Abstract
Background
Antimicrobial resistance among pneumococci has greatly increased over the past two to three decades. Resistance to tetracycline (tet(M)), chloramphenicol (cat) and macrolides (erm(B) and/or mef(A/E)) is generally conferred by acquisition of specific genes that are associated with mobile genetic elements, including those of the Tn916 and Tn5252 families. The first tetracycline-, chloramphenicol- and macrolide-resistant pneumococci were detected between 1962 and 1970; however, until now the oldest pneumococcus shown to harbour Tn916 and/or Tn5252 was isolated in 1974. In this study the genomes of 38 pneumococci isolated prior to 1974 were probed for the presence of tet(M), cat, erm(B), mef(A/E) and int (integrase) to indicate the presence of Tn916/Tn5252-like elements.
Results
Two Tn916-like, tet(M)-containing, elements were identified among pneumococci dated 1967 and 1968. The former element was highly similar to that of the PMEN1 multidrug-resistant, globally-distributed pneumococcal reference strain, which was isolated in 1984. The latter element was associated with a streptococcal phage. A third, novel genetic element, designated ICESpPN1, was identified in the genome of an isolate dated 1972. ICESpPN1 contained a region of similarity to Tn5252, a region of similarity to a pneumococcal pathogenicity island and novel lantibiotic synthesis/export-associated genes.
Conclusions
These data confirm the existence of pneumococcal Tn916 elements in the first decade within which pneumococcal tetracycline resistance was described. Furthermore, the discovery of ICESpPN1 demonstrates the dynamic variability of pneumococcal genetic elements and is contrasted with the evidence for Tn916 stability.
Keywords: Streptococcus pneumoniae, Antimicrobial resistance, Mobile genetic elements, Tetracycline resistance, ICE elements
Background
Pneumococci (Streptococcus pneumoniae) are a major bacterial cause of disease for which antimicrobial resistance is of increasing concern. A recent report estimated that in 2009 >24% of disease-causing pneumococci in the USA were tetracycline-nonsusceptible and 8.2% were chloramphenicol-nonsusceptible [1]. In some European countries, such as Spain and France, these rates were higher: 35.1% and 42.3% of pneumococci isolated in these countries, respectively, were tetracycline-nonsusceptible, while 26.1% and 20.3%, respectively were chloramphenicol-nonsusceptible [2]. Similarly, 39.2% of pneumococci in the USA [1], 26.7% of those in Spain and 30.0% of those in France were macrolide-nonsusceptible [2]. In some regions of Asia rates were even higher: 84.8%, 66.8% and 96.4% of pneumococcal isolates were tetracycline-, chloramphenicol- and erythromycin-nonsusceptible, respectively [3].
Among pneumococci, chloramphenicol and tetracycline nonsusceptibility are conferred through acquisition of the cat and tet genes (most commonly tet(M)). The cat gene encodes a chloramphenicol acetyltransferase, which catalyses the conversion of chloramphenicol into non-active derivatives. tet(M) encodes a ribosomal protection protein shown to initiate reversal of tetracycline-ribosome binding, an event which would otherwise inhibit protein synthesis [4].
Pneumococcal nonsusceptibility to erythromycin and other macrolides is most commonly conferred by acquisition of the erm(B) and/or mef(A/E) genes [5,6]. erm(B) encodes an N-methyltransferase which mediates macrolide-target site alteration [7]. mef(A/E) encodes macrolide efflux pumps [8] allowing the resistant cell to reduce its internal macrolide concentration and thereby eliminate the detrimental effects of these drugs.
In recent years it has become apparent that the majority of tet(M) and/or cat genes among tetracycline- and/or chloramphenicol-nonsusceptible pneumococci are contained within genetic elements known as integrative conjugative elements (ICEs) [9]. tet(M) is associated with the Tn916 family of ICEs, the first of which was described in 1981 from an Enterococcus faecalis isolate [10]. cat is associated with the Tn5252 ICE family, first described in 1991 [11], where, besides the typical intTn5252 integrase, a different integrase (intICESp23FST81) has recently been described [12].
To date, Tn916-like ICEs have been identified among >35 bacterial genera [13]. Tn916 itself is ~18 kb in length and contains 24 genes arranged into functional modules associated with conjugal transfer, recombination, transcriptional regulation and accessory functions (e.g. tetracycline resistance [14]). The recombination module contains the integrase (int) and excisionase (xis) genes. The int gene encodes a tyrosine recombinase responsible for integration and excision of the element from the host chromosome. The product of xis drives the directionality of the recombination process, most commonly by promoting excision [14]. Tn916 has been shown to have the ability to transpose intracellularly and to mediate its own intercellular transfer via conjugation [14]. Successful conjugal transfer between pneumococci and into pneumococci from other species is a strain-dependent property [15,16]; transfer between pneumococci by transformation has also been documented [17]. Aside from Tn916, other members of this family may contain alternate accessory genes and/or feature a smaller, independent genetic element integrated within the Tn916 genetic background, e.g. the erm(B)-containing Tn917 or Omega elements, or the mef(E)-containing MEGA (macrolide efflux genetic assembly) element [17-19].
