Abstract
Background
Telomere maintenance in Drosophila relies on the targeted transposition of three very special non-LTR retrotransposons, HeT-A, TART, and TAHRE (HTT). The sequences of the retrotransposon array build up the telomere chromatin in this organism. We have recently reported the role of the chromosomal protein Putzig/Z4 in maintaining a proper chromatin structure at the telomere domain of Drosophila. Because the Putzig protein has been found in different cellular complexes related with cell proliferation, development, and immunity, we decided to investigate whether the previously described Putzig partners, DREF/TRF2 and KEN, could also be involved in the telomere function in this organism.
Results
We have found that mutant alleles for Dref/Trf2 and Ken show alterations in HeT-A and TART expression, suggesting a possible role of these protein complexes in the regulation of the telomere retrotransposons. In agreement, both HeT-A and TART contain the specific DNA binding sequences for the DREF and the KEN protein proteins.
Conclusions
We have identified three new negative regulators involved in the control of the expression of the telomeric retrotransposons, Dref, Trf2, and Ken. Our results offer some clues on which other chromatin-related proteins might be involved in telomere regulation and retrotransposon control.
Background
The telomeres in Drosophila are constituted by an array of three specialized non-LTR retrotransposons, HeT-A, TART, and TAHRE (HTT array), whose targeted transpositions at the end of the chromosomes are analogous to the telomere replication performed by the holoenzyme telomerase in most eukaryotes [1-3]. Since the main genes involved in telomere elongation in Drosophila are embedded at the telomere chromatin, a study of the regulation and structure of the chromatin at this domain is important in understanding the telomere function in this organism. The chromatin at the telomere domain, the HTT array, attracts a different set of proteins from the subtelomeric domain, telomere associated sequences (TAS), and nucleates a specific class of chromatin with mixed characteristics of heterochromatin and euchromatin [4-6], and RSS, unpublished observations).
The chromosomal protein Z4/Putzig (Pzg) is a seven zinc-finger protein known to localize at polytene chromosome interbands and necessary to maintain the band-interband structure in these chromosomes [7]. A study using a Drosophila mutant line, tel1 characterized by the presence of telomeres ten times longer than the average wild-type Drosophila telomeres [8], identified Pzg as a component of the telomere domain [4]. These findings led us to investigate the role of Pzg at Drosophila telomeres. We found that the lack of Pzg disturbs the structure of the telomeric chromatin affecting the stability of the telomeres and causing telomere fusions (TFs) [6]. The telomere function of Pzg is coordinated with other proteins present at the HTT array, such as JIL-1 or HP1a [6]. The equilibrium between these proteins is one of the keys to obtaining a precise level of expression of the telomere retrotransposons, HeT-A and TART. A recent study has also confirmed the presence of Pzg at the telomeres when screening for proteins that interact with another component of the HTT array, the Prod protein [9,10].
Pzg is not a telomere-specific protein and has been shown to be an important cofactor in at least three different pathways related with chromatin remodeling. In most cases, Pzg exerts its effects by mediating chromatin changes and acts as an activator; these are the cases of the nucleosome remodeling factor (NURF) and the DREF/TRF2 complexes [11,12]. The role of Pzg in the DREF/TRF2 complex is related with the necessary remodeling of the chromatin around the promoters of replication-related genes. The DREF homo-dimer binds specifically to the DRE sequence and, together with TRF2, is required for the cellular shift from the resting state into the proliferating state [13]. Nevertheless, Pzg can also negatively regulate the expression, as, for example, when it directly binds the co-repressor KEN in the JAK/STAT pathway [14]. The identification of Pzg in a protein complex composed of KEN and NURF in immunoprecipitation experiments, together with the observation of melanotic tumors in pzg mutant flies, which was due to an overexpression of defense response genes, strongly suggested the involvement of Pzg and NURF in the transcriptional repression of the JAK/STAT pathway genes [14,15].
Understanding whether any of these mechanisms involving Pzg could be linked to its telomere role is relevant to a better understanding of both telomere biology in Drosophila and how the regulation of the non-LTR retrotransposons HeT-A and TART could be related to the replication or defense mechanism of the organism.
