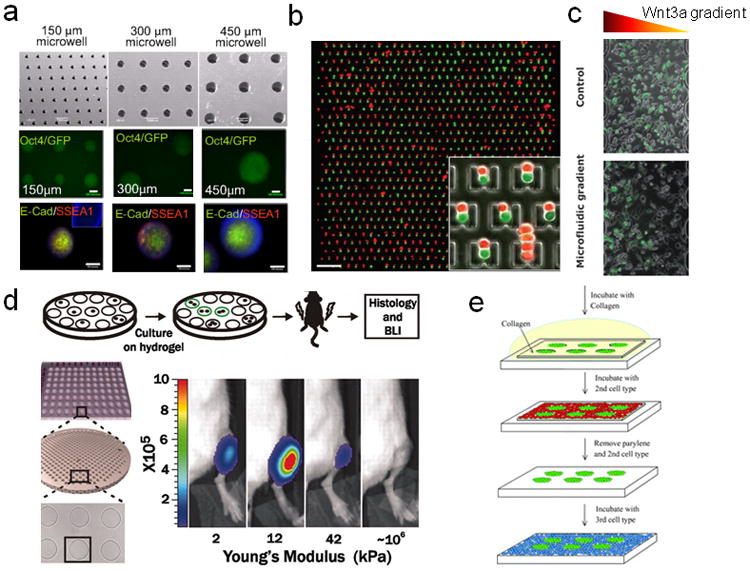
Examples illustrating the diverse capabilities of micro- and nanoengineered platforms used for control and analysis of stem cells. (a) Microwell-mediated control of embryoid body size in 3D. Oct4, ES cell pluripotency markers, SSEA1 and E-cadherin in Ebs within microwells are shown. (b) Microfluidic control of cell pairing and stem cell fusion. Red and green fluorescence image overlay image of CellTracker-labeled 3T3s loaded into the 2 mm 2 mm device. Scale bar: 200 um. (c) Case study: activation of the canonical Wnt3a/β-catenin pathway in cells cultured in microbioreactors. Panel A: cells were exposed for 12 h either to uniform Wnt3a concentration (control, top row) or to a microfluidic-generated gradient of Wnt3a concentration (microfluidic gradient, bottom row). Bright field images (left column), fluorescent images of the activated Venus-expressing cells (middle column) and merged images of Venus and bright field (right column) are shown. Images were taken in the mid sections of the culture channel [73]. (d) Culture on pliant hydrogel promotes muscle stem cell self-renewal. Hydrogel arrays with hundreds of microwells containing single MSCs were followed by time-lapse microscopy for 3 days. Videos were automatically processed and analyzed [74]. e) Microfabricated co-culture systems to pattern multiple cells types. Parylene-C stencils are pretreated with hyaluronic acid (HA) to reduce non-specific binding and then ES cells are allowed to attach to the substrate through the holes. Thereafter the entire area is treated with collagen to now encourage cell attachment and seeded with a second cell type, followed by a repetition of the process to co-culture a third cell type81 (reprinted with permission from the Royal Chemical Society).
On the flip side, experience of many researchers indicate that maintaining long term gradients in microfluidics has been a significant challenge necessitating a significant amount of fluid flow, and consequently of usage of large amounts of reagents [4, 23]. Stem cells tend to be sensitive to shear stress, and continuous fluid flow over days can affect them adversely [24, 25]. Though novel approaches exist to create “static” or low flow conditions to contain shear stress, most of the methods still require complex microfluidics circuitry, or relatively cumbersome standardization procedures [23, 24, 26]. Therefore, unless methods are optimized and standardized, we opine that microfluidics is suitable for maintenance of gradients for a few hours to a day. Therefore unless more specific devices and solutions are used, we suggest short term perturbation to study cell viability, signal transduction pathways, migration etc. when gradients are required for experiments. Possible solutions to address these disadvantages are listed in Table 1.
Table 1.
List of advantages and potential pitfalls of various micro- and nanoengineered tools, and possible solutions to avoid them.