Among pneumococci, Tn5252-like ICEs are most commonly identified in association with Tn916-like ICEs [9,11,12,18] whereby the resulting composite element is known as Tn5253 (or Tn5253-like) and may be highly variable [9,18]. In addition to the cat gene, Tn5252 (~48 kb in length) contains an independent int gene and the umuDC gene, shown to provide protection from UV damage [20]. In some cases Tn5252-like ICEs also contain a lantibiotic synthesis gene cluster [12,18]. Lantibiotics, or lanthionine-containing antibiotics, are antimicrobial compounds with activity against a range of gram-positive bacterial species [21].
There is evidence to suggest that Tn916-like and Tn5252-like elements were present among pneumococci in the 1970s [22,23] and to our knowledge the oldest pneumococcus for which such an element has been described was isolated in 1974 [11,22]. However, tetracycline-nonsusceptible pneumococci were first identified in 1962 [24], while pneumococcal resistance to erythromycin and chloramphenicol were recognised in 1967 [25] and 1970 [26], respectively. Given the important role of these ICEs in the dissemination of resistance determinants, it therefore seems likely that they, or similar genetic elements, were present among pneumococcal populations from at least as early as the 1960s. The aim of this study was to use our historical genome collection [27] to search for, characterise, and compare such older elements among pneumococci isolated prior to 1974, thus pre-dating the earliest known pneumococcal Tn916 and Tn5252 representatives [10,11,22].
Results
Nucleotide comparison of pneumococcal ICE int genes
Ten uniquely designated pneumococcal ICEs were identified from Genbank. Four elements (Tn1131, Tn5253, ICESpn11876 and ICESpn11930) each contained two independent int genes. Six elements each contained only a single int gene. Alignment of all 14 int nucleotide sequences showed four clearly defined clusters; sequences assigned to the same cluster shared >98% nucleotide identity (Table 1).
Table 1.
Integrase genes identified in specific pneumococcal genetic elements, grouped by nucleotide sequence similarity
| Group |
Integrase |
Genetic elementa |
Genbank |
|---|---|---|---|
| gene type | representativea | accession no. | |
| 1 |
intTn916 |
Tn1311(a) |
FN667862.2 |
| |
|
Tn1545 |
X61025.1 |
| |
|
Tn2010 |
AB426620.1 |
| |
|
Tn5251 |
FJ711160.1 |
| |
|
Tn5253(a) |
EU351020.1 |
| |
|
Tn6003 |
AM410044.5 |
| |
|
ICESpn11876(a) |
FR671404.1 |
| |
|
ICESpn11930(a) |
FR671403.1 |
| 2 |
intICESp23FST81 |
ICESpn11876(b) |
FR671404.1 |
| |
|
ICESpn6094 |
FR670347.2 |
| 3 |
intICESpn8140 |
ICESpn8140 |
FR671412.1 |
| 4 |
intTn5252 |
Tn1131(b) |
FN667862.2 |
| |
|
Tn5253(b) |
EU351020.1 |
| ICESpn11930(b) | FR671403.1 |
a. (a) and (b) refer to pairs of different integrase genes found in elements Tn1131, Tn5253, ICESpn11876 and ICESpn11930. Other representatives of genetic elements in these groups may be found in the ICEberg database (http://db-mml.sjtu.edu.cn/ICEberg/).
Identification of resistance determinants and transposon int genes among pneumococci isolated prior to 1974
In total, 19 unique CCs and 23 unique serotypes/groups were represented by the 38 isolates included in this study (Table 2). The BIGS database BLASTn tool [28] was used to screen the genomes of these isolates for the presence of each of the tet(M), cat, erm(B) and mef(A/E) resistance determinants, and one representative of each of the int nucleotide sequence clusters. Two isolates, 14/5 (1967) and 18C/3 (1968), were positive for both intTn916 and tet(M). Isolate PN1 (1972) did not possess tet(M), cat, erm(B) or mef(A/E) resistance determinant genes, although it had intTn5252. No other isolates were positive for any of the tet(M), cat, erm(B) or mef(A/E) resistance determinant genes, or any int genes.
Table 2.
