Abstract
A genetically modified recombinant gH625-c-prune was prepared through conjugation of c-prune with gH625, a peptide encompassing 625–644 residues of the glycoprotein H of herpes simplex virus 1, which has been proved to possess the ability to carry cargo molecules across cell membranes. C-prune is the C-terminal domain of h-prune, overexpressed in breast, colorectal, and gastric cancers, interacting with multiple partners, and representing an ideal target for inhibition of cancer development. Its C-terminal domain results in an intrinsically disordered domain (IDD), and the peculiar properties of gH625 render it an optimal candidate to act as a carrier for this net negatively charged molecule by comparison with the positively charged TAT. A characterization of the recombinant gH625-c-prune fusion protein was conducted by biochemical, cellular biology and confocal microscopy means in comparison with TAT-c-prune. The results showed that the gH625-c-prune exhibited the ability to cross biomembranes, opening a new scenario on the use of gH625 as a novel multifunctional carrier.
Keywords: delivery, IDP
Introduction
Cell compartmentalization and impermeability of membranes represent a major obstacle for delivery of therapeutic molecules; in fact, many bioactive molecules are incapable of overcoming the membrane permeability barrier, and this represents the major impediment for gene and drug targeting in theranostics. Although a wide variety of biodrugs, including peptides and proteins, are now produced on a commercial scale, one challenging task for pharmaceutical researchers is to devise ways to deliver these drugs effectively and safely via noninvasive, patient-friendly routes.1,2
Cell-penetrating peptides (CPPs) are a valuable class of natural short peptide sequences which have been recently considered very promising for cellular uptake and membrane interaction studies.3 While individual CPPs differ in length and sequence, they share some common features, which include their amphipathic nature, net positive charge, hydrophobicity and helical moment, the ability to interact with lipidic membranes, and to adopt a distinct secondary structure upon association with lipids.4 These short cationic peptides (nanomolecules), either by covalent or noncovalent binding, have been exploited for the delivery of a range of macromolecules in the biologically active form, such as proteins, plasmids, siRNA (short interfering ribonucleic acids), nanoparticles, peptide nucleic acids, and liposomes, that are otherwise impermeable to the plasma membrane. Unfortunately, CPP application in cellular delivery has been hampered by the controversy regarding their uptake mechanism; in fact, no defined picture of the mechanism has yet emerged.3–7
Among the CPPs, the human immunodeficiency virus type 1 (HIV-1) TAT basic domain has been used extensively for macromolecule delivery.7–9 TAT is a nuclear transcriptional activator protein consisting of 101 amino acids. This protein is required for viral replication by HIV-1. The truncated sequence of HIV-1 TAT 49–57 (RKKRRQRRR) is a highly basic region typically involved in cellular translocation,7 and proteins as small as green fluorescent proteins (30 kDa) and as large as immunoglobulin G (150 kDa) have been delivered efficiently by TAT peptide.10
Problems in the application of CPPs arise from the unfavorable physicochemical and biological properties of therapeutic peptides and proteins to deliver, which may affect their absorption. In fact, the use of a charged delivery peptide may represent a disadvantage in some cases. Therefore, it is fundamental to exploit novel molecules which present different structural characteristics and use different internalization mechanisms.11 For this reason, great attention has recently been devoted to the study of hydrophobic peptides that efficiently traverse biological membranes, promoting lipid membrane-reorganizing processes, such as fusion or pore formation, and thus involving temporary membrane destabilization and subsequent reorganization.12 Viral-derived peptides, in particular those derived from viral entry proteins, may be useful as delivery vehicles due to their intrinsic properties of inducing membrane perturbation.13–15 Delivery across cellular membranes involves several mechanisms such as direct transfer through cell surface membrane by lipid membrane fusion or transient permeabilization of the cell membrane; or after endocytosis, transfer across vesicular membranes by lipid disruption, pore formation, or fusion.
The nineteen-residues peptide gH625 was previously identified as a membrane-perturbing domain in the glycoprotein H (gH) of herpes simplex virus 1; gH625 interacts with model membranes, contributing to their merging12,16 and is able to traverse the membrane bilayer and transport a cargo into the cytoplasm and across the blood–brain barrier.1,2,17,18 In particular, we recently reported the ability of gH625 peptide to transport quantum dots inside the cytoplasm in an efficient way and only partially involving endocytic pathways;18 moreover, we reported its ability to enhance intracellular penetration of liposomes functionalized with the peptide on the external leaflet,17 nanoparticles,2 and dendrimers.19 Compared with the TAT peptide, which mainly exploits the endocytic pathway, the viral membranotropic peptide gH625 crosses membrane bilayers mainly through a translocation mechanism.
The hydrophobicity of gH625 compared with the positively charged TAT can represent an advantage for the delivery of an acidic and natively unfolded protein. This study has indeed been focused on the different delivery of an intrinsically disordered C-terminal domain of h-prune20 fused to TAT and gH625. H-prune is a member of the DHH protein super-family, overexpressed in breast, colorectal, and gastric cancers.21–24 H-prune possesses a hitherto phosphodiesterase (cAMP-PDE) activity, and inhibition of PDE activity with dipyridamole suppresses cell motility in breast cell lines.25,26 H-prune contains two DHH domains at its N-terminus, followed by a third C-terminal domain, which was previously shown to be responsible for the interaction with different partners such as GSK-3beta and with Nm23-H1.27,28 The conformational analysis of the C-terminal domain of h-prune performed by nuclear magnetic resonance showed that it assumes a random coil conformation with the exception of some protein regions with helical structural propensity and a disulfide bridge as unique rigid linkage.20 In particular, the recombinant C-terminal domain of h-prune (residues 354–453, c-prune) was sufficient to bind the characterized interacting Nm23-H1 endogenous protein as verified by Western blotting analysis and nuclear magnetic resonance spectroscopy. In addition, further recent studies using a mimetic peptide, derived from the minimal interaction region of Nm23-H1 with h-prune, showed the impairment of neuroblastoma in vivo and elected it as possible therapeutic strategy.20
Here, we report the use of gH625 for delivery of c-prune as representative of a class of intrinsically disordered proteins (IDPs). The peculiar properties of gH625 render it an optimal candidate to act as a carrier for net negatively charged molecules by comparison with the positively charged TAT. A characterization of the recombinant gH625-c-prune fusion protein was conducted by biochemical, cellular biology, and confocal microscopy means. The results showed that the gH625-c-prune exhibited the ability to cross biomembranes, opening a new scenario for the use of gH625 as a novel multifunctional carrier.