Results
Mutations in Dref, Trf2, and Ken affect the telomeric retrotransposons HeT-A and TART
We investigated whether mutations in Dref, Trf2, and Ken affected the expression of the telomeric retrotransposons HeT-A and TART. As mentioned, Pzg has been found in the same protein complexes as DREF and KEN. TRF2 (TATA-box-binding protein (TBP) related factor 2) has been found in a complex with NURF and DREF. It was demonstrated that the recognition of promoters of replication-related genes by TRF2 depended on the presence of DREF that directly binds to specific DNA motifs [13]. If the function of Pzg at the telomeres depends on the action of the DREF complex, we would expect similar effects of mutant alleles of Trf2 and Dref over the telomere retrotransposons. Therefore, the mutant alleles included in this study are DrefKG0994, Trf260071, and Ken1 mutants.
We have previously observed that null mutants of pzg do not affect the expression of the HeT-A retrotransposon [6], although the hypomorph mutant Z47.1 affects its expression and genomic copy number [6]. We included two different pzg mutant alleles in these studies, the pzg hypomorph (Z47.1) and a pzg null mutant (pzg66), in order to investigate whether they affected the expression of the TART retrotransposon. For all mutants, we analyzed the levels of HeT-A and TART mRNA by quantitative real-time PCR. Because the number of copies of the telomeric retrotransposons varies among stocks, we normalized the HeT-A and TART mRNA data by the number of copies of the retrotransposon in each stock, to obtain the level of expression of each copy of HeT-A and TART. Moreover, the presence of a higher copy number of one of the telomere retrotransposons in a mutant allele could be indicative of an increased expression and rate of terminal transposition.
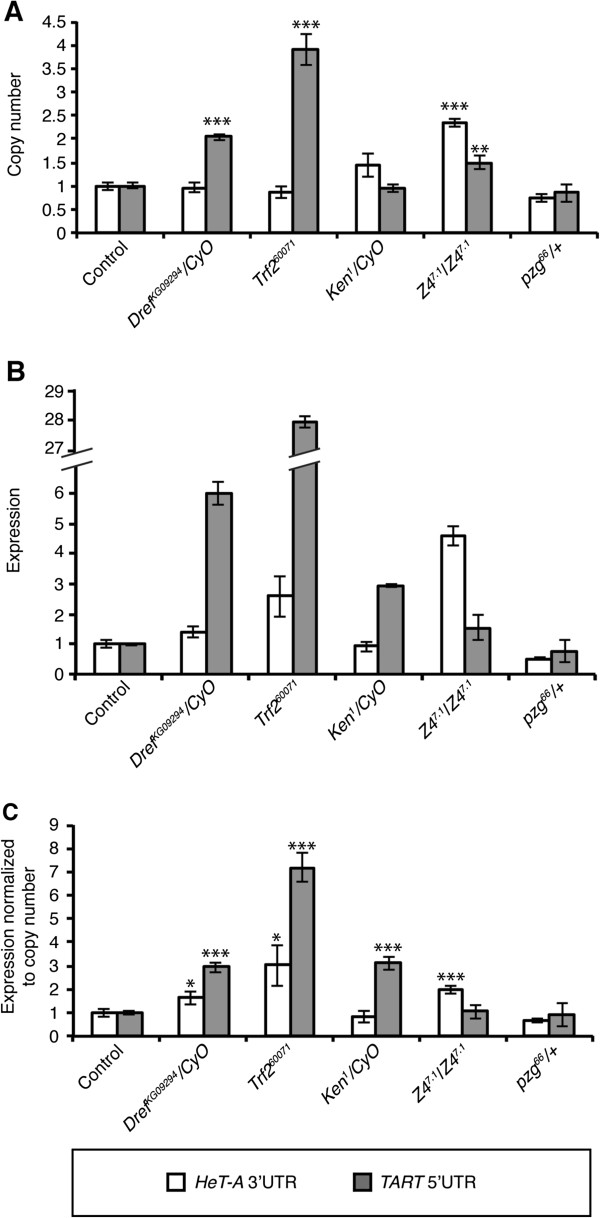
To determine the number of copies of HeT-A and TART in each stock, we extracted genomic DNA from third-instar larvae without salivary glands. DrefKG0994, Trf260071, and Ken1 mutant alleles did not show differences in HeT-A copy number (Figure 1A), but in the Z47.1/Z47.1 hypomorph mutant, an increase in HeT-A copy number was observed, as we have previously demonstrated [6]. Results for the TART element are different; DrefKG0994, Trf26007,1 and the Z47.1/Z47.1 alleles show a significant increase of TART copies in their genomes, while Ken1 and the null allele of pzg, pzg66, do not show a significant change in the number of copies of TART in their genomes (Figure 1A).