| Tools | Advantages | Potential Pitfalls and Limitations | Solutions for the Specific Pitfalls | Complexity of Solutions |
|---|---|---|---|---|
| Microfluidics Gradient Generators | Pneumatic valve controls can quickly become unmanageable with increasing complexity | Keep number of conditions limited | ++ | |
| Automate pneumatic valve control [34] | ++++ | |||
| Absorption of biochemicals due to high surface area to volume ratio | Check for absorption profile of biochemical factor in pdms [71] | ++++ | ||
| Long term differentiation assays with perfusion | Shear stress | Use H-chip designs to avoid direct flow of fluid over cells | ++ | |
| Dampen flow by specialized perfusion[24, 26] | +++ | |||
| Culture cells in protected etched channels or microwells[25, 63] | ++ | |||
| Loss of autocrine paracrine signaling[62] | Maintain pulsatile flow with long periods (5–6 hours) with no flow [24] | ++++ | ||
| Use no flow conditions, and culture cells in high throughput microwells with media replenishment every 6–12 hours [46, 76] | +++ | |||
| Co-culture |
|
Sophisticated requirements for temporal control[49] | +++++ | |
| Control of shear and rigidity |
|
Difficult to ascertain effect on cell viability by shear alone [60] | Ex-chip control always suggested with comparable cell density and media | ++ |
|
Temporal control of rigidity difficult and require sophisticated set ups[59] | +++++ | ||
| Control of topography |
|
Expensive to fabricate [77–79] | CFL [80] and electrospinning[60] are cheaper but difficult to fabricate | +++++ |
| Small surface area prohibiting biochemical experiments [70] | Use CFL substrata from commercial or expert laboratories to minimize optimizations | + | ||
| 3D control not available in all topographical features | Electrospinning allows 3D control. CFL allows 2.5D control | ++ |
A significant advantage of small experimental chamber size in microfluidics chips increasingly harnessed by cell biologists is experimental miniaturization leading to a much greater analysis throughput requiring smaller amounts of cells and reagents [13, 27–30]. However, as a note of caution, high throughput screening of stem cells for differentiation has met only with mixed success. In a particularly memorable feat, Quake and colleagues have demonstrated a microfluidic device with 2056 valves, and 256 observation chambers containing cells stimulated with different stimuli [31], and an automated cell culture system for long term observation of stem cell differentiation [32]. In a considerably less massive application example, we have demonstrated an immunostaining based multiplexed device that allows temporal control of multiple biochemical stimuli to deduce the responses of various signaling effector molecules in tens of thousands of cells [33]. The device is completely automated providing consistency in cell seeding, stimulation profiles, immunostaining, data acquisition and analyses across experiments. Overall, such microfluidic experiments provide an interesting complement to other high-throughput assays, such as DNA microarray-based analysis. While microarray experiments normally allow for testing a very limited number of conditions with a very high number of read-outs (e.g., genes whose expression is assayed), whereas high-throughput microfluidic experimentation allows analysis of a very high number of conditions with a relatively small but still very informative number of read-outs. These platforms can be used for hypothesis generation, narrowing down conditions, and for drug screening for both research and industrial purposes. High throughput screening of ECM molecules can be obtained by micropatterning, and protein microarray technology.
Microfluidics can also be used to create more biomimetic niche-like conditions for stem cells in a high throughput manner [34, 35] or used for single cell profiling of stem cells in chemically defined conditions [36]. Similarly, microfluidics can allow a heterogeneous stem cell population or heterogeneous embryoid bodies [37] to be counted, and sorted [38], while allowing microscopy at the same time. However, many of the “proof of concept” devices may be difficult to operate and require skill acquirement limiting large scale use in conventional biological laboratories.
Building neighborhood with selection
Adult stem cells reside in specialized microenvironments called stem cell niches that allow both homotypic and heterotypic juxtracrine interactions [1], and paracrine signaling between stem cells and other somatic cell types (Fig. 1). However, these cell-cell interactions cannot be replicated in a controlled fashion using conventional co-culture experiments. Paracrine and autocrine signaling influencing stem cells can be studied using specialized microfluidic devices [39]. Interestingly, cells accord a higher sensitivity to paracrine signaling in microculture presenting a more sensitive method to probe the cells [40].
Figure 1.
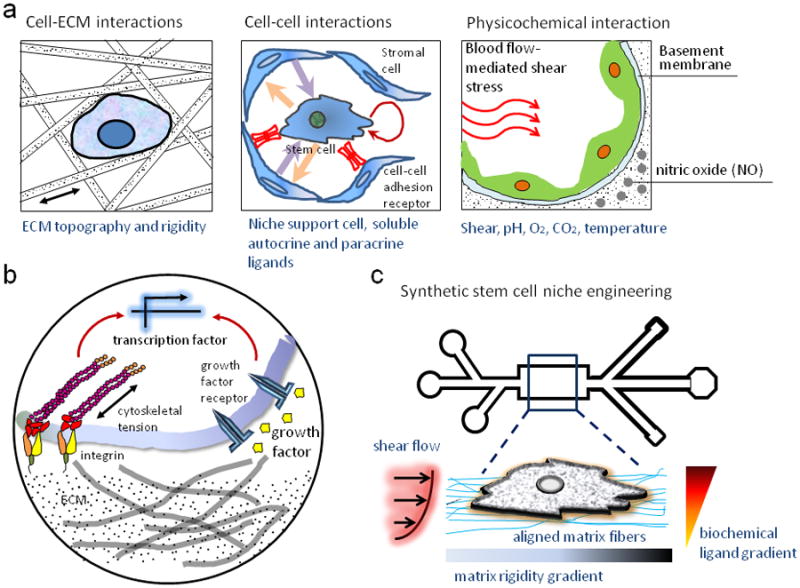
The nesting cell: A stem cell is exposed to multivariate cues including cell-cell interactions, cell-ECM interaction, soluble factors and biophysical factors like substratum rigidity, topography, shear stress, oxygen, and pH (A); Novel techniques like microfluidics, and micro-nanoengineering can allow mimicking the microenvironmental condition a cell experiences in vivo, and allow more precisely control of experimental parameters like shear stress, biochemical gradients, substrate rigidity and nanotopography, and cell positioning (B).