Experimental procedures
Materials
Escherichia coli strains DH5α (Life Technologies, Carlsbad, CA, USA) and BL21 (DE3) STAR (Life Technologies) were used for cloning and expression of the recombinant proteins, respectively. pET28b+ and pETM11 (Novagen Inc, Madison, WI, USA) were used as expression vectors. HeLa cells (American Type Culture Collection [ATCC], Manassas, VA, USA) were grown in DMEM (Dulbecco’s Modified Eagle’s Medium) supplemented with 10% fetal bovine serum, 1% glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies) at 37°C in humidified air with 5% CO2.
Cloning strategy of gH625-c-prune and TAT-c-prune
The coding sequence of gH 625–644 (60 bp) was amplified by polymerase chain reaction (PCR) using specific primers from genomic DNA of herpes simplex virus 1. To ensure correct orientation into the multiple cloning sites of the vector, the forward primer (5′-GGAATTCCATATGCACG-GCCTCGCCTCG-3′) incorporated an NdeI restriction enzyme site, whereas the reverse primer (5′-CATGCCATG-GGGAAGGCGCGGATCAG-3′) incorporated an NcoI site. The digested fragment was ligated to the gene encoding the C-terminal domain of h-prune (amino acid sequence 353–454, pETM11-c-prune) previously digested with the restriction enzymes NcoI and XhoI.20 The fused segment was finally ligated to the expression vector pET28b+, which provided a cleavable N-terminal His6 tag. The coding sequence of TAT-c-prune was obtained through two consecutive PCR reactions. The first PCR reaction was carried out using pETM11-c-prune as a template and the forward primer (5′-CCGCCA-GCGCCGCCGCCCCCTGCTCCAGGAAGC-3′) and the reverse primer (oligo rev454) incorporated XhoI restriction enzyme site (5′-CCGCTCGAGTTACTTCTTGGACAG-GGAGGCTG-3′). The product of this PCR was used as a template for a second PCR reaction using the forward primer incorporated NcoI restriction enzyme site (5′-CATGCCA-TGG GCTACGGCCGCAAGAAACGCCGCCAGCGC-CGCC-3′) and the oligo rev454.
The coding sequence of TAT-c-prune was ligated to the expression vector pETM11, which provided a cleavable N-terminal His6 tag. The resulting positive plasmids were used to transform E. coli BL21 (DE3) STAR strain. The expression of the recombinant proteins (gH625-c-prune and TAT-c-prune) was carried out using the transformed cells grown at 37°C in 0.5 L of LB (Luria-Bertani broth) containing 50 μg/mL kanamycin and then inducing them overnight with 0.5 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) at 22°C.
Production of recombinant gH625-c-prune and TAT-c-prune
C-prune was expressed and purified as elsewhere described.20 The gH625-c-prune and TAT-c-prune were purified from inclusion bodies. Briefly, the cells were resuspended incubated for 16 hours at room temperature in 30 mL of 50 mM Tris-HCl (pH 7.5), 8 M urea, 300 mM NaCl, and 5% glycerol (buffer A), and then sonicated to promote cell lysis. The soluble and insoluble fractions of the lysate were then separated by centrifugation (17,000 rpm for 20 minutes at 4°C). The soluble fraction of the lysate was mixed with 4 mL of Ni-NTA resin (Qiagen, Hilden, Germany) equilibrated with the same buffer and then incubated for 1 hour at room temperature. The column was washed with ten volumes of denaturing buffer (buffer A) plus 50 mM imidazole. The denatured proteins were then eluted by increasing imidazole concentration (300 mM). The purity and yield of the protein were analyzed by 15% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis), and the protein concentration was determined by the Bradford assay using bovine serum albumin (BSA) as protein standard. The refolding procedure was carried out by dilution of the denatured proteins at a concentration of 0.2–0.4 mg/mL in 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5% glycerol, 0.5M L-arginine and 5 mM dithiothreitol (DTT). After an overnight mild stirring at 4°C, the insoluble proteins were removed by centrifugation at 12,000 rpm for 30 minutes at 4°C, and the refolded proteins were subjected to proteolytic digestion, with thrombin for gH625-c-prune and TEV protease for TAT-c-prune, to remove the His-tag. The concentration of refolded proteins was estimated by Bradford assay. The proteins were then loaded onto a Superdex 75 gel filtration column (HiLoad 10/30; GE Healthcare Bio-Sciences AB, Uppsala, Sweden) pre-equilibrated with 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5% glycerol, 1.5% D-glucose and 5 mM DTT.