Pneumococcal isolates included in this study
| Isolate | Serotype | Year | Country | ST | CC | Genome accession no.b |
|---|---|---|---|---|---|---|
| 7A/2 |
7A |
1937 |
Denmark |
191 |
191 |
ERR025824 |
| 14/3 |
14 |
1939 |
Denmark |
875 |
1106 |
ERR026719 |
| 17 F/2 |
17 F |
1939 |
Denmark |
123 |
113 |
ERR026724 |
| 18C/2 |
18C |
1939 |
Denmark |
4706 |
113 |
ERR026186 |
| 22A/2 |
22A |
1939 |
Denmark |
7181 |
490 |
ERR025822 |
| 18B/2 |
18B |
1941 |
Denmark |
4706 |
113 |
ERR026193 |
| 35C/2 |
35C |
1941 |
Denmark |
7196 |
113 |
ERR026192 |
| 2/3 |
2 |
1943 |
Denmark |
3744 |
490 |
ERR026715 |
| 35C/3 |
35C |
1943 |
Denmark |
5989 |
113 |
ERR025823 |
| 23A/2 |
23A |
1945 |
Denmark |
439 |
439 |
ERR025821 |
| 7B/2 |
7B |
1952 |
USA |
7180 |
66 |
ERR025825 |
| 7 F/2 |
7 F |
1952 |
USA |
7210 |
218 |
ERR026172 |
| 9 L/2 |
9 L |
1952 |
USA |
5979 |
124 |
ERR026178 |
| 9 N/2 |
9 N |
1952 |
USA |
7205 |
None3782 |
ERR026179 |
| 14/2 |
14 |
1952 |
USA |
124 |
124 |
ERR025830 |
| 17 F/1 |
17 F |
1952 |
USA |
574 |
574 |
ERR025831 |
| 19 F/8 |
19 F |
1952 |
Denmark |
7229 |
4399 |
ERR026190 |
| 2/2 |
2 |
1956 |
USA |
574 |
574 |
ERR025820 |
| 10 F/2 |
10 F |
1956 |
USA |
7186 |
490 |
ERR025828 |
| 11C/1 |
11C |
1957 |
USA |
7201 |
124 |
ERR025829 |
| 9 N/6 |
9 N |
1960 |
Denmark |
71 |
66 |
ERR026180 |
| 12 F/3 |
12 F |
1961 |
Denmark |
7228 |
4399 |
ERR026195 |
| 14/4 |
14 |
1961 |
Denmark |
134 |
124 |
ERR026188 |
| 18 F/1 |
18 F |
1961 |
USA |
7182 |
490 |
ERR026173 |
| 7 F/3 |
7 F |
1962 |
Denmark |
191 |
191 |
ERR026176 |
| 9A/1 |
9A |
1962 |
USA |
312 |
156/162 |
ERR025826 |
| 12 F/2 |
12 F |
1962 |
USA |
7194 |
Singleton7194 |
ERR026183 |
| 17 F/3 |
17 F |
1962 |
Denmark |
392 |
392 |
ERR026725 |
| 17 F/4 |
17 F |
1962 |
Denmark |
392 |
392 |
ERR026726 |
| 19 F/5 |
19 F |
1962 |
Denmark |
7229 |
4399 |
ERR026717 |
| 19 F/10 |
19 F |
1963 |
Denmark |
7230 |
Singleton7230 |
ERR026174 |
| 14/5a |
14 |
1967 |
Denmark |
15 |
15 |
ERR026720 |
| 23 F/4 |
23 F |
1967 |
Australia |
7184 |
Singleton7184 |
ERR026191 |
| 9 V/4 |
9 V |
1968 |
Denmark |
123 |
113 |
ERR025827 |
| 18C/3a |
18C |
1968 |
Denmark |
7195 |
113 |
ERR026716 |
| PN2 |
4 |
1969 |
PNG |
7152 |
Singleton7152 |
ERR163219 |
| 19 F/11 |
19 F |
1972 |
Denmark |
71 |
66 |
ERR026175 |
| PN1a | 6 | 1972 | PNG | 7151 | 277 | ERR163220 |
Note: PNG = Papua New Guinea; ST = Sequence Type; CC = Clonal Complex.
a. Isolates that possess the mobile elements discussed in this manuscript and are depicted in Figure 1.
b. European Nucleotide Archive accession number.
Within the 14/5 genome, the intTn916 and tet(M) genes were located on a Tn916 element which was structurally identical (i.e. contained the same putative genes in the same order and was a complete BLASTn match) to the Tn916 region of ICESp23FST81 (the PMEN1 pneumococcal reference isolate ICE, Figure 1). In the 14/5 genome this Tn916 ICE was inserted upstream of the pspA locus, which encodes pneumococcal surface protein A.
Figure 1.