Again, microfabrication-based approaches can come to the rescue in the form of micro-stamps and micro-stencils [41–44]. These are simple ready-to-use devices that allow sequential seeding of each cell type, in predefined patterns that can be designed to control for the extent, and type of cell-cell interactions in a very precise manner (Fig. 2e). Stem cells are frequently maintained as non adherent sphere cultures (e.g. neurospheres, cardiospheres, embryoid bodies etc.). Microwells created using photolithography can also allow controlling the size and uniformity of the sphere cultures (Fig. 2a) [45, 46]. Recently, the laminar flow in microfluidic devices has also been used to define cell-cell interactions conditions by using microfabricated “capture-cups” to immobilize cells of one type, and then reversing the flow to capture cells of another type [47]. Cell-cell interactions and fusion can be effectively controlled by combination of microfluidics and microfabrication technology (Fig. 2b). Even temporal control of cell-cell interactions, potentially useful to understand the kinetics of stem cell-somatic cell interactions [48], has been demonstrated using micro-machined movable silicon parts [49]. Dielectrophoresis (DEP) can be used to spatially localize live cells by electrical current. Achieving such precise control of cell-cell interactions both spatially and temporally is nearly impossible using conventional techniques, presenting stem cell biologists with an array of tools to ask questions that were difficult to answer previously. Microstencils are relatively easy to use and handle, and require few optimizations (e.g. cell seeding density). In our opinion, these stencils can easily be included as standard co-culture platforms with defined interacting area between cells of various types. Many more designs to obtain more precise, novel, and interesting methods to get cells to interact at the experimentalist’s whim should be in the offing.
Engineering the mechanics
The physiological microenvironment of the stem cells not only consists of the biochemical factors, but also the physical properties of the microenvironment in which the cells reside [50–52]. ECM exists in vivo in a complex arrangement of fiber and sheet like structures, with nanoscale features that extend over centimeters [52, 53]. While ECM can interact biochemically with the cells, the mechanics of the stem cell-ECM interactions is probably equally, if not more important. For example, human mesenchymal stem cells cultured on substrata of rigidities mimicking various tissue types in vivo differentiate into somatic cells of the respective tissue type [54]. Similarly, stem cells are sensitive to the topography of the ECM substrata [53], and other mechanical forces [52]. Despite clear evidence supporting considerable role of the mechanical cell environment, commonly employed culture conditions involve growing stem cells on flat plates or cover slips with rigidity orders of magnitude higher than the ones observed in mammalian tissue Such culture conditions can considerably influence cellular phenotypes, altering cell stemness, differentiation potential, and migration properties [55]. Even though protocols to “maintain” stem cells on plastic surfaces have been standardized, we believe that it is essential to contrast the phenotype of cells when cultured on microenvironment more closely mimicking the physiological one for the given stem cell type.
Controlling certain mechanical perturbations is straightforward. For example control of shear stress can be achieved quite simply by microfluidics, either by controlling the input pressure of fluid flow, or by controlling the resistance in the channels [24, 56–58]. Similarly, gradients of substrate elasticity can also be created using microfluidics, though more interesting methods exist for specific perturbations (e.g. temporal and spatial control). To control substratum elasticity, for instance, in one interesting example, microfabricated post array made of a polymer polydimethylsiloxane (PDMS) with magnetic control allow application of precise elastic force to the cell at subcellular localization, and also for sensing the force applied by cells [59]. In a recent study, muscle stem cells were time lapsed on pliable hydrogel surfaces in arrays of microwells to study their differentiation into skeletal muscle cells (Fig. 2d). In contrast, creation of platforms to precisely control substrate topography remains a difficult process, and protocols to develop them exist only in a few university environments. Consequently, the role of topography of the ECM in controlling stem cell functions is still poorly understood, and frequently underestimated. Various methods have been developed and used for presentation of topographical cues to cultured cells include colloidal lithography [35], polymer demixing [36], and nanoimprinting [37]. These techniques employ creative means to define features of extremely small sizes (tens to hundreds of nanometers) on flat or 3-dimensional substrata, but are difficult to establish; therefore collaboration with laboratories with established setups or commercial procurement is the norm.
However, most of the platforms to present topographical cues remain expensive, and can be fabricated in selective engineering labs. Another concern with many of the current techniques is that owing to their small surface areas, they are not amenable to large biochemical experiments (e.g. western blot, RT-PCR, immunoprecipitation, microarrays etc) within reasonable cost constraints. While most of these techniques can be quite expensive, techniques like electrospinning [60] and capillary force lithography (CFL) [61] are becoming popular with stem cell biologists for their lower costs, and capability to produce large surface area samples with precisely controlled topography.
