Abstract
Many gene families in higher plants have expanded in number, giving rise to diverse protein paralogs with specialized biochemical functions. For instance, plant general transcription factors such as TFIIB have expanded in number and in some cases perform specialized transcriptional functions in the plant cell. To date, no comprehensive genome-wide identification of the TFIIB gene family has been conducted in the plant kingdom. To determine the extent of TFIIB expansion in plants, I used the remote homology program HHPred to search for TFIIB homologs in the plant kingdom and performed a comprehensive analysis of eukaryotic TFIIB gene families. I discovered that higher plants encode more than 10 different TFIIB-like proteins. In particular, Arabidopsis thaliana encodes 14 different TFIIB-like proteins and predicted domain architectures of the newly identified TFIIB-like proteins revealed that they have unique modular domain structures that are divergent in sequence and size. Phylogenetic analysis of selected eukaryotic organisms showed that most life forms encode three major TFIIB subfamilies that include TFIIB, Brf, Rrn7/TAF1B/MEE12 subfamilies, while all plants and some algae species encode one or two additional TFIIB-related protein subfamilies. A subset of A. thaliana GTFs have also expanded in number, indicating that GTF diversification and expansion is a general phenomenon in higher plants. Together, these findings were used to generate a model for the evolutionary history of TFIIB-like proteins in eukaryotes.
Keywords: Transcription, Gene family, Phylogenetics, Arabidopsis thaliana, Evolution
1. Introduction
All eukaryotic life forms contain at least three types of multi-subunit nuclear RNA polymerases (Pols) I, II, and III that transcribe DNA into different forms of RNA1-3. For instance, Pol I synthesizes most ribosomal RNAs (rRNAs), Pol II synthesizes mRNAs, small nuclear RNAs (snRNAs), and microRNAs, and Pol III synthesizes transfer RNAs (tRNAs), the 5S rRNA and other snRNAs4-11. Eukaryotic transcription initiation by Pol I, II, and III each require the action of several general transcription factors (GTFs) for accurate gene transcription2,3. The well-studied Pol II transcription system utilizes up to seven different GTFs that include TATA binding protein (TBP), TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH 7,12. The GTFs work together to help Pol II find the promoter, open doublestranded DNA, select a start site, initiate RNA synthesis, and facilitate Pol II clearance from the promoter 7,12. Like the Pol II-specific GTFs, Pol I and III utilize paralogous GTFs to initiate transcription2,3,13,14.
TFIIB and TBP are two of the best-studied GTFs among the eukaryotic and archeal transcription initiation factors. TFIIB-like factors are universally required for eukaryotic and archael transcription initiation 2,14-18. For example, archaea and eukaryotic Pol I and III require paralogous TFIIB-like factors called TFB, Rrn7/TAF1B/MEE12, and TFIIB-related factor 1 (Brf1), respectively 3,17. In the Pol II preinitiation complex, TFIIB simultaneously interacts with Rpb1 Dock and Rpb2 Wall domains of Pol II, TBP, and DNA upstream and downstream of TBP 7,12,19-22. TFIIB also plays roles at several key steps during the transcription cycle that include transcription start site selection (TSS), promoter opening, abortive initiation, promoter clearance, and termination 20,23-30. TBP is also universally required for transcription initiation by archaeal Pol and each of the three major eukaryotic Pols 2,3,14,31-33. TBP binds specifically to TATA or TATA-like DNA sequences upstream of the transcription start site of promoter DNA, and induces a sharp 90° bend in DNA7,33,34.
Plants are unique because they contain two additional multi-subunit nuclear Pols (IV-V) that perform specialized and distinct transcriptional functions. For instance, Pol IV is required for small interfering RNA (siRNA) biogenesis whereas Pol V is required for siRNA targeting and RNA-directed DNA methylation 35-45. Current evolutionary models suggest that Pol IV and V evolved from Pol II due to their sequence homology, shared subunit composition, and overlapping functions 46-49. Besides Pols, plants also contain multiple copies of TBP and TFIIB family proteins. For example, Arabidopsis thaliana encodes two distinct forms of TBP50 and up to eight different TFIIB-like proteins that consist of two TFIIBs, three TFIIB-related factors (Brf), one Rrn7/TAF1B-like protein, and two plant specific TFIIB-related proteins (Brp)17,51-55.
Gene family expansion can result in proteins with redundant, specialized, and diversified functions. To determine the extent of TFIIB expansion and their emergence in the plant kingdom, I preformed a simple computational approach using the remote homology detection search program HHpred56. I examined various eukaryotic genomes including several plant, mosses, algae, fungi, and metazoan genomes to identify TFIIB homologs, determine their phylogenetic relationships, and compare structural homology with their well-characterized yeast and mammalian counterparts. The present study identified a new TFIIB-like subfamily and examined the evolutionary history of TFIIB family proteins in the eukaryotic kingdom. The A. thaliana genome was also searched for GTF homologs for each of the three eukaryotic Pols, showing that most higher plant GTF gene families have also expanded in number.
2. Materials and Method
2.1. HHpred protein similarity search
Sequence and structure similarity searches were performed by HHpred (http://toolkit.tuebingen.mpg.de/hhpred) to search against the A. thaliana genome database of Hidden Markov Models (HMM) under default settings and thresholds. The A. thaliana genome database used for this study only contains annotated protein coding genes, while annotated pseudogenes are not included in the database.
2.2. TFB protein mining from complete genome sequences
A. thaliana TFB protein sequences listed in Table S1 were used as the comment for PSI-BLAST and TBLASTN against selected genomes using the National Center for Biotechnology and Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST). It was expected that all TFB-like proteins would contain an N-terminal zinc ribbon, a variable linker segment, and a cyclin fold domain. A total of 20 different eukaryotic genomes listed in Table 1 were searched for these protein sequence signatures. The E-value threshold was increased to 10 for most searches to increase search sensitivity. Reiterative PSI-BLAST searches were preformed until no new protein matches were detected. When searching for Rrn7/TAF1B/MEE12 homologs, threshold settings were relaxed to an E-value threshold of 100, since these proteins exhibit significant sequence divergence between species. Potential TFB homologs were further filtered by PHI-BLAST or screened manually for a zinc ribbon consensus sequence in the form of the following regular expression: (CxxC/Hx14-17CxxC/HG). From this pool, protein sequences were individually searched by HHpred against the A. thaliana genome database as above. At total of 101 protein sequences were identified and their protein sequences are listed in Table S1 Orthologous groups were identified on the basis of phylogenetic clustering, HHpred probability, and percent identity scores.