Figure 1.
HeT-A and TART expression and copy number in Dref, Trf2, Ken, and pzg mutants. (A)Dref and Trf2 mutants have more TART copies than control flies but no difference in HeT-A copies is observed. Ken and pzg66/+ mutants do not affect HeT-A or TART copy number. Z47.1/Z47.1 mutants have more HeT-A and TART copies. (B,C)HeT-A transcripts increase in Dref, Trf2, and Z47.1/Z47.1 mutants but no effect is observed in Ken mutants. TART transcripts increase in Dref, Trf2, and Ken mutants and do not change in pzg mutants. HeT-A is represented in white bars and TART in grey bars. Error bars represent standard deviations of three independent experiments. Asterisks indicate statistically significant differences (* P < 0.05 to 0.01; ** P < 0.01 to 0.001; ***, P < 0.001) in HeT-A and TART expression and copy number of each mutant compared with respective controls. For all the analyzed stocks, the number of copies and level of expression were normalized to their respective controls, w1118 and ry506, which in turn were normalized to 1, to simplify the interpretation of the data.
Next, we analyzed the mRNA levels of HeT-A and TART in the same mutants. To obtain the expression data we extracted mRNA from whole third-instar larvae and analyzed them by quantitative real-time PCR. Unlike HeT-A, the TART element does not show an increase in transcription in any of the pzg mutant alleles after normalizing the data. On the other hand, in accordance with the genomic copy number obtained, a significant increase in TART expression was observed for the DrefKG0994 and the Trf260071 alleles. These same alleles show a similar behavior for the expression of the HeT-A retrotransposon although less accentuated than for its telomere partner (Figure 1B,C). These observations are in accordance with a possible link between the role of Pzg and the DREF/TRF2 protein complexes in the control of the telomere retrotransposons. Finally, the Ken1 allele also shows an increase in TART expression although no effect in HeT-A expression was observed (Figure 1B,C). These results indicate that DrefKG0994, Trf260071, and Ken1 mutant alleles affect gene expression differently at the telomere in Drosophila.
The TART promoter contains DREF binding sequences
After showing that the DREF/TRF2 complex had a role in controlling the expression of the telomeric retrotransposons, HeT-A, TART, and TAHRE, we searched for the presence of the DREF binding sequence (5′-TATCGATA) along the sequence of the telomeric retrotransposons [16]. We did not find the DREF motif in the sequence of HeT-A and TAHRE. However, we were able to identify two DREF binding sequences in TART; one in the 5′ UTR around 170 bp downstream of the transcription start site and another around 600 bp upstream the end of the 3′ UTR. To investigate whether these binding sites were conserved among the different TART subfamilies we performed a nucleotide sequence alignment using ClustalW software with the available copies in the databases (Figure 2). The sequence alignment revealed that the DREF binding site at the 5′ UTR was highly conserved among all TART subfamilies (Figure 2A) while the 3′ UTR sequence was only present in the TART A subfamily (Figure 2B). The DREF binding site at the TART 5′ UTR is located at the TART promoter that drives sense transcription, while the 3′ UTR binding site lies around the area where a putative TART anti-sense promoter has been proposed [17].
Figure 2.

DREF binding sites. Nucleotide sequence alignment of TART A, B, and C subfamilies using ClustalW software. (A)TART 5′ UTR alignment; DREF binding site is 100% conserved in all TART subfamilies. (B)TART 3′ UTR alignment; DREF binding site is only conserved in the TART A subfamily. The alignment was performed for each domain, 5′ UTR, open reading frame, and 3′ UTR, but only the region corresponding to the DREF binding site is shown. Numbers on the right of the alignment refer to the position corresponding to each sequence.
