Abstract
Background
Integrin beta-5 (ITGB5) and mucin 13 (MUC13) genes are highly expressed on the apical surface of intestinal epithelia and are thought to be candidate genes for controlling the expression of the receptor for enterotoxigenic Escherichia coli (ETEC) F4ac. Human MUC13 protein has an expected role in protecting intestinal mucosal surfaces and porcine ITGB5 is a newly identified potential receptor for ETEC F4ac.
Methodology/Principal Findings
To test the hypothesis that ITGB5 and MUC13 both play key roles in protection of the intestinal mucosa against pathogenic bacterium, porcine intestinal epithelial cells (IPEC-J2) were transfected with ITGB5-targeting, MUC13-targeting or negative control small interfering RNA (siRNA), respectively. Firstly, we measured mRNA expression levels of mucin genes (MUC4, MUC20), pro-inflammatory genes (IL8, IL1A, IL6, CXCL2), anti-inflammatory mediator SLPI, and PLAU after RNAi treatments with and without ETEC infection. Secondly, we compared the adhesions of ETEC to the pre- and post-knockdown IPEC-J2 cells of ITGB5 and MUC13, respectively. We found that ITGB5 and MUC13 knockdown both had small but significant effects in attenuating the inflammation induced by ETEC infection, and both increased bacterial adhesion in response to F4ac ETEC exposure.
Conclusions/Significance
Our current study first reported that ITGB5 and MUC13 are important adhesion molecules of mucosal epithelial signaling in response to Escherichia coli in pigs. These data suggest that both ITGB5 and MUC13 play key roles in defending the attachment and adhesion of ETEC to porcine jejunal cells and in maintaining epithelial barrier and immunity function.
Introduction
Enterotoxigenic Escherichia coli (ETEC) is recognized as a common cause of diarrhoea in humans and swine, their adherence to intestinal cells is mediated by proteinaceous, species-specific colonization factors [1]. F4 fimbriae are the major colonization factors associated with porcine neonatal and postweaning diarrhoea caused by ETEC [2], [3]. Among the three different antigenic variants (F4ab, F4ac, and F4ad) of F4 fimbriae, F4ac is the most prevalent [4], [5]. ETEC having F4ac fimbriae inducing severe diarrhoea is dependent on the presence of the specific receptors for F4ac (F4acR) [6], which is encoded by the responsible gene [7]. To date, the candidate genes of F4acR having been investigated in several specific pig populations by association study between genetic markers with in vitro F4ac adhesion phenotypes include: mucin 4 (MUC4) [8], MUC13 [7], [9], MUC20 [10] and integrin beta-5 (ITGB5) [11].
Besides action as a physical barrier limiting access of microbes and toxins to the underlying tissues, the contiguous lining intestinal mucosal epithelial cells secrete long filamentous cell-surface mucins which are a major constituent of the mucus barrier [12], [13]. For these membrane anchored cell-surface mucins, in addition to protection, they play an important role in the survival of mucosal epithelial cells, and participate in intracellular signal transduction [12]. Mucin 13 is a transmembrane mucin glycoprotein highly expressed in the jejunum of pig [9]. More recently, the mRNA expression level of MUC13 is shown to be down-regulated in the inflamed intestine cells of porcine induced by F4ac ETEC strain infection [5]. In human, MUC13 was reported to regulate epithelial inflammation in response to inflammatory and infectious stimuli [12], [13]. Being adjacent to the MUC13 gene on porcine chromosome 13 (SSC13), ITGB5 was our newly identified candidate gene for F4acR via genome wide association study [11]. As one member of the “focal adhesion” family, ITGB5 is an attractive candidate to be tested as biomarker and/or new drug target in human pancreatic cancer [14]. In the mouse, the ITGB5 subunit was found to be expressed on both the apical and basal surface of endometrial epithelium [11], [15].
In this study, we aimed to detect the effects of deficiency of MUC13 or ITGB5 in porcine intestinal cells upon F4ac ETEC infection and their mucosal immunogenicity.
Results
Silencing of the Target Genes
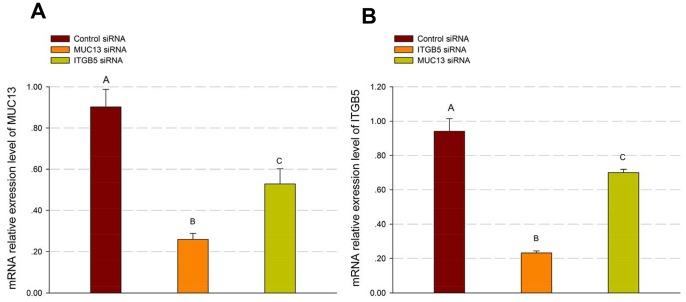
RNA interference was used to investigate the function of MUC13 and ITGB5 in porcine intestinal cells. Firstly, we designed and synthesized siRNAs specifically targeting porcine MUC13 and ITGB5, respectively (Table 1). Subsequently, the siRNAs were transfected into IPEC-J2 cells by using Lipofectamine® RNAiMAX Reagent. In Figure 1A, we show the mRNA expression levels of MUC13 in cells transfected with negative siRNA duplexes (negative control) and with MUC13-targeting siRNA, respectively, at 44 h post-transfection. The results indicated that mRNA expression of MUC13 was significantly suppressed by the MUC13-targeting siRNA (Fold-change = −3.48, P<0.001). Compared with negative control, at 44 h post-transfection, the mRNA expression of ITGB5 was significantly attenuated by the ITGB5-targeting siRNA (Fold-change = −4.05, P<0.001; Figure 1B).
