Abstract
TrkH belongs to a superfamily of K+ transport proteins required for growth of bacteria in low external K+ concentrations. The crystal structure of TrkH from Vibrio parahaemolyticus showed that TrkH resembles a K+ channel, and may have a gating mechanism substantially different from K+ channels. TrkH assembles with TrkA, a cytosolic protein comprising two Regulate-the-Conductance-of-K+, or RCK domains, which are found in certain K+ channels and control their gating. However, fundamental questions on whether TrkH is an ion channel and how it is regulated by TrkA remain unresolved. Here we show single-channel activity of TrkH that is upregulated by ATP via TrkA. We report two structures of the tetrameric TrkA ring, one in complex with TrkH and one in isolation, in which the ring assumes two dramatically different conformations. These results suggest a mechanism for how ATP increases TrkH activity by inducing conformational changes in TrkA.
K+ is concentrated in all living cells and is essential for many physiological processes. In bacteria, homeostasis of K+ is largely mediated by specialized K+ transport proteins known as the Superfamily of K+ Transporters, or SKT proteins1. The bacterial TrkH/TrkG/KtrB proteins form the largest sub-family of SKT proteins, and the crystal structure of TrkH from Vibrio parahaemolyticus, VpTrkH, was reported recently2. Each of the four connected homologous domains of TrkH resembles a subunit of the KcsA K+ channel, comprising two transmembrane helices connected by a re-entrant P-loop (M1-P-M2)3, and the four domains encircle a central ion permeation pathway. In addition, TrkH has two structural features atypical of K+ channels: a dimeric quaternary structure, with each protomer containing its own pore; and a long membrane-embedded loop, inserted into the middle of a pore-lining helix, that occludes the ion permeation pathway. The loop is conserved across the TrkH/TrkG/KtrB family, and previous studies have shown that deletion of this intramembrane loop increases KtrB-mediated K+ uptake, suggesting that it may act as a gate4,5.
The TrkH/TrkG/KtrB integral membrane proteins form a complex with an intracellular regulatory protein, and the genes encoding the two proteins reside in the same operon in many bacteria. The protein that assembles with TrkH/TrkG is TrkA, while KtrB interacts with KtrA6-8. Both TrkA and KtrA are composed of RCK domains9-16, which can be classified into two types based on their cognate ligands: bacterial RCK domains possessing conserved GXGXXG motifs that bind to nucleotides8,12,14,17,18, sometimes referred to as KTN domains, and the bacterial and mammalian RCKs that lack a nucleotide binding motif and bind to ions such as Ca2+ (11,15,16,19). The organization of RCK domains varies considerably. For example, the KtrA and TrkA protomers contain one and two RCK domains, respectively, expressed as separate cytoplasmic proteins, while for K+ channels the RCK domains are directly connected to the termini of their associated channels. Previous RCK structures suggest that regardless of gene architecture, a total of eight RCK domains associate in a configuration known as a gating ring9,11,14-16,19.
The TrkH/TrkG/KtrB proteins were initially thought to be either H+- or Na+-dependent K+ transporters20-22, but their sequence and structural similarity to K+ channels suggests that these proteins could function as ion channels. However, ion channel activity has never been demonstrated for any member of the TrkH/TrkG/KtrB family of proteins. Furthermore, if TrkH is an ion channel, what effect does its associated protein TrkA have on gating of the channel? Current models of gating in K+ channels with RCK domains propose that a dilation in the diameter of the gating ring directly translates into movement of the four pore-lining helices and opening of the permeation pathway9,11,15,16,19,23. However, this model of a four-fold symmetric gating ring expansion appears incompatible with the dimeric architecture of TrkH, and does not account for the intramembrane loop. In this study, we attempt to address these unresolved issues by characterizing the structure and function of the TrkH-TrkA complex.