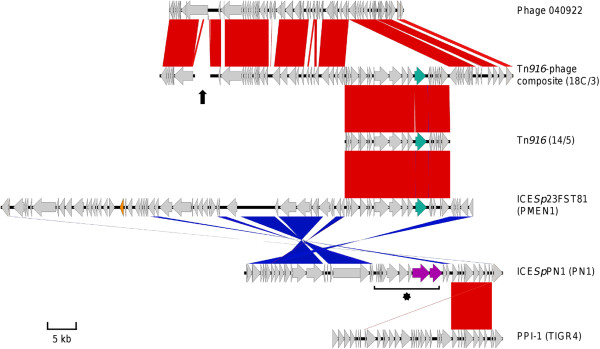
Comparison of mobile genetic elements identified in this study and those previously described. Elements are named as in text, and isolates in which these elements were identified are named in parentheses. Predicted genes are depicted by horizontal arrows; cyan arrows represent tet(M) resistance genes, the orange arrow indicates cat, and purple arrows represent lantibiotic-associated genes. Red bars represent BLASTn matches between sequences. Blue bars represent reverse-oriented BLASTn matches. The region of missing sequence within the Tn916-(bacterio)phage composite element is indicated by a vertical black arrow; the asterisk marks an ~11.3 kb insertion in ICESpPN1 (as described in Results). See Additional file 1 for details about the genes found within the Tn916-phage composite and ICESpPN1 elements.
Within the 18C/3 genome, intTn916 and tet(M) were located on a Tn916 element, itself associated with a putative bacteriophage showing similarity to the Streptococcus phage 040922 (Genbank accession no. FR671406.1; Figure 1) and inserted in the genome downstream of the trxB locus, which encodes a pyridine nucleotide-disulphide oxidoreductase. The genome of the 18C/3 bacteriophage contained 48 genes, 31 of which were predicted to encode hypothetical proteins. A further 13 genes were predicted to encode functional phage proteins such as the phage integrase, lytic amidase, holin and structural phage proteins (Additional file 1; Genbank accession no. KC488256). It should be noted here that contiguous sequence could not be obtained across the full length of the 18C/3 Tn916-bacteriophage composite element, despite repeated attempts using PCR and conventional Sanger sequencing techniques. The position of the missing region is shown in Figure 1 and likely included part of the pblB gene, which may play a role in adhesion [29]. Relative to those of 14/5 and ICESp23FST81, the 18C/3 Tn916-like sequence contained a 155 bp deletion between orf12 and tet(M).
Comparative analyses indicated that PN1 harboured a novel, composite ICE which was inserted in the genome upstream of the rbgA locus, which encodes a ribosomal biogenesis GTPase. This novel ICE, designated ICESpPN1, contained 45 genes and regions of similarity to Tn5252 and PPI-1 (Pneumococcal Pathogenicity Island 1) as described below (Figure 1 and Additional file 1; Genbank accession no. KC488257).
The 15 5’-most genes of ICESpPN1 showed similarity to those of the Tn5252-like region of ICESp23FST81 and included repA, which encodes replication initiation factor A. A further four and two putative genes also showed similarity to those of Tn5252 and were separated by a ~11.3 kb insertion (Figure 1). This insertion contained 10 putative genes, including four with ≥96% nucleotide sequence identity to those of a two-component signalling system described for Streptococcus mitis strain B6 [30] and S. pneumoniae ICESpn8140 [18]. These genes were predicted to encode an ABC-type antimicrobial peptide transporter, an ABC-type transporter permease, a sensor histidine kinase and a response regulator. Directly 3’ of the two-component system cluster were two genes for which the predicted peptide products showed 64% amino acid identity to the LanM lantibiotic and 76% amino acid identity to a lantibiotic transporter, respectively.
The 3’-most ICESpPN1 gene represented intTn5252 and was preceded by a putative excisionase gene, predicted to encode a protein with 83% amino acid identity to a putative Streptococcus sp. excisionase. Directly upstream of the putative excisionase gene was a region which was structurally identical to part of PPI-1 of TIGR4, another pneumococcal reference isolate (Genbank accession no. NC_003028.3, see Figure 1). This region spanned nine predicted genes, the predicted protein products of which included the PlcR putative transcriptional regulator, an IS200 family transposase, a putative protein kinase, and both a putative ABC-transporter ATP-binding protein and permease.
Comparison of Tn916 sequence regions
The 14/5 Tn916-like nucleotide sequence differed from that of ICESp23FST81 by only 101 substitutions and a 5 bp insertion within orf14. 94 of the nucleotide differences between these sequences were located within the tet(M) gene, which is consistent with the previous observation that tet(M) possesses a mosaic nucleotide sequence [31]. Additionally, this Tn916-like sequence shared >99% nucleotide identity with Tn916-like ICEs identified among contemporary bacterial isolates including Streptococcus suis, Streptococcus parauberis and Staphylococcus aureus[32-34]. The 18C/3 Tn916-like nucleotide sequence was highly similar to that of the 14/5, differing by only two nucleotide substitutions (and the deletion described above).
Discussion
This study of an historical collection of pneumococci has led to the discovery of two of the earliest known representatives of pneumococcal Tn916-like ICEs and a novel composite ICE. The Tn916-like ICEs from pneumococci isolated in 1967 and 1968, respectively, were highly similar to the Tn916-like region of ICESp23FST81 from the PMEN1 reference strain dated 1984 [12], and contemporary Tn916-like ICEs. Both of these ICEs contained the tetracycline-resistance determinant tet(M), but did not contain any other resistance determinant genes. Identification of such elements among pneumococci isolated in the late 1960s confirms their existence from the first decade within which tetracycline resistance was reported among pneumococci [24]. (Note that tetracycline was released for use in 1948, chloramphenicol in 1947 and erythromycin in 1952 [35].)