Table 1. Small RNA sequences.
| Target genes | GenBank Accession Number | Type | Sequence | Quantity |
| MUC13 | NM_001105293.1 | RNA | CCAGCUUGUUGAGGUAGAAGUAGUA | Stealth |
| RNA | UACUACUUCUACCUCAACAAGCUGG | Stealth | ||
| ITGB5 | NM_001246669.1 | RNA | AAUCCGUGCAUUGGCUACAAGUUAU | Stealth |
| RNA | AUAACUUGUAGCCAAUGCACGGAUU | Stealth |
Figure 1. MUC13-targeting and ITGB5-targeting siRNA reduced the expression of MUC13 and ITGB5 in IPEC-J2 cells.
IPEC-J2 cells were transfected with MUC13-targeting, ITGB5-targeting or negative control siRNA for 44 h, and the expression levels of MUC13 (A) and ITGB5 (B) were measured by real-time RT-PCR, respectively. For (A and B), results are the mean ± SE (standard error) from IPEC-J2 cells of three independent replicates (n = 3). In each group, values without a common letter are significantly different (P<0.01).
Because MUC13 and ITGB5 are located next to each other in porcine genome and can be expressed in the same cell, to examine whether there are correlation [16]/compensation [13] between the expression levels of the two adjacent genes in IPEC-J2 cells, we measured MUC13 mRNA expression in ITGB5 knockdown (ITGB5-KD) cells as well as the mRNA expression of ITGB5 in MUC13 knockdown (MUC13-KD) cells. There was a small significant decrease in the expression of MUC13 in ITGB5-KD cells (Fold-change = −1.71, P<0.005; Figure 1A), and in the expression of ITGB5 in MUC13-KD cells (Fold-change = −1.34, P<0.01; Figure 1B), respectively. These results demonstrated that there are potential correlation between the expressions of ITGB5 and MUC13 in IPEC-J2 cells.
When MUC13 or ITGB5 was silenced, the mRNA expression of the two genes was measured in silenced cells infected with F4ac ETEC. We observed that silence of MUC13 decreased the expression of MUC13 and ITGB5, while silence of ITGB5 only decreased the expression of itself in the infected cells (Figure S1).
ITGB5 and MUC13 Similarly Regulate Cell-surface Mucin Expression in Response to ETEC
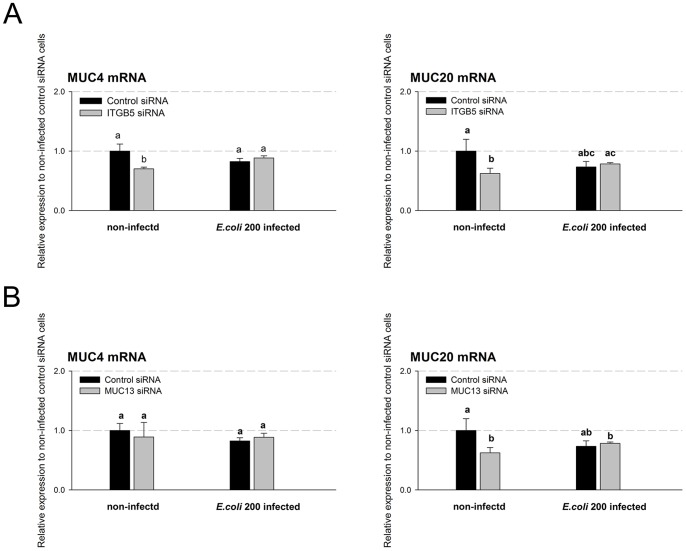
In addition to MUC13, two other genes, encoding structurally similar cell surface mucins and also located on Chr13, are known to be expressed in porcine jejunum: MUC4 and MUC20 [3]. To identify whether there is interaction between MUC13 or ITGB5 with each of the two mucin genes, we firstly separately compared the mRNA expression levels of MUC4 and MUC20 in ITGB5- or MUC13-deficient cells with that in the negative control cells at 44 h post-transfection of siRNAs. There were small significant decreases in the expression of MUC4 (∼30%, P<0.05) and of MUC20 (∼38%, P<0.05) in ITGB5-deficient cells (Figure 2A). When MUC13 was knocked down there was a ∼37% significant decrease (P<0.05) in MUC20 mRNA expression (Figure 2B), but no significant change in MUC4 expression. These results indicated that the expressions of MUC4 and MUC20 might be correlated with ITGB5 expression and that MUC20 might be also related to MUC13 in IPEC-J2 cells.
Figure 2. Expression of cell surface mucin genes in control and ITGB5-silencing/MUC13-silencing IPEC-J2 cells infected or non-infected with F4ac ETEC.
(A) IPEC-J2 cells were transfected with ITGB5-targetingor negative control siRNA for 44 h and infected with or without F4ac ETEC for 3 h. Total RNA from non-infected or infected cells were used to measure the mucin genes mRNA expression by real-time PCR. (B) IPEC-J2 cells were transfected with MUC13-targeting or negative control siRNA for 44 h and infected and assessed as in A. Statistics: mean ± SE; n = 3; In each group, values without a common letter are significantly different (P<0.05).
