Abstract
Betapapillomavirus is a genus of papillomaviruses (PVs) commonly found in human skin and associated with both benign and malignant skin lesions. Only 2 previous beta-PVs have been fully characterized in nonhuman species. This report describes a novel beta-PV, named Macaca fascicularis PV type 2 (MfPV2), isolated from exophytic skin papillomas on the hands and feet of a 2-year-old male cynomolgus monkey (M. fascicularis). On histology the papillomas were composed of diffusely thickened epidermis with superficial foci of cytomegaly, cytoplasmic pallor, marginalized chromatin, and rare eosinophilic intranuclear inclusion bodies. Positive immunostaining for p16 and the proliferation marker Ki67 was present multifocally within affected epidermis, most prominently within basal-type cells. Complete sequence identity (100%) was noted between PV genomes fully sequenced from hand and foot lesions. The MfPV2 genome was 7632 base pairs in length and included putative open reading frames (ORFs) for E1, E2, E4, E6, E7, L1, and L2 genes, similar to other PVs. The closest relatives to MfPV2 based on the L1 ORF sequence were all beta-PVs. These included human PV (HPV) 9, HPV115, HPV76, HPV75, and MfPV1 (60–70% pairwise identity for all), the latter of which was also isolated from hand and foot papillomas in a cynomolgus macaque. Phylogenetic analysis placed MfPV2 in a new species group (beta-6), distinct from HPVs (beta-1 to beta-5) and MfPV1 (beta-1). These findings characterize a new nonhuman beta-PV and provide additional support for the idea that tissue tropism among ancestral primate PVs developed prior to divergence of certain Old World primate lineages.
Keywords: papilloma, papillomavirus, Betapapillomavirus, primate, Macaca, wart
Papillomaviruses (PVs) are a diverse family of small nonenveloped DNA viruses associated with a wide spectrum of proliferative epithelial and fibroepithelial lesions ranging from benign skin and mucosal papillomas to cervical, oral, and other epithelial cancers.2,21,35 A recent analysis of PV taxonomy identified more than 180 types of animal PVs within 29 genera.3 The majority of known PVs come from primates, including more than 120 HPVs, 15 nonhuman primate (NHP) PVs, and numerous other putative primate PVs partially sequenced but not fully described and typed.1–4,6,7 Primate PVs have been detected in a variety of Old World NHP species, including macaques, common and pygmy chimpanzees, colobus monkeys, and gorillas.3 The best-studied NHP PVs are from cynomolgus and rhesus macaques, which carry numerous types of genital mucosal PVs (possibly rivaling the diversity of genital HPVs)4–6 and develop PV-associated genital neoplasia similar to that seen in human populations.20,31,32 Descriptions of NHP PVs from skin lesions are rare, however, with only a few reports of PV-associated cutaneous papillomas in colobus monkeys5,17,23 and a cynomolgus macaque.15 In this report we describe features of hand and foot papillomas associated with a novel cutaneous PV in a cynomolgus macaque.
Methods
During routine physical examination, approximately 20 to 30 papillomas were observed on the palmar and plantar surfaces of a juvenile male cynomolgus macaque (Macaca fascicularis) approximately 2 years old. This captive-bred monkey was Mauritius-origin and spent time at a Covance facility and Bristol-Myers Squibb primate quarantine and research sites before the masses were noted. The masses had been present for approximately 1 month prior to removal.
Biopsies of hand and foot papillomas were divided and portions of each were fixed in 10% neutral buffered formalin and snap-frozen upon removal. Fixed papillomas were embedded in paraffin and sectioned to 5 µm in thickness for hematoxylin and eosin (HE) and immunohistochemical staining. HE-stained slides were evaluated qualitatively for histologic changes by multiple pathologists (SHTG, CEW, JMC). Immunostaining procedures were performed on fixed, paraffin-embedded tissues using commercially available primary monoclonal antibodies for Ki67 (Ki67-MIB1, Dako, Carpinteria, CA) and p16 (p16 INK4a) (CMC811, Cell Marque, Hot Springs, AR); macaque PV antiserum (polyclonal rabbit antibodies raised to virus-like particles for MfPV3, a genital alpha-genus PV); and a commercial PV antibody (rabbit polyclonal antibody NCL-PVp raised to bovine PV type 1 (BPV1) antigens; Novacastra; Newcastle, UK).32 The proliferation marker Ki67 and the tumor suppressor protein p16 are widely used markers of PV-associated lesions.19 Rabbit MfPV3 antiserum was generated by Covance Laboratories (Princeton, NJ). Primary antibodies were diluted 1:50 (Ki67), 1:25 (p16), or 1:100 (MfPV3 and NCL-PVp) in Tris buffer, pH 7.4, containing 0.5% casein as blocking reagent. Staining methods included antigen-retrieval with citrate buffer (pH 6.0), biotinylated rabbit anti-mouse Fc antibody as a linking reagent, alkaline phosphatase-conjugated streptavidin as the label, hematoxylin as the counterstain, and Vector Red as the chromogen (Vector Laboratories, Burlingame, CA). All immunostaining batches included negative staining control slides treated with species-matched nonimmune serum in place of the primary antibody. Normal cynomolgus macaque footpad and skin sections were used as reference control tissues.
Total DNA was isolated from frozen portions of hand and foot lesions, and PV DNA was initially identified by polymerase chain reaction (PCR) amplification using consensus FAP primers within the L1 open reading frame (ORF).11 The 480-base pair (bp) FAP sequence revealed maximum similarity of 70% to HPV9 (a beta-genus HPV) with a BLAST search against GenBank. Hence, type-specific primer sets based on the sequence of the FAP product and a consensus region from the E1 ORF were designed to amplify the complete genomes in 2 overlapping fragments (3621 bp and 4111 bp, respectively). For overlapping PCR, an equal mixture of AmpliTaq Gold Taq DNA polymerase (Applied Biosystems, Foster City, CA) and Pwo Platinum Taq DNA Polymerase (High Fidelity, Invitrogen, Carlsbad, CA) were used as previously described.30 PCR products of appropriate size were cloned into the TOPO TA pCR2.1 vector (Invitrogen) and sequenced in the Einstein Sequencing Facility (Bronx, NY). Subsequent sequencing was performed using primer walking. Complete genomic sequences were compiled from the sequences of overlapping cloned PCR products and deposited into NCBI/GenBank (accession GU014531).