Recent advances in ECM biology has confirmed that cellular behavior is different on ECM coated 2D surfaces as compared to the native 3D environment [3]. Though 3D cultures (collagen gels, Matrigel etc.) have been in use, the role of mechanical cues in a 3D context (topography, and rigidity) has also come under increasing investigation. Electrospinning allows formation of nano-threads with ECM proteins, or with other polymers that are coated with ECM proteins, and provide a more natural 3D microenvironment to the stem cells than possible through other techniques [60]. Further, the most common platform to grow 3D cultures, Matrigel, can now be created within microfluidics allowing precise perturbations in the 3D [43]. Both the cost and the size of experimental platforms to present nanotopographical cues are coming down in recent years. Most of these platforms are also relatively easy to use, though they require highly sophisticated protocols to fabricate. Therefore, it is recommended that the standardized platforms either available commercially, or through nanoengineering laboratories are used to minimize difficult to attain optimizations in the desired feature sizes. These developments are quite promising, and in the near future commercial substrata with defined nanotopographical features that mimic different tissue ECM may become available.
Avoiding pitfalls
Stem cells present many unique problems for experimentation, including high susceptibility to shear stress, the need for prolonged experimentation to allow differentiation events to occur, and dependence on complex liquid and solid microenvironments that mimic the stem cell niche. These requirements make experimentation with stem cells particularly challenging, and if one is not careful in understanding potential pitfalls of micro- and nano-fabricated platforms, the results can be very disappointing. For example, if not carefully controlled, continuous perfusion can result in the shear stress harmful to some cell types, or in increased loss of molecules mediating paracrine and autocrine signaling molecules necessary for cell growth, survival or differentiation [62]. Therefore, it is preferable to use devices that do not depend on continuous flow over the cells, at least in long term, to maintain the desired liquid micro-environment, including environments with gradients of signaling inputs or drugs [23–25, 63, 64]. However, it should be noted that not all stem cell types are negatively sensitive to shear stress, which may even be required to induce targeted differentiation. Microfluidics can be useful, therefore, in understanding the role of shear stress on various stem cell phenotype, including differentiation. For non-adherent or semi-adherent cell types, e.g. embryonic stem cells and spheroids, one must choose the devices that do not involve continuous perfusion of media above cells, or minimize shear stress. If however, cell perfusion is important to maintain, e.g., to define more complex signaling input distribution over the cell population analyzed, one can modify the device design to decrease the adverse effects of flow, e.g., the shear stress. A simple method to significantly reduce shear stress is to increase the chamber height (shear stress reduces 4 times for each 2-fold increase in height). Another method is to create microwells in the cell substratum that protect cell by locally reducing the effects of flow shear [25, 63]. Ideally, flow based control of cell medium should be only used for short term perturbations, for instance, to understand signaling response to a specific growth factor, cytokine, or a drug. For long term cultures, H-type devices [65, 66], pulsating flow using a microcontroller [67], or flow dampeners, e.g., osmotic pumps [18] or more sophisticated flow controls [23, 24, 64] can be used. An additional current hurdle is that all these devices require training, more advanced equipment (especially the fluid control interfacing), and significant optimization relevant to the use of particular stem cell types. The standardization and convenience provided by commercialization of these platforms may ultimately require less time investment, but at present remain elusive.
As has been mentioned earlier, culture in microfluidics can result in large absorption of chemicals due to high surface area: volume ratio in the chambers. Further, biologists should seriously consider the logistics of how cells/explants/large spheroids will be introduced within the microfluidic platforms. Towards solving these issues, novel devices have been designed more recently that allow conducting a large portion of experimentation in an open, unsealed environment, which can be sealed at a desired time point, for more precise cell stimulation, using magnetic force [26], or vacuum [68]. These devices can be very useful to conduct experiments with large explants, or for interventional microfluidics experiments. In spite of less restrictive demands, these platforms still require considerable optimization to control for cell viability in response to shear stress, loss of paracrine signaling, as well as depending on more advanced interfacing set-ups.
The requirement for complex fabrication techniques can be partially alleviated by recent advances in stereolithography based tools allowing one to obviate the need for silicon-based microlithography [69]. These tools also permit creating of larger and more geometrically complex microfluidics devices that are easier to use, introduce significantly smaller amount of shear stress, and also facilitate hydrogel cultures within the device. We envision that these efforts will be enormously useful in making the microfluidic control less disruptive for stem cell experimentation. This may also help address the commonly occurring concern with microfluidic device of the large surface area to volume ratio, that might result in significant absorption of the active ingredients in the media (e.g. growth factors, drugs, antibodies etc.) Another minor concern which biologists should consider is the absorption of biochemicals in microfluidic devices. Regehr et al. detail the absorption rates of various biochemicals in PDMS devices [70, 71].
Cell substratum nanopatterning techniques have now been progressively optimized and characterized for stem cell based experiments. However, their limitations should be considered before using the corresponding technologies. For example, electrospinning can produce 3D gels presenting nanocues to the cultured cells, but their anisotropy is less controllable than that obtained using other techniques. In contrast, CFL can allow designing arbitrary nanocues, and is relatively inexpensive to use, allowing however cells to obtain topographical cues only from the 2D substratum. Another useful technique that can be employed to present nanocues to cultured cells without requiring expensive equipment is self-assembly based co-polymer patterning. This method can be used to create a wide variety of nanopatterns, but one should be careful to use only those chemicals that have been demonstrated to self-assemble in the desired topographical structures [72].