Secondly, we assessed the relative effects of ITGB5 and MUC13 on mRNA expression of the two mucin genes (MUC4 and MUC20) in response to 3 h infection with ETEC strain 200 (MOI = 8∶1) at 44 h after siRNAs transfection. After exposure to ETEC strain 200 for 3 h, no marked differences were observed in the mRNA expressions of MUC4 and MUC20 between negative control and ITGB5-deficient/MUC13-deficient cells, respectively.
ITGB5 and MUC13 Similarly Regulate mRNA Expression of Markers of Inflammation in Response to ETEC Infection
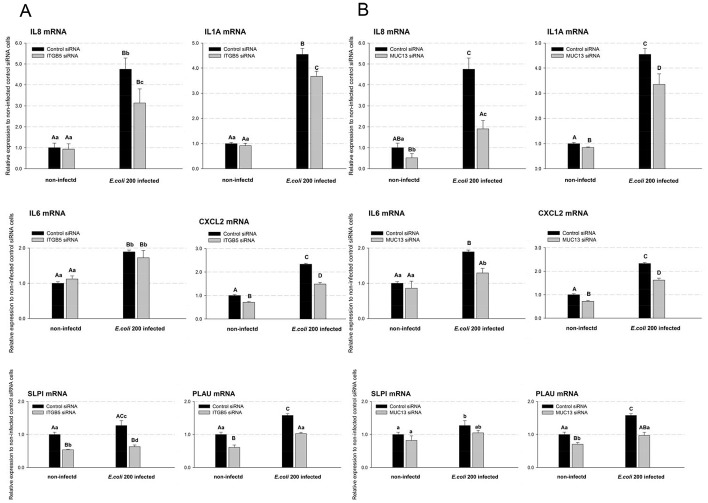
To detect the effects of ITGB5 and MUC13 on the expressions of pro-inflammatory cytokines/chemokines, the mRNA levels of IL8, IL1A, IL6 and CXCL2 were measured in F4ac ETEC infected and non-infected ITGB5-KD or MUC13-KD cells, respectively. In the non-infected cells, knockdown of ITGB5 only slightly reduced CXCL2 mRNA expression (∼29%, P<0.01; Figure 3A). As a comparison, knockdown of MUC13 significantly reduced the mRNA expression levels of IL8, IL1A and CXCL2 (P<0.01; Figure 3B).
Figure 3. ITGB5 and MUC13 modulated intestinal epithelial cell inflammatory reaction in response to F4ac ETEC.
IPEC-J2 cells were transfected with ITGB5-targeting and MUC13-targeting individually or negative control siRNA for 44 h and co-cultured with F4ac ETEC for 3 h or not. The mRNA levels of the inflammatory markers were measured by real-time PCR. Statistics: mean ± SE; n = 3; In each group, values without a common capital letter are significantly different (P<0.01), without a common lower case letter are significantly different (P<0.05).
As we expected, exposure to the bacteria strikingly induced the mRNA expression of all the four important pro-inflammatory cytokines/chemokines (IL8, IL1A, IL6 and CXCL2) [17], [18] in the negative control cells. Intriguingly, knockdown of ITGB5 and MUC13 in IPEC-J2 cells both negatively regulated the expressions of IL8, IL1A and CXCL2 mRNA in response to ETEC infection, and the decrease of IL6 mRNA expression was also observed in MUC13-deficient cells (Figure 3).
In addition to the pro-inflammatory cytokines, in the F4ac ETEC infected negative control cells, there were significantly higher mRNA levels of SLPI (anti-inflammatory mediator) [19] and PLAU (encodes a serine protease which can in turn activate plasminogen into plasmin, and the latter is involved in degradation of the extracellular matrix) [20] than in the non-infected negative control cells (Figure 3). Highlighting the significance of ITGB5 and MUC13, after exposure to ETEC ITGB5−/MUC13-deficient cells produced no more SLPI and PLAU than the non-infected negative control cells.
These results primarily revealed the pro-inflammatory activity of MUC13 and ITGB5 in porcine intestinal epithelial cells.
ITGB5 Depletion and MUC13 Depletion Similarly Enhanced F4ac ETEC Adhesion to the IPEC-J2 Cells
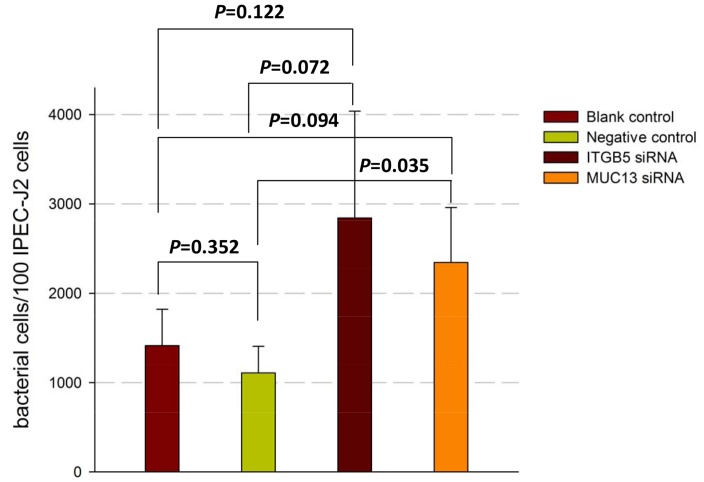
As both ITGB5 and MUC13 are candidate genes for ETEC F4ac receptor [3], [11], we finally assessed the influences of these two genes on adhesion of F4ac ETEC to the piglet small intestinal epithelial cells (Figure 4). IPEC-J2 cells were separately transfected with ITGB5- or MUC13-targeting siRNA for 48 h, and the cells were co-cultured with live bacteria for a further 3 h. Quantitative real-time PCR was used to measure the counts of adherent bacteria as described previously [5].