Single-channel activities of the TrkH-TrkA complex
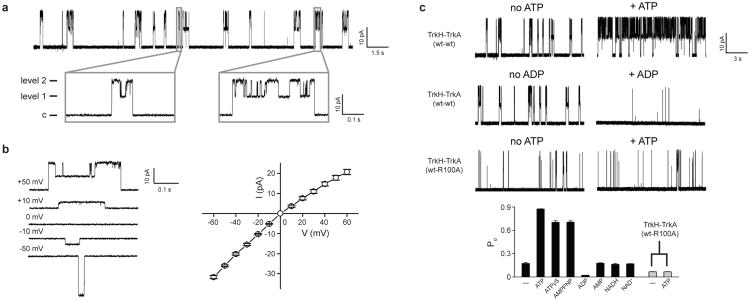
Patch clamp was performed on Escherichia coli spheroplasts heterologously expressing TrkH and TrkA from V. parahaemolyticus using an inside-out configuration. A single-channel current trace in 200 mM symmetrical K+ solution is shown in Fig. 1a. Occasional bursts of channel activities were observed, and these bursts were interrupted by prolonged closures. Within each burst of openings there are two current levels, level 1 and 2, with level 2 approximately double the size of level 1 in amplitude (Supplementary Table 1). Every burst of openings starts with a sojourn from the baseline directly to level 2 followed by openings alternating between levels 1 and 2, and terminates >91% of the time directly from level 2. This behavior is inconsistent with two entirely independent channels, but suggests a single TrkH dimer whose two pores open and close in a mostly concerted fashion.
Figure 1. Single-channel activities of the TrkH-TrkA complex.
a. Single-channel currents through the TrkH-TrkA complex were recorded in symmetrical 200 mM KCl. Current traces are shown at a holding potential of +50 mV. The closed state is indicated as “c”. The gray rectangles show two individual bursts of channel openings. b. The open-channel current-voltage relationship. Current traces recorded at different voltages are shown in the left panel, and current amplitudes are plotted in the right panel. c. Current traces of the TrkH-TrkA (wt-wt) complex and the TrkH-TrkA (wt-R100A) complex in the absence (left) and presence (right) of 5 mM ATP or ADP. The Popen after perfusion of different ligands is plotted in the bar graph. Added ligand concentrations are as follows: ATP, 5 mM; ATPγS, 1 mM; AMP-PNP, 1 mM; ADP, 5 mM; AMP, 5 mM; NADH, 1 mM; NAD+, 1 mM. Error bars are s.e.m. of 3-9 independent measurements.
As a control, currents from spheroplasts expressing only TrkH were also recorded. Similar bursts of channel activities with two levels of amplitudes were observed (Supplementary Table 1 and Supplementary Fig. 1a). However, the overall open probability of TrkH without TrkA is higher than that of the complex (0.65 ± 0.03 vs. 0.17 ± 0.01), suggesting that TrkA affects activity of TrkH. To verify that the single-channel currents were from TrkH, a cysteine substitution was made to a pore-lining residue (I220C) based on the known TrkH structure2. Perfusion of the cysteine modifying reagent [2-(trimethylammonium) ethyl] methanethiosulfonate (MTSET)24 reversibly reduced current amplitudes as well as the overall open probability for the I220C mutant for either TrkH alone or the TrkH-TrkA complex (Supplementary Fig. 1b and c). Wild type TrkH was not affected by MTSET (Supplementary Fig. 1c).
The open channel current-voltage (I-V plot) relationship is plotted in Fig. 1b and showed a slight inward rectification: level 2 has a chord conductance of 350 pS at +60 mV and 550 pS at -60 mV. Selectivity of the TrkH ion channel was examined by changing the intracellular side of the solution. Although TrkH does not discriminate Rb+ and Cs+ from K+, it conducts Na+ and Li+ at a significantly slower rate (Supplementary Table 1 and Supplementary Fig. 2). Given that the K+ channel signature sequence, TVGYG, which lines the selectivity filter, is not highly conserved in TrkH, the weak selectivity of TrkH is not surprising. In addition, substituting most of the intracellular Cl- with a large organic anion, gluconate, does not affect the reversal potential, indicating that the TrkH channel does not conduct Cl- (Supplementary Fig. 2d).