Phylogenetic analysis was performed as previously described.6 Briefly, the nucleotide sequences of complete L1 ORFs from representative PV types were used to evaluate the phylogenetic position of MfPV2. MrBayes v3.1.2 with 10,000,000 cycles for the Markov chain Monte Carlo (MCMC) algorithm was used to generate a phylogenetic tree.14,27
Results
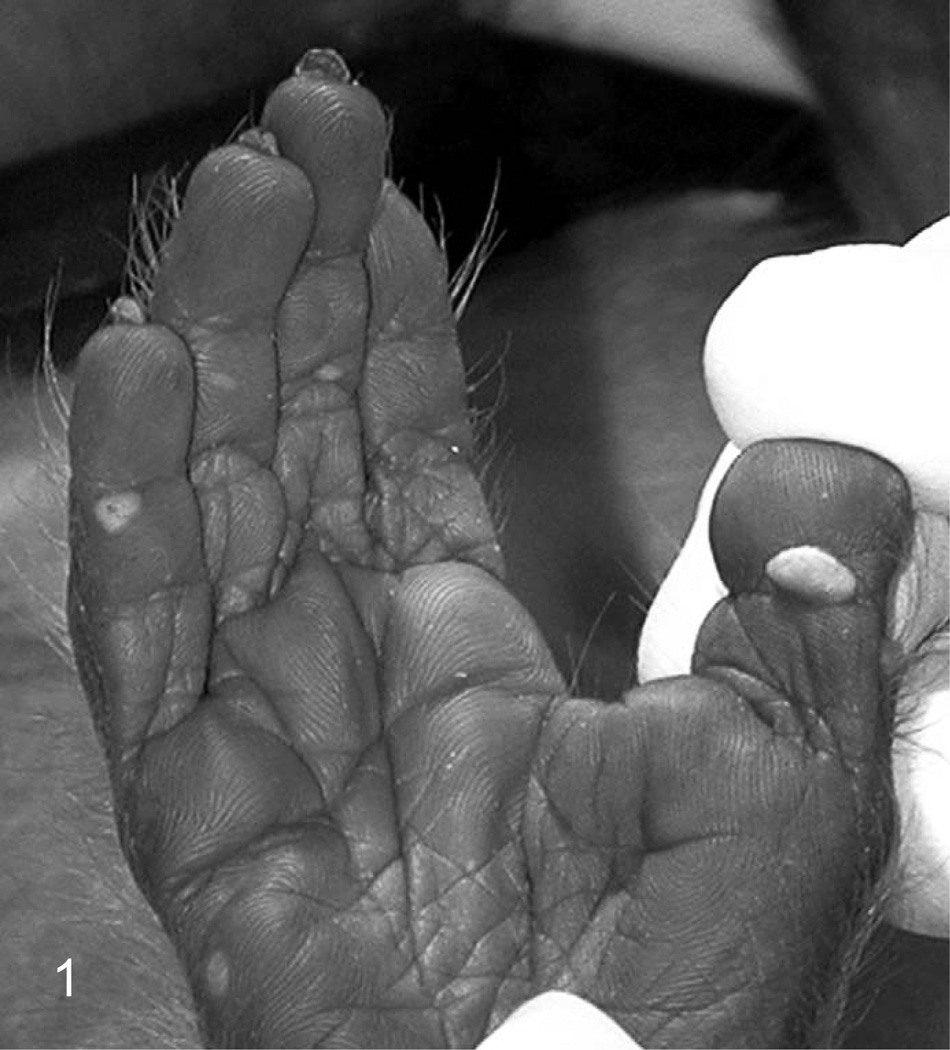
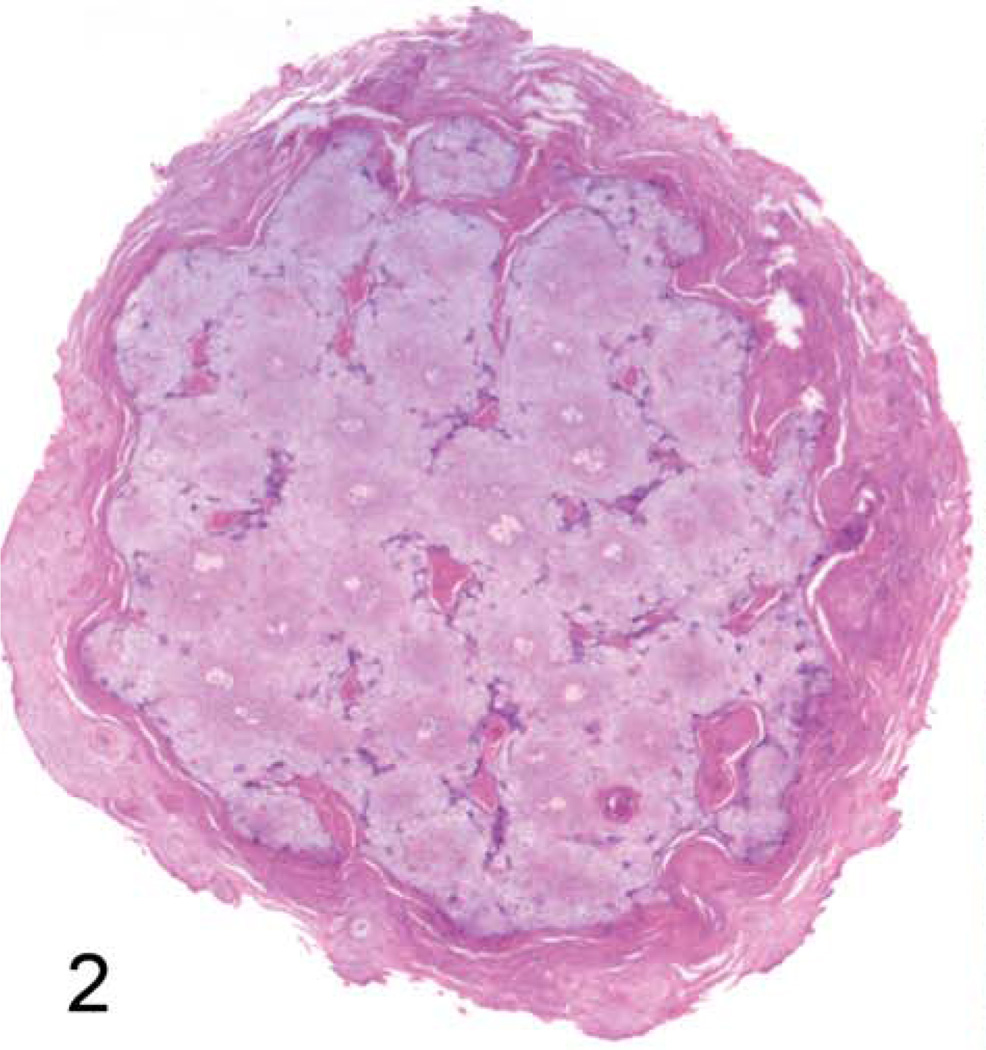
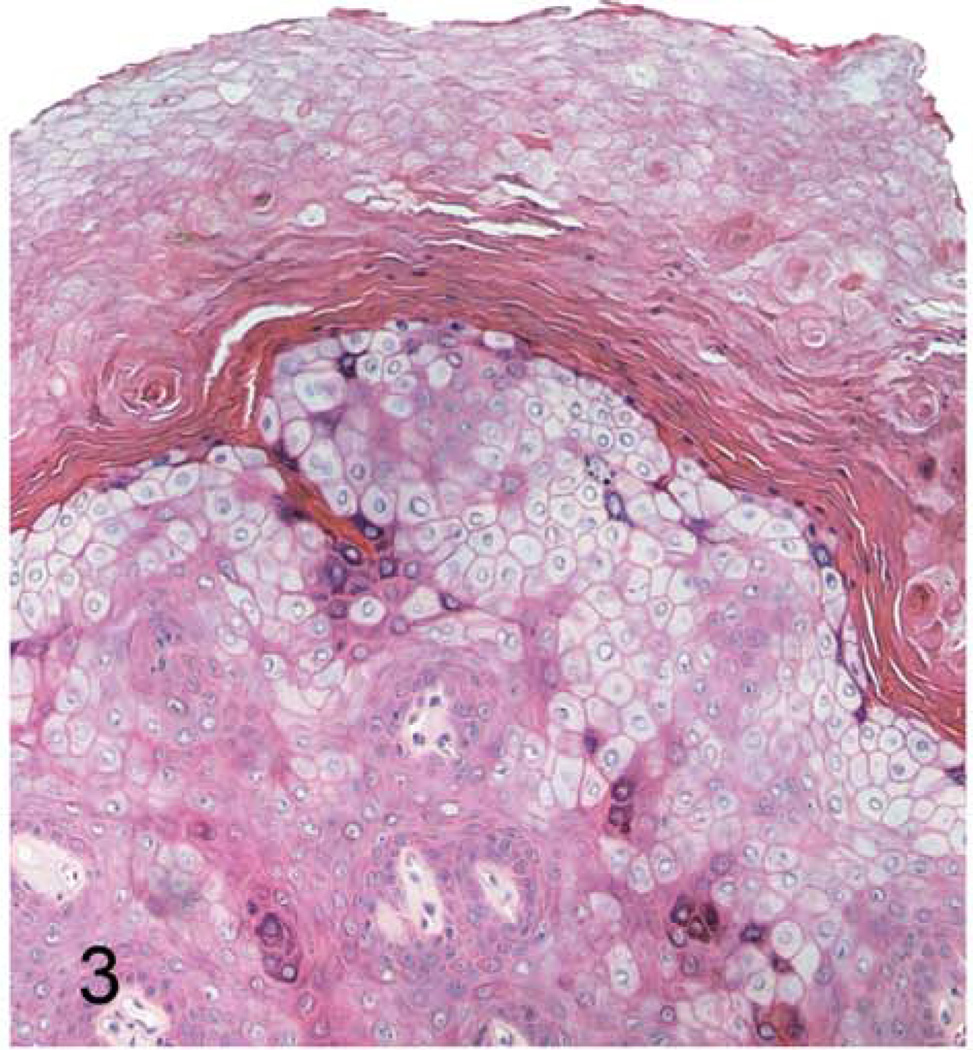
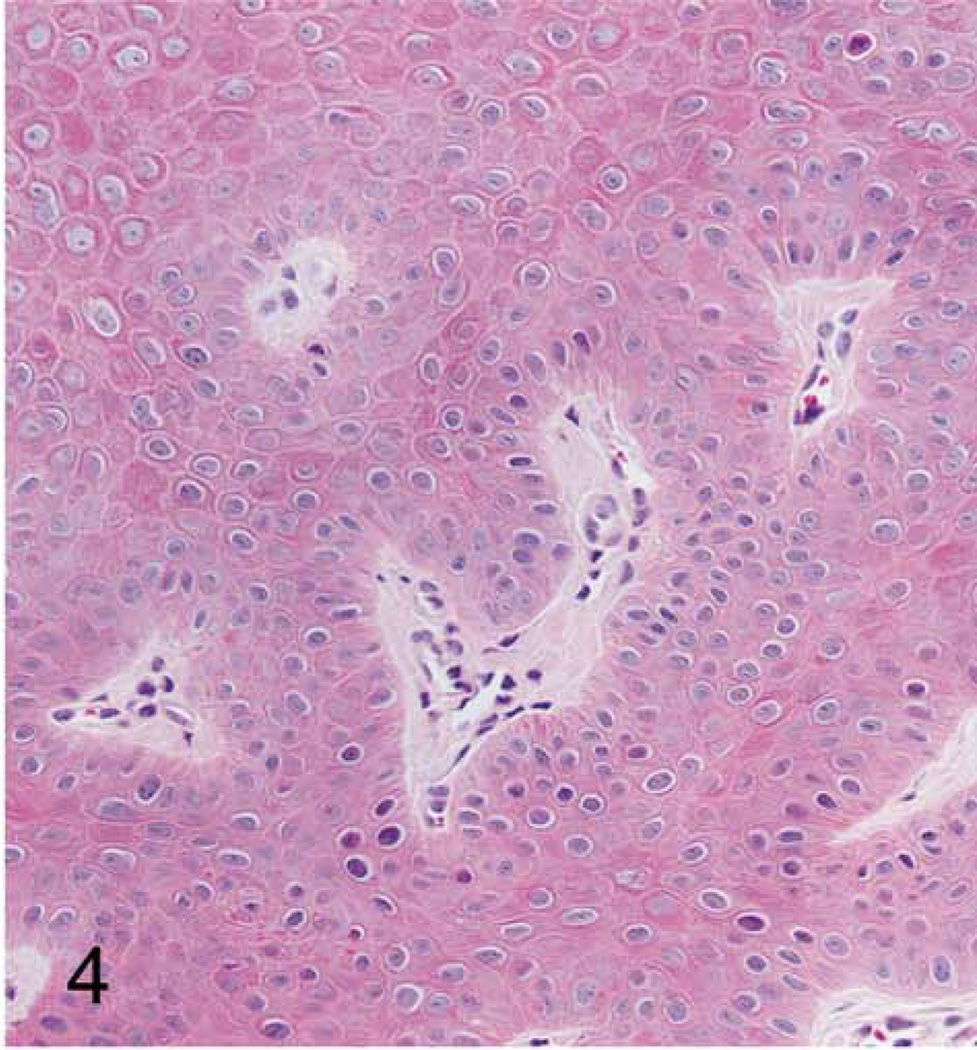
Grossly, the hand and foot papillomas were exophytic, nonpigmented, and variably raised (Fig. 1). Papillomas were moveable with the skin and ranged in size from 0.1 × 0.1 × 0.1 to 0.3 × 0.5 × 0.7 cm. Hand and foot masses appeared similar on histology. Features included diffusely thickened epidermis separated by attenuated islands of dermis and covered by a layer of keratin up to ~1 mm in thickness (Figs. 2 and 3). Pockets of superficial dermis