Figure 4. Overview of relative adhesion of F4ac ETEC to IPEC-J2 cells transfected with ITGB5-targeting/MUC13-targeting siRNA.
IPEC-J2 cells were transfected with ITGB5-targeting and MUC13-targeting individually or negative control siRNA (Negative control) for 48 h and co-cultured with F4ac ETEC for 3 h. Adhesion of F4ac ETEC to the four group (including three transfected groups and one non-transfected group (Blank control)) cells were measured by real-time PCR. Statistics: mean ± SE; n = 3.
We observed that, compared with the negative control (transfected with control siRNA), knockdown of ITGB5 and MUC13 both increased the number of F4ac ETEC adhering to the porcine intestinal epithelial cells at the P value of 0.072 and 0.035, respectively. The enhancement of F4ac ETEC adhesion by deficiency of ITGB5 or MUC13 in IPEC-J2 cells is a novel observation.
Discussion
In the present study, we primarily identified the effects of deficiency of MUC13 or ITGB5 in porcine intestinal cells upon F4ac ETEC infection and their mucosal immunogenicity. In human studies, gastric MUC13 is found to be down-regulated after H. pylori infection [21] and has been shown to be aberrantly expressed in gastrointestinal cancers [22]. ITGB5 is up-regulated in pancreatic adenocarcinoma [14],[23], and its repression impairs angiogenesis both in vitro and in vivo [24]. We have previously found that the expression of porcine MUC13 and MUC4 was down-regulated in IPEC-J2 cells post infection with the same F4ac ETEC strain, and the expression of ITGB5 was not significantly decreased [5]. Indeed, our present study provided evidence for the first time that decreased endogenous ITGB5 mRNA levels were associated with decreased mRNA levels of MUC13 (Figure 1B), MUC4, MUC20 (Figure 2A), CXCL2, SLPI and PLAU (Figure 3A), while decreased endogenous MUC13 mRNA levels was associated with decreased ITGB5 (Figure 1A), MUC20 (Figure 2B), IL8, IL1A, CXCL2 and PLAU mRNA (Figure 3B), in the in vitro porcine intestinal epithelial cells.
Up to now, the relationship between MUC13 and ITGB5 in swine is still undefined. Based on our study results, the deficiency of MUC13 may suppress the expression of ITGB5 (Figure S1). On the other hand, ITGB5 might regulate the expression of mucin genes (including MUC13), since Bianchi-Smiraglia et al found that integrin β5 deficiency induced both inhibitions of the Src-FAK and MEK-ERK signaling pathways [25], as the latter is essential for the activation of mucin genes transcription factor CREB (cAMP response element-binding protein) [26], . Moreover, in the current study we observed that silence of ITGB5 decreased the expression of MUC13 (Fold-change = −1.71, P<0.005; Figure 1B) while there was a small decrease in the expression of ITGB5 in MUC13-KD cells (Fold-change = −1.34, P<0.01; Figure 1B). In our previous study [5], when the expression of MUC13 was intensively reduced by F4ac ETEC infection, the mRNA expression level of ITGB5 did not significantly decrease. The exact relationship of ITGB5 and MUC13 should be further tested by applying gene transfection of ITGB5/MUC13 into IPEC-J2 cells or gene knockout of ITGB5/MUC13 to confirm their effects on each other’s expression.
The relationships between MUC13 and other paralogs such as MUC4 and MUC20 in porcine are unknown too, but no CNV (copy number variation) was detected in this region of pig chromosome 13 based on our Porcine SNP60 genotyping data [29] and copy number assay by quantitative real time PCR (our unpublished data). Studies have found strong correlation (be consistently up- or down-regulated) between the expression levels of genes that are located close to each other on the genome in mammals [16], [30]. Thus, the low co-expression of these genes could also partially and potentially be explained by ITGB5-targeting or MUC13-targeting RNA interference.
Our current study provides the first in vitro evidence that porcine ITGB5 and MUC13 have similar functions in modulating intestinal epithelial resistance to ETEC-induced inflammation. In human, Sheng et al [31] have proved that MUC13 has modulation effect on chemokine (IL8 and CXCL2) secretion in LS513 intestinal epithelial cells through a nuclear factor-κB-dependent pathway. For human ITGB5, the pathways it is involved in include IL-8 signaling (INGENUITY TARGET EXPLORER), phagosome (conserved biosystem, from KEGG), immune system (organism-specific biosystem, from REACTOME) and antigen processing-cross presentation (organism-specific biosystem, from REACTOME). Published reports have shown that monocytes from three patients lacking of integrin β3 (ITGB3) expression were unable to produce interleukin 8 in response to BLP (SK4) [32]. Additionally, human ITGB5 over-expression promotes the angiogenic properties of circulating angiogenic cells via Srckinase-mediated activation of STAT3, followed by increased transcription of CXCL8 and CCL2 [33]. Similar to the effect of human ITGB3 and ITGB5, our data showed that the genetic ablation of ITGB5 impaired responses of porcine intestinal epithelial cells to ETEC. A pathway including porcine MUC13 and ITGB5 has still not been defined. Based on the almost identical effect of the independent knockdown of the two adjacent genes in the present study, it implied that porcine MUC13 and ITGB5 may be involved in the same pathway which includes IL8, CXCL2 and other immune related genes.
