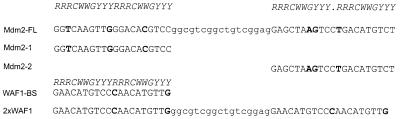Abstract
Genotoxic stress activation of the tumor suppressor transcription factor p53 involves post-translational C-terminal modifications that increase both protein stability and DNA binding activity. We compared the requirement for p53 protein activation of p53 target sequences in two major p53-regulated genes, p21/WAF1 (encoding a cell cycle inhibitory protein) and Mdm2 (encoding a ubiquitin ligase that targets p53 for proteolytic degradation). The p53 binding site in the proximal p21/WAF1 promoter contains a single p53 binding consensus sequence, while the p53 binding site in the Mdm2 promoter contains two consensus sequences linked by a 17 bp spacer. Binding of recombinant p53 protein to the p21/WAF1 binding site required monoclonal antibody PAb421, which can mimic activating phosphorylation and/or acetylation events at the C-terminus. In contrast, recombinant p53 bound strongly to the Mdm2 binding site in the absence of PAb421 antibody. Separate binding to each consensus sequence of the Mdm2 binding site still required PAb421, indicating that p53 binding was not simply due to greater affinity to the Mdm2 consensus sequences. Linking two p21/WAF1 binding sites with the 17 bp spacer region from the Mdm2 gene eliminated the PAb421 requirement for p53 binding to the p21/WAF1 site. These results suggest a mechanism for regulation of Mdm2 gene transcription that differs from that other p53-induced genes by its lack of a requirement for C-terminal activation of p53 protein. A steady induction of Mdm2 protein would maintain p53 protein at low levels until post-translational modifications following DNA damage increased p53 activity towards other genes, mediating p53 growth inhibitory and apoptotic activities.
INTRODUCTION
The p53 tumor suppressor protein transactivates cell cycle arrest and apoptosis genes. p53 gene inactivation by mutation is one of the most common alterations in human tumors (1). p53 activation is mediated by post-translational modifications that increase both the p53 protein half-life and affinity for its DNA binding sequence. The increase in p53 half-life is mainly due to reduced affinity for Mdm2, a p53 binding protein that mediates p53 ubiquitination and degradation through the proteasome pathway (2,3). The Mdm2 gene itself is transcriptionally activated by p53, closing an autoregulatory feedback loop (4). Besides protein stabilization, p53 is thought to require activation of its sequence-specific DNA binding function to transactivate its target genes. The p53 protein binds to a consensus binding motif made up of two decameric palindromes separated by 0–13 bp (5), represented in Figure 1. The binding of recombinant or mammalian expressed p53 protein to this sequence is weak but can be activated in vitro by agents that interact with or modify the last 30 amino acids in the p53 C-terminus. Phosphorylation within this 30 amino acid region by casein kinase II, binding of PAb421, alternative splicing, deletion, or incubation with peptides corresponding to this region can activate the p53 DNA binding function (6–8). Furthermore, in vitro translated p53 requires incubation with either PAb421 or nuclear extracts for significant binding to the consensus sequence (9). Models have been proposed in which the p53 C-terminus folds and interacts with the central core of the p53 protein, maintaining the molecule in a latent form (inhibited for DNA binding) (8). Post-translational modifications of the C-terminus after stress or genotoxic stimuli could diminish this interaction, resulting in activation of the p53 protein–DNA binding function. These post-translational modifications include serine and threonine phosphorylations and lysine acetylations (10).
Figure 1.
p53 binding sequences used in these experiments. Residues matching the p53 binding sequence consensus are in upper case italics at the top. In the consensus sequence, R stands for any purine, Y stands for any pyrimidine and W stands for A or T. The 17 nt separating the p53 binding sequences in Mdm2-FL and 2×WAF1 are in lower case. Those residues within the p53 binding sequence that deviate from the consensus sequence are in bold. Mdm2-FL and WAF1-BS are natural p53 binding sites present in the promoters of the Mdm2 and WAF1 genes, respectively. Mdm2-FL is a dimer of p53 binding sites; Mdm2-1 and Mdm2-2 are the isolated p53 binding sites from Mdm2-FL. 2×WAF1 is composed of two WAF1-BS binding sites linked by the same 17 bp sequence separating the single p53 binding sites in Mdm2-FL.