Circular dichroism (CD) measurements
CD spectra were recorded with a J-810 spectropolarimeter equipped with a Peltier temperature control system (Model PTC-423-S, Jasco Europe, Cremella (LC), Italy). Far-ultraviolet (far-UV) measurements (195–260 nm) were carried out at 20°C using a 0.1 cm optical path length cell and proteins (8 μM) and peptides (40 μM) were dissolved in a buffer containing 10 mM Tris-HCl (pH 7.5), NaCl 100 mM, 1.25% glycerol, and DTT 1 mM. CD spectra, recorded with a time constant of 4 seconds, a 2 nm bandwidth, and a scan rate of 10 nm min1, were signal-averaged over at least three scans. The baseline was corrected by subtracting the complete buffer spectrum. Thermal denaturation curves were recorded over the 20°C–90°C temperature interval, following the CD signal at 222 nm. The curve was registered using a 0.1 cm path length cell, a protein concentration of 25 μM, and a scan rate of 1.0°C min−1.
Western blot analysis
His-tagged gH625-c-prune (2 mg) was immobilized on Ni-NTA resin (the capacity of the resin is 10 mg/mL of tagged protein). HeLa cells were resuspended in phosphate buffered saline (PBS) supplemented with 1% Triton X-100 and protease inhibitors. After 30 minutes on ice, cells were centrifuged at 13,500 rpm for 30 minutes. The amount of total proteins present in the extract was determined by Bradford assay. Total extract of HeLa cells (4 mg of total protein) was incubated with the resin at 4°C overnight. After extensive washing, the elution was carried out with increasing imidazole concentration (300 mM). The collected fractions were analyzed by Western blotting using an anti-Nm23-H1 antibody and an anti-His antibody as control of immobilized protein.
Preparation of small unilamellar vesicles (SUVs)
SUVs consisting of dioleoylphosphatidylcholine (DOPC)/cholesterol (Chol) (1:1) and 25% of Br-PC (1-palmitoyl-2-dibromo-stearoylphosphatidylcholine) were prepared according to the extrusion method of Hope et al29 in 5 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 100 mM NaCl, pH 7.4. Lipids were dried from a chloroform solution under a stream of nitrogen gas and lyophilized overnight. For fluorescence experiments, dry lipid films were suspended in buffer by vortexing for 1 hour and sonicated for 30 minutes.
Tryptophan fluorescence experiments
Br-PC employed as quencher of tryptophan fluorescence is suitable for probing membrane insertion of gH625-c-prune, since it acts over a short distance and does not drastically perturb the membrane.30,31 gH625-c-prune, containing the tryptophan residue in the peptide sequence, was added (final concentration of 4 μM) to 2 mL of buffer (5 mM HEPES, 100 mM NaCl, pH 7.4) containing 100 μM of Br-PC/Chol SUV and a total lipid concentration of 400 μM, thus establishing a lipid/peptide molar ratio of 100:1. Emission spectrum of the tryptophan was recorded with excitation set at 295 nm. SUV composed of DOPC/Chol (1:1) and which contained 25% of either 6,7-Br-PC, 9,10-Br-PC, or 11,12-Br-PC were used. Three separate experiments were conducted for each condition. In control experiments, gH625-c-prune in DOPC/Chol (1:1) SUVs without Br-PC was used.
Labeling of gH625-c-prune and TAT-c-prune
The gH625-c-prune, TAT-c-prune, and c-prune were labeled with N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-1,3-diazole (IANBD; Life Technologies) a fluorophore that interacts specifically with reduced cysteines.16 The reaction was conducted in a buffer containing 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5% glycerol, 1.5% D-glucose, and 5 mM DTT with a tenfold molar excess of IANBD added as a stock solution (10 mg/mL in dimethylformamide) for 4 hours at 4°C in slow agitation. The excess of IANBD was removed by extensive dialysis at 4°C in the reaction buffer.
Flow cytometry of cell association of gH625-c-prune
Cell association was measured by flow cytometry, equipped with a 488 nm argon laser (FACSCalibur, Becton Dickinson, CA, USA). NBD-labeled gH625-c-prune (5 μM) was added to 5 × 105 HeLa cells in a final volume of 0.5 mL in Opti-MEM medium (Life Technologies) at 37°C in slow agitation for 10 minutes. Thereafter, the cells were washed in PBS, 0.1% BSA and then resuspended in 0.5 mL of PBS, 0.1% BSA for flow cytometric analysis. The sample was excited at 488 nm, fluorescence was measured at 525 nm, and a total of 20,000 events for sample were collected. A stock solution of dithionite (1 M) was freshly prepared in 1 M Tris-HCl (pH 10). Following flow cytometric analysis, 5 μL of dithionite stock solution was added to cells maintained at 4°C, and the fluorescence of internalized gH625-c-prune was measured after 5 minutes. Experiments were performed in triplicate.
Cellular uptake kinetics and endocytosis inhibition of gH625-c-prune and TAT-c-prune by confocal microscopy
For protein uptake-kinetics experiments on nonfixed living cells, 105 HeLa cells were incubated with 30 μM of NBD-protein solutions at 37°C for 10 and 30 minutes. After incubation, cells were rinsed twice with PBS, and fresh cell culture medium without phenol red was added.16
To inhibit the endocytic pathways, HeLa cells were treated with sodium azide, cytochalasin D, and low temperature. To be exact, 40 μM sodium azide was added to the cell culture medium for 30 minutes, followed by 30 minutes of incubation with proteins. Moreover, to disassemble actin microfilaments, HeLa cells were treated with 30 μM cytochalasin D in cell culture medium for 30 minutes at 37°C before protein incubation. After cytochalasin D treatment, cells were incubated with protein solution for 30 minutes. Furthermore, 4°C blocking experiments were carried out by incubating cells with protein solutions at 4°C for 30 minutes.
All the samples were observed by a confocal laser scanning microscope (CLSM) (LSM510; Carl Zeiss Meditec AG, Jena, Germany) equipped with a 488 nm Argon laser line and with a 63× water-immersion objective. Confocal images of all samples were analyzed by ImageJ (National Institutes of Health, Bethesda, MD, USA) software to evaluate the protein uptake in terms of intra cellular fluorescence intensity. Fifty cells per sample were analyzed. Protein uptake inhibition data were reported as the percentages of the intracellular fluorescence intensity of the samples normalized to intracellular fluorescence intensity of control nontreated cells.