To date, only three well characterized porcine intestinal epithelial cell lines are available: IPEC-1, IPEC-J2 and IPI-2I [34]. Three major points demonstrate that IPEC-J2 represents a better model for this study than the two other cell lines: (I) Non-immortalized cell line IPEC-J2 can serve as a better model of the normal host intestinal epithelium than the transformed IPI-2I cell line [34], (II) Pavlova et al. showed that six F4 ETEC strains all failed to enhance IL-8 and TNFα mRNA expression in the IPI-2I cell line [35], and (III) porcine ileum is a site of minor importance for F4+ ETEC pathogenesis [36]. Furthermore, There are some RNAi studies performed in one cell line [37], [38], [39]. In order to prevent the time-caused conflict to harvest the cells for RNA extraction and for adhesion assay, respectively, we set the time point of harvesting cells (infected or non- infected with F4ac ETEC) for adhesion assay 4 h later than that harvesting cells (infected or non- infected with F4ac ETEC) for RNA extraction.
Adhesion of the F4ac ETEC to IPEC-J2 cells has been reported [5], [40]. Since ITGB5 and MUC13 are striking candidate genes for receptor of F4ac ETEC [7], [9], [11], the effects of them on ETEC-IPEC-J2 adhesion were firstly assessed. Surprisingly, knockdown of ITGB5 and MUC13 in IPEC-J2 cells both increased F4ac ETEC adhesion (Figure 4). Integrins, a structurally elaborate family of adhesion molecules, was reported to participate in a wide range of biological processes, including tissue repair, angiogenesis and inflammation [41]. The observation about ITGB5 was in line with a published study showing that the absence of ITGB5 in Hela cells increased Salmonella invasion [42]. On the basis of published reports that bacterial pathogens often induce host cell apoptosis to promote their survival and dissemination [1], [43], [44], we speculated that the potential reason for increase of F4ac ETEC adhesion is that ITGB5-or MUC13-deficiency induced cell apoptosis and impair metabolic activity of IPEC-J2 cells.
In summary, our work demonstrated that the deficiency of endogenous ITGB5 and MUC13 can: (I) influence mRNA expression mutually; (II) affect the RNA levels of MUC20, CXCL2 and PLAU etc; (III) inhibit the normal responses of intestinal epithelial cells to ETEC infection; (IV) promote F4ac ETEC to adhere to porcine intestinal epithelial cells in vitro.
Materials and Methods
Cell Culture and Bacterial Strain
Porcine intestinal cells (IPEC-J2) were grown in Dulbecco’s modified eagle medium (DMEM)/Ham’s F-12 (1∶1) medium (DMEM/F12, GIBCO, Invitrogen, Beijing) containing 5% fetal bovine serum, and were maintained in a 95% air-5% CO2 humidified atmosphere at 37°C and subcultured at 4-day intervals. F4ac ETEC strain 200 (O149:K91:F4ac, LT+, STa+, STb+, EAST1+) were removed from cryo-storage and cultured in Ordinary Broth Agar at 37°C for three generations (24 h per generation) [5].
SiRNA Transfection
Small interfering RNAs (siRNAs) specifically targeting MUC13 and ITGB5 and the corresponding negative control (Stealth RNAi Negative Control Low GC Duplex) were chemically synthesized by Invitrogen (Carlsbad, CA). The sequences of small interfering RNAs were shown in Table 1. Porcine intestinal cells (IPEC-J2 cells) were transfected with siRNAs using Lipofectamine® RNAiMAX Transfection Reagent and Reverse Transfection protocol (Invitrogen) according to the manufacturer’s protocol. Briefly, after digesting IPEC-J2 cells in the logarithmic phase, cell suspension was prepared with a concentration of 100 cells/µl in complete growth medium without antibiotics. Diluted 3 µl gene-specific siRNA oligomers (20 µM) in 500 µl Opti-MEM®I reduced serum medium (Opti-MEM, Invitrogen) and added 5µl Lipofectamine® RNAiMAX to each well containing the diluted RNAi molecules. After 20 min incubation at room temperature, the complexes were added to one cell of the 6-well plate. To each well with RNAi duplex-Lipofectamine® RNAiMAX complexes, added 2500 µl of the diluted cell suspension. The IPEC-J2 cells incubated with transfection medium (500 µl Opti-MEM®I reduced serum medium +2500 µl complete growth medium) only were used as Blank control and that transfected with a non-target control siRNA were used as Negative control. Each transfection repeated three times.
Infection of the Cell Lines
For enterotoxigenic Escherichia coli infection post siRNA transfection:
At 44 hours post-transfection, monolayers of the cells in 6-well tissue culture plates were washed twice with PBS, and 1 ml of DMEM/F12 was added. A total of 20 µl of bacterial suspension of F4ac ETEC strain 200 (108 CFU/ml, MOI (multiplicity of infection) = 8∶1) was added. Then, the cells and bacteria were co-incubated at 37°C in a 5% CO2- 95% air atmosphere for a further 3 h. These samples were used to RNA isolation.