HeT-A, TART and TAHRE contain KEN binding sites
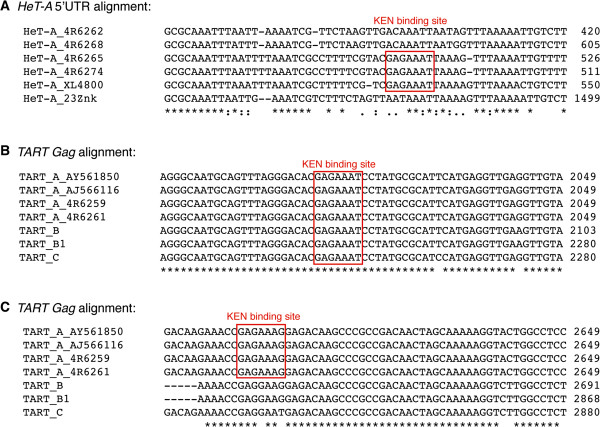
Similarly, we searched for KEN binding sites (5′-GAGAAAK, K = G/T) [18] in HeT-A, TART, and TAHRE. We found that this sequence was present in the three telomeric retrotransposons. HeT-A has a KEN binding site at the 5′ UTR that is conserved in three of the six analyzed sequences (Figure 3A). For this analysis, we only used the complete HeT-A sequences available in the databases (for more information, see [19]). TART has two KEN binding motifs inside the Gag coding sequence (ORF1); one is present in the three TART subfamilies, while the other is only present in the TART A subfamily (Figure 3B,C). Finally, in TAHRE, we found six copies of the KEN binding site (data not shown), three at the 5′ UTR, one at the reverse transcriptase domain (ORF 2), and two at the 3′ UTR. We could not perform a sequence multi-alignment analysis for TAHRE, since only one full sequence is available in databases [20].
Figure 3.
KEN binding sites. (A)HeT-A 5′ UTR alignment; KEN binding site is conserved in three of the six HeT-A sequences analyzed. (B)TART Gag alignment; KEN binding site is conserved in the three TART subfamilies. (C)TART Gag alignment; the second KEN binding site is only conserved in the TART A subfamily. Nucleotide sequence alignment was performed using ClustalW software. The alignment was performed for each domain, 5′ UTR, open reading frame, and 3′ UTR, but only the region corresponding to the DREF binding site is shown. Numbers on the right of the alignment refer to the position corresponding to each sequence.
Both these observations, the alterations in the levels of expression of the telomere retrotransposons in mutant alleles of Dref, Trf2, and Ken and the presence of both DREF and KEN motifs in the sequences of the different telomere retrotransposons, suggest that the complexes that these proteins nucleate are able to bind to the telomeric array, and therefore that they are susceptible to regulate the expression of the telomeric retrotransposons. In future, in vivo evidence for these bindings will confirm the regulatory role of the DREF/TRF2 and Ken protein complexes at the telomere array.
Discussion
We have previously demonstrated that the product encoded by the Z4/putzig gene is involved in maintaining telomere stability in Drosophila[6]. Because Pzg is not a telomere-specific protein and has been related with different protein complexes involved in diverse cellular functions, we decided to investigate whether the role of Pzg at the telomeres could involve any of the previously defined Pzg partners from other protein complexes [11,13-15].
Dref and Trf2 control the expression of HeT-A and TART
Although a previous study had reported that no DREF binding sites were present at the HTT array [10], we identified two DREF binding sites in the TART element of D. melanogaster; one at the 5′UTR, highly conserved among TART subfamilies, and another at the 3′UTR, only conserved in the TART A subfamily (Figure 2). The high conservation of the 5′UTR DREF binding site, suggests the existence of selective pressure acting at this DNA binding motif. Interestingly, the TART A subfamily, the one that contains both DREF binding motifs, is the more abundant of the TART subfamilies in D. melanogaster[21]. In addition to the presence of the DREF DNA binding motif in the sequence of the TART element, the recruitment of the DREF/TRF2 complex at the telomeres has been recently demonstrated [10].