Results
Expression and purification of c-prune, gH625-c-prune, and TAT-c-prune
Recombinant gH625-c-prune and TAT-c-prune were purified from inclusion body using a nickel affinity chromatography in denaturing conditions. C-prune (molecular weight [MW] 11528 Da) was purified as already described.20 The collected fractions were subjected to a refolding process via direct dilution to prevent aggregation and precipitation of the protein. The refolded proteins were subjected to proteolytic digestion with thrombin (gH625-c-prune, MW 14190 Da composed by GSHMHGLASTL TRWAHYNALIRAFPM and c-prune) and TEV protease (TAT-c-prune, MW 12736 Da composed by GAMGYGRKKRRQRRR and c-prune), in the optimized conditions, to remove the His-tag with an efficiency of about 60%. The digested sample was loaded onto a Superdex 75 gel filtration column pre-equilibrated with an appropriate buffer (see Methods) to separate the protein aggregates from the properly folded forms. The purity and yield of the proteins obtained were analyzed by 15% SDS-PAGE and Bradford assay respectively (Figure 1). Accordingly to the intrinsically disordered nature of c-prune, it presents an abnormal migratory behavior with an apparent molecular weight of about 15 kDa.20,32
Figure 1.
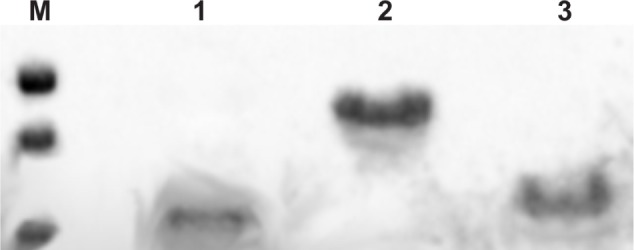
Electrophoretic analysis. 15% SDS-PAGE of molecular weight markers: 25 kDa, 20 kDa, and 14 kDa (M), TAT-c-prune (1), gH625-c-prune (2), and c-prune (3) purified to homogeneity.
Abbreviation: SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
gH625-c-prune and TAT-c-prune both show an anomalous electrophoretic mobility. The yields of the proteins are 2 mg/L for TAT-c-prune and 6 mg/L for gH625-c-prune.
CD measurements
The secondary structures of gH625-c-prune, TAT-c-prune, c-prune, and gH625 peptide were determined by CD spectroscopy in the far-UV spectral region (195–260 nm). CD spectra show that gH625-c-prune protein has a high proportion of secondary structure compared with c-prune. In fact, CD spectra performed on c-prune,20 TAT-c-prune, and gH625 peptide revealed spectra with minima at ~200–205 nm, which were characteristic of disordered polypeptides (Figure 2A). Instead, the CD spectrum of the recombinant protein gH625-c-prune, in the same conditions, is characterized by the presence of two minima (at 208 and 222 nm) and one maximum (at 195 nm), which are characteristic of α-helical proteins (Figure 2A).33 The thermal denaturation curve of gH625-c-prune presents a sigmoidal shape with a single inflection point, corresponding to Td values of about 52°C (Figure 2B). The thermal denaturation of gH625-c-prune is not a reversible process due to the protein precipitation.
Figure 2.
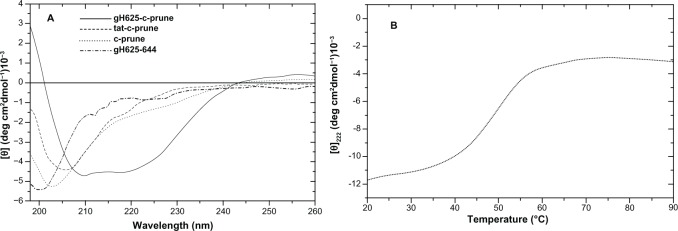
Far-UV CD spectra. (A) Far-UV CD spectra of gH625-c-prune (solid line), TAT-c-prune (dashed line), c-prune (dotted line), and gH625 (dashed-dotted line) in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1.25% glycerol, and 1 mM DTT. The spectra were recorded using 0.1 cm path length cells. The protein and peptide concentrations were 8 μM and 40 μM, respectively. (B) Temperature-induced denaturation curves of gH625-c-prune (25 μM) obtained by recording the molar ellipticity at 222 nm in 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5% glycerol, and 1 mM DTT.
Abbreviations: CD, circular dichroism; DTT, dithiothreitol; UV, ultraviolet.
Analysis of gH625-c-prune biological function
To verify whether c-prune retained its biological activity after conjugation with the peptide gH625, we verified the complex formation with Nm23-H1 by Western blotting, as previously demonstrated for isolated c-prune.20 His-tagged gH625-c-prune was immobilized on the resin, and total extract of HeLa cells was loaded onto it (see Methods). The Western blotting analysis showed that gH625-c-prune is still able to interact with Nm23-H1 in HeLa extract (Figure 3), suggesting that the folding of the protein does not affect its biological function.
Figure 3.
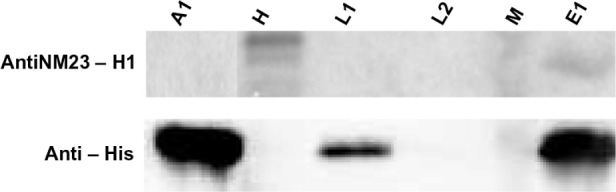
Recombinant gH625-c-prune retains the ability to bind to intracellular Nm23-H1. Lane A1: His-tagged gH625-c-prune (2 μg). Lane H: HeLa total extract (50 μg). Lane L1: wash with 50 mM imidazole (2 μg). Lane L2: wash with 50 mM imidazole of resin without gH625-c-prune immobilized (50 μg). Lane M: molecular weight marker. Lane E1: elution with 300 mM imidazole (2 μg).