At 48 hours post-transfection, monolayers of cells in 6-well tissue culture plates were washed twice with PBS, and 1 ml of DMEM/F12 was added. A total of 30 µl of bacterial suspension of F4ac ETEC strain 200 (108 CFU/ml, MOI = 10∶1) was added. Then, the cells and bacteria were co-incubated at 37°C in a 5% CO2-95% air atmosphere for a further 3 h. These samples were used to measure the adhesion values of the ETEC.
RNA Preparation and Real-time RT-PCR
The IPEC-J2 cells used to isolate RNA were washed twice (E.coli non-infected) or four times (E.coli infected) with PBS, then lysed with TRIZOL Reagent (Life technologies, Carlsbad, CA, USA) directly in the culture dishes. Isolation of RNA was performed using TRIZOL Reagent following the manufacturer’s instructions.
According to the manufacturer’s instructions, complementary DNA (cDNA) was synthesized from the RNA using the Prime Script® RT reagent Kit with gDNA Eraser (Perfect Real Time) (TAKARA BIO, Dalian, China). The real time RT-PCR reactions were performed in a final volume of 20 µl with the Roche SYBR Green PCR Kit (Roche, Hercules, CA, USA) using a LightCycler® 480 Real-Time PCR System (Roche, Hercules, CA, USA). The relative RNA expression levels of siRNA targeting genes (ITGB5, MUC13), mucin genes (MUC4, MUC20), pro-inflammatory genes (IL8, IL1A, IL6, CXCL2), anti-inflammatory mediator SLPI, and PLAU were analyzed against housekeeping gene ACTB using the 2−ΔΔCt (the gene primers were listed in Table 2). Duplicate qRT-PCRs were conducted on each sample and the average Ct was used for the analysis.
Table 2. Primers for quantitative real time PCR and RT-PCR.
| Genes | GenBank Accession Number | Primers | Sequence |
| MUC13 [5] | NM_001105293 | F | 5′- GAGACTGGCTTTAGCAACCT-3′ |
| R | 5′- AGTCTATCAAACCCTCACAC-3′ | ||
| ITGB5 [5] | DQ786571 | F | 5′-GACTGTCTGCTTATCCACCC-3′ |
| R | 5′-CCATTCTTGACCAGGTTTGT-3′ | ||
| MUC4 [5] | NM_001206344 | F | 5′- AGGATGCCCAATGGCTCTACT-3′ |
| R | 5′- AAGGAGGCTGGTTCCGTTGAT-3′ | ||
| MUC20 [5] | NM_001113440 | F | 5′- CAGCAAAGACCTCTAAGATGG-3′ |
| R | 5′- CAGCAGGGAGACTTGGATGG-3′ | ||
| IL8 [5] | NM_213867 | F | 5′-CAAGCAAAAACCCATTCTCCG-3′ |
| R | 5′-CCAGCACAGGAATGAGGCATA-3′ | ||
| IL1A | NM_214029 | F | 5′-TAAGAATCTCAGAAACCCGAC-3′ |
| R | 5′-GGCTGATTTGAAGTAGTCCAT-3′ | ||
| CXCL2 [5] | NM_001001861 | F | 5′- TGCAGACCGTGCAAGGAATT-3′ |
| R | 5′- TGGCTATGACTTCCGTTTGGT-3′ | ||
| IL6 | NM_214399 | F | 5′-GAGAGCAATAAGGGAAATGTC-3′ |
| R | 5′-TCTTCATCCACTCGTTCTGT-3′ | ||
| SLPI | NM_213870 | F | 5′- CTGGGTGACTTAAAATGCTG-3′ |
| R | 5′- CAAAGTAGATGGTGGTGGTA-3′ | ||
| PLAU | NM_213945 | F | 5′-AAACCCTTCACTCCAGCACT-3′ |
| R | 5′-TTGTCGGTACGGATCTTCAG-3′ | ||
| ACTB [5] | AY550069 | F | 5′- GCTCTTCCAGCCCTCCTTCC-3′ |
| R | 5′- ACAGCACCGTGTTGGCGTAG-3′ |
[5] Reference No. 5.
Adhesion Assay
The adhesion values of the F4ac ETEC strain 200 to IPEC-J2 cells were evaluated using a real-time quantitative PCR assay of STa gene as described previously [5]. As internal standards in this study we amplified serial dilutions of the respective bacteria in PBS ranging from 1×105 to 1×102 CFU/µl.
Supporting Information
Infection with F4ac ETEC did not influence the expression of MUC13 or ITGB5 in MUC13 -KD and ITGB5 -KD IPEC-J2 cells.
(TIF)
Acknowledgments
The authors thank Prof. Dr. Michael F.G. Schmidt, Freie Universität Berlin, Germany and Dr. Junjun Wang, China Agricultural University, China for kindly providing the IPEC-J2 cell line [5], [34]. Meanwhile, grateful acknowledgements are made to the reviewers for their constructive suggestions for our manuscript.