We have observed an increase of transcription of both HeT-A and TART retrotransposons in the DrefKG09294 and Trf260071 mutant alleles (Figure 1C). In agreement with the presence of DREF binding sites in the TART element, we find that the difference in TART transcription for the DrefKG09294 and Trf260071 mutant alleles is especially significant. Moreover, both the DrefKG09294 and Trf260071 mutant alleles contain a higher copy number of TART elements, suggesting that this increase in transcription is translated in an increase in terminal transposition. In a DrefKG09294 and Trf260071 mutant background, the intermingled nature of HeT-A and TART at the HTT array [22] could help to spread the possible change in chromatin structure caused by the lack of binding of DREF/TRF2 at the TART sequences towards the HeT-A promoter, provoking an indirect increase in the transcription of HeT-A despite not containing DREF binding sites (Figure 1C).
The increase in expression from the HTT array in the mutant alleles of Dref and Trf2 indicates that in a wild-type situation this protein complex behaves as a repressor of the telomere retrotransposons. A priori, we were expecting DREF/TRF2 to activate the telomere expression in synchrony with cell proliferation [11,13]. Nevertheless, in accordance with our results, the DREF/TRF2 complex has also been reported to contribute to gene repression.
The DREF/TRF2 complex competes for DNA binding with the BEAF (boundary element-associated factor) because both DREF and BEAF have overlapping binding sites (BEAF binds to 5′-CGATA motifs) [23]. Moreover, it was shown that both proteins are able to target the same promoter [24]. Boundary elements have the ability to insulate a transgene from its chromosomal context by blocking enhancer-promoter interactions and heterochromatin spreading [25]. In D. melanogaster, BEAF associates preferentially with active transcribed genes [26]. The presence of BEAF at DREF-regulated genes leads to a decrease in the deposition of the heterochromatic mark H3K9me3, de-repressing those genes from the surrounding heterochromatin [24,27,28]. Our results are compatible with this scenario, in which equilibrium between the binding of both complexes would be necessary to obtain a fine-tuned regulation of the expression of the telomere retrotransposons. The binding of BEAF to the DREF binding sites at the TART promoter would protect the telomere retrotransposon from a repressive environment, and the binding of DREF would protect the genome from an excessive transcription, transposition, and telomere elongation. In a DREF mutant background, a major occupancy of BEAF at the HTT array would create an opening of the surrounding chromatin and, consequently, also increase the levels of HeT-A expression.
Our initial hypothesis was that Pzg could be exerting its role at the telomeres through its interaction with the DREF/TRF2 complex. In contrast with the increase in TART transcription observed in the DrefKG09294 and Trf260071 mutant alleles, the results obtained with two pzg mutant alleles, Z47.1 hypomorph and pzg66 null, show no change in the expression level of the TART retrotransposon. This result suggests that although the DREF/TRF2 has been related with Pzg in other situations, in this case they might be acting independently.
More chromatin modifiers have been related with the DREF/TRF2 complex, like the linker Histone H1, which is also involved in heterochromatin and transposable element gene expression as well as the NURF subunit ISWI (involved in ATP dependent nucleosome sliding) [29-32]. The loss of Trf2 could also affect the telomere chromatin through a secondary effect on these other heterochromatin components. Future studies involving additional mutant alleles of the mentioned chromatin components will help to elucidate the molecular mechanism by which the DREF/TRF2 complex affects the telomeres.