Table 1. Number of TFIIB related proteins in selected species.
| Species | Taxonomic group | TFIIB | Brf1 | Rrn7 | Brp1 | Brp2 | Brp3 | Brp4 | Brp5 | Brp6 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | Humans | 1 | 2 | 1 | - | - | - | - | - | - | 4 |
| Drosophita metanogaster | Fruit fly | 1 | 1 | 1 | - | - | - | - | - | - | 3 |
| Caenorhabditis elegans | Nematode | 1 | 1 | 1 | - | - | - | - | - | - | 3 |
| Monosiga brevicoHis | Simple metazoan | 1 | 1 | 1 | - | - | - | - | - | - | 3 |
| Dictyostelium discoideum | Slime Mold | 1 | 1 | 1 | - | - | - | - | - | - | 3 |
| Saccharomyces cerevisiae | Fungi | 1 | 1 | 1 | - | - | - | - | - | - | 3 |
| Phaeodactylum tricornuttim | Diatom | 2 | 1 | 1 | - | - | - | - | - | - | 4 |
| Ectocarpus siliculosus | Brown algae | 2 | 1 | 1 | - | - | - | - | - | - | 4 |
| Hemiselmis andersenii | Red algae, Nucleomorph | 1 | 1 | - | - | - | - | - | - | - | 2 |
| Gwllardta theta | Red algae. Nuclear | 1 | 1 | 1 | 1 | - | - | - | - | - | 4 |
| Guillardia theta | Red algae, Nucleomorph | 1 | 1 | - | - | - | - | - | - | - | 2 |
| Cyanidioschyzon merolae | Red algae | 1 | 1 | 1 | 1 | - | - | - | - | - | 4 |
| Ostreococcus tatiri | Green algae | 1 | 1 | 1 | - | - | - | - | 1 | - | 4 |
| Micromonas pussitla | Green algae | 1 | 1 | 1 | - | - | - | - | 1 | - | 4 |
| Volvox carteri | Green algae | 1 | 1 | 1 | - | - | - | - | 1 | - | 4 |
| Chlamydomonas reinhardtii | Green algae | 1 | 1 | 1 | - | - | - | - | 1 | - | 4 |
| Physcomitrelta patens | Bryophytes | 2 | 1 | 1 | - | - | - | - | 1 | - | 5 |
| Selaginelta moellendorffii | Lycophytes | 2 | 3 | 1 | 2 | - | - | - | 2 | - | 10 |
| Oryza sativa | Angiosperms, monocots | 3 | 1 | 1 | 1 | 1 | - | - | 1 | - | 8 |
| Populus trichocarpa | Angiosperms, dicots | 3 | 2 | 1 | 1 | 1 | - | - | 1 | - | 9 |
| Arabidopsis thaliana | Angiosperms, dicots | 2 | 4 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 14 |
2.3 Phylogenetic analysis and tree construction
Maximum likelihood phylogenetic trees of selected protein sequences were constructed with the PhyML 3.0 online program (http://www.phylogeny.fr)57 using advanced mode bootstrapping (100 bootstraps) and one of three substitution models (JTT, LG, and Dayhoff). Each matrix model generated trees with similar clustering. A total of 98 eukaryotic TFB protein sequences were aligned by their TFIIB homology domains (BHD). Multiple sequence alignments were combined from HHpred alignments and converted to FASTA format with minor manual manipulation to remove gaps in the comment protein sequence.
3. Results
3.1. The A. thaliana genome contains 14 different TFB-like genes
To search genome-wide for TFIIB-like (TFB) protein coding genes in the A. thaliana genome, I used HHpred to search various TFIIB family proteins against the A. thaliana genome database. Three different TFB proteins were used as queries that include AtMEE12 (Pol I), AtTFIIB1 (Pol II), and AtBrf1 (Pol III). Using AtMEE12 and AtTFIIB1 as queries each detected 11 matches (Fig.1A,B), while AtBrf1 detected 13 matches (Fig.1C). In total, 14 different A. thaliana proteins matched with high probability (>97%) to the three AtTFB family proteins. Among the 8 TFB-like proteins described in the literature, 6 additional TFB-like proteins were identified (Unk1-6). Four of the newly identified TFB-like proteins were assigned new numerical AtBrp names that include AtBrp3 (Unk3), AtBrp4 (Unk2), AtBrp5 (Unk4), and AtBrp6 (Unk5). The remaining two new AtTFB proteins were homologous to either the AtMEE12 or AtBrf1 C-terminal domain (CTD) and assigned names that include AtMEE12CTD (Unk1) and AtBrfCTD (Unk6).
Figure 1. HHpred identification of A. thaliana TFB proteins.
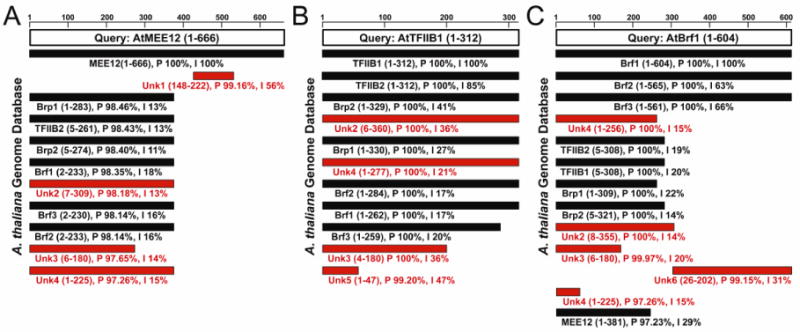
A. thaliana queries are AtMEE12 (A), AtTFIIB1 (B), and AtBrf1 (C) depicted as open boxes, and matches to the queries are shown as bars below. Black bars denote known TFB family members and red bars indicate newly identified TFB members. Below each bar is the name of the protein match and region of homology between the comment and match. The percent probability (P) and identity (I) are also indicated. The scale bar represents length of amino acids.