Overall, as in other new and rapidly developing area of analysis, one should exercise sufficient care in recognizing the inherent limitations of the novel techniques. Their novelty might also be a barrier in itself, based on the illusion of arduous learning curves required. In the end, the new micro- and nano-fabrication techniques can be invaluable for all the reasons listed above, if the cells and stimuli of interests are carefully tested and optimized, and the appropriate controls performed. These axioms of experimental science will remain true during this and other revolutionary changes in the techniques and methods used. The stem cell biologists therefore must be aware of the limitations of each new experimental platform before designing specific experiments.
Why should the stem cell biologists pay attention?
Recent advances in stem cell biology have underlined the importance of the role of microenvironment in influencing nearly all cellular behaviors, including morphology, migration, and fate. Cells are exposed to a combination of cues including biochemical growth factors and cytokines, ECM matrix proteins, substratum rigidity, oxygen tension, shear stress, and tissue nanotopography in a spatially and temporally varying fashion. Presentation of these cues to the cells in a consistent, precise, and combinatorial manner is one of the most significant challenges facing stem cell biologists. Stem cells also offer many unique challenges to the biologists. For example, precise and consistent characterization techniques for stem cells that are also inexpensive to use for cross laboratory comparison of stem cells is still a challenge. Methods are required to study very small subpopulation of primary stem cells. Microfluidics and microfabricated devices have provided a solution to these challenges, and are embraced by an increasing number of research laboratories working in the area of stem cells and other cell biology disciplines. The microfabrication techniques have been perfected in the microelectronic industry over decades, presenting to the biologists a highly consistent design paradigm that can be readily used to develop stem cell-specific platforms. These novel tools bring many biochemical techniques to a single chip, allowing sequential integration of otherwise independent experimental steps on a single platform. With the advantage of multiplexing that pneumatic valves provide, and parallelization offered by the miniaturized experimental systems, experiments can now be performed on cells with a dramatically increased precision, reliability, and consistency, while reducing cost and experimental errors to a significant extent.
Consequently, screening of stem cells with a combination of factors has become simpler, less expensive, and many folds faster. However, challenges remain. Stem cells are frequently finicky, and concerns regarding their high sensitivity to shear, viability in microfluidic devices during long term cultures, and high absorption of proteins by the walls continue to remain. Microfluidic devices can quickly become unmanageable with increasing complexity of design, requiring advanced skill and optimization. In addition, the nanofabricated platforms tend to be expensive and small in surface area prohibiting biochemical experiments. Many of these problems are now being addressed by simpler designs, and stem-cell specific changes in design of devices (e.g. low perfusion, deeper chambers, large input/output reservoirs to avoid handling of tubes), but it would be prudent to wait for such optimizations to be completed before large scale experiments can be brought completely to these devices. However, we opine, that for experiments involving short time frame (hours to a day), and for few conditions, microfluidics offers many advantages that can be employed to perform more careful and precise experimentations. A good rule of thumb may be to always test the viability of cells in the device for the length of experiments against a conventional tissue-culture dish. Arguably, these smart Petri dishes can and should revolutionize the modern biology further, making it truly quantitative, systems science, as well as much approximating the conditions present in live organisms.
Table 2.
A few important and relevant parameters for choosing (or to avoid choosing) the appropriate micro- nanofabricated platform for specific stem cell based experiments.