The Tn916-like element dated 1968 in isolate 18C/3 was inserted within a phage showing similarity to the Streptococcus phage 040922 (Genbank accession no. FR671406). Bacteriophages are known to mediate horizontal gene transfer between bacteria through the process of transduction [36], although it is not clear to what extent this process occurs amongst pneumococci, or whether the 040922 phage has retained this ability. Pneumococcal Tn916-like ICEs have not generally been associated with phage [9,16,17,19] and no such other phage-associated representatives are present in the Genbank database. Thus it seems unlikely that phage-mediated transduction has played an important role in the dissemination of Tn916-like ICEs, although it is possible that the 18C/3 ICE was acquired in this way.
Aside from the tet(M) genes described above, no other tet(M), cat, erm(B) or mef(A/E) resistance determinants were identified among the pneumococcal genomes studied here. There are other resistance mechanisms such as erm(A), erm(TR) or tet(O) that have been found in pneumococci but are believed to be rare [37,38]. We searched for these genes among our genome collection and found no evidence for them, thus in this study we focussed on the common tetracycline, chloramphenicol and erythromycin resistance mechanisms among pneumococci.
In addition to those described above, an ICE-associated int gene was identified among one further isolate, PN1, dated 1972. It is also worth noting that this strain was among the earliest penicillin-nonsusceptible pneumococci to be identified, by virtue of the possession of altered penicillin-binding protein genes [27]. Further analyses indicated that this pneumococcus harboured a novel composite ICE, designated ICESpPN1, which contained regions of similarity to the previously described Tn5252-like ICEs and PPI-1 of the TIGR4 reference isolate. Additionally this ICE contained a two-component signalling system gene cluster and two putative lantibiotic biosynthesis-/export- associated genes.
Lantibiotics are small, lanthionine-containing antimicrobial peptides. Synthesis and secretion follow detection of extracellular signals and are auto-regulated by two-component signal systems such as that putatively described above for ICESpPN1. Antimicrobial activity is achieved following post-translational modification. Production of immunity proteins is required to protect the host cell [21]; however, no such post-translational modification enzyme- or immunity protein- associated genes were identified within ICESpPN1, calling into question the ability of PN1 to produce a functional version of this lantibiotic.
PPI-1 contains a 5’ region which is highly conserved among pneumococci and a 3’ region which is not conserved (with the exception of the most 3’ gene) [12,39]. ICESpPN1 contained a region which was structurally identical to the non-conserved region of TIGR4 PPI-1. Previously it was noted that loci within the conserved regions of PPI-1 showed similarity to those of Tn5252[39]. Subsequently it was suggested that sequences may be exchanged between these elements via homologous recombination [12]. Such a process could explain the patterns of shared sequence structure described here.
Previous authors have demonstrated the capacity for diversity among pneumococcal ICEs [9,18], and the description of ICESpPN1 reiterates this dynamic variability. The finding that two pneumococci isolated in the late 1960s harboured Tn916-like ICEs, and that these ICEs were highly similar to that of the epidemiologically-successful PMEN1 reference strain and contemporary Tn916-like elements, demonstrates the ability of these ICEs to persist, almost unchanged (≥99% nucleotide sequence identity), within bacterial populations for many years. The phenotypic effects associated with possession of such ICEs have an important impact on our ability to treat pneumococcal disease. In this context, understanding the processes driving the spread, maintenance and/or diversification of these elements is of the utmost importance.
Conclusions
In this study we discovered the oldest known examples of tetracycline resistance-conferring pneumococcal genetic elements: two different Tn916-like, tet(M)-containing, elements identified among pneumococci dated 1967 and 1968. The former element was highly similar to that of the PMEN1 multidrug-resistant, globally-distributed pneumococcal reference strain isolated in 1984. The latter element was uniquely associated with a streptococcal phage. We also described a novel ICE element in a pneumococcal isolate recovered in Papua New Guinea in 1972, and interestingly, this isolate was also one of the earliest penicillin-nonsusceptible pneumococci. This novel element, designated ICESpPN1, contained a region of similarity to Tn5252, a region of similarity to a pneumococcal pathogenicity island and novel lantibiotic synthesis/export-associated genes. The importance of antimicrobial resistance among pneumococci is unequivocal, and our work sheds further light on how these particular resistance determinants have evolved.