3.2. Predicted domain architecture of A. thaliana TFB-like proteins
Using the HHpred alignments and the known domain architectures of TFB family proteins as a guide, I assigned the domains of all 14 A. thaliana TFIIB family proteins (Fig.2A). TFIIB-like proteins contain three domains beginning with a zinc ribbon domain at the N-terminus followed by a linker domain and two cyclin fold repeats. Brf- and Rrn7/TAF1B/MEE12-like proteins consist of the same three TFIIB-like domains, but also contain an extended CTD following the cyclin fold repeats. Among the newly identified AtTFB proteins, four are missing one or more TFIIB-like domains. For instance, the domain architecture of AtBrp3 is similar to TFIIB by containing a zinc ribbon and linker domain, but only contains a single cyclin fold. Likewise, the domain architecture of AtBrp6 is missing the entire cyclin fold repeat domain and only consists of a zinc ribbon and linker domain.
Figure 2. Domain architecture and phylogenetic relationships of A. thaliana TFB family proteins.
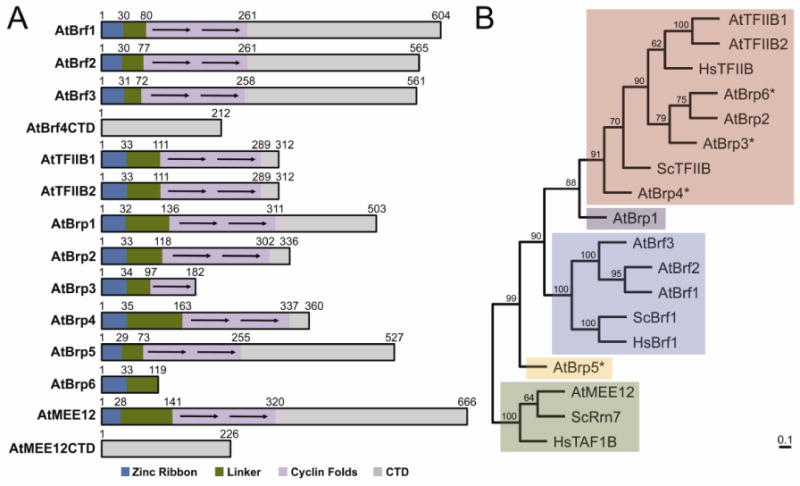
(A) Schematic representation of TFB family protein domain architecture. Each domain is represented by a different color. Arrows indicate individual cyclin fold repeats. (B) Evolutionary relationship of A. thaliana TFB-like proteins. The phylogenetic tree was created by comparing the TFIIB Homology Domain (BHD) domain that consists of the zinc ribbon, linker, and cyclin fold domains for each TFB-like protein. AtBrf4CTD and AtMEE12CTD were excluded from this analysis since neither proteins contain a BHD. Each clade is shaded a different color based on their closest evolutionary relationship. The numbers at each node represent the percentage of trees supporting the specific branching pattern in the bootstrap analysis. The scale bar represents the number of estimated changes per position for a unit of branch length.
The two remaining AtTFB proteins detected by HHpred contain familiar domain architectures. For example, AtBrp4 consists of a TFIIB-like domain architecture with the exception that the linker domain is 50 residues longer than compared to AtTFIIB1. AtBrp5 also contains a similar domain organization as AtBrfs and AtMEE12 where it contains a large CTD. However, the AtBrp5 CTD lacks any apparent sequence similarity with the AtBrf and AtMEE12 CTDs, indicating that it likely has a unique structure and function in transcription. The DNA sequences proximal to the truncated AtTFBs also lack any similarity TFIIB-like domains, ensuring the truncation of these genes is not the result of incorrect annotation and gene boundary assignment, but it is possible that these truncated AtTFB family proteins may represent uncharacterized pseudogenes. None of the AtTFB family protein identified above are listed in available A. thaliana pseudogene datasets58,59, but four pseudogenes associated with the AtBrf2, AtBrf3, AtMEE12, and AtMEE12CTD genes were previously identified and are listed in Table S2
3.3. Phylogenetic relationships of A. thaliana TFB-like proteins
To further compare and classify each AtTFB protein, I used phylogenetics to group the AtTFB family proteins based on their evolutionary relationships with respect to their human and yeast counterparts. An unrooted phylogenetic tree was constructed from a multiple HHpred sequence alignments of AtTFB protein family members, encompassing the zinc ribbon, linker, and cyclin fold domains. AtBrp2 and AtBrp3 cluster together with TFIIB orthologs, while AtBrp4, AtBrp1, and AtBrp6 cluster with the TFIIB clade, but are progressively more divergent (Fig.2B). AtBrfs and AtMEE12 each group well with their human and yeast counterparts. Notably, AtBrp5 is very divergent and does not cluster well with any of the three major TFB clades, indicating that it may represent a new and distinct TFB protein clade in plants.
3.4. Sequence conservation of A. thaliana TFB-like protein domains
To examine the protein sequence conservation of AtBrp domains, I determined the protein sequence identity between AtBrps protein domains relative to three reference AtTFB family proteins that include AtTFIIB1, AtBrf1, and AtMEE12. In general, the AtBrp zinc ribbon domains are more similar to AtTFIIB with the exception of the zinc ribbon domains of AtBrp1 and AtBrp5 (Fig.3A,D), possibly suggesting these zinc ribbons may contact Pol II given their similarity. The AtBrp1 and AtBrp5 zinc ribbon domains lack significant sequence identity to any of the reference proteins, indicating they may contact unique Pol domains.
Figure 3. Protein identity comparisons of A. thaliana TFB family protein domains.