Mdm2- and cyclin-dependent kinase inhibitor WAF1 (11) genes are transcriptional targets of the p53 protein, encoding proteins that participate in p53 stability regulation and p53-mediated G1 arrest, respectively. Mdm2 differs from other p53 regulated genes in that its promoter contains a double p53 binding site composed of two p53 consensus sequences separated by a 17 bp spacer (4). In contrast, the p53 binding sites proximal to the promoters of other known p53 regulated genes, such as WAF1, contain a single p53 consensus sequence (11). A critical question is whether differences in the particular p53 binding elements in target genes may alter the requirement for p53 post-translational modification and activation. Such differences could modulate the transcriptional activation kinetics of different p53 target genes and regulate the p53 response. To begin to answer this question, we analyzed the relative DNA binding activities of recombinant human p53 proteins to the p53 binding sites present in the WAF1 and Mdm2 genes. Either monoclonal antibody PAb421 or deletion of the last 30 amino acids in the p53 sequence was used to mimic the activation events at the p53 C-terminus. The results indicate that the requirement for p53 C-terminal activation depends on the particular binding element; binding to the single p53 site present in the WAF1 promoter requires activation, while binding to the double p53 binding site present in the Mdm2 gene promoter is constitutive. This suggests a mechanism of differential regulation of p53 target genes responsive to stress or genotoxic stimuli.
MATERIALS AND METHODS
Recombinant p53 protein production in insect cells
The recombinant human wild-type p53 and C-terminally deleted p53Δ30 proteins were expressed in Sf9 insect cells and purified as previously described (12).
Electrophoretic mobility shift assay (EMSA)
Double-stranded oligonucleotides for the Mdm2-P2 promoter and WAF1 promoters (detailed in Fig. 1) were synthesized by the Biopolymer Core Facility of Roswell Park Cancer Institute (Buffalo, NY) and by Takara Biomedicals (Kyoto, Japan). The oligonucleotides were end-labeled with polynucleotide kinase and [γ-32P]ATP. EMSA was carried out in a total volume of 20 ml containing 20–200 ng of recombinant p53 protein and a molar excess of the radiolabeled oligonucleotide in binding buffer [20 mM HEPES–NaOH pH 7.9, 25 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 2 mM spermidine, 10% glycerol, 60 ng poly(dI-dC) and 20 mg BSA]. The reaction mixture was incubated for 30 min at 4°C, followed by electrophoresis in 5% native gel for 30 min at 200 V. The gel was dried and exposed to a phosphorimaging plate and analyzed using a BAS2000 system (Fuji Film, Tokyo, Japan).
Determination of the binding affinity constant (Kd) of p53 to the Mdm2 promoter
A fixed amount of recombinant p53 protein (100 ng) was incubated with increasing concentrations of radioactively labeled Mdm2-FL double-stranded oligonucleotide (Fig. 1), under the same conditions used for EMSA assays. Bound and free oligonucleotide was determined by EMSA and quantified by phosphorimaging. The Kd value was estimated by simple regression analysis (SAS/INSIGHT, SAS Institute Inc.). Data are expressed as the mean of three independent experiments, including the standard deviation of the mean.