KEN is a repressor of TART
The JAK/STAT signaling pathway is responsible for the activation of the immune response genes [18,33]. KEN is a well-defined repressor of the JAK/STAT pathway and competes with STAT for the binding to a target gene [33]. Recently, NURF301 and Pzg were also found to be recruited by KEN and repress the JAK/STAT pathway [14,15]. When we analyzed the effect of the Ken mutant allele Ken1 in Drosophila telomeres, an increase in TART expression was observed but no effects on HeT-A expression were detected (Figure 1C). Accordingly, we found that the KEN binding sequences are not conserved in all the analyzed HeT-A sequences in opposition to the high conservation of at least one of the KEN binding sites in the TART sequence (Figure 3). A possible explanation could be that KEN might be involved in the recruitment of the chromatin-remodeling complex NURF to TART sequences repressing its transcription. In a Ken mutant background, the NURF complex is not recruited to the telomeres, leading to a relaxation of the chromatin. The fact that no effect on HeT-A expression is observed in Ken mutants suggests that the degree of chromatin relaxation is weaker than the one observed in Dref and Trf2 mutants. As with the DREF/TRF2 complex, we have found that the pzg alleles here analyzed do not show a change in transcription equivalent to the one observed in the Ken1 mutant. This result suggests a possible independent role of pzg and the Ken at Drosophila telomeres. We do not know whether the role of Ken controlling the expression of the TART element keeps any relationship with the JAK/STAT pathway, but it is possible that independent roles of Ken from the JAK/STAT pathway exist and that the control of telomere transcription could be the first one to be described.
Conclusions
We have identified three new genes involved in the regulation of the expression of the telomeric retrotransposons, Dref, Trf2, and Ken. Our results offer new insights in the composition and regulation of the telomere chromatin, pointing to unexpected relationships with other chromatin-related proteins and different pathways of the cell that had not been previously related with either telomere or retrotransposon regulation.
Methods
Fly stocks and crosses
Fly stocks were maintained and crosses preformed at 25°C on standard Drosophila corn meal medium. w1118 and ry506 were used as control, depending on the genetic background of each strain. w1118; Trf260071/FM7c, ry506; DrefKG09294/CyO and ry506; Ken1/CyO were obtained from Bloomington Stock Center. w1118; Trf260071/FM7c balancer was changed to FM7c-GFP to allow the selection of hemizygous males to perform the experiments. The three mutations correspond to P-element insertions inside the coding region. The hypomorph line Z47.1/TM3Sb, lacking the promoter region, came from Harald Eggert and Harald Saumweber and the line pzg66/TM6 was provided by Anja Nagel, this line is a null mutant that results in embryonic and early larval lethality.
Sequence alignments
The sequence alignments were carried out using ClustalW software. For the accession number of the HeT-A sequences used in the alignments see [19] and for the accession number of TART and TAHRE sequences see [21].
Genomic DNA extraction
Genomic DNA was extracted from adult flies to quantify the number of HeT-A and TART copies in each strain. Ten third-instar larvae without salivary glands were homogenized in 200 μl solution A (0.1 M Tris-HCl pH9.0, 0.1 M EDTA and 1% SDS) and incubated at 70°C for 30 min. 28 μl 8 M KAc were added and the samples incubated for 30 min on ice. Cell debris was harvested at maximum speed for 15 min at 4°C. The supernatant was transferred to a new tube and the DNA precipitated by adding 0.5 volumes isopropanol and centrifuging at 15.000 rpm for 5 min. Pelleted DNA was washed with 1 volume 70% ethanol and centrifuged. Finally, the DNA pellet was air-dried, and re-suspended in 50 μl 1× TE by rotating overnight at 4°C. After genomic DNA extraction, the number of copies was determined by quantitative real-time PCR using 2 ng of DNA per reaction.
Because certain endoreplication of the telomeric sequences exists in salivary glands, we depleted the samples from this tissue in order to have a copy number that would reflect the real copy number of the adult organism.
Primers used for TART amplification were, for TART_5′ UTR_F, GATAATAGCCGATAAGCCCGCCA, and for TART_5′ UTR_R, AAGACACAGCGGTTGATCGATATG. Primers used for HeT-A amplification were HeT-A_3′ UTR_F (CCCCGCCAGAAGGACGGA) and HeT-A_3′ UTR_R (TGTTGCAAGTGGCGCGCA). Primers used for actin amplification were Actin_F (GCGCCCTTACTCTTTCACCA) and Actin_R (ATGTCACGGACGATTTCACG).