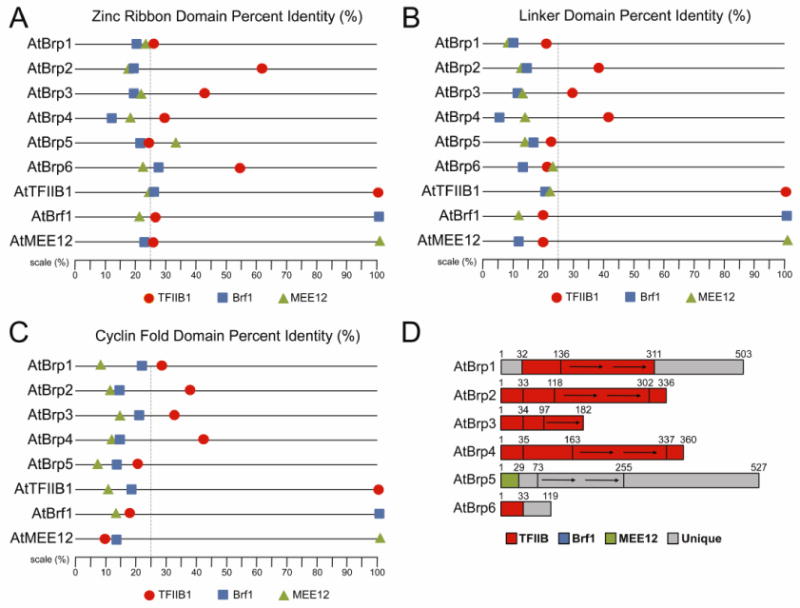
Shown for each protein is the percent identity of the zinc ribbon (A), linker (B), and cyclin folds (C) for the list proteins relative to the three major A.thaliana TFB representatives that include AtTFIIB1 (red circle), AtBrf1 (blue square), and AtMEE12 (green triangle). For each protein, the percent identity is plotted on the horizontal line following the protein name. The gray dashed line indicates the 25% threshold. (D) Schematic of the AtBrp family proteins. Domains are colored according to the AtTFB representative they share highest sequence identity above the 25% threshold.
The AtTFB linker and cyclin fold domains were also compared (Fig.3B,C,D). AtBrp1-4 each contain linker and cyclin domains most similar to TFIIB, possibly indicating they preform TFIIB-like functions in Pol II and TBP binding. The linker domain of AtBrp5 and AtBrp6, and the AtBrp5 cyclin domains lack significant sequence similarity to any of the three AtTFB reference proteins. The TFIIB linker domain or connecting region contains a segment called the B-reader that immediate follows the zinc ribbon domain20. The B-reader contains a conserved helical sequence patch (EWRTF) that is positioned near the TSS that reads the template to find a suitable start site20,60. Among the AtBrp proteins, the conserved TFIIB B-reader motif is found only in AtBrp2, while all the remaining AtBrps lack this motif, possibly indicating they recognize unique TSSs.
3.5. Expression profiles of A.thaliana TFB family genes
The expression patterns of all the A.thaliana TFB proteins were also investigated using the AtGenExpress Tileviz developmental dataset61 to determine whether the TFB genes are transcribed during plant development. The AtTFB genes were clustered into four distinct groups based on the their expression profiles and compared with representative Pol I-V genes of the two largest subunits (Fig.S1). All the AtTFB genes were widely expressed in the different plants tissues, indicating that each of these genes are transcribed in the plant cell and possibly translated into proteins. Notably, the AtBrp5 and AtTFIIB1 expression profiles clustered well together, indicating that AtBrp5 may perform a general transcriptional role like AtTFIIB1 in all tissues and developmental stages.
3.6. Comparative analysis of eukaryotic TFB-like proteins
To identify TFB gene family protein members in other eukaryotic genomes, I used reiterative PSI-BLAST to search various AtTFB protein sequence against selected eukaryotic genome databases. PHI-Blast was also performed on various proteins containing zinc ribbon and/or cyclin domains using a simple AtTFB zinc ribbon regular expression. Potential TFB-like protein matches were independently searched by HHpred against the A. thaliana genome database to determine if they were homologous to any of the AtTFB family proteins. Each TFB-like proteins identified among the various eukaryotic genome contained zinc ribbon and cyclin domain sequence signatures. Names for each of the TFB proteins were assigned based on the HHpred and phylogenetic analysis. Using this approach, a total of 19 different eukaryotic genomes were analyzed that include several metazoan species, fungi, algae, and plant species. In general, I expected to find that each eukaryotic genome contained at least one TFIIB-, Brf-, and Rrn7/TAF1B/MEE12-like protein. The total number of TFB-like proteins identified is summarized in Table 1. Among the 101 different TFB-like proteins detected, three or more TFB-like proteins were found in each of the genomes searched with the exception of the nucleomorph genomes of the red algae Cryptophytes Hemi andersenii and Guillardia theta. Only two TFB-like proteins orthologous to TFIIB and Brf1 were detected in the nucleomorph genomes, and lacked an Rrn7/TAF1B/MEE12-like protein. Given the compact nature of nucleomorph genomes in red algae and their unique biology, the Rrn7/TAF1B/MEE12-like gene may have been lost62-64.
To examine the phylogenetic relationship among the eukaryotic TFB protein family members, I generated a phylogenetic tree from TFB protein sequence alignments using the Jones-Taylor-Thornton (JTT) amino acid replacement method65,66 The phylogenetic tree divided the TFB proteins into five distinct subfamilies where each member of the same subfamily clustered together (Fig.4). In addition to the three expected clades that include TFIIB, Rrn7/TAF1B/MEE12, and Brf, two additional clades were formed that represent the Brp1-like and Brp5-like proteins only found in plant and algae species. Alternate matrices such as LG67 and Dayhoff 68 resulted in genetic trees with similar branching and overall topologies (Fig.S2).
Figure 4. Phylogenetic analysis of TFB protein families in selected eukaryotic species.