| Platform | Techniques | Some Useful Numbers | Notes |
|---|---|---|---|
| Microfluidics | Photolithography | Chamber height1: 10–500μm Minimum feature size2: 5–10μm Length of culture: 2 days 3D culture: difficult Ease of handling: moderate High throughput: yes | 1. High chamber height reduces shear.2. Small feature size allow efficient packing of device. |
| Stereolithography | Chamber height1: 100–1000μm Minimum feature size2: 25μm Length of culture: 7–10 days 3D culture: easy Ease of handling: easy High throughput: difficult | ||
| Microstencils | Ease of handling1: very easy Size of “holes”: 5–500μm Cell types: 2–3 Shape of cell-cell interface: arbitrary Incorporation in microfluidics: difficult | 1. Small size of holes present difficulty in introducing cells. | |
| Nanofabrication | Capillary force lithography | Feature size1: >10nm Sample size2: 5cm Shape of features: arbitrary Material compatibility: UV assisted polymerization Length of culture: 1–21 days Ease of handling: very easy Cost: low Microscopy: any Incorporation in microfluidics: easy | 1. Possibility to obtain smaller feature size allow a higher flexibility to create a more biomimetic ECM microenvironment.2. Large sample size allow biochemical experiments. |
| Electrospinning | Feature size: 10–100nm Sample size1: >1cm3 Shape of features: grooves Material compatibility: limited Length of culture: 1–15 days Ease of handling: moderately easy Cost: moderate Microscopy: confocal | 1. Electrospinning allows only 3D samples. | |
| Nanoimprinting | Feature size: 10–100nm Sample size1: >1cm3 Shape of features: grooves, pillars Material compatibility: UV or thermal curable materials Microscopy: any Cost: moderate Incorporation in microfluidics: easy | Nanoimprinting equipment is required. | |
| Mechanical Gradients in Microfluidics | Polyethylene glycol (PEG) gradients | FDA approved: yes Range of rigidity1,86: 50–500kPa Mode of polymerization: UV/chemical Ease of preparation: moderately difficult Ease of ECM coating: moderately easy | 1. Range of rigidity achieved by change in cross-linker concentration. |
| Polyacrylamide gradients | FDA approved: no Range of rigidity1,86: 0.01–100kPa Mode of polymerization: UV/chemical Ease of preparation: moderately easy Ease of ECM coating1: difficult | 1. ECM coating on PAAM presents complications. | |
| PDMS gradients | FDA approved: no Range of rigidity86: 10–1000kPa Mode of polymerization: UV/chemical Ease of ECM coating: easy Ease of preparation: moderately difficult Ease of gradients: difficult | ||
| Nanofabrication of polymers | Ease of preparation1: difficult Ease of handling: easy Gradient shape: arbitrary Ease of ECM coating: easy | 1. Unlike microfluidics based gradient generation, nanofabricated samples require complex fabrication procedures. |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441(7097):1075–9. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 2.Blank U, Karlsson G, Karlsson S. Signaling pathways governing stem-cell fate. Blood. 2008;111(2):492–503. doi: 10.1182/blood-2007-07-075168. [DOI] [PubMed] [Google Scholar]
- 3.Even-Ram S, Artym V, Yamada KM. Matrix control of stem cell fate. Cell. 2006;126(4):645–7. doi: 10.1016/j.cell.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Salieb-Beugelaar GB, et al. Latest Developments in Microfluidic Cell Biology and Analysis Systems. Analytical Chemistry. 2010;82(12):4848–4864. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 5.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442(7101):403–11. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 6.Beebe DJ, Mensing GA, Walker GM. PHYSICS AND APPLICATIONS OF MICROFLUIDICS IN BIOLOGY. Annual Review of Biomedical Engineering. 2002;4(1):261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 7.Meyvantsson I, Beebe DJ. Cell Culture Models in Microfluidic Systems. Annual Review of Analytical Chemistry. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- 8.Shimada H, et al. Concentration gradient of vascular endothelial growth factor in the vitreous of eyes with diabetic macular edema. Invest Ophthalmol Vis Sci. 2009;50(6):2953–5. doi: 10.1167/iovs.08-2870. [DOI] [PubMed] [Google Scholar]
- 9.Wang SJ, et al. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300(1):180–9. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 10.Moore KA, I, Lemischka R. Stem cells and their niches. Science. 2006;311(5769):1880–5. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 11.Raymond K, et al. Adhesion within the stem cell niches. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22(7):863–6. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 13.Yang F, et al. High throughput optimization of stem cell microenvironments. Comb Chem High Throughput Screen. 2009;12(6):554–61. doi: 10.2174/138620709788681916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceradini DJ, Gurtner GC. Homing to hypoxia: HIF-1 as a mediator of progenitor cell recruitment to injured tissue. Trends Cardiovasc Med. 2005;15(2):57–63. doi: 10.1016/j.tcm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Carmona G, et al. Activation of Epac stimulates integrin-dependent homing of progenitor cells. Blood. 2008;111(5):2640–6. doi: 10.1182/blood-2007-04-086231. [DOI] [PubMed] [Google Scholar]
- 16.Burns CE, Zon LI. Homing sweet homing: odyssey of hematopoietic stem cells. Immunity. 2006;25(6):859–62. doi: 10.1016/j.immuni.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Dertinger SKW, et al. Generation of Gradients Having Complex Shapes Using Microfluidic Networks. Analytical Chemistry. 2001;73(6):1240–1246. [Google Scholar]
- 18.Park JY, et al. Differentiation of Neural Progenitor Cells in a Microfluidic Chip-generated Cytokine Gradient. Stem Cells. 2009 doi: 10.1002/stem.202. [DOI] [PubMed] [Google Scholar]