were filled with thin-walled vascular channels and lined by relatively monomorphic basal-type epidermal cells with dense chromatin (Fig. 4). Basal-type cells extended 4 to 8 cell layers from the basal lamina (compared with 1 to 2 layers in normal epidermis). More superficial epidermal cells exhibited varying degrees of cytomegaly and nuclear pleomorphism. Features of these cells included cytoplasmic pallor, nuclear clearing with peripheralized chromatin, eccentric basophilic to magenta nucleoli 2 to 3 µm in diameter, and occasional pale eosinophilic intranuclear inclusions (Fig. 5). Intranuclear inclusions were round to ovoid, up to ~7 µm in diameter, and typically surrounded by clear haloes of cleared chromatin (Fig. 5, inset). Cells within the granulosum layer often contained 2 to 3 nuclei and variable amounts of deeply basophilic clumped granular material (keratohyalin presumed).
Figure 1.
Foot papillomas; macaque. Raised nonpigmented papillomas on the footpads of amale cynomolgus monkey. Papillomas with a similar gross appearance were also observed on the handpads.
Figure 2. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Hand papilloma; macaque. Transverse section of papilloma showing epidermal expansion with scattered pale foci of intervening dermis. 20×. HE.
Figure 3. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Hand papilloma; macaque. Superficial keratin and epidermal hyperplasia. 100×. HE.
Figure 4. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Foot papilloma; macaque. Islands of vascularized dermis surrounded by monomorphic basal-type epidermal cells. 200×. HE.
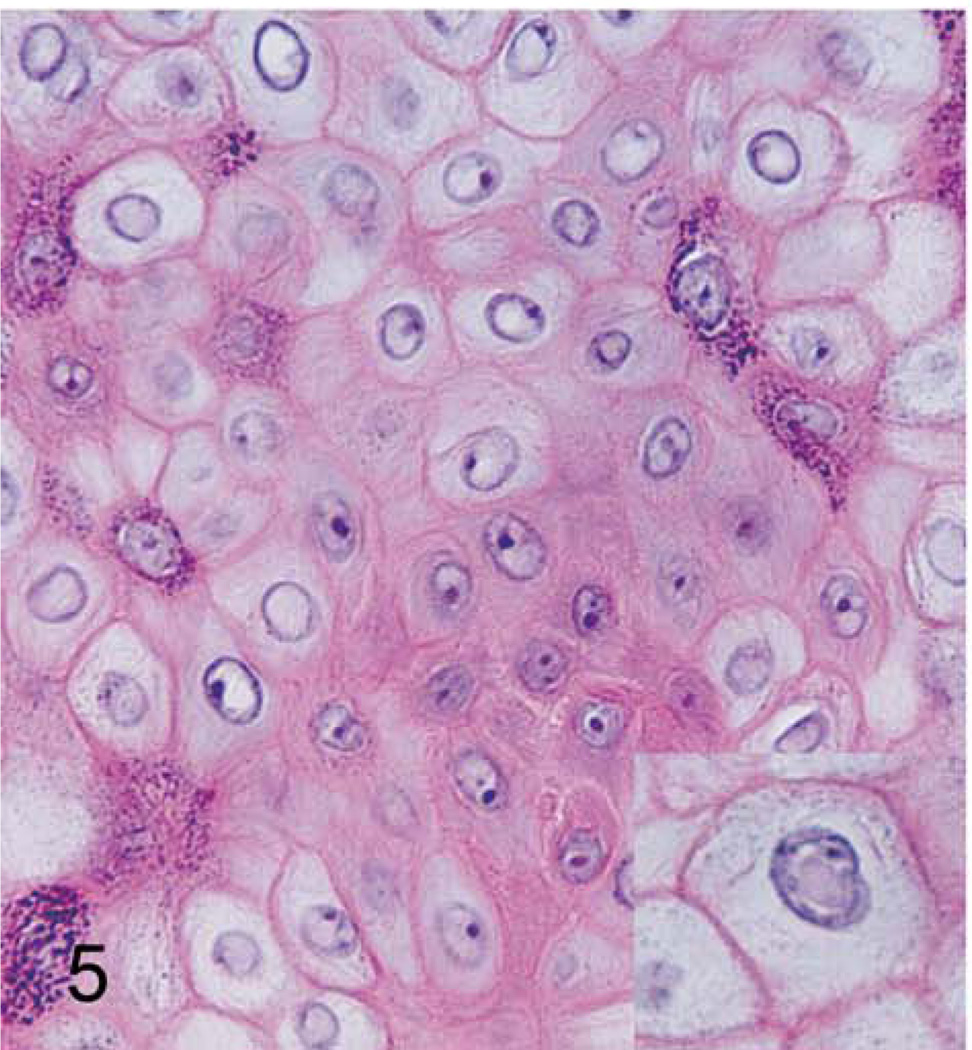
Figure 5. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Hand papilloma; macaque. Cytomegaly, cytoplasmic pallor, central nuclear clearing, and occasional pale eosinophilic intranuclear inclusion bodies (inset) within superficial epidermal cells. 400×. HE.