Funding Statement
This work was supported by the Program of New Breed Development via Transgenic Technology (2009ZX08009-146B), the Project for Science and Technological Innovation of Yunnan Province (2010AB001), Beijing Municipal Bureau of Agricultural pilot demonstration projects (20100222), the Earmarked Fund for Modern Agro-Industry Technology System (CARS-37-04B) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT1191). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Johnson AM, Kaushik RS, Rotella NJ, Hardwidge PR (2009) Enterotoxigenic Escherichia coli modulates host intestinal cell membrane asymmetry and metabolic activity. Infect Immun 77: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Molle I, Joensuu JJ, Buts L, Panjikar S, Kotiaho M, et al. (2007) Chloroplasts assemble the major subunit FaeG of Escherichia coli F4 (K88) fimbriae to strand-swapped dimers. J Mol Biol 368: 791–799. [DOI] [PubMed] [Google Scholar]
- 3. Jacobsen M, Cirera S, Joller D, Esteso G, Kracht SS, et al. (2011) Characterisation of five candidate genes within the ETEC F4ab/ac candidate region in pigs. BMC Res Notes 4: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rampoldi A, Jacobsen MJ, Bertschinger HU, Joller D, Burgi E, et al. (2011) The receptor locus for Escherichia coli F4ab/F4ac in the pig maps distal to the MUC4-LMLN region. Mamm Genome 22: 122–129. [DOI] [PubMed] [Google Scholar]
- 5. Zhou C, Liu Z, Jiang J, Yu Y, Zhang Q (2012) Differential gene expression profiling of porcine epithelial cells infected with three enterotoxigenic Escherichia coli strains. BMC Genomics 13: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Melkebeek V, Rasschaert K, Bellot P, Tilleman K, Favoreel H, et al. (2012) Targeting aminopeptidase N, a newly identified receptor for F4ac fimbriae, enhances the intestinal mucosal immune response. Mucosal Immunol 5: 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren J, Yan X, Ai H, Zhang Z, Huang X, et al. (2012) Susceptibility towards enterotoxigenic Escherichia coli F4ac diarrhea is governed by the MUC13 gene in pigs. PLoS One 7: e44573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng QL, Ren J, Yan XM, Huang X, Tang H, et al. (2007) The g.243A>G mutation in intron 17 of MUC4 is significantly associated with susceptibility/resistance to ETEC F4ab/ac infection in pigs. Anim Genet 38: 397–400. [DOI] [PubMed] [Google Scholar]
- 9. Zhang B, Ren J, Yan X, Huang X, Ji H, et al. (2008) Investigation of the porcine MUC13 gene: isolation, expression, polymorphisms and strong association with susceptibility to enterotoxigenic Escherichia coli F4ab/ac. Anim Genet 39: 258–266. [DOI] [PubMed] [Google Scholar]
- 10. Ji H, Ren J, Yan X, Huang X, Zhang B, et al. (2011) The porcine MUC20 gene: molecular characterization and its association with susceptibility to enterotoxigenic Escherichia coli F4ab/ac. Mol Biol Rep 38: 1593–1601. [DOI] [PubMed] [Google Scholar]
- 11. Fu WX, Liu Y, Lu X, Niu XY, Ding XD, et al. (2012) A genome-wide association study identifies two novel promising candidate genes affecting Escherichia coli F4ab/F4ac susceptibility in swine. PLoS One 7: e32127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheng YH, Lourie R, Linden SK, Jeffery PL, Roche D, et al. (2011) The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis. Gut 60: 1661–1670. [DOI] [PubMed] [Google Scholar]
- 13. Sheng YH, Triyana S, Wang R, Das I, Gerloff K, et al. (2012) MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal Immunol 6: 557–568. [DOI] [PubMed] [Google Scholar]
- 14. Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, et al. (2011) Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 10: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aplin JD, Spanswick C, Behzad F, Kimber SJ, Vicovac L (1996) Integrins beta 5, beta 3 and alpha v are apically distributed in endometrial epithelium. Mol Hum Reprod 2: 527–534. [DOI] [PubMed] [Google Scholar]
- 16. Thygesen HH, Zwinderman AH (2005) Modelling the correlation between the activities of adjacent genes in Drosophila. BMC Bioinformatics 6: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shirkey TW, Siggers RH, Goldade BG, Marshall JK, Drew MD, et al. (2006) Effects of commensal bacteria on intestinal morphology and expression of proinflammatory cytokines in the gnotobiotic pig. Exp Biol Med (Maywood) 231: 1333–1345. [DOI] [PubMed] [Google Scholar]
- 18. Gao Y, Flori L, Lecardonnel J, Esquerre D, Hu ZL, et al. (2010) Transcriptome analysis of porcine PBMCs after in vitro stimulation by LPS or PMA/ionomycin using an expression array targeting the pig immune response. BMC Genomics 11: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adapala VJ, Buhman KK, Ajuwon KM (2011) Novel anti-inflammatory role of SLPI in adipose tissue and its regulation by high fat diet. J Inflamm (Lond) 8: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ismail HA, Quan JH, Wei Z, Choi IW, Cha GH, et al. (2012) Gene expression profiles in genetically different mice infected with Toxoplasma gondii: ALDH1A2, BEX2, EGR2, CCL3 and PLAU. Korean J Parasitol 50: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jonckheere N, Van Seuningen I (2010) The membrane-bound mucins: From cell signalling to transcriptional regulation and expression in epithelial cancers. Biochimie 92: 1–11. [DOI] [PubMed] [Google Scholar]