- 19.Melin J, Quake SR. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annual Review of Biophysics and Biomolecular Structure. 2007;36(1):213–231. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 20.Cheung YK, et al. Microscale control of stiffness in a cell-adhesive substrate using microfluidics-based lithography. Angew Chem Int Ed Engl. 2009;48(39):7188–92. doi: 10.1002/anie.200900807. [DOI] [PubMed] [Google Scholar]
- 21.Kim DH, et al. Microengineered platforms for cell mechanobiology. Annu Rev Biomed Eng. 2009;11:203–33. doi: 10.1146/annurev-bioeng-061008-124915. [DOI] [PubMed] [Google Scholar]
- 22.Polinkovsky M, et al. Fine temporal control of the medium gas content and acidity and on-chip generation of series of oxygen concentrations for cell cultures. Lab Chip. 2009;9(8):1073–84. doi: 10.1039/b816191g. [DOI] [PubMed] [Google Scholar]
- 23.Liu K, et al. Cell culture chip using low-shear mass transport. Langmuir. 2008;24(11):5955–5960. doi: 10.1021/la8003917. [DOI] [PubMed] [Google Scholar]
- 24.Korin N, et al. Periodic “flow-stop” perfusion microchannel bioreactors for mammalian and human embryonic stem cell long-term culture. Biomed Microdevices. 2009;11(1):87–94. doi: 10.1007/s10544-008-9212-5. [DOI] [PubMed] [Google Scholar]
- 25.Joanne Wang C, et al. A microfluidics-based turning assay reveals complex growth cone responses to integrated gradients of substrate-bound ECM molecules and diffusible guidance cues. Lab Chip. 2008;8(2):227–37. doi: 10.1039/b713945d. [DOI] [PubMed] [Google Scholar]
- 26.Tkachenko E, et al. An easy to assemble microfluidic perfusion device with a magnetic clamp. Lab Chip. 2009;9(8):1085–95. doi: 10.1039/b812184b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong J, Edel JB, deMello AJ. Micro- and nanofluidic systems for high-throughput biological screening. Drug Discovery Today. 2009;14(3–4):134–146. doi: 10.1016/j.drudis.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Kamei KI, et al. An integrated microfluidic culture device for quantitative analysis of human embryonic stem cells. Lab on a Chip. 2009;9(4):555–563. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]
- 29.Meyvantsson I, et al. Automated cell culture in high density tubeless microfluidic device arrays. Lab on a Chip. 2008;8(5):717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 30.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nature Biotechnology. 2008;26(1):120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 31.Hong JW, Quake SR. Integrated nanoliter systems. Nat Biotechnol. 2003;21(10):1179–83. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Sjoberg R, et al. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79(22):8557–63. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 33.Cheong R, Wang CJ, Levchenko A. Using a microfluidic device for high-content analysis of cell signaling. Sci Signal. 2009;2(75):pl2. doi: 10.1126/scisignal.275pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carrion B, et al. Recreating the perivascular niche ex vivo using a microfluidic approach. Biotechnol Bioeng. doi: 10.1002/bit.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobel S, Lutolf M. High-throughput methods to define complex stem cell niches. Biotechniques. 48(4):ix–xxii. doi: 10.2144/000113401. [DOI] [PubMed] [Google Scholar]
- 36.Kamei K, et al. Microfluidic image cytometry for quantitative single-cell profiling of human pluripotent stem cells in chemically defined conditions. Lab Chip. 10(9):1113–9. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lillehoj PB, et al. Continuous sorting of heterogeneous-sized embryoid bodies. Lab Chip. 10(13):1678–82. doi: 10.1039/c000163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu HW, et al. An integrated microfluidic system for isolation, counting, and sorting of hematopoietic stem cells. Biomicrofluidics. 4(2) doi: 10.1063/1.3454767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung KE, et al. Transition to invasion in breast cancer: a microfluidic in vitro model. Integrative Biology. 2010 doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domenech M, et al. Cellular observations enabled by microculture: paracrine signaling and population demographics. Integrative Biology. 2009;1(3):267–274. doi: 10.1039/b823059e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright D, et al. Generation of static and dynamic patterned co-cultures using microfabricated parylene-C stencils. Lab Chip. 2007;7(10):1272–9. doi: 10.1039/b706081e. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Hegde M, Jayaraman A. Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions. Lab on a Chip. 2010;10(1):43–50. doi: 10.1039/b911367c. [DOI] [PubMed] [Google Scholar]
- 43.Huang CP, et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab on a Chip. 2009;9(12):1740–1748. doi: 10.1039/b818401a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen ZL, et al. Patterning Mammalian Cells for Modeling Three Types of Naturally Occurring Cell-Cell Interactions. Angewandte Chemie-International Edition. 2009;48(44):8303–8305. doi: 10.1002/anie.200902708. [DOI] [PubMed] [Google Scholar]
- 45.Hwang YS, et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci U S A. 2009;106(40):16978–83. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi YY, et al. Controlled-size embryoid body formation in concave microwell arrays. Biomaterials. 2010;31(15):4296–4303. doi: 10.1016/j.biomaterials.2010.01.115. [DOI] [PubMed] [Google Scholar]