Since TrkA contains a conserved nucleotide-binding motif, we examined channel activity in the presence of NAD+, NADH, and ATP. While perfusion of NADs to the intracellular side of the channel did not produce significant changes in the currents, perfusion of 5 mM ATP substantially increased the open probability (Popen) of the TrkH-TrkA complex (Fig. 1c, Supplementary Fig. 3). The increase in Popen is the result of both a reduced time between bursts and a prolonged burst duration (Fig. 1c). To verify that this effect was mediated by TrkA, we mutated R100 on TrkA, which forms part of the ATP binding site (see Structure of the isolated TrkA tetramer below), to alanine, and the mutant no longer responds to ATP with an increase in Popen (Fig 1c). This increase in Popen does not require ATP hydrolysis, because two non-hydrolysable ATP analogs, adenosine 5′-O-(3-thiotriphosphate) (ATPγS) and adenosine 5′-(β,γ-imidotriphosphate) (AMP-PNP), both produced a similar increase in Popen (Fig. 1c). This result is consistent with earlier studies indicating that the Trk system in bacteria requires ATP25 but is not an ATPase26. Interestingly, ADP had the opposite effect to that of ATP and significantly reduced the Popen, while AMP has a negligible effect (Fig. 1c and Supplementary Fig. 3).
Structure of the TrkA-TrkH complex
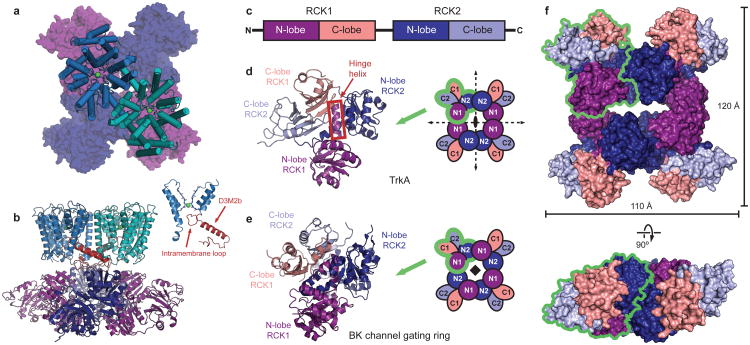
To understand how TrkH and TrkA interact, we co-expressed the two proteins, and crystallized the stable purified complex (Supplementary Fig. 4). Although NADH does not affect channel activity (Fig. 1c), inclusion of NADH in the crystallization solution improved crystal size and quality. The structure of the complex was solved at a resolution of 3.8 Å (Supplementary Fig. 5, Supplementary Table 2). In the asymmetric unit, two TrkH dimers interact with a ring of four TrkA protomers; this probably occurs because the internal symmetry of the TrkA tetramer creates two equivalent interfaces on both sides of the ring, as discussed in detail below. In the membrane, one TrkH dimer most likely interacts with one TrkA tetramer (Supplementary Fig. 6).
The tetrameric TrkA gating ring associates with the cytoplasmic face of the TrkH dimer (Fig. 2a, b). The structure of TrkH in the complex is almost identical to the previously solved structure of TrkH in isolation (Supplementary Fig. 7a). In particular, the pore of the ion conduction pathway is still obstructed by the intramembrane loop (Fig. 2b inset, Supplementary Fig. 7b). Each TrkH monomer interacts with TrkA through a cytosolic loop following the second half of the broken M2 helix in domain 3, D3M2b (Fig. 2b and Supplementary Fig. 4), which is tilted to lie nearly parallel to the bilayer and is connected directly to the intramembrane loop. The interface includes a number of salt bridges between positively charged residues on the D3M2b-D4 loop and negatively charged residues on TrkA (Supplementary Fig. 8b). A number of residues on the cytosolic side of TrkH are missing from the electron density, including an eighteen-residue stretch on the loop connecting domains 1 and 2 (Supplementary Fig. 8a). Although this cytosolic loop is not highly conserved among the TrkG/TrkH/KtrB proteins, because of its proximity to TrkA it may be involved in additional contacts not resolved in the crystal structure.