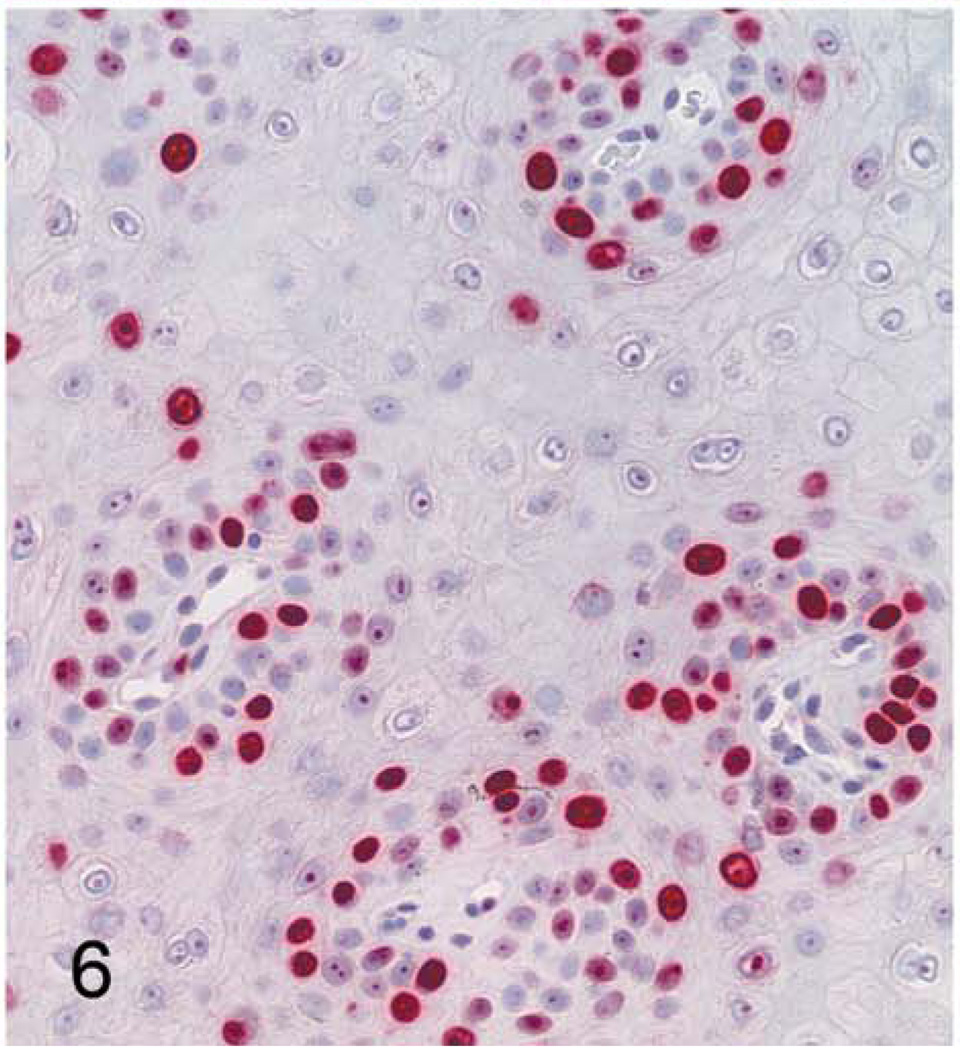
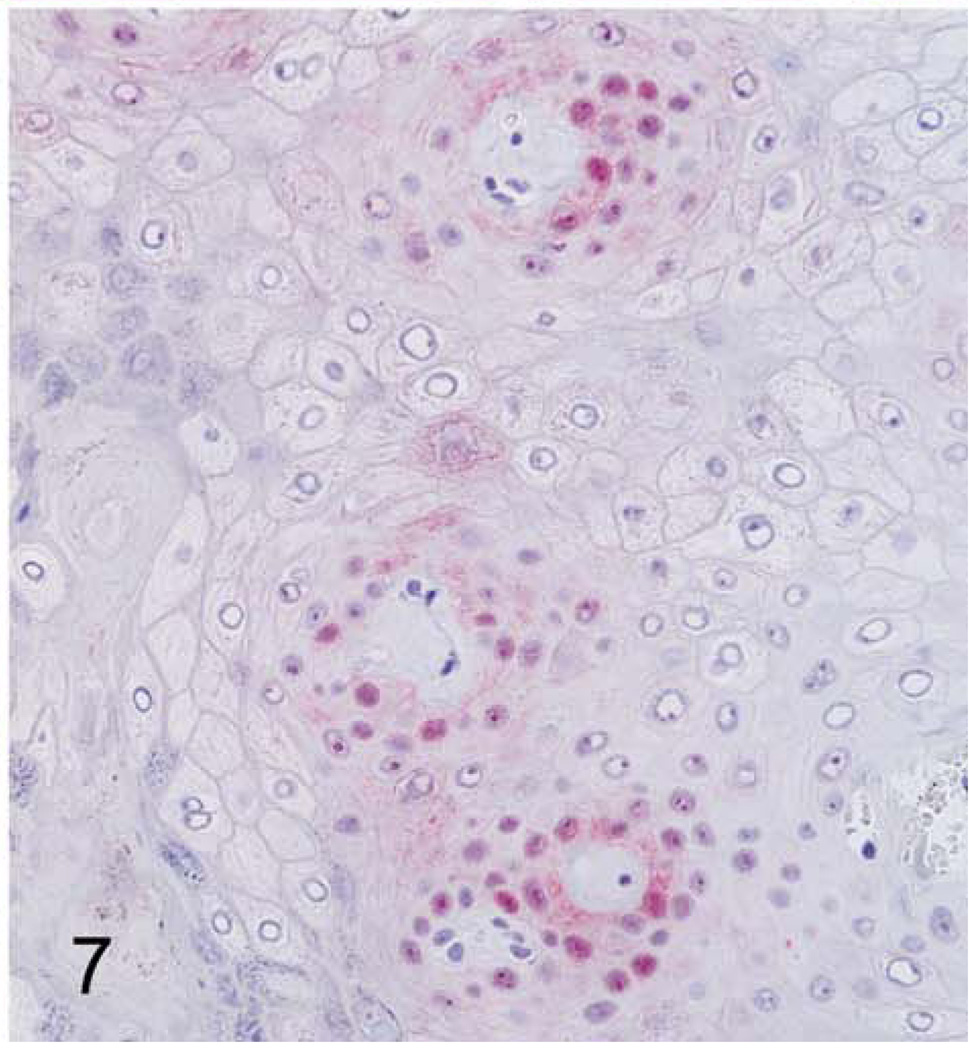
With immunohistochemistry, both hand and foot papillomas showed strong nuclear labeling for the proliferation marker Ki67 within basal-type epidermal cells, particularly in regions adjacent to intervening islands of dermis (Fig. 6); more scattered Ki67 labeling was present within more superficial epidermal layers. Cytoplasmic and nuclear immunolabeling for p16 was also most prominent within basal-type cells, although staining intensity was weaker than with Ki67 (Fig. 7). In normal control footpad epidermis, Ki67 labeling was limited to 1 to 2 layers of basal cells, whereas p16 staining was completely absent. Cytoplasmic and membranous immunostaining for MfPV3 antigen was present within more superficial epidermal cells and multifocally within the keratin layer (interpreted as nonspecific staining). Immunostaining for anti-BPV1 antigen (NCL-PVp) was limited to scattered foci within the keratin layer (interpreted as artifactual staining of dying corneal cells).