Fluorescence measurements
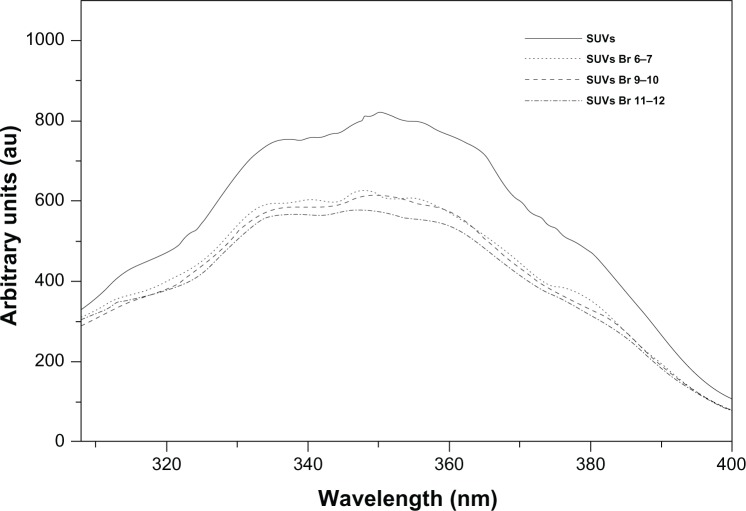
A tryptophan residue naturally present in the sequence of a protein or a peptide can serve as an intrinsic probe for the localization of the peptide within a membrane; in fact, the fluorescence emission of a tryptophan residue increases when the amino acid enters a more hydrophobic environment, and together with an increase in quantum yield, the maximal spectral position will be shifted toward shorter wavelengths (blue shift). The peptide gH625 contains a tryptophan residue in the middle of the sequence; Figure 4 shows the fluorescence emission spectra of the peptide gH625 upon interaction with Br-DOPC/Chol vesicles. In this experiment the gH625-c-prune location inside the bilayer was investigated by measuring the relative quenching of the gH625 tryptophan fluorescence by the probes 11,12-Br-PC, 9,10-Br-PC, and 6,7-Br-PC, which differ in the position of the quencher moiety along the hydrocarbon chain and allow to establish the depth of the peptide insertion into the membrane. For all the conditions tested, a quenching of the tryptophan emission is evident. These results indicate that, upon binding to vesicles, the peptide is deeply inserted into the membrane bilayer.
Figure 4.
Tryptophan fluorescence emission spectra. Tryptophan fluorescence emission spectra of the gH625-c-prune upon interaction with Br-DOPC/Chol vesicles. gH625-c-prune incubated in buffer without SUV (solid line); with SUV that contained 6,7-Br-PC (dashed line), with SUV that contained 9,10-Br-PC (dotted line), and with SUV that contained 11,12-Br-PC (dashed-dotted line).
Abbreviations: Br-PC, 1-palmitoyl-2-dibromo-stearoylphosphatidylcholine; Chol, cholesterol; DOPC, dioleoylphosphatidylcholine; SUV, small unilamellar vesicle.
Determination of NBD-gH625-c-prune cellular uptake
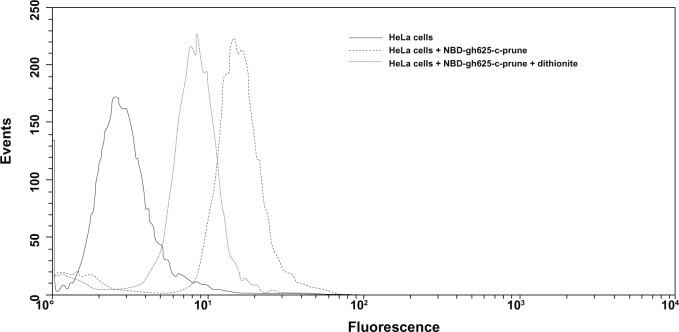
We have determined the fraction of NBD-labeled gH625-c-prune taken up into HeLa cells by flow cytometry. This fraction is determined by comparing the fluorescence intensity before and after addition of sodium dithionite, an essentially membrane-impermeant molecule, which suppresses irreversibly the fluorescence of the accessible NBD-moiety localized on the external cell surface. We incubated NBD-gH625-c-prune (5 μM) with HeLa cells at 37°C. After 10 minutes of incubation, we measured the quenching of the NBD-gH625-c-prune by dithionite treatment. The dithionite reaction was performed at low temperature (4°C) because the dithionite diffusion in the biological membranes is strongly reduced in comparison with high temperatures.34 After the addition of dithionite, there is an increase of fluorescence compared with the intrinsic fluorescence of the cells (Figure 5). This shows that a percentage of protein (~50%) has been internalized in 10 minutes. Increasing protein concentration, time of incubation, or dithionite concentration did not result in an increase of yield’s internalization. NBD was used in the same experiment to exclude that the fluorescence signal was due to internalization of the fluorophore alone, and no cellular fluorescence was observed at concentration up to 15 μM (data not shown).
Figure 5.
Flow cytometric analysis of HeLa cells treated with NBD-gH625-c-prune. Cells are treated for 10 minutes with 5 μM NBD-gH625-c-prune in the presence or absence of dithionite. Untreated cells (solid line), cells treated with NBD-gH625-c-prune (dashed line), cells treated with NBD-gH625-c-prune and dithionite (dotted line). Single result representative of three similar experiments.