RESULTS
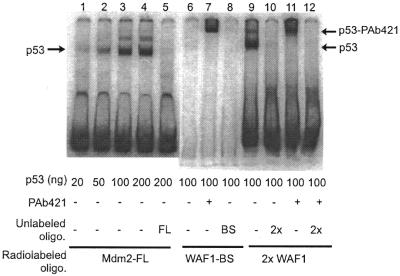
The binding of human wild-type p53 to the WAF1 binding site and full-length Mdm2 binding site (WAF1-BS and Mdm2-FL in Fig. 1, respectively) is shown in Figure 2. As expected, binding to the WAF1 binding site required C-terminal activation by antibody PAb421; a retardation complex was formed only in the presence of PAb421 antibody in the reaction (Fig. 2, lane 7) and not in its absence (Fig. 2, lane 6). However, under the same conditions, binding to the native Mdm2 site occurred in the absence of PAb421 and could be detected even with 20 ng of p53 protein (Fig. 2, lanes 1–4), while in the absence of PAb421, no binding to WAF1-BS could be detected even with 100 ng of p53 protein (lane 6). When two WAF1 binding sites were linked by the same 17 bp spacer sequence found in the double Mdm2 binding site (to produce the 2×WAF1 oligonucleotide, Fig. 1), p53 protein could bind to the new oligonucleotide in the absence of PAb421 antibody (Fig. 2, lane 9). There was no increase in signal of the more slowly migrating 2×WAF1–p53 complex in the presence of PAb421 antibody (Fig. 2, lane 11 compared to lane 9), suggesting that the p53 protein was fully capable of binding to the 2×WAF1 template.
Figure 2.
Binding activity of human wild-type p53 protein to the Mdm2 and WAF1 gene p53 binding sites. Recombinant human wild-type p53 protein was produced and purified from insect cells, and its binding activity was analyzed by EMSA as described in the text. The indicated radiolabeled oligonucleotides (Mdm2-FL, WAF1-BS and 2×WAF1, described in Fig. 1) were incubated with 20–200 ng of purified p53 protein, and the radiolabeled oligonucleotide–p53 bound complexes were separated in a non-denaturating polyacrylamide gel. Where indicated, 200 ng of PAb421 antibody, which binds to the C-terminal domain of p53, were also included in the reaction. The positions of the p53-radiolabeled oligonucleotide complexes and the PAb421–p53-radiolabeled supershifted complexes are indicated by arrows. In indicated lanes, binding to the radiolabeled oligonucleotide was competed by a 100-fold molar excess of unlabeled oligonucleotide, indicated by FL (Mdm2-FL), BS (WAF1-BS) or 2× (WAF12×).
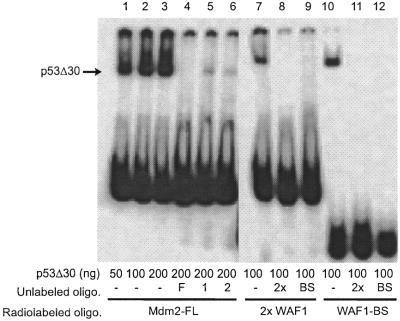
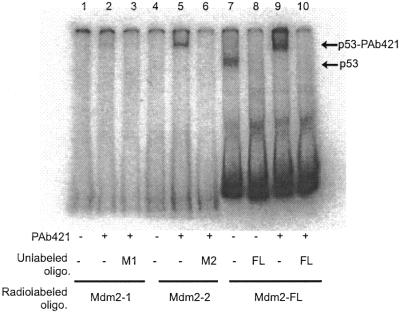
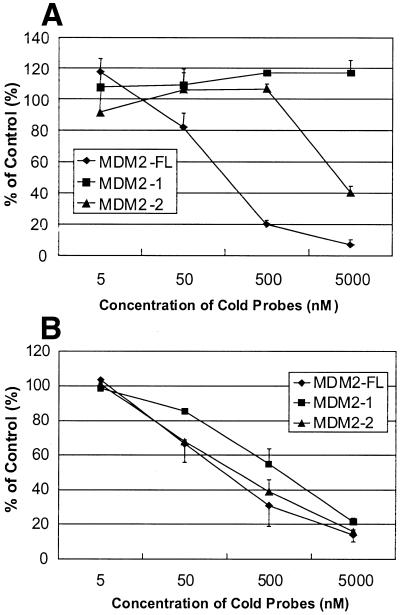
Deletion of the C-terminal 30 amino acids in p53, which contain the PAb421 epitope, has been shown to activate p53 binding to its consensus sequence equally as well as addition of PAb421 antibody (8). Thus, a p53 protein without the last 30 amino acids (p53Δ30) should bind equally well to single WAF1, double Mdm2-FL and double 2×WAF1 oligonucleotides. The p53Δ30 protein was expressed in insect cells, and the binding of the purified p53Δ30 protein to these oligonucleotides was tested (Fig. 3). No significant differences were seen between the binding activity of p53Δ30 to