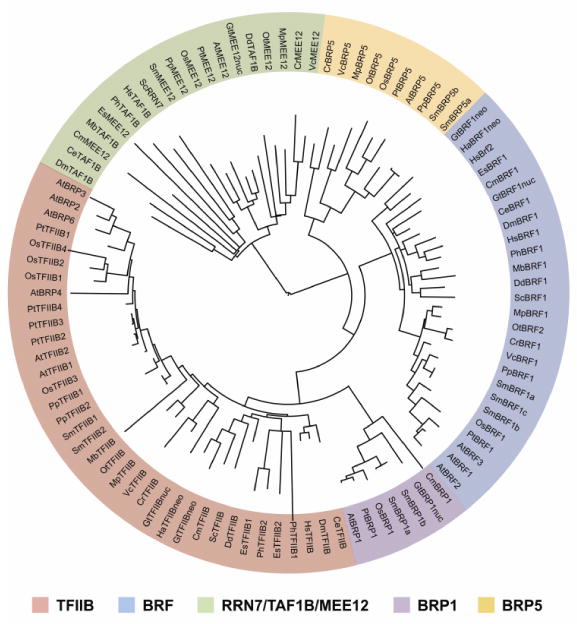
The phylogenetic tree was created by comparing TFIIB homology domains that consist of the zinc ribbon, linker, and cyclin fold domains in each of the selected eukaryotic organisms. Saccharomyces cerevisiae (Sc), Monosiga brevicollis (Mb), Dictyostelium discoideum (Dd), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Phaeodactylum tricornutum (Ph), Ectocarpus siliculosus (Es), Hemiselmis andersenii (Ha), Guillardia theta (Gt), Cyanidioschyzon merolae (Cm), Ostreococcus tauri (Ot), Volvox carteri (Vc), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Populus trichocarpa (Pt), Arabidopsis thaliana (At). Each major TFB protein subfamily is shaded by a unique color. The TFB subfamilies include TFIIB, Brf, Rrn7/TAF1B/MEE12, Brp1, and Brp5. Nuclear genome, nuc; Nucleomorph genome, neo. The tree was visualized with iTOL80,81.
A model that describes a hypothetical evolutionary history of the five major TFB protein subfamilies is depicted in Figure 5. Both the Brp1 and Brp5 subfamilies are specific to algae and plants, but Brp1 subfamily is not found in any of the green algae genomes examined in this study. The Brp1 subfamily first emerges in red algae, but then is missing in green algae and Bryophyte mosses. It is not clear why members of the Brp1 subfamily would be missing in these species, but the Brp1 subfamily clearly reemerges in the genomes of Lycophyte mosses and higher plants. The Brp5 subfamily has a more simple evolutionary history were it first emerges in green algae and is continuously found in mosses and higher plant species. Finally, there is also a clear expansion in the number of TFB proteins beginning with lycophtyes and continuing with the higher plant species.
Figure 5. Model for the evolutionary history of the TFB protein family in eukaryotes.
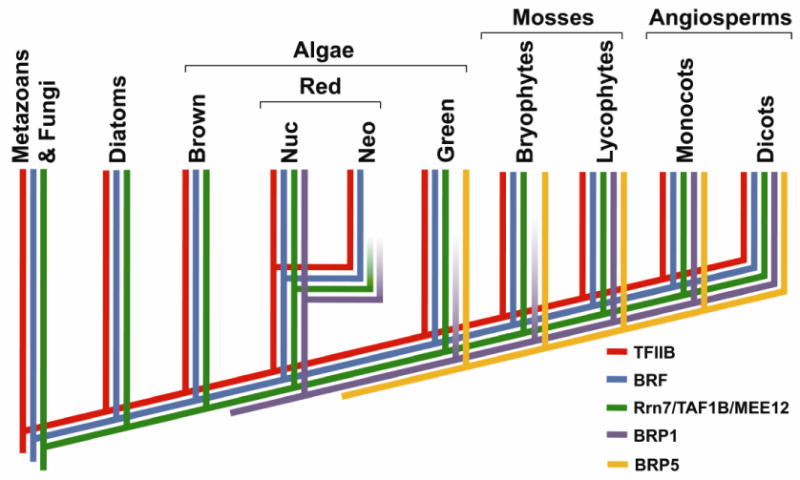
The hypothetical phylogenetic tree depicting the presence or absence of TFB protein family genes in a range of green plants, algae, diatoms, fungi and metazoan species. The five major TFB protein families are depicted by a colored line. The taxonomic names for each group are indicated above each branch.
3.7. Expansion of A. thaliana GTF genes
To determine the extent of GTF expansion in plants, I searched the A. thaliana genome for other plant GTF homologs. Using a similar strategy as described for AtTFBs, I used HHpred to search 23 representative GTF family proteins among the three Pols against the A. thaliana genome database. The search results are summarized in Table 2. For most plant GTFs, multiple copies were detected, indicating that GTF expansion is system-wide and possibly pair up in unique combinations to control transcription. For a few GTFs, it was difficult to identify the most direct ortholog since the proteins contained common repeated motifs that match with high probability to numerous proteins that are clearly not transcription factors. Besides these cases, multiple copies of GTFs were detected for more than half of the GTFs analyzed, clearly indicating GTF expansion is a general feature of A. thaliana and higher plants.