Figure 2. Structure of the TrkH-TrkA complex.
a.Structure of the complex between the TrkH dimer (light blue and teal) and the TrkA tetramer (dark blue and purple), viewed from the periplasmic side. The TrKA protomer is shown as a surface representation. b. The TrkH-TrkA complex viewed from within the plane of the membrane. Helix D3M2b in each TrkH subunit is highlighted in red. The inset on right shows homologous domains 1 and 3 of TrkH, with the intramembrane loop and helix D3M2b highlighted. c. Schematic showing the organization of RCK domains in a TrkA subunit. d-e. Cartoon representation of a TrkA protomer (d) or a protomer from the BK channel gating ring (e), colored by subdomain according to the color scheme in panel c. Schematics illustrating the organization of the tetrameric gating ring with the two-fold or four-fold symmetry axes marked are shown on the right. The green outlines delineate an individual subunit in each tetramer. f. Surface representation of the TrkA tetramer in the TrkH-TrkA complex, viewed from the periplasmic side (top) or parallel to the membrane (bottom). The green outline delineates an individual subunit.
Fold and quaternary structure of the TrkA RCK gating ring
The TrkA protomer contains two tandem RCK domains, RCK1 and RCK2, so that the gating ring contains a total of eight RCK domains. Each RCK domain comprises an N-terminal lobe assuming a Rossman fold, connected to a smaller C-terminal lobe by a “hinge” helix (Fig. 2c, d). A key difference between the TrkA structure and the BK and MthK channel-associated RCK structures11,15,16 is that the TrkA tetramer has D2 point group symmetry, involving three perpendicular, intersecting, two-fold rotational symmetry axes (Fig. 2d, f), whereas the BK gating ring has C4 symmetry (Fig. 2e). As a consequence of its “dimer of dimers” arrangement, the TrkA tetramer provides a two-fold symmetric interaction surface to match the two-fold symmetry of the TrkH dimer. Additionally, the tetramer contains two distinct types of subunit interfaces: N1-N1 interfaces, formed between the N-terminal lobes of the RCK1 domains; and N2-N2 interfaces, formed by the N-terminal lobes of the RCK2 domains. In contrast, the four-fold symmetric BK gating ring contains four identical N1-N2 interfaces.
The TrkA protomer contains two GXGXXG nucleotide-binding motifs27, one in the N-lobe of each RCK domain. The N2 lobe binding site in each TrkA protomer is occupied by a large electron density consistent with NADH, which was included as an additive to facilitate crystallization. At this low resolution the oxidized and reduced forms cannot be distinguished (Supplementary Fig. 9). The other binding site is unoccupied.
Structure of the isolated TrkA tetramer
We were also able to obtain crystals of TrkA without TrkH and in the presence of 10 mM ATPγS that diffracted to a resolution of 3.05 Å, and solved the structure by molecular replacement (Fig. 3a, Supplementary Table 2). Isolated TrkA forms a tetramer in solution (Supplementary Fig. 10) as well as in the crystal lattice (Supplementary Fig. 11). Each protomer contains two large densities consistent with ATPγS molecules, with one bound to the GXGXXG motif in the N lobe of each RCK domain, so that the gating ring contains a total of eight bound nucleotides (Supplementary Fig 12).