Methods
Genomic sequencing, serotype and genotype data
Whole-genome sequence data for pneumococci sampled from an historical isolate collection were previously generated and described [27]. Thirty-six of these genome sequences represented pneumococci isolated prior to 1974 and were thus included in this study (Table 2). The genomes of two additional pneumococci, PN2 and PN1, isolated in 1969 and 1972 respectively (Table 2), had recently been added to our historical isolate collection and were included because they were also isolated before 1974. These genome sequences were generated on the Illumina Hi-seq platform; production of 200 bp insert libraries was followed by 100 nucleotide paired-end sequencing using standard protocols. Illumina reads were assembled to consensus contigs using Velvet [40]. Data were deposited in a BIGS database [28]. Serotype/group and genotype data (as defined by multilocus sequence typing [41]) for all isolates were previously described. Isolates were assigned to clonal complexes (CCs – clusters of genotypes descended from a recent common ancestor) by a modified goeBURST method [27,42].
Identification of putative ICEs
Reference nucleotide sequences for each of the tet(M), cat, erm(B) and mef(A/E) resistance determinants were retrieved from Genbank. int gene sequences were retrieved from each uniquely designated pneumococcal ICE, identified by interrogation of Genbank using the following search term combinations: Organism = Streptococcus pneumoniae + Title = Transposon, Organism = Streptococcus pneumoniae + Title = Conjug*, Organism = Streptococcus pneumoniae + Title = element. int nucleotide sequences were aligned by MUSCLE [43] and imported to MEGA5 [44] for assignment to clusters (by visual comparison), and sequence identity calculation. The BIGS database BLASTn tool [28] was used to search isolate genomes for the tet(M), cat, erm(B) and mef(A/E) resistance determinants, and a single representative of each int gene cluster.
Isolates positive for any resistance determinant or int gene were further studied by extraction of the relevant Velvet contig from the BIGS database and comparison to the genomes of the pneumococcal reference strains R6 and PMEN1 (Genbank accession no. AE007317.1 and NC_011900.1, respectively) using the Artemis Comparison Tool (ACT) [45]. Candidate genetic elements were identified as those which contained one or more of the resistance determinants of interest (i.e. tet(M), cat, erm(B) and mef(A/E)) and/or int genes, and were not similar to any region of the R6 genome, which does not contain any Tn916/Tn5252-like ICEs. Candidate Tn916-like and/or Tn5252-like elements were further identified as those which showed similarity to the Tn916 and/or Tn5252-like regions of the PMEN1 reference ICE, ICESp23FST81 [12]. Regions of the resistant determinant/int-containing contigs that did not show similarity to any part of the R6 genome or PMEN1 ICE were extracted and queried against the Genbank database by BLASTn. Sequences representing the best BLAST matches were retrieved from the database for further comparison using ACT.
Identification of additional ICE regions and genome integration sites
When a single Velvet contig contained putative ICE sequences plus regions showing similarity to the R6 reference genome, the latter were considered to represent the genomic flanking regions of the ICE. Consequently putative ICE genomic integration sites were identified by reference to the R6 genome annotation.
When genomic flanking sequences were not contiguous to the putative ICE sequences, SMALT [46] was used to map Illumina sequence reads to the Velvet consensus assembly. Mapped assemblies were converted to a gap5 database [47]. Illumina reads that mapped to the ends of the putative ICE-containing contigs were checked for the location of their corresponding paired reads. This enabled the identification of consensus assembly contigs representing the adjacent region(s) of the genome. Comparison of these contigs to the R6 genome using ACT identified the putative ICE integration sites. Sequences which were not similar to the R6 genome were considered additional ICE regions and were queried against the Genbank database by BLAST.
Association of the original ICE assembly contigs, additional putative ICE contigs and/or the putative genomic flanking region contigs was confirmed by standard PCR amplification in 25 μl reaction volumes followed by agarose gel electrophoresis (primers available upon request). Conventional Sanger sequencing was used to close sequence gaps within putative ICEs. PCR products to be used for Sanger sequencing were precipitated with 60 μl of 20% PEG (polyethylene glycolate) / 2.5 M NaCl and washed with 70% ETOH. Sequencing was completed as described previously [27].
Prediction of genes and Tn916 sequence comparison
Putative genes were predicted using Prodigal [48]. Where possible, putative functions were assigned by BLAST match to sequences deposited in Genbank. Tn916 sequence regions were aligned by MUSCLE [43] and imported to MEGA5 [44] for visual inspection / sequence identity calculation.
Abbreviations
ABC: ATP-binding cassette; ACT: Artemis Comparison Tool; BIGS: Bacterial Isolate Genome Sequence; cat: Chloramphenicol resistance gene; CC: Clonal complex; erm(B) and/or mef(A/E): Erythromycin resistance genes; ICE: Integrative Conjugative Element; int: Integrase gene; MEGA: Macrolide efflux genetic assembly; MEGA5: Molecular Evolutionary Genetics Analysis 5; PCR: Polymerase chain reaction; PMEN: Pneumococcal Molecular Epidemiology Network; PPI-1: Pneumococcal Pathogenicity Island 1; ST: Sequence type; tet(M): Tetracycline resistance gene; UV: Ultraviolet; xis: Excisionase gene.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KLW and ABB designed the study, analysed data, wrote the paper; LML and RH provided bacterial isolates; JP and SDP provided whole genome sequencing data; KLW and AVT assembled whole genome sequence data and performed bioinformatics analyses to close sequence gaps; KLW and ABB performed PCR and Sanger sequencing reactions to manually close sequence gaps. All authors discussed the results, read and approved the manuscript for publication.