Table 2. Mumber of A.thafiana GTF family proteins.
| GTF Family | Subunit | Pol | Total |
|---|---|---|---|
| TBP | - | l,II,III | 2 |
|
| |||
| Rrn3 | - | I | 3 |
| CF | Rrn11 | I | N.A. |
| Rrn7 | I | 2 | |
| Rrn6 | I | N.A. | |
| TFIF | A34.5 | I | 1 |
| A49 | I | 1 | |
|
| |||
| TFIIA | Toa1 | II | 3 |
| Toa2 | II | 1 | |
| TFIIB | - | II | 5 |
| TFIIF | Tfg1 | II | 1 |
| Tfg2 | II | 2 | |
| TFIIE | Tfa1 | II | 3 |
| Tfa2 | II | 2 | |
|
| |||
| TFIIIB | Brf1 | III | 4 |
| Bdp1 | III | 1 | |
| TFIIIF | C37 | III | 1 |
| C53 | III | 2 | |
| TFIIIE | C34 | III | 1 |
| C82 | III | 1 | |
| C31 | III | 2 | |
| TFIIIC | Tfc1 | III | 2 |
| Tfc3 | III | 2 | |
4. Discussion
TFB family proteins play integral and fundamental roles during the transcription cycle. In the present study, the A. thaliana genome was comprehensively searched for new members of the TFB protein family, resulting in the discovery of a new Brp5 subfamily. The Brp5 subfamily is particularly interesting given its sequence divergence from other TFB family proteins. The emergence of the Brp5 subfamily also correlates well with the emergence of Pol IV in green algae and land plant species46,48,49. Since the Pol IV-V GTF requirements remain unclear, an attractive although speculative model is that Brp5 may act as a Pol IV-V GTF. It seems plausible that Pol IV-V would require their own specific TFIIB-like factor as shown for archeal and eukaryotic Pols I-III. Clearly future genetic and biochemical studies will be necessary to determine the transcriptional role of the Brp5 subfamily and its potential protein interaction partners.
The nucleomorph genomes of the unicellular cryptophyte algae species apparently lack an Rrn7/TAF1B/MEE12-like gene. Nucleomorph genomes consist of three small and compact chromosomes that encode significantly fewer genes than their larger nuclear counterparts in the same red algae cell62,69-71. Likewise, the nucleomorph rDNA locus is also reduced in size, consisting of a single rDNA cistron at the ends of each chromosome, whereas most eukaryotes contain multiple rDNA copies69,70,72. If nucleomorph genomes lost an Rrn7/TAF1B/MEE12-like factor they may have also lost some or all of the genes encoding the Pol I transcription machinery. Available genome analysis suggests otherwise, since genes encoding the Pol I subunits are found in the H.andersenii and G.theta genomes62,73,74. Assuming nucleomorph rRNA synthesis is essential, it remains unclear how Pol I would initiate rDNA transcription without a Pol I-specific TFB-like protein. An intriguing model is that one of the two remaining TFB subfamily members could potentially take over this role. Another possibility is that that the nuclear Rrn7/TAF1B/MEE12-like factor may be imported into the nucleomorph nucleus, as suggested for the nucleopore complex genes encoded only in the nuclear genome64,70. More detailed molecular and biochemical studies may shed light on these intriguing possibilities.
Red algae and some plants encode two evolutionary distinct Pol I TFBs, while most other eukaryotic cells encode a single Pol I GTF most similar to the Rrn7/TAF1B/MEE12 subfamily. In A. thaliana cells, gene expression patterns of AtMEE12 and AtBRP1 indicate that they are widely expressed in plant cells and tissues, as expected for general Pol I factors, but protein localization studies confound their proposed roles in rDNA transcription. For instance, AtBRP1 protein localizes to the cytosolic face of the outer plastid envelope of wild-type plant cells rather than the expected nucleolar localization for a Pol I factor as shown for CmBrp152,54. AtBrp1 shuttles to the nucleus upon treatment with proteosome inhibitors54 and possibly upon Agrobacterium tumefaciens75 infection through it interaction with viral effector protein VirE3, suggesting that the Pol I activity of the BRP1 subfamily transcriptional functions has become more specialized in higher plants. Together, it seems that both AtBRP1 and AtMEE12 have unique functions in Pol I transcription, and future studies will be necessary to ascertain their precise role in Pol I transcription.
A.thaliana encodes an expansive repertoire of GTFs, but the roles of these expanded GTF family members remain unclear. Available evidence from the few A.thaliana TFB proteins that have been genetically and biochemically characterized, suggest that they perform specialized functions in the transcription process. Archaea are a clear biological example of GTF diversification and specialization, where species such as Halobacterium NRC-1 encodes six TBP-like proteins and seven TFIIB-like proteins that pair up in different combinations to coordinate transcription76-79. A similar phenomenon is possible for these different GTF homologs in A.thaliana. Further exploration of their potential roles may uncover unique properties of the GTFs and Pols in plant and eukaryotic transcription.
Supplementary Material
Supplementary Figure 1. Developmental expression profiles for A. thaliana TFB genes. Expression profiles of all AtTFB genes and the two largest subunits of all five plant Pols indicated on the right were compared in various developmental stages and plant organs indicated along the top of the each column. Expression values were complied from the A. thaliana Tiling array express (AtTAX) database61. The expression data was visualized and analyzed using Multiple experiment viewer (MeV)82,83. Hierarchical clustering was also used to group the expression profiles and is indicated on the left84. The color scale representing log signal values is shown at the bottom.
Supplementary Figure 2. Additional phylogenetic analysis of TFB protein families in selected eukaryotic species. The phylogenetic tree was created by comparing TFIIB homology domains that consist of the zinc ribbon, linker, and cyclin fold domains in each of the selected eukaryotic organisms. Saccharomyces cerevisiae (Sc), Monosiga brevicollis (Mb), Dictyostelium discoideum (Dd), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Phaeodactylum tricornutum (Ph), Ectocarpus siliculosus (Es), Hemiselmis andersenii (Ha), Guillardia theta (Gt), Cyanidioschyzon merolae (Cm), Ostreococcus tauri (Ot), Volvox carteri (Vc), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Populus trichocarpa (Pt), Arabidopsis thaliana (At). LG (A) and Dayhoff (B) substitution matrices were used to generate the phylogenetic trees. Each major TFB protein subfamily is shaded by a unique color. The TFB subfamilies include TFIIB, Brf, Rrn7/TAF1B/MEE12, Brp1, and Brp5. Nuclear genome, nuc; Nucleomorph genome, neo. The tree was visualized with iTOL80,81.
Highlights.