WAF1-BS and 2×WAF1 oligonucleotides (Fig. 3, lanes 7 and 10). Competition between radiolabeled and unlabeled WAF1-BS and 2×WAF1 oligonucleotides showed no difference in affinity of p53Δ30 to any of these oligonucleotides: both the unlabeled single and double WAF1 oligonucleotides could compete with equal efficiency for binding to radiolabeled WAF1-BS and 2×WAF1 (Fig. 3, lanes 8, 9 and 11, 12). This suggests that the double p53 binding site configuration can fully substitute for the C-terminal activation of p53 protein, at least when using the WAF1 binding site. However, the single sites Mdm2-1 and Mdm2-2, each representing half of the full-length Mdm2 binding site as indicated in Figure 1, were not able to fully compete for binding to the double Mdm2 binding site (Fig. 3, lanes 3–6). This could be due to a lower intrinsic affinity of the p53 protein for the single Mdm2, as opposed to the single WAF1 site. The relative affinities of full-length p53 protein for the single or double Mdm2 DNA binding sites were compared, as shown in Figures 4 and 5. As expected from the results in Figure 2, the p53 protein bound to the naturally occurring double Mdm2 site in the absence of PAb421 antibody (Fig. 4, lane 7) and was supershifted in its presence (Fig. 4, lane 9). The p53 protein failed to bind monomers of the Mdm2 binding site (Fig. 4, lanes 1 and 4) unless activated by PAb421 (Fig. 4, lanes 2 and 5). Competition experiments, in which increasing concentrations of cold Mdm2-1, Mdm2-2 and Mdm2-FL displaced bound radiolabeled Mdm2-FL, confirmed that p53 protein has a higher affinity for Mdm2-FL than for Mdm2-1 or Mdm2-2 (Fig. 5A). The affinity for Mdm2-2 was higher than for Mdm2-1, since at the highest concentration (5000 nM), Mdm2-2 could compete for ∼60% of the labeled Mdm2-FL oligonucleotide, while at the same concentration, Mdm2-1 still was unable to compete for Mdm2-FL binding to any degree. Because the results shown in Figure 4 suggested that p53 protein may have still higher affinity for Mdm2-FL in the presence of PAb421 than in its absence (Fig. 4, compare lanes 7 and 9), the same competition studies were carried out in the presence of PAb421 (Fig. 5B). As expected, in the presence of PAb421 the affinity of p53 protein for the radioactively labeled Mdm2-FL and each of the unlabeled competitors were similar, but with Mdm2-1 still the least effective competitor. Because there was an indication in Figure 4 (lane 9 compared to lane 7) that while not required, PAb421 could still increase the binding to the Mdm2-FL, affinity constants (Kd) were measured. The Kd of p53 protein binding to Mdm2-FL was 21.28 ± 1.19 nM in the absence of PAb421 and 3.12 ± 0.62 nM in the presence of PAb421. Thus, the double binding site in the Mdm2 promoter alleviates the requirement for C-terminal activation, but C-terminal activation may still be required for full DNA binding activity of the p53 protein.
Figure 3.