Supplementary Material
(Genes identified in the phage-Tn916 composite element found in pneumococcal isolate 18C/3) and Table S2. (Genes identified in the novel ICE element, ICESpPN1, which was found in the pneumococcal isolate PN1).
Contributor Information
Kelly L Wyres, Email: kelly.wyres@googlemail.com.
Andries van Tonder, Email: andries.vantonder@zoo.ox.ac.uk.
Lotte M Lambertsen, Email: lotte_lambertsen@yahoo.dk.
Regine Hakenbeck, Email: hakenb@rhrk.uni-kl.de.
Julian Parkhill, Email: parkhill@sanger.ac.uk.
Stephen D Bentley, Email: sdb@sanger.ac.uk.
Angela B Brueggemann, Email: angela.brueggemann@zoo.ox.ac.uk.
Acknowledgements
This work was supported by the Wellcome Trust through grants 083511 (awarded to ABB, a Wellcome Trust Career Development Fellow) and 098051 (awarded to the Sanger Institute core sequencing and informatics teams). Figure 1 was produced using the Easyfig software (http://sourceforge.net/projects/easyfig/).
References
- Jones RN, Sader HS. Declining antimicrobial susceptibility of Streptococcus pneumoniae in the United States: report from the SENTRY antimicrobial surveillance program (1998–2009) Diagn Microbiol Infect Dis. 2010;68:334–336. doi: 10.1016/j.diagmicrobio.2010.08.024. [DOI] [PubMed] [Google Scholar]
- European Antimicrobial Resistance Surveillance Network (EARS-Net) database. [ http://www.ecdc.europa.eu]
- Kim SH, Song J, Chung DR, Thamlikitkul V, Yang Y, Wang H, Lu M, So TM, Hsueh PR, Yasin RM. et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56(3):1418–1426. doi: 10.1128/AAC.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose K, Stephens DS. In: The Pneumococcus. Tuomanen EI, editor. Washington D. C: ASM Press; 2004. Macrolide, quinolone, and other non-β-lactam antibiotic resistance in Streptococcus pneumoniae; pp. 351–366. [Google Scholar]
- Farrell DJ, Klugman KP, Pichichero M. Increased antimicrobial resistance among nonvaccine serotypes of Streptococcus pneumoniae in the pediatric population after the introduction of 7-valent pneumococcal vaccine in the United States. Pediatr Infect Dis J. 2007;26(2):123–128. doi: 10.1097/01.inf.0000253059.84602.c3. [DOI] [PubMed] [Google Scholar]
- Calatayud L, Ardanuy C, Cercenado E, Fenoll A, Bouza E, Pallares R, Martin R, Liñares J. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob Agents Chemother. 2007;51(9):3240–3246. doi: 10.1128/AAC.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39(3):577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41(10):2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson-Begg SK, Roberts AP, Hall LMC. Diversity of putative Tn5253-like elements in Streptococcus pneumoniae. Int J Antimicrob Agents. 2009;33(4):364–367. doi: 10.1016/j.ijantimicag.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoubi P, Kilic AO, Vijayakumar MN. Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol. 1991;173(5):1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Walker D, Romero P, Lennard N, Paterson GK, Bason NC, Mitchell AM, Quail MA, Andrew PW, Parkhill J. et al. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniaeSpain23F ST81. J Bacteriol. 2009;191(5):1480–1489. doi: 10.1128/JB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. Fems Microbiol Rev. 2011;35(5):856–871. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17(6):251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Celli J, Poyart C, Trieu-Cuot P. Use of an excision reporter plasmid to study the intracellular mobility of the conjugative transposon Tn916 in gram-positive bacteria. Microbiology. 1997;143:1253–1261. doi: 10.1099/00221287-143-4-1253. [DOI] [PubMed] [Google Scholar]
- Santoro F, Oggioni MR, Pozzi G, Iannelli F. Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 2010;308:150–158. doi: 10.1111/j.1574-6968.2010.02002.x. [DOI] [PubMed] [Google Scholar]
- Del Grosso M, Scotto d'Abusco A, Iannelli F, Pozzi G, Pantosti A. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2004;48(6):2037–2042. doi: 10.1128/AAC.48.6.2037-2042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS. et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Grosso M, Camilli R, Iannelli F, Pozzi G, Pantosti A. The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: Insertion sites and association with other genetic elements. Antimicrob Agents Chemother. 2006;50(10):3361–3366. doi: 10.1128/AAC.00277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Najar U, Vijayakumar MN. An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J Bacteriol. 1999;181(9):2782–2788. doi: 10.1128/jb.181.9.2782-2788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey JM, van der Donk WA. Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- Dang-Van A, Tiraby G, Acar JF, Shaw WV, Bouanchaud DH. Chloramphenicol resistance in Streptococcus pneumoniae: enzymatic acetylation and possible plasmid linkage. Antimicrob Agents Chemother. 1978;13(4):577–583. doi: 10.1128/AAC.13.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey RC, Swenson JM, Clark NC, Thornsberry C. DNA hybridization studies of a nucleotide sequence homologous to transposon Tn1545 in the "Minnesota" strain of multiresistant Streptococcus pneumoniae isolated in 1977. Diagn Microbiol Infect Dis. 1989;12(1):13–16. doi: 10.1016/0732-8893(89)90038-2. [DOI] [PubMed] [Google Scholar]