Most life forms encode three TFIIB gene subfamilies
Plants encode two additional TFIIB subfamilies
Most higher plants encode 10 or more TFIIB-like factors
Arabidopsis thaliana encodes 14 different TFIIB-like proteins
GTF expansion is a general phenomenon in plants
Acknowledgments
I would like to thank members of the Hahn lab at the FHCRC and Ted Young at University of Washington for their helpful discussions and encouragement. I also thank Mia Levine of the Malik lab at the FHCRC for critical reading of the manuscript and insightful comments. BAK was supported by grant R01GM075114 to Steven Hahn.
Abbreviations
- Pol
RNA polymerase
- rRNA
ribosomal RNA
- snRNA
small nuclear RNA
- tRNA
transfer RNA
- GTF
general transcription factor
- TBP
TATA-binding protein
- Brf1
TFIIB-related factor 1
- siRNA
small interfering RNA
- BRP
plant specific TFIIB-related protein
- TFB
TFIIB-like protein
- CTD
C-terminal domain
- At
Arabidopsis thaliana
- JTT
Jones-Taylor-Thornton
- TSS
Transcription Start Site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Critical reviews in biochemistry and molecular biology. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Molecular cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nature reviews Microbiology. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 4.Acker J, Conesa C, Lefebvre O. Yeast RNA polymerase III transcription factors and effectors. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annual review of pharmacology and toxicology. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 6.Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochemical Society transactions. 2008;36:590–594. doi: 10.1042/bst0360590. [DOI] [PubMed] [Google Scholar]
- 7.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung KH, Stumph WE. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc) Critical reviews in biochemistry and molecular biology. 2011;46:11–26. doi: 10.3109/10409238.2010.518136. [DOI] [PubMed] [Google Scholar]
- 9.Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene. 2012;493:185–194. doi: 10.1016/j.gene.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Schanen BC, Li X. Transcriptional regulation of mammalian miRNA genes. Genomics. 2011;97:1–6. doi: 10.1016/j.ygeno.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider DA. RNA polymerase I activity is regulated at multiple steps in the transcription cycle: recent insights into factors that influence transcription elongation. Gene. 2012;493:176–184. doi: 10.1016/j.gene.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Bushnell DA, Kornberg RD. RNA polymerase II transcription: Structure and mechanism. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CC et al. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannini A. A structural perspective on RNA polymerase I and RNA polymerase III transcription machineries. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Blattner C et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes & development. 2011;25:2093–2105. doi: 10.1101/gad.17363311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science (New York, NY) 2011;333:1637–1640. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutson BA, Hahn S. TFIIB-related factors in RNA polymerase I transcription. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naidu S, Friedrich JK, Russell J, Zomerdijk JC. TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science (New York, NY) 2011;333:1640–1642. doi: 10.1126/science.1207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Kostrewa D et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science (New York, NY) 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science (New York, NY) 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 23.Grunberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nature structural & molecular biology. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mischo HE, Proudfoot NJ. Disengaging polymerase: Terminating RNA polymerase II transcription in budding yeast. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luse DS. Promoter clearance by RNA polymerase II. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan CD. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagrm.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goel S, Krishnamurthy S, Hampsey M. Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH. The Journal of biological chemistry. 2012;287:557–567. doi: 10.1074/jbc.M111.281576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiesler SC, Weinzierl RO. The linker domain of basal transcription factor TFIIB controls distinct recruitment and transcription stimulation functions. Nucleic acids research. 2011;39:464–474. doi: 10.1093/nar/gkq809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Roberts SG. New insights into the role of TFIIB in transcription initiation. Transcription. 2010;1:126–129. doi: 10.4161/trns.1.3.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Molecular cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 31.De Carlo S, Lin SC, Taatjes DJ, Hoenger A. Molecular basis of transcription initiation in Archaea. Transcription. 2010;1:103–111. doi: 10.4161/trns.1.2.13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiduschek EP, Ouhammouch M. Archaeal transcription and its regulators. Molecular microbiology. 2005;56:1397–1407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- 33.Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- 34.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wierzbicki AT et al. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes & development. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan EH, Blevins T, Ream TS, Pikaard CS. Functional consequences of subunit diversity in RNA polymerases II and V. Cell reports. 2012;1:208–214. doi: 10.1016/j.celrep.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nature reviews Molecular cell biology. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 38.Lahmy S, Bies-Etheve N, Lagrange T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics: official journal of the DNA Methylation Society. 2010;5:4–8. doi: 10.4161/epi.5.1.10435. [DOI] [PubMed] [Google Scholar]
- 39.Zheng B et al. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes & development. 2009;23:2850–2860. doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pontes O, Costa-Nunes P, Vithayathil P, Pikaard CS. RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Molecular plant. 2009;2:700–710. doi: 10.1093/mp/ssp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He XJ et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes & development. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PloS one. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends in plant science. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harbor symposia on quantitative biology. 2006;71:449–459. doi: 10.1101/sqb.2006.71.028. [DOI] [PubMed] [Google Scholar]
- 45.Onodera Y et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Tucker SL, Reece J, Ream TS, Pikaard CS. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genomewide and segmental gene duplications, retrotransposition, and lineagespecific subfunctionalization. Cold Spring Harbor symposia on quantitative biology. 2010;75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- 47.Ream TS et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Molecular cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo J, Yoshikawa N, Hodson MC, Hall BD. Duplication and paralog sorting of RPB2 and RPB1 genes in core eudicots. Molecular phylogenetics and evolution. 2007;44:850–862. doi: 10.1016/j.ympev.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Hall BD. A multistep process gave rise to RNA polymerase IV of land plants. Journal of molecular evolution. 2007;64:101–112. doi: 10.1007/s00239-006-0093-z. [DOI] [PubMed] [Google Scholar]