RNA extraction and cDNA synthesis
Total RNA was isolated from ten whole third-instar larvae and extracted using RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNase Free DNase Set (Qiagen) was used to remove genomic DNA contaminations as follows: one on column during the extraction accordingly to manufacturer’s protocol, and two in solution for 2 hours at 37°C. RNA was cleaned by precipitation and its quality assessed using NanoDrop spectrophotometry.
One microgram of RNA was reverse transcribed into cDNA using Transcriptor First Strand cDNA Synthesis Kit (Roche) with oligo(dT) primers, and the expression of the different transcripts analyzed by quantitative real-time PCR. For each fly strain, two independent RNA extractions were prepared and analyzed independently three times. Primers used for TART amplification were, for TART_5′ UTR_F, GATAATAGCCGATAAGCCCGCCA, and for TART_5′ UTR_R, AAGACACAGCGGTTGATCGATATG. Primers used for HeT-A amplification were HeT-A_3′ UTR_F (CCCCGCCAGAAGGACGGA) and HeT-A_3′ UTR_R (TGTTGCAAGTGGCGCGCA). Primers used for actin amplification were Actin_F (GCGCCCTTACTCTTTCACCA) and Actin_R (ATGTCACGGACGATTTCACG).
We did not deplete the samples of salivary glands in the expression analysis, since the telomere retrotransposons are not expressed in this tissue [34].
Quantitative real-time PCR
Quantitative Real Time-PCR was performed to determine HeT-A and TART copy number and expression. The iQ5 Multicolor Real-Time PCR Detection System was used and the iQ™ SYBR® Green Supermix (BioRad) was used to prepare the reactions. Relative levels of HeT-A and TART expression were determined using the threshold cycle and normalized to actin levels. Three independent experiments of two samples each strain were performed.
Abbreviations
BEAF: Boundary element-associated factor; DREF: DNA replication-related element-binding factor; HP1a: Heterochromatin protein 1a; HTT: HeT-A, TART and TAHRE array; LTR: Long terminal repeats; NURF: Nucleosome remodeling factor; PCR: Polymerase chain reaction; TAS: Telomere associated sequences; TBP: TATA-box-binding protein; TF: Telomere fusion; TRF2: TATA-box-binding protein related factor 2.
Competing interests
The authors declare no competing interests.
Authors’ contributions
RS-S participated in the design of the study, performed research, analyzed data, and helped in drafting the manuscript. MDV performed research and analyzed data. EC conceived the study, analyzed data, and wrote the manuscript. All authors approved the final manuscript.
Contributor Information
Rute Silva-Sousa, Email: rute.sousa@ibe.upf-csic.es.
Míriam Díaz Varela, Email: miriam.diaz.varela@gmail.com.
Elena Casacuberta, Email: elena.casacuberta@ibe.upf-csic.es.
Acknowledgements
This work was supported by a grant from the Spanish Ministry of Science and Innovation BFU2009-08318/BMC to EC, a PhD Fellowship Fundação para a Ciência e Tecnologia, Portugal SFRH / BD / 36291 / 2007 to RS-S, and a JAEintro (JAEINT_11_01262) from the CSIC to MDV.