- Evans W, Hansman D. Tetracycline-resistant pneumococcus. Lancet. 1963;281:451. doi: 10.1016/s0140-6736(75)91547-0. [DOI] [PubMed] [Google Scholar]
- Dixon JMS. Pneumococcus resistant to erythromycin and lincomycin. Lancet. 1967;289:573. doi: 10.1016/s0140-6736(67)92803-6. [DOI] [PubMed] [Google Scholar]
- Cybulska J, Jeljaszewicz J, Lund E, Munksgaard A. Prevalence of types of Diplococcus pneumoniae and their susceptibility to 30 antibiotics. Chemotherapy. 1970;15:304–316. doi: 10.1159/000220695. [DOI] [PubMed] [Google Scholar]
- Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Liñares J, JM R, Kristinsson KG, Beall BW, Klugman KP. et al. The multidrug-resistant PMEN1 pneumococcus is a paradigm for genetic success. Genome Biol. 2012;13(11):R103. doi: 10.1186/gb-2012-13-11-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;10(11):595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Croucher NJ, Hiller NL, Hu FZ, Ehrlich GD, Bentley SD, García E, Mitchell TJ. Comparative genomic analysis of ten Streptococcus pneumoniae temperate bacteriophages. J Bacteriol. 2009;191(15):4854–4862. doi: 10.1128/JB.01272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denapaite D, Brückner R, Nuhn M, Reichmann P, Henrich B, Maurer P, Schähle Y, Selbmann P, Zimmermann W, Wambutt R. et al. The genome of Streptococcus mitis B6 - what is a commensal? PLoS One. 2010;5(2):e9426. doi: 10.1371/journal.pone.0009426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oggioni MR, Dowson CG, Smith JM, Provvedi R, Pozzi G. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid. 1996;35(3):156–163. doi: 10.1006/plas.1996.0018. [DOI] [PubMed] [Google Scholar]
- Holden MTG, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C. et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS ONE. 2009;4(7):e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Kanai K, Yoshikoshi K. Structural characterization of Tn916-like element in Streptococcus parauberis serotype II strains isolated from diseased Japanese flounder. Lett Appl Microbiol. 2009;48(6):770–776. doi: 10.1111/j.1472-765X.2009.02609.x. [DOI] [PubMed] [Google Scholar]
- Schijffelen MJ, Boel CH, van Strijp JA, Fluit AC. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics. 2010;11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR. Humans as the world's greatest evolutionary force. Science. 2001;293:1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt W-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna VA, Roberts MC. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J Antimicrob Chemother. 1998;42(5):613–619. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- Lambertsen LM, Ekelund K, Hansen DS, Kaltoft M, Christensen JJ, Hammerum AM. Erythromycin resistance caused by erm(A) subclass erm(TR) in a Danish invasive pneumococcal isolate: are erm(A) pneumococcal isolates overlooked? Scand J Infect Dis. 2008;40:6–7. doi: 10.1080/00365540701854717. [DOI] [PubMed] [Google Scholar]
- Brown JS, Gilliland SM, Holden DW. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol. 2001;40(3):572–585. doi: 10.1046/j.1365-2958.2001.02414.x. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- Francisco AP, Bugalho M, Ramirez M, Carriço JA. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics. 2009;10:152. doi: 10.1186/1471-2105-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, MA R, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21(16):3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Sanger Institute - SMALT. [ http://www.sanger.ac.uk/resources/software/smalt/]
- Bonfield JK, Whitwham A. Gap5–editing the billion fragment sequence assembly. Bioinformatics. 2010;26(14):1699–1703. doi: 10.1093/bioinformatics/btq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Genes identified in the phage-Tn916 composite element found in pneumococcal isolate 18C/3) and Table S2. (Genes identified in the novel ICE element, ICESpPN1, which was found in the pneumococcal isolate PN1).