- 50.Li YF, Dubois F, Zhou DX. Ectopic expression of TATA box-binding protein induces shoot proliferation in Arabidopsis. FEBS letters. 2001;489:187–191. doi: 10.1016/s0014-5793(01)02101-9. [DOI] [PubMed] [Google Scholar]
- 51.Cavel E et al. A plant-specific transcription factor IIB-related protein, pBRP2, is involved in endosperm growth control. PloS one. 2011;6:e17216. doi: 10.1371/journal.pone.0017216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imamura S, Hanaoka M, Tanaka K. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. The EMBO journal. 2008;27:2317–2327. doi: 10.1038/emboj.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen YH et al. The central cell plays a critical role in pollen tube guidance in Arabidopsis. The Plant cell. 2007;19:3563–3577. doi: 10.1105/tpc.107.053967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lagrange T, et al. Transcription factor IIB (TFIIB)-related protein (pBrp), a plant-specific member of the TFIIB-related protein family. Molecular and cellular biology. 2003;23:3274–3286. doi: 10.1128/MCB.23.9.3274-3286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldwin DA, Gurley WB. Isolation and characterization of cDNAs encoding transcription factor IIB from Arabidopsis and soybean. The Plant journal: for cell and molecular biology. 1996;10:561–568. doi: 10.1046/j.1365-313x.1996.10030561.x. [DOI] [PubMed] [Google Scholar]
- 56.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dereeper A et al. Phylogeny.fr: robust phylogenetic analysis for the nonspecialist. Nucleic acids research. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karro JE et al. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic acids research. 2007;35:D55–60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou C et al. Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant physiology. 2009;151:3–15. doi: 10.1104/pp.109.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2012 doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 61.Laubinger S et al. At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome biology. 2008;9:R112. doi: 10.1186/gb-2008-9-7-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Douglas S et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 63.Gilson PR, McFadden GI. Jam packed genomes--a preliminary, comparative analysis of nucleomorphs. Genetica. 2002;115:13–28. doi: 10.1023/a:1016011812442. [DOI] [PubMed] [Google Scholar]
- 64.Neumann N, Jeffares DC, Poole AM. Outsourcing the nucleus: nuclear pore complex genes are no longer encoded in nucleomorph genomes. Evolutionary bioinformatics online. 2006;2:23–34. [PMC free article] [PubMed] [Google Scholar]
- 65.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences: CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 66.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 67.Le SQ, Gascuel O. Animproved general amino acid replacement matrix. Molecular biology and evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 68.Kosiol C, Goldman N. Different versions of the Dayhoff rate matrix. Molecular biology and evolution. 2005;22:193–199. doi: 10.1093/molbev/msi005. [DOI] [PubMed] [Google Scholar]
- 69.Moore CE, Archibald JM. Nucleomorph genomes. Annual review of genetics. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- 70.Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. BioEssays: news and reviews in molecular, cellular and developmental biology. 2007;29:392–402. doi: 10.1002/bies.20551. [DOI] [PubMed] [Google Scholar]
- 71.Cavalier-Smith T. Nucleomorphs: enslaved algal nuclei. Current opinion in microbiology. 2002;5:612–619. doi: 10.1016/s1369-5274(02)00373-9. [DOI] [PubMed] [Google Scholar]
- 72.Silver TD, Moore CE, Archibald JM. Nucleomorph ribosomal DNA and telomere dynamics in chlorarachniophyte algae. The Journal of eukaryotic microbiology. 2010;57:453–459. doi: 10.1111/j.1550-7408.2010.00511.x. [DOI] [PubMed] [Google Scholar]
- 73.Lane CE et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lane CE, Archibald JM. Novel nucleomorph genome architecture in the cryptomonad genus hemiselmis. The Journal of eukaryotic microbiology. 2006;53:515–521. doi: 10.1111/j.1550-7408.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Rodriguez FM, Schrammeijer B, Hooykaas PJ. The Agrobacterium VirE3 effector protein: a potential plant transcriptional activator. Nucleic acids research. 2006;34:6496–6504. doi: 10.1093/nar/gkl877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bleiholder A, Frommherz R, Teufel K, Pfeifer F. Expression of multiple tfb genes in different Halobacterium salinarum strains and interaction of TFB with transcriptional activator GvpE. Archives of microbiology. 2012;194:269–279. doi: 10.1007/s00203-011-0756-z. [DOI] [PubMed] [Google Scholar]
- 77.Turkarslan S et al. Niche adaptation by expansion and reprogramming of general transcription factors. Molecular systems biology. 2011;7:554. doi: 10.1038/msb.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coker JA, DasSarma S. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC genetics. 2007;8:61. doi: 10.1186/1471-2156-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baliga NS, et al. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Molecular microbiology. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 80.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics (Oxford, England) 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 81.Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic acids research. 2011;39:W475–478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saeed AI et al. TM4 microarray software suite. Methods in enzymology. 2006;411:134–193. doi: 10.1016/s0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 83.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 84.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Developmental expression profiles for A. thaliana TFB genes. Expression profiles of all AtTFB genes and the two largest subunits of all five plant Pols indicated on the right were compared in various developmental stages and plant organs indicated along the top of the each column. Expression values were complied from the A. thaliana Tiling array express (AtTAX) database61. The expression data was visualized and analyzed using Multiple experiment viewer (MeV)82,83. Hierarchical clustering was also used to group the expression profiles and is indicated on the left84. The color scale representing log signal values is shown at the bottom.
Supplementary Figure 2. Additional phylogenetic analysis of TFB protein families in selected eukaryotic species. The phylogenetic tree was created by comparing TFIIB homology domains that consist of the zinc ribbon, linker, and cyclin fold domains in each of the selected eukaryotic organisms. Saccharomyces cerevisiae (Sc), Monosiga brevicollis (Mb), Dictyostelium discoideum (Dd), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Phaeodactylum tricornutum (Ph), Ectocarpus siliculosus (Es), Hemiselmis andersenii (Ha), Guillardia theta (Gt), Cyanidioschyzon merolae (Cm), Ostreococcus tauri (Ot), Volvox carteri (Vc), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Populus trichocarpa (Pt), Arabidopsis thaliana (At). LG (A) and Dayhoff (B) substitution matrices were used to generate the phylogenetic trees. Each major TFB protein subfamily is shaded by a unique color. The TFB subfamilies include TFIIB, Brf, Rrn7/TAF1B/MEE12, Brp1, and Brp5. Nuclear genome, nuc; Nucleomorph genome, neo. The tree was visualized with iTOL80,81.