Binding activity of human C-terminally deleted p53Δ30 protein to the binding sites from Mdm2 and WAF1 genes. Recombinant p53Δ30 was produced and purified from insect cells, and binding activity was analyzed by EMSA, as described in the text. The amount of p53Δ30 protein added to each reaction is indicated. The location of the p53Δ30-radiolabeled oligonucleotide complexes is indicated by arrows. Where indicated, a 100-fold molar excess of unlabeled oligonucleotide Mdm2-FL (F), Mdm2-1 (1), Mdm2-2 (2), WAF1-BS (BS) and 2×WAF1 (2×) was added.
Figure 4.
Relative binding of human wild-type p53 protein to the naturally occurring double Mdm2 binding site (Mdm2-FL) or each single site (Mdm2-1 or Mdm2-2). Binding was assayed by EMSA using 100 ng of recombinant human wild-type p53 protein produced in insect cells, as described in the text. Monoclonal antibody PAb421 (200 ng) was included in the reaction where indicated, as well as a 100-fold molar excess of unlabeled oligonucleotides Mdm2-1 (M1), Mdm2-2 (M2) or Mdm2-FL (FL). The locations of p53–oligonucleotide complexes and PAb421–p53–oligonucleotide complexes are indicated by arrows.
Figure 5.
Effect of competitor sequences from the Mdm2 P2 promoter on binding of p53 to [32P]Mdm2-FL. Binding was analyzed in the absence (A) or presence (B) of monoclonal antibody PAb421. In all cases, 100 ng of purified p53 protein was incubated with 2.5 nM [32P]Mdm2-FL for 30 min at 4°C, and DNA–protein complexes detected by EMSA. Binding to the radioactively labeled double-stranded oligonucleotide was competed with cold Mdm2-1, Mdm2-2 and Mdm2-FL oligonucleotides, at the indicated concentrations. The percentage of bound 32P-Mdm2-FL, compared to the control without cold oligonucleotide, was determined by phosphorimaging. In a single experiment each reaction was done in duplicate, and the results shown are the mean of three independent experiments. Error bars indicate the standard deviation of the mean.
DISCUSSION
Taken together, our results indicate that the p53 protein has constitutive DNA binding activity for the tandem sites present in the Mdm2 gene promoter, and the requirement for p53 C-terminal activation depends on the particular DNA binding sequence. These results provide a potential mechanism for differential regulation of p53 target genes after genotoxic stress. This is particularly relevant for p53 steady-state regulation and for response to DNA damage, considering the major role of Mdm2 protein in regulating p53 protein stability. The Mdm2 protein binds to the p53 N-terminal domain and promotes p53 protein ubiquitination and proteolytic degradation. The Mdm2 protein is also responsible for the low levels of p53 protein found in normal cells in the absence of genotoxic stimuli. DNA damage induces p53 phosphorylation at the N-terminus, diminishing the interaction with Mdm2 protein and increasing p53 protein stability (13). The proposed regulatory feedback loop (4), through induction of Mdm2 gene transcription by activated p53, is a critical mechanism to maintain low levels of p53 protein in the absence of a genotoxic stimulus and to control the extent of p53 protein induction after DNA damage. That the double nature of the Mdm2 gene p53 binding site alleviates the requirement of p53 C-terminal activation for effective DNA binding is likely to contribute to the efficiency of the Mdm2–p53 self-regulatory loop. Because no other activation events are required to induce Mdm2, p53 protein would be maintained at low steady state levels by Mdm2 protein. After genotoxic damage, p53 is post-translationally modified in the C- and N-termini (10). The N-terminal post-translational modifications inhibit Mdm2 binding, resulting in accumulation of p53 proteins activated for inducing WAF1 and other p53 downstream genes. That p53 binding activity to the Mdm2 promoter still increases after C-terminal activation may contribute to the efficiency of the Mdm2–p53 regulatory loop after genotoxic damage.