Uptake kinetics and endocytosis inhibition
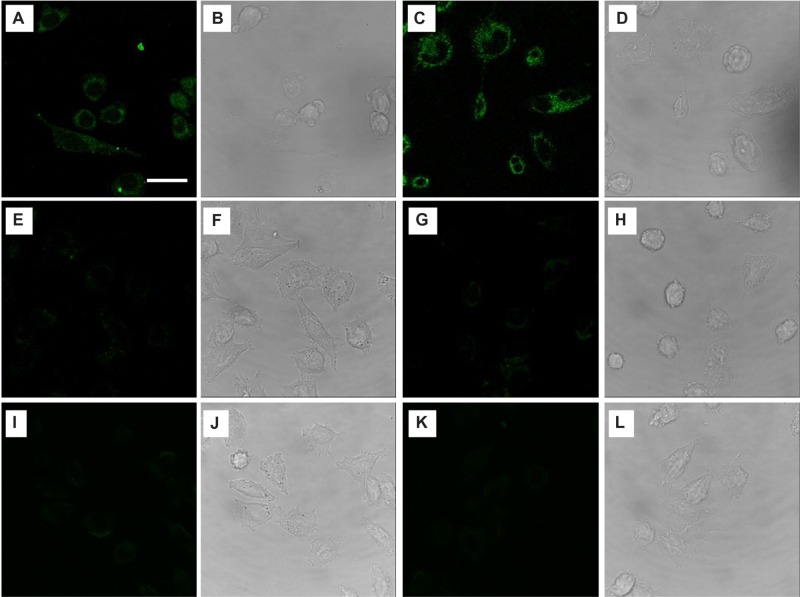
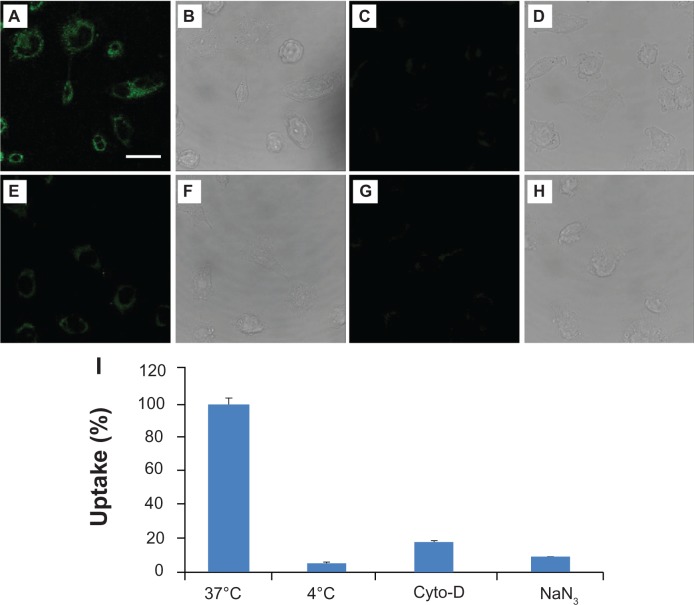
To verify whether gH625 peptide could promote prune internalization, we examined uptake in HeLa cells by confocal microscopy. We incubated 30 μM gH625-c-prune solution in cell culture medium for 10 and 30 minutes at 37°C. Images show a fluorescence signal in HeLa cells already after 10 minutes incubation for gH625-c-prune (Figure 6A). Moreover, by increasing incubation time, the intracellular fluorescence intensity increased (Figure 6C). Conversely, no fluorescence was observed in case of c-prune (Figure 6I and K), and only a slight intracellular fluorescence was detectable after incubation with TAT-c-prune (Figure 6E and G). Transmitted light images in all conditions tested are reported in Figure 6B, D, F, H, J, and L. gH625-c-prune uptake was partially due to endocytic mechanisms as demonstrated by endocytosis inhibition experiments. In particular, gH625-c-prune uptake was significantly inhibited by NaN3 and 4°C treatments (Figure 7A, C, and G). Indeed, more than 90% reduction in gH625-c-prune uptake was observed (Figure 7I). In addition, 80% uptake inhibition was observed after incubation with cytochalasin D (Figure 7E). The transmitted light images in all conditions tested are reported in Figure 7B, D, F, and H. These data demonstrate that gH625-c-prune internalization significantly depends on active, ATP (adenosine triphosphate)-dependent mechanisms and suggest that there is a small amount of gH625-c-prune that use a different, probably passive, mechanism to cross the cell membrane.
Figure 6.
Uptake kinetics of HeLa cells incubated at 37°C with gH625-c-prune (A–D), TAT-c-prune (E–H), and c-prune (I–L) for 10 (left panels) and 30 (right panels) minutes. Fluorescence (A, C, E, G, I, and K) and transmitted light (B, D, F, H, J, and L) images. Magnification bar: 20 μm.
Figure 7.
Uptake inhibition experiments. Confocal laser scanning microscope images of HeLa cells incubated with gH625-c-prune protein at 37°C (A and B), NaN3 (C and D), and cytochalasin D (E and F) at 4°C (G and H). Fluorescence (A, C, E, and G) and transmitted light (B, D, F, and H) images. Percentage of uptake as a function of inhibition treatments (I). Magnification bar: 20 μm.
Discussion
A peptide encompassing 625–644 residues of the gH of herpes simplex virus 1 represents a very attractive peptide for studies on membrane interactions.11 Recent studies indicated also that gH625 cellular uptake is associated with its ability to interact with membrane lipids and to form a transient helical structure that temporarily affects membrane organization, thereby facilitating insertion into the membrane and translocation bypassing the endocytic pathway.16 Structural studies highlighted that when gH625 is in helical conformation, the polar residues concentrate on one face of the helix, giving it an amphiphilic character common to fusion peptides of most fusion glycoproteins of class I enveloped viruses. In addition, gH625 has been used to efficiently deliver quantum dots in cells, that do not significantly traverse the membrane bilayer on their own;18 liposomes;17 dendrimers;1 or nanoparticles through the blood–brain barrier.2
Therefore, gH625 can be considered a promising tool to efficiently perform intracellular delivery and may represent an alternative strategy to the use of the well characterized positively charged TAT peptide.