References
- Silva-Sousa R, López-Panadѐs E, Casacuberta E. Drosophila telomeres: an example of co-evolution with transposable elements. Genome Dyn. 2010;7:46–67. doi: 10.1159/000337127. [DOI] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Drosophila telomeres: a variation on the telomerase theme. Fly (Austin) 2008;2:101–110. doi: 10.4161/fly.6393. [DOI] [PubMed] [Google Scholar]
- Capkova Frydrychova R, Biessmann H, Mason JM. Regulation of telomere length in Drosophila. Cytogenet Genome Res. 2008;122:356–364. doi: 10.1159/000167823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF. Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J Cell Sci. 2005;118:5465–5477. doi: 10.1242/jcs.02654. [DOI] [PubMed] [Google Scholar]
- Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, Walter MF, Mason JM. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171:1767–1777. doi: 10.1534/genetics.105.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Sousa R, López-Panadès E, Piñeyro D, Casacuberta E. The chromosomal proteins JIL-1 and Z4/Putzig regulate the telomeric chromatin in Drosophila melanogaster. PLoS Genet. 2012;8:e1003153. doi: 10.1371/journal.pgen.1003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert H, Gortchakov A, Saumweber H. Identification of the Drosophila interband-specific protein Z4 as a DNA-binding zinc-finger protein determining chromosomal structure. J Cell Sci. 2004;117:4253–4264. doi: 10.1242/jcs.01292. [DOI] [PubMed] [Google Scholar]
- Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, Gatti M, Mason JM. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török T, Benitez C, Takács S, Biessmann H. The protein encoded by the gene proliferation disrupter (prod) is associated with the telomeric retrotransposon array in Drosophila melanogaster. Chromosoma. 2007;116:185–195. doi: 10.1007/s00412-006-0090-4. [DOI] [PubMed] [Google Scholar]
- Takács S, Biessmann H, Reddy HM, Mason JM, Török T. Protein interactions on telomeric retrotransposons in Drosophila. Int J Biol Sci. 2012;8:1055–1061. doi: 10.7150/ijbs.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler SJ, Nagel AC. Putzig is required for cell proliferation and regulates Notch activity in Drosophila. Mol Biol Cell. 2007;18:3733–3740. doi: 10.1091/mbc.E07-03-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler SJ, Nagel AC. A novel Pzg-NURF complex regulates Notch target gene activity. Mol Biol Cell. 2010;21:3443–3448. doi: 10.1091/mbc.E10-03-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- Kugler SJ, Gehring EM, Wallkamm V, Krüger V, Nagel AC. The Putzig-NURF nucleosome remodeling complex is required for ecdysone receptor signaling and innate immunity in Drosophila melanogaster. Genetics. 2011;188:127–139. doi: 10.1534/genetics.111.127795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SY, Xiao H, Glover BP, Tjian R, Wu C, Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta. 2008;1779:81–89. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Belote JM, Levis RW. Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons. Nucleic Acids Res. 2006;34:5498–5507. doi: 10.1093/nar/gkl709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Piñeyro D, López-Panadès E, Lucena-Pérez M, Casacuberta E. Transcriptional analysis of the HeT-A retrotransposon in mutant and wild type stocks reveals high sequence variability at Drosophila telomeres and other unusual features. BMC Genomics. 2011;12:573. doi: 10.1186/1471-2164-12-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martín-Gallardo A, Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21:1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu Rev Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–383. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- Emberly E, Blattes R, Schuettengruber B, Hennion M, Jiang N, Hart CM, Käs E, Cuvier O. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008;6:2896–2910. doi: 10.1371/journal.pbio.0060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkess G, West AG. Chromatin insulator elements: establishing barriers to set heterochromatin boundaries. Epigenomics. 2012;4:67–80. doi: 10.2217/epi.11.112. [DOI] [PubMed] [Google Scholar]
- Vogelmann J, Valeri A, Guillou E, Cuvier O, Nollmann M. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus. 2011;2:358–369. doi: 10.4161/nucl.2.5.17860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Zhao K, Laemmli UK. The scs′ boundary element: characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Käs E, Laemmli UK. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma. 2002;110:519–531. doi: 10.1007/s00412-001-0181-1. [DOI] [PubMed] [Google Scholar]
- Bouazoune K, Brehm A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006;14:433–449. doi: 10.1007/s10577-006-1067-0. [DOI] [PubMed] [Google Scholar]
- Siriaco G, Deuring R, Chioda M, Becker PB, Tamkun JW. Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics. 2009;182:661–669. doi: 10.1534/genetics.109.102053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, Fyodorov DV, Skoultchi AI. Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 2009;23:452–465. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujatovic O, Zaragoza K, Vaquero A, Reina O, Bernués J, Azorín F. Drosophila melanogaster linker histone dH1 is required for transposon silencing and to preserve genome integrity. Nucleic Acids Res. 2012;40:5402–5414. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombría JC, Sotillos S. JAK/STAT signalling: STAT cannot play with Ken and Barbie. Curr Biol. 2006;16:R98–R100. doi: 10.1016/j.cub.2006.01.021. [DOI] [PubMed] [Google Scholar]
- George JA, Pardue M-L. The promoter of the Heterochromatic Drosophila Telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2002;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]