Figure 6. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Foot papilloma; macaque. Nuclear immunostaining for the proliferation marker Ki67 (MIB1). 400×.
Figure 7. Cutaneous hand and foot papillomas in a cynomolgus macaque associated with MfPV2.
Hand papilloma; macaque. Positive nuclear and cytoplasmic immunostaining for the tumor suppressor protein p16. 400×.
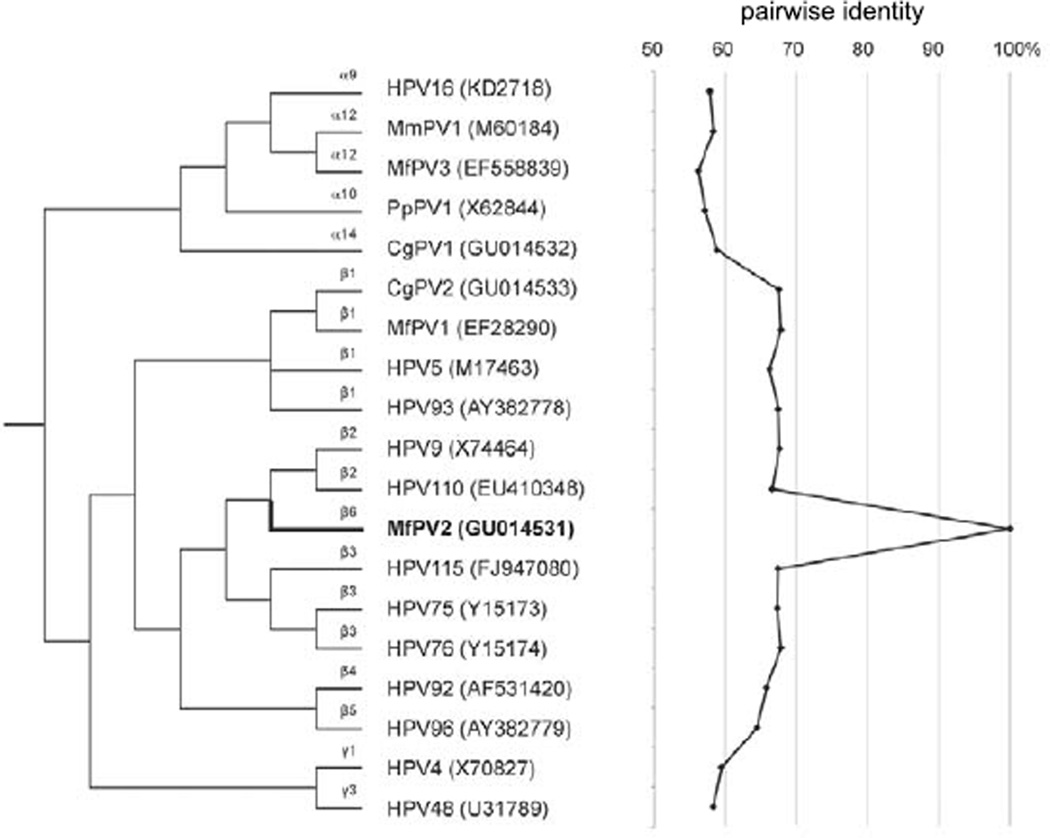
Papillomavirus DNA isolated from both hand and foot lesions contained the identical viral genomic sequence (100% identity). The PV genome was 7632 bp in length and included putative ORFs for early (E6, E7, E1, E2, and E4) and late (L2 and L1) PV genes. The upstream regulatory region (URR) containing many cis-acting regulatory sequences that control viral transcription and replication was identified between the stop codon of L1 and the first start codon of the E6 ORF. The full L1 ORF nucleotide sequence was less than 70% similar to other PVs (Fig. 8), indicating a novel species of MfPV. Phylogenetic analysis inferred from the L1 ORF sequence placed this virus in the beta-PV genus within a new species group (beta-6).3,7 The virus was named MfPV2 (based on actual date of sequencing, which came prior to that of alpha-PVs MfPV3 to MfPV11). The full L1 sequence of MfPV2 (1542 bp) was most similar to that of beta-3 and beta-2 HPVs (~67.2% pairwise similarities). The MfPV2 L1 gene sequence also showed similarities to MfPV1 (67.8% pairwise similarity), a beta-1 PV also identified from hand and foot papillomas in a cynomolgus macaque,15 and Colobus monkey PV type 2 (CgPV2) (67.5% pairwise similarity), isolated from cutaneous papillomas in a colobus monkey (Colobus guereza),5,17 but was relatively distant to NHP alpha-PVs (genital) such as MfPV3 and CgPV1 (56.2% and 58.8% pairwise similarities, respectively). The latter findings indicate closer relation of primate PVs infecting similar tissues (eg, mucosal or cutaneous) than PVs isolated from the same Old World NHP host species.
Figure 8.
Phylogenetic tree showing that MfPV2 forms a distinct species group (beta-6) within the beta-PV genus. The L1 ORF nucleotide sequence pairwise similarities of MfPV2 are shown in the figure to the right of dendrogram. Species groups (alpha, beta, and gamma) are shown to the left of PV types. NCBI/Genbank identifiers are shown in parentheses.