In this paper, we report the validation of gH625 as carrier of an intrinsically disordered domain, the C-terminal domain of h-prune. We designed, cloned, expressed, and purified the recombinant gH625-c-prune as a soluble product. Far-UV CD spectra revealed an unexpected α-helical conformation of the protein when compared with the CD spectra of gH625 or c-prune alone, both typical of unfolded species. Nonetheless, the folding of gH625-c-prune retains an anomalous electrophoretic mobility. This behavior could be justified by the preponderance of a rather high content. In spite of this change in structure, gH625-c-prune retained its activity binding to its partner Nm23-H1 as verified by Western blot analysis. The quenching of tryptophan fluorescence by brominated phospholipid probed the membrane insertion of gH625-c-prune, indicating that the protein bound to the peptide is located inside the membrane bilayer. This experiments may indicate that in the cell, gH625-c-prune may be able to cross the cell membrane. In addition, flow cytometric analyses revealed a significant signal of fluorescence corresponding to the internalization of gH625-c-prune NBD-labeled, confirmed by the quenching of external fluorescence following dithionite treatment. Finally confocal microscopy data performed on gH625-c-prune showed that gH625 is able to deliver c-prune in cells, and data obtained with different inhibitors showed that the entrance to the cell is strongly dependent on endocytic pathway, although a fraction of the protein is able to cross the bilayer through a translocation mechanism, which has been hypothesized to be the main crossing mechanism used by the peptide gH625. It is worth mentioning that not only is this the first time that gH625 has been shown to be able to deliver a protein intracellularly, but this is also the first time that an IDP was shown to be easily transported across the membrane bilayer, retaining its biological activity.
In comparison with gH625-c-prune, we started a characterization also of TAT-c-prune. The typical negative charge of an intrinsically disordered domain,35,36 such as c-prune, could represent a problem to obtain a fusion construct with TAT CPP. Indeed, TAT-c-prune has been obtained as an unstable product in comparison with the stable gH625-c-prune, achieving rather high levels of yield. Furthermore, CD analyses of TAT-c-prune showed a spectrum typical of an unstructured protein. In addition, confocal microscopy data performed at the same concentration and incubation time for both proteins, gH625-c-prune and TAT-c-prune, revealed that only gH625 plays a role for c-prune delivery. We can hypothesize that the folding of gH625-c-prune, as monitored by CD, can represent an improvement and so facilitate the insertion into the membrane and subsequent translocation of the protein.
There is a natural abundance of IDPs involved in the pathogenesis of numerous human diseases, which may suggest that in the future these proteins may represent a huge reservoir of novel drug targets. A key role is thus played by the availability of delivery tools for IDP. Altogether, our results open a wider scenario on the potential applications of this membranotropic peptide fused with an IDP. One of these could be, once delivered in the cell, to perform functional and/or structural characterization of the protein in physiological conditions.37 In particular, this approach could give the opportunity to generate novel therapeutic molecules by drug discovery to find out new strategies for the treatment, in the case of c-prune, of h-prune overexpressing tumors.
Acknowledgments
This work was funded by PON-1_2388 – Verso la medicina personalizzata: nuovi sistemi molecolari per la diagnosi e la terapia di patologie oncologiche ad alto impatto sociale and by the Italian MIUR for financial support (PRIN2009 and FIRB RBAP114AMK_006).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Carberry TP, Tarallo R, Falanga A, et al. Dendrimer functionalization with a membrane-interacting domain of herpes simplex virus type 1: towards intracellular delivery. Chemistry. 2012;18(43):13678–13685. doi: 10.1002/chem.201202358. [DOI] [PubMed] [Google Scholar]
- 2.Guarnieri D, Falanga A, Muscetti O, et al. Shuttle-mediated nanoparticle delivery to the blood brain barrier. Small. 2013;9(6):853–862. doi: 10.1002/smll.201201870. [DOI] [PubMed] [Google Scholar]
- 3.Zorko M, Langel U. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57(4):529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Deshayes S, Morris MC, Divita G, Heitz F. Cell-penetrating peptides: tools for intracellular delivery of therapeutics. Cell Mol Life Sci. 2005;62(16):1839–1849. doi: 10.1007/s00018-005-5109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang M, Chou JC, Lee HJ. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant Cell Physiol. 2005;46(3):482–488. doi: 10.1093/pcp/pci046. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J Biol Chem. 2002;277(4):2437–2443. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 7.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272(25):16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 8.Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21(2):45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- 9.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8(7):848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 10.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285(5433):1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 11.Galdiero S, Vitiello M, Falanga A, Cantisani M, Incoronato N, Galdiero M. Intracellular delivery: exploiting viral membranotropic peptides. Curr Drug Metab. 2012;13(1):93–104. doi: 10.2174/138920012798356961. [DOI] [PubMed] [Google Scholar]