- 47.Skelley AM, et al. Microfluidic control of cell pairing and fusion. Nat Methods. 2009;6(2):147–52. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao DL, Ma L, Shen K. Transient cell-cell interactions in neural circuit formation. Nature Reviews Neuroscience. 2009;10(4):262–271. doi: 10.1038/nrn2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hui EE, Bhatia SN. Micromechanical control of cell-cell interactions. Proc Natl Acad Sci U S A. 2007;104(14):5722–6. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adamo L, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459(7250):1131–5. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stolberg S, McCloskey KE. Can shear stress direct stem cell fate? Biotechnol Prog. 2009;25(1):10–9. doi: 10.1002/btpr.124. [DOI] [PubMed] [Google Scholar]
- 52.Wang JH, Thampatty BP. Mechanobiology of adult and stem cells. Int Rev Cell Mol Biol. 2008;271:301–46. doi: 10.1016/S1937-6448(08)01207-0. [DOI] [PubMed] [Google Scholar]
- 53.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5(1):17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 55.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462(7272):433–41. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim L, et al. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip. 2006;6(3):394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 57.Cimetta E, et al. Micro-bioreactor arrays for controlling cellular environments: design principles for human embryonic stem cell applications. Methods. 2009;47(2):81–9. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park JY, et al. Simultaneous generation of chemical concentration and mechanical shear stress gradients using microfluidic osmotic flow comparable to interstitial flow. Lab Chip. 2009;9(15):2194–202. doi: 10.1039/b822006a. [DOI] [PubMed] [Google Scholar]
- 59.Sniadecki NJ, et al. Magnetic microposts as an approach to apply forces to living cells. Proc Natl Acad Sci U S A. 2007;104(37):14553–8. doi: 10.1073/pnas.0611613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlberg B, et al. Electrospun polyurethane scaffolds for proliferation and neuronal differentiation of human embryonic stem cells. Biomed Mater. 2009;4(4):45004. doi: 10.1088/1748-6041/4/4/045004. [DOI] [PubMed] [Google Scholar]
- 61.Kim DH, et al. Guided Cell Migration on Microtextured Substrates with Variable Local Density and Anisotropy. Adv Funct Mater. 2009;19(10):1579–1586. doi: 10.1002/adfm.200990041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellison D, Munden A, Levchenko A. Computational model and microfluidic platform for the investigation of paracrine and autocrine signaling in mouse embryonic stem cells. Mol Biosyst. 2009;5(9):1004–12. doi: 10.1039/b905602e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korin N, et al. Design of well and groove microchannel bioreactors for cell culture. Biotechnol Bioeng. 2009;102(4):1222–30. doi: 10.1002/bit.22153. [DOI] [PubMed] [Google Scholar]
- 64.Luo CX, et al. A fast cell loading and high-throughput microfluidic system for long-term cell culture in zero-flow environments. Biotechnology and Bioengineering. 2008;101(1):190–195. doi: 10.1002/bit.21877. [DOI] [PubMed] [Google Scholar]
- 65.Abhyankar VV, et al. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6(3):389–93. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 66.Paliwal S, et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446(7131):46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 67.Kim JY, et al. A cell culturing system that integrates the cell loading function on a single platform and evaluation of the pulsatile pumping effect on cells. Biomed Microdevices. 2008;10(1):11–20. doi: 10.1007/s10544-007-9105-z. [DOI] [PubMed] [Google Scholar]
- 68.Chung BG, et al. A hybrid microfluidic-vacuum device for direct interfacing with conventional cell culture methods. BMC Biotechnol. 2007;7:60. doi: 10.1186/1472-6750-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han A, et al. Multi-layer plastic/glass microfluidic systems containing electrical and mechanical functionality. Lab Chip. 2003;3(3):150–7. doi: 10.1039/b302118a. [DOI] [PubMed] [Google Scholar]
- 70.Regehr KJ, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9(15):2132–9. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6(12):1484–6. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 72.Bita I, et al. Graphoepitaxy of self-assembled block copolymers on two-dimensional periodic patterned templates. Science. 2008;321(5891):939–43. doi: 10.1126/science.1159352. [DOI] [PubMed] [Google Scholar]
- 73.Cimetta E, et al. Microfluidic device generating stable concentration gradients for long term cell culture: application to Wnt3a regulation of beta-catenin signaling. Lab Chip. 10(23):3277–83. doi: 10.1039/c0lc00033g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 5995;329:1078–81. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang F, et al. Combinatorial extracellular matrices for human embryonic stem cell differentiation in 3D. Biomacromolecules. 11(8):1909–14. doi: 10.1021/bm100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukuda J, et al. Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials. 2006;27(30):5259–67. doi: 10.1016/j.biomaterials.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 77.Chua KN, et al. Functional nanofiber scaffolds with different spacers modulate adhesion and expansion of cryopreserved umbilical cord blood hematopoietic stem/progenitor cells. Exp Hematol. 2007;35(5):771–81. doi: 10.1016/j.exphem.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goto M, et al. Micro- and nanometer-scale patterned surface in a microchannel for cell culture in microfluidic devices. Anal Bioanal Chem. 2008;390(3):817–23. doi: 10.1007/s00216-007-1496-4. [DOI] [PubMed] [Google Scholar]
- 79.Luo W, Yousaf MN. Tailored electroactive nanorods for biospecific cell adhesion and differentiation. Chem Commun (Camb) 2009;(10):1237–9. doi: 10.1039/b817212a. [DOI] [PubMed] [Google Scholar]
- 80.Kim DH, et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A. 2010;107(2):565–70. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]