Discussion
Here, we describe skin papillomas associated with a novel beta-PV named MfPV2. This virus represents the third typed nonhuman beta-PV and the first member of the beta-6 species group.7 Related beta-PVs include HPV9, HPV75, HPV76, MfPV1, and CgPV2, all of which are also associated with cutaneous papillomas.5,8,15,17,23 Histologic features included cytomegaly, cytoplasmic and nuclear clearing, and intranuclear inclusions, distinct from the karyomegaly, nuclear atypia, and koilocytosis seen in alpha-PV–associated cervical intraepithelial neoplasia.19,20,34 This information adds to current knowledge of beta-PV–associated lesions and NHP tumor virus diversity.
Nongenital cutaneous HPVs are associated with a diverse array of dermatologic conditions, including common, mosaic, flat, and filiform papillomas (warts); epidermal dysplasia; and squamous cell carcinomas (SCCs).16 Beta-HPVs in particular are associated with epidermodysplasia verruciformis (a hereditary skin condition of florid papillomatosis linked to HPV types 5 and 8), cutaneous papillomas, and various premalignant and malignant skin lesions, including SCCs.8,9,12,13 Often cited as pathogens primarily of immunosuppressed patients, beta-HPVs (especially group 2 species) have recently been identified in healthy skin as well as nonmelanoma skin cancers from immunocompetent patients.12,13 Human PVs specifically associated with palmar and plantar papillomas include types 1, 2, 4, and 63.7 These HPVs also cause common skin warts elsewhere and represent diverse alpha (HPV2), gamma (HPV4), and mu (HPV1 and HPV63) genera,7 suggesting that palmoplantar infections may be related to mechanical factors as well as specific virus/host cell interactions.
A variety of benign and malignant PV-associated skin neoplasms have been reported in the veterinary literature.18 In addition to NHP tumors noted previously,5,15,17,23 reported lesions include fibropapillomas in cattle, sheep, elk, and deer (delta and epsilon PVs); sarcoids in horses (delta PVs); keratinaceous papillomas and SCCs in rabbits (kappa PVs); in situ and invasive SCCs in cats (lambda and dyotheta PVs) and dogs (tau and chi PVs); papillomas, keratoacanthomas, and in situ SCCs in multimammate mice (iota PV); trichoepitheliomas and sebaceous carcinomas in harvest mice (pi PV); cutaneous papillomas in manatees (rho PV); and other miscellaneous skin tumors in a porcupine (sigma PV), grey parrot (theta PV), raccoon (lambda PV), ferret (novel PV), and fruit bat (psi PV).3,18,10,24,25,26,29,33 Despite the recent expansion of this list, the exact role of human and nonhuman PVs in the pathogenesis of skin malignancies is unclear.
Papillomaviruses target specific host cell types and sites and exhibit specialized life cycle dynamics adaptive to these ecological microniches.22,28 The anatomical preferences of HPVs can be predicted based in part on the sequence of the L1 gene, which codes for the major PV capsid protein.7 Alpha-HPVs primarily infect genital/mucosal sites and include the more oncogenic genital HPVs, whereas beta- and gamma-HPVs generally inhabit nongenital skin and include HPVs associated with more exophytic warty lesions.3,7 As seen with MfPV2, NHP PV sequences sort alongside HPVs within these same genera,2–5,7 indicating that phylogenetic separation between genital/mucosal and skin PVs also occurs in nonhuman PVs and that niche specialization of ancestral primate PVs may have occurred prior to divergence of certain Old World primate lineages.5,6 Future identification and comparative analysis of additional NHP beta-PVs should be useful in the study of primate PV evolution and viral genetic determinants of skin tumorigenesis.
Supplementary Material
Acknowledgements
We thank Hermina Borgerink, Jean Gardin, Anne Dunn, Andrew Prior, and Suzanne Leanza for their technical contributions.
Financial Disclosure/Funding
This work was supported by the National Institutes of Health (NIH) grants R21 AI083962-01 from the National Institute of Allergy and Infectious Diseases (NIAID) and K01 RR21322-05 from the National Center for Research Resources (NCRR), center grants to the Einstein Cancer Research Center and the Center for AIDS Research, and the Wake Forest University School of Medicine Translational Science Institute. The contents are solely the responsibility of the authors and do not necessarily represent the view of the NIAID, NCRR, or NIH.
Footnotes
Declaration of Conflicting Interests
The authors declared that they had no conflicts of interest with respect to their authorship or the publication of this article.