- 12.Galdiero S, Falanga A, Vitiello G, et al. Role of membranotropic sequences from herpes simplex virus type I glycoproteins B and H in the fusion process. Biochim Biophys Acta. 2010;1798(3):579–591. doi: 10.1016/j.bbamem.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Galdiero S, Falanga A, Vitiello M, Browne H, Pedone C, Galdiero M. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2005;280(31):28632–28643. doi: 10.1074/jbc.M505196200. [DOI] [PubMed] [Google Scholar]
- 14.Galdiero S, Falanga A, Vitiello M, et al. The presence of a single N-terminal histidine residue enhances the fusogenic properties of a membranotropic peptide derived from herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2010;285(22):17123–17136. doi: 10.1074/jbc.M110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galdiero S, Falanga A, Vitiello M, et al. Analysis of a membrane interacting region of herpes simplex virus type 1 glycoprotein H. J Biol Chem. 2008;283(44):29993–30009. doi: 10.1074/jbc.M803092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galdiero S, Russo L, Falanga A, et al. Structure and orientation of the gH625–644 membrane interacting region of herpes simplex virus type 1 in a membrane mimetic system. Biochemistry. 2012;51(14):3121–3128. doi: 10.1021/bi201589m. [DOI] [PubMed] [Google Scholar]
- 17.Tarallo R, Accardo A, Falanga A, et al. Clickable functionalization of liposomes with the gH625 peptide from Herpes simplex virus type I for intracellular drug delivery. Chemistry. 2011;17(45):12659–12668. doi: 10.1002/chem.201101425. [DOI] [PubMed] [Google Scholar]
- 18.Falanga A, Vitiello MT, Cantisani M, et al. A peptide derived from herpes simplex virus type 1 glycoprotein H: membrane translocation and applications to the delivery of quantum dots. Nanomedicine. 2011;7(6):925–934. doi: 10.1016/j.nano.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Falanga A, Tarallo R, Galdiero E, Cantisani M, Galdiero M, Galdiero S. Review of a viral peptide nanosystem for intracellular delivery. J Nanophoton. 2013;7(1):071599. [Google Scholar]
- 20.Carotenuto M, Pedone E, Diana D, et al. Neuroblastoma tumorigenesis is regulated through the Nm23-H1/h-Prune C-terminal interaction. Sci Rep. 2013;3:1351. doi: 10.1038/srep01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi T, Hino S, Oue N, et al. Glycogen synthase kinase 3 and h-prune regulate cell migration by modulating focal adhesions. Mol Cell Biol. 2006;26(3):898–911. doi: 10.1128/MCB.26.3.898-911.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi T, Oue N, Wada S, et al. h-Prune is an independent prognostic marker for survival in esophageal squamous cell carcinoma. Ann Surg Oncol. 2009;16(5):1390–1396. doi: 10.1245/s10434-007-9585-3. [DOI] [PubMed] [Google Scholar]
- 23.Oue N, Yoshida K, Noguchi T, Sentani K, Kikuchi A, Yasui W. Increased expression of h-prune is associated with tumor progression and poor survival in gastric cancer. Cancer Sci. 2007;98(8):1198–1205. doi: 10.1111/j.1349-7006.2007.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massidda B, Sini M, Budroni M, et al. Molecular alterations in key-regulator genes among patients with T4 breast carcinoma. BMC Cancer. 2010;10:458. doi: 10.1186/1471-2407-10-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Angelo A, Garzia L, Andre A, et al. Prune cAMP phosphodiesterase binds nm23-H1 and promotes cancer metastasis. Cancer Cell. 2004;5(2):137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 26.D’Angelo A, Zollo M. Unraveling genes and pathways influenced by H-prune PDE overexpression: a model to study cellular motility. Cell Cycle. 2004;3(6):758–761. [PubMed] [Google Scholar]
- 27.Marino N, Zollo M. Understanding h-prune biology in the fight against cancer. Clin Exp Metastasis. 2007;24(8):637–645. doi: 10.1007/s10585-007-9109-3. [DOI] [PubMed] [Google Scholar]
- 28.Galasso A, Zollo M. The Nm23-H1-h-Prune complex in cellular physiology: a ‘tip of the iceberg’ protein network perspective. Mol Cell Biochem. 2009;329(1–2):149–159. doi: 10.1007/s11010-009-0115-4. [DOI] [PubMed] [Google Scholar]
- 29.Hope MJ, Bally MB, Webb G, Cullis PR. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812(1):55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 30.Bolen EJ, Holloway PW. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 1990;29(41):9638–9643. doi: 10.1021/bi00493a019. [DOI] [PubMed] [Google Scholar]
- 31.De Kroon AI, Soekarjo MW, De Gier J, De Kruijff B. The role of charge and hydrophobicity in peptide-lipid interaction: a comparative study based on tryptophan fluorescence measurements combined with the use of aqueous and hydrophobic quenchers. Biochemistry. 1990;29(36):8229–8240. doi: 10.1021/bi00488a006. [DOI] [PubMed] [Google Scholar]
- 32.Bourhis JM, Receveur-Brechot V, Oglesbee M, et al. The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein Sci. 2005;14(8):1975–1992. doi: 10.1110/ps.051411805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmad E, Rabbani G, Zaidi N, Ahmad B, Khan RH. Pollutant-induced modulation in conformation and beta-lactamase activity of human serum albumin. PLoS One. 2012;7(6):e38372. doi: 10.1371/journal.pone.0038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angeletti C, Nichols JW. Dithionite quenching rate measurement of the inside-outside membrane bilayer distribution of 7-nitrobenz-2-oxa-1,3-diazol-4-yl-labeled phospholipids. Biochemistry. 1998;37(43):15114–15119. doi: 10.1021/bi9810104. [DOI] [PubMed] [Google Scholar]
- 35.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62(1):24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 36.Ahmad E, Kamranur Rahman S, Masood Khan J, Varshney A, Hasan Khan R. Phytolacca americana lectin (Pa-2; pokeweed mitogen): an intrinsically unordered protein and its conversion into partial order at low pH. Biosci Rep. 2010;30(2):125–134. doi: 10.1042/BSR20090035. [DOI] [PubMed] [Google Scholar]
- 37.Ito Y, Selenko P. Cellular structural biology. Curr Opin Struct Biol. 2010;20(5):640–648. doi: 10.1016/j.sbi.2010.07.006. [DOI] [PubMed] [Google Scholar]