- 22. Chauhan SC, Ebeling MC, Maher DM, Koch MD, Watanabe A, et al. (2012) MUC13 mucin augments pancreatic tumorigenesis. Mol Cancer Ther 11: 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van den Broeck A, Vankelecom H, Van Eijsden R, Govaere O, Topal B (2012) Molecular markers associated with outcome and metastasis in human pancreatic cancer. J Exp Clin Cancer Res 31: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caporali A, Emanueli C (2011) MicroRNA regulation in angiogenesis. Vascul Pharmacol 55: 79–86. [DOI] [PubMed] [Google Scholar]
- 25. Bianchi-Smiraglia A, Paesante S, Bakin AV (2012) Integrin beta5 contributes to the tumorigenic potential of breast cancer cells through the Src-FAK and MEK-ERK signaling pathways. Oncogene 32: 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song KS, Seong JK, Chung KC, Lee WJ, Kim CH, et al. (2003) Induction of MUC8 gene expression by interleukin-1 beta is mediated by a sequential ERK MAPK/RSK1/CREB cascade pathway in human airway epithelial cells. J Biol Chem 278: 34890–34896. [DOI] [PubMed] [Google Scholar]
- 27. Kim SW, Hong JS, Ryu SH, Chung WC, Yoon JH, et al. (2007) Regulation of mucin gene expression by CREB via a nonclassical retinoic acid signaling pathway. Mol Cell Biol 27: 6933–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC (2011) Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol Cancer Res 9: 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J, Jiang J, Fu W, Jiang L, Ding X, et al. (2012) A genome-wide detection of copy number variations using SNP genotyping arrays in swine. BMC Genomics 13: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hurst LD, Pal C, Lercher MJ (2004) The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet 5: 299–310. [DOI] [PubMed] [Google Scholar]
- 31. Sheng YH, Triyana S, Wang R, Das I, Gerloff K, et al. (2012) MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal Immunol 6: 557–568. [DOI] [PubMed] [Google Scholar]
- 32. Gerold G, Abu Ajaj K, Bienert M, Laws HJ, Zychlinsky A, et al. (2008) A Toll-like receptor 2-integrin beta3 complex senses bacterial lipopeptides via vitronectin. Nat Immunol 9: 761–768. [DOI] [PubMed] [Google Scholar]
- 33. Leifheit-Nestler M, Conrad G, Heida NM, Limbourg A, Limbourg FP, et al. (2010) Overexpression of integrin beta 5 enhances the paracrine properties of circulating angiogenic cells via Src kinase-mediated activation of STAT3. Arterioscler Thromb Vasc Biol 30: 1398–1406. [DOI] [PubMed] [Google Scholar]
- 34. Arce C, Ramirez-Boo M, Lucena C, Garrido JJ (2010) Innate immune activation of swine intestinal epithelial cell lines (IPEC-J2 and IPI-2I) in response to LPS from Salmonella typhimurium. Comp Immunol Microbiol Infect Dis 33: 161–174. [DOI] [PubMed] [Google Scholar]
- 35. Pavlova B, Volf J, Alexa P, Rychlik I, Matiasovic J, et al. (2008) Cytokine mRNA expression in porcine cell lines stimulated by enterotoxigenic Escherichia coli. Vet Microbiol 132: 105–110. [DOI] [PubMed] [Google Scholar]
- 36. Devriendt B, Stuyven E, Verdonck F, Goddeeris BM, Cox E (2010) Enterotoxigenic Escherichia coli (K88) induce proinflammatory responses in porcine intestinal epithelial cells. Dev Comp Immunol 34: 1175–1182. [DOI] [PubMed] [Google Scholar]
- 37. Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, et al. (2004) Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene 23: 4681–4689. [DOI] [PubMed] [Google Scholar]
- 38. Goshima G, Vale RD (2003) The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 162: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H, Mu X, Chen J, Liu H, Shi W, et al. (2013) RNAi silencing targeting RNF8 enhances radiosensitivity of a non-small cell lung cancer cell line A549. Int J Radiat Biol. [DOI] [PubMed]
- 40. Koh SY, George S, Brozel V, Moxley R, Francis D, et al. (2008) Porcine intestinal epithelial cell lines as a new in vitro model for studying adherence and pathogenesis of enterotoxigenic Escherichia coli. Vet Microbiol 130: 191–197. [DOI] [PubMed] [Google Scholar]
- 41. Shimaoka M, Springer TA (2003) Therapeutic antagonists and conformational regulation of integrin function. Nat Rev Drug Discov 2: 703–716. [DOI] [PubMed] [Google Scholar]
- 42. Misselwitz B, Dilling S, Vonaesch P, Sacher R, Snijder B, et al. (2011) RNAi screen of Salmonella invasion shows role of COPI in membrane targeting of cholesterol and Cdc42. Mol Syst Biol 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ashida H, Mimuro H, Ogawa M, Kobayashi T, Sanada T, et al. (2011) Cell death and infection: a double-edged sword for host and pathogen survival. J Cell Biol 195: 931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thi EP, Lambertz U, Reiner NE (2012) Sleeping with the enemy: how intracellular pathogens cope with a macrophage lifestyle. PLoS Pathog 8: e1002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Infection with F4ac ETEC did not influence the expression of MUC13 or ITGB5 in MUC13 -KD and ITGB5 -KD IPEC-J2 cells.
(TIF)