References
- 1.Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J Virol. 2000;74:11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonsson A, Hansson BG. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J Virol. 2002;76:12537–12542. doi: 10.1128/JVI.76.24.12537-12542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen HZ, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan SY, Bernard HU, Ratterree M, Birkebak TA, Faras AJ, Ostrow RS. Genomic diversity and evolution of papillomaviruses in rhesus monkeys. J Virol. 1997;71:4938–4943. doi: 10.1128/jvi.71.7.4938-4943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan SY, Ostrow RS, Faras AJ, Bernard HU. Genital papillomaviruses (PVs) and epidermodysplasia verruciformis PVs occur in the same monkey species: implications for PV evolution. Virology. 1997;228:213–217. doi: 10.1006/viro.1996.8400. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, van Doorslaer K, DeSalle R, Wood CE, Kaplan JR, Wagner JD, Burk RD. Genomic diversity and interspecies host infection of Macaca fascicularis papillomaviruses (MfPVs) within the alpha papillomavirus α12 species. Virol. 2009;393:304–310. doi: 10.1016/j.virol.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 8.Delius H, Saegling B, Bergmann K, Shamanin V, de Villiers EM. The genomes of three of four novel HPV types, defined by differences of their L1 genes, show high conservation of the E7 gene and the URR. Virology. 1998;240:359–365. doi: 10.1006/viro.1997.8943. [DOI] [PubMed] [Google Scholar]
- 9.Dell’Oste V, Azzimonti B, De Andrea M, Mondini M, Zavattaro E, Leigheb G, Weissenborn SJ, Pfister H, Michael KM, Waterboer T, Pawlita M, Amantea A, Landolfo S, Gariglio M. High beta-HPV DNA loads and strong seroreactivity are present in epidermodysplasia verruciformis. J Invest Dermatol. 2009;129:1026–1034. doi: 10.1038/jid.2008.317. [DOI] [PubMed] [Google Scholar]
- 10.Erdélyi K, Bálint A, Dencso L, Dán A, Ursu K. Characterisation of the first complete genome sequence of the roe deer (Capreolus capreolus) papillomavirus. Virus Res. 2008;135:307–311. doi: 10.1016/j.virusres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J Gen Virol. 1999;80(Pt 9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 12.Forslund O. Genetic diversity of cutaneous human papillomaviruses. J Gen Virol. 2007;88:2662–2669. doi: 10.1099/vir.0.82911-0. [DOI] [PubMed] [Google Scholar]
- 13.Forslund O, Iftner T, Andersson K, Lindelof B, Hradil E, Nordin P, Stenquist B, Kirnbauer R, Dillner J, de Villiers EM Viraskin Study Group. Cutaneous human papillomaviruses found in sun-exposed skin: beta-papillomavirus species 2 predominates in squamous cell carcinoma. J Infect Dis. 2007;196:876–883. doi: 10.1086/521031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 15.Joh J, Hopper K, Van Doorslaer K, Sundberg JP, Jenson AB, Ghim SJ. Macaca fascicularis papillomavirus type 1: a non-human primate betapapillomavirus causing rapidly progressive hand and foot papillomatosis. J Gen Virol. 2009;90:987–994. doi: 10.1099/vir.0.006544-0. [DOI] [PubMed] [Google Scholar]
- 16.Karagas MR, Nelson HH, Sehr P, Waterboer T, Stukel TA, Andrew A, Green AC, Bavinck JN, Perry A, Spencer S, Rees JR, Mott LA, Pawlita M. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst. 2006;98:389–395. doi: 10.1093/jnci/djj092. [DOI] [PubMed] [Google Scholar]
- 17.Kloster BE, Manias DA, Ostrow RS, Shaver MK, McPherson SW, Rangen SR, Uno H, Faras AJ. Molecular cloning and characterization of the DNA of two papillomaviruses from monkeys. Virology. 1988;166:30–40. doi: 10.1016/0042-6822(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 18.Munday JS, Kiupel M. Papillomavirus-associated cutaneous neoplasia in mammals. Vet Pathol. 2010;47:254–264. doi: 10.1177/0300985809358604. [DOI] [PubMed] [Google Scholar]
- 19.Nam EJ, Kim JW, Hong JW, Jang HS, Lee SY, Jang SY, Lee DW, Kim SW, Kim JH, Kim YT, Kim S, Kim JW. Expression of the p16 and Ki-67 in relation to the grade of cervical intraepithelial neoplasia and high-risk human papillomavirus infection. J Gynecol Oncol. 2008;19:162–168. doi: 10.3802/jgo.2008.19.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrow RS, McGlennen RC, Shaver MK, Kloster BE, Houser D, Faras AJ. A rhesus monkey model for sexual transmission of a papillomavirus isolated from a squamous cell carcinoma. Proc Natl Acad Sci USA. 1990;87:8170–8174. doi: 10.1073/pnas.87.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkin DM, Bray F. Chapter 2: the burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S11–S25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 22.Peh WL, Middleton K, Christenson N, Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu JW, Boorbar J. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol. 2002;76:10401–10416. doi: 10.1128/JVI.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangan SR, Gutter A, Baskin GB, Anderson D. Virus associated papillomas in colobus monkeys (Colobus guereza) Lab Anim Sci. 1980;30:885–889. [PubMed] [Google Scholar]
- 24.Rector A, Tachezy R, Van Doorslaer K, MacNamara T, Burk RD, Sundberg JP, Van Ranst M. Isolation and cloning of a papillomavirus from a North American porcupine by using multiply primed rolling-circle amplification: the Erethizon dorsatum papillomavirus type 1. Virology. 2005;331:449–456. doi: 10.1016/j.virol.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 25.Rector A, Van Doorslaer K, Bertelsen M, Barker IK, Olberg RA, Lemey P, Sundberg JP, Van Ranst M. Isolation and cloning of the raccoon (Procyon lotor) papillomavirus type 1 by using degenerate papillomavirus-specific primers. J Gen Virol. 2005;86:2029–2033. doi: 10.1099/vir.0.80874-0. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues A, Gates L, Payne HR, Kiupel M, Mansell J. Multicentric squamous cell carcinoma in situ associated with papillomavirus in a ferret. Vet Pathol. 2010;47:964–968. doi: 10.1177/0300985810369899. [Epub 2010 May 13]. [DOI] [PubMed] [Google Scholar]
- 27.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 28.Schiffman M, Herrero R, Desalle R, Hildesheim A, Wacholder S, Rodriguez AC, Bratti MC, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Wright TC, Solomon D, Chen Z, Schussler J, Castle PE, Burk RD. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Tachezy R, Rector A, Havelkova M, Wollants E, Fiten P, Opdenakker G, Jenson B, Sundberg J, Van Ranst M. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002;2:19. doi: 10.1186/1471-2180-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terai M, Burk RD. Identification and characterization of 3 novel genital human papillomaviruses by overlapping polymerase chain reaction: candHPV89, candHPV90, and candHPV91. J Infect Dis. 2002;185:1794–1797. doi: 10.1086/340824. [DOI] [PubMed] [Google Scholar]
- 31.Wood CE, Borgerink H, Register TC, Scott L, Cline JM. Cervical and vaginal epithelial neoplasms in cynomolgus macaques. Vet Pathol. 2004;41:108–115. doi: 10.1354/vp.41-2-108. [DOI] [PubMed] [Google Scholar]
- 32.Wood CE, Chen Z, Cline JM, Miller BE, Burk RD. Characterization and experimental transmission of an oncogenic papillomavirus in female macaques. J Virol. 2007;81:6339–6345. doi: 10.1128/JVI.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodruff RA, Bonde RK, Bonilla JA, Romero CH. Molecular identification of a papilloma virus from cutaneous lesions of captive and free-ranging Florida manatees. J Wildl Dis. 2005;41:437–441. doi: 10.7589/0090-3558-41.2.437. [DOI] [PubMed] [Google Scholar]
- 34.Wright TC, Kurman RJ, Ferenczy AF. Precancerous lesions of the cervix. In: Kurman RJ, editor. Blaustein’s Pathology of the Female Genital Tract. 5th ed. New York, NY: Springer-Verlag; 2002. pp. 253–324. [Google Scholar]
- 35.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.