Protein interactions between p53 molecules binding to individual consensus sequences within the double site are probably responsible for the increased affinity towards double sites. The p53 protein contains an oligomerization domain in the C-terminal third of the molecule that mediates the formation of p53 tetramers (14). Further stabilization can occur through stacking of these tetramers into higher order aggregates through the hydrophobic central domain (15). Cooperative binding to double consensus sites would increase the affinity of p53 for its DNA binding sequence without the need for C-terminal activation. To our knowledge, the Mdm2 gene is the only p53 responsive gene described with a double p53 binding sequence capable of binding by latent p53. The current results suggest a mechanism whereby transcription of Mdm2 in cells could be differentially regulated compared to other p53 regulated genes. However, they do not rule out the possibility that additional modifications of the p53 protein can further modulate DNA binding specificity for other p53 responsive gene promoters.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Danny Reinberg and Dr Ron Drapkin for supplying the plasmids used to construct the p53-dependent transcription templates and Dr Enrico Mihich for helpful discussions. We also thank Jennifer Carlson and Michelle Bryant for assistance with preparation of this manuscript. This work was supported by the Taisho Pharmaceutical Co., Ltd, RPCI Institute Core grant CA16056 and Oregon Cancer Center grant P30CA69533.
References
- 1.Greenblatt M.S., Bennett,W.P., Hollstein,M. and Harris,C.C. (1994) Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res., 54, 4855–4878. [PubMed] [Google Scholar]
- 2.Fuchs S.Y., Adler,V., Buschmann,T., Wu,X. and Ronai,Z. (1998) Mdm2 association with p53 targets its ubiquitination. Oncogene, 17, 2543–2547. [DOI] [PubMed] [Google Scholar]
- 3.Honda R. and Yasuda,H. (1999) Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J., 18, 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X.W., Bayle,J.H., Olson,D. and Levine,A.J. (1993) The p53–mdm-2 autoregulatory feedback loop. Genes Dev., 7, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 5.El-Deiry W.S., Kern,S.E., Pietenpol,J.A., Kinzler,K.W. and Vogelstein,B. (1992) Definition of a consensus binding site for p53. Nat. Genet., 1, 45–49. [DOI] [PubMed] [Google Scholar]
- 6.Hupp T.R., Meek,D.W., Midgley,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y., Liu,Y., Lee,L., Miner,Z. and Kulesz-Martin,M. (1994) Wild-type alternatively spliced p53: binding to DNA and interaction with the major p53 protein in vitro and in cells. EMBO J., 13, 4823–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hupp T.R., Sparks,A. and Lane,D.P. (1995) Regulation of the specific DNA binding function of p53. Cell, 83, 237–245. [DOI] [PubMed] [Google Scholar]
- 9.Funk W.D., Pak,D.T., Karas,R.H., Wright,W.E. and Shay,J.W. (1992) A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol., 12, 2866–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giaccia A.J. and Kastan,M.B. (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev., 12, 2973–2983. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry W.S., Tokino,T., Velculescu,V.E., Levy,D.B., Parsons,R., Trent,J.M., Lin,D., Mercer,W.E., Kinzler,K.W. and Vogelstein,B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell, 75, 817–825. [DOI] [PubMed] [Google Scholar]
- 12.Albor A., Kaku,S. and Kulesz-Martin,M.F. (1998) Wild type and mutant forms of p53 activate human topoisomerase I: a possible mechanism for the gain of function in mutants. Cancer Res., 58, 2091–2094. [PubMed] [Google Scholar]
- 13.Unger T., Juven-Gershon,T., Moallem,E., Berger,M., Sionov,R.V., Lozano,G., Oren,M. and Haupt,Y. (1999) Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J., 18, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P., Reed,M., Wang,Y., Mayr,G., Stenger,J.E., Anderson,M.E., Schwedes,J.F. and Tegtmeyer,P. (1994) p53 domains: structure, oligomerization, and transformation. Mol. Cell. Biol., 14, 5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenger J.E., Tegtmeyer,P., Mayr,G.A., Reed,M., Wang,Y., Wang,P., Hough,P.V. and Mastrangelo,I.A. (1994) p53 oligomerization and DNA looping are linked with transcriptional activation. EMBO J., 13, 6011–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]