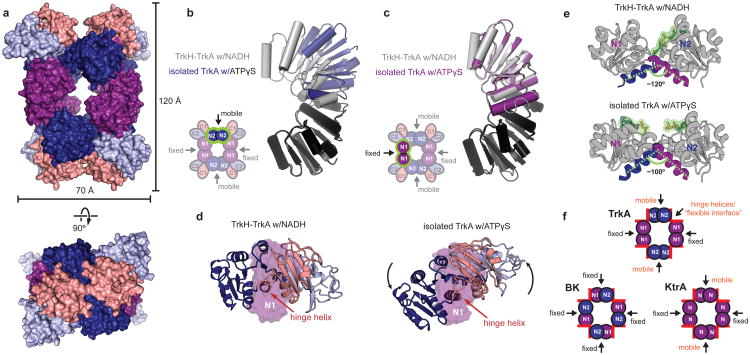
Figure 3. Comparison of TrkA in the TrkA-only and TrkH-TrkA complex structures.
a. Surface representation of the isolated TrkA tetramer, viewed from the periplamic side (top) or parallel to the membrane (bottom), colored by subdomain according to the color scheme in 2c. b-c. Superposition of the N2-N2 interface (b) and N1-N1 interface (c) in the TrkH-TrkA (gray) and TrkA-only (blue or purple) structures. The alignment was based on the domains colored black in both structures. d. A single TrkA protomer from the TrkH-TrkA complex structure (left) and a protomer from the TrkA-only structure (right) viewed from the same orientation after superposition by their N1 subdomains. N1 is shown as a surface, while the other three lobes are shown as cartoon. e. The N1 and N2 lobes in a subunit of TrkA from the NADH-bound TrkH-TrkA complex (top) and the ATPγS-bound TrkA (bottom) structures are shown with their hinge helices highlighted. NADH or ATPγS are shown in green. f. Cartoons of the TrkA, BK, and KtrA gating rings showing only the N lobes. The hinge helices are marked with red rectangles. Description of interfaces as “fixed” or “rotating” is based on comparison of structures 2HMS and 2HMW for KtrA, and 3NAF and 3U6N for BK.
Comparison of the structure of isolated TrkA to the structure from the TrkH-TrkA complex shows a substantial conformational change (Fig. 3a). This change is generated by markedly different behavior of the N1-N1 and N2-N2 interfaces. The N1-N1 interface shows no change between the two structures (Fig. 3b, Supplementary Video 1). In contrast, the N-lobes of the N2-N2 interface rotate and slide past each other (Fig. 3c, Supplementary Video 2), with one N2-N2 interface undergoing a larger translation than the other (Supplementary Fig. 13, Supplementary Video 3). The different behavior of the N1-N1 and N2-N2 interfaces converts TrkA from a roughly planar conformation to a twisted conformation (Fig. 3a).
The conformational change within individual protomers driving the “flat” to “twisted” transition in the gating ring can be visualized by aligning protomers from the ATPγS-bound structure and the complex by their immobile N1 subdomains only, which reveals rigid-body rotations of the N2, C1, and C2 lobes about the hinge helix connecting N1 to C1 (Fig. 3d, Supplementary Video 4). Given that the two structures were obtained at low resolution and bound to non-physiological ligands, it is difficult to identify specific protein-ligand interactions that may drive this conformational change. However, we note that the γ-phosphate groups on the bound ATPγS molecules are located very close to the cleft between the two N-lobes (Fig. 3e, Supplementary Fig. 13). The width of this cleft changes between the two structures, as measured by a change in the intersection angle of the hinge helices from 120° in the TrkA-TrkH structure to approximately 100° in the ATPγS-bound TrkA structure (Fig. 3e).
The change in the angle of intersection between hinge helices (Fig. 3e) is not unique to TrkA; similar movements have been observed in the KefC12,13, KtrA14, and MthK19 gating rings. In contrast, the interfaces corresponding to the intersubunit interfaces in TrkA (referred to as “assembly” interfaces in MthK and BK channels) appear to vary in their behavior between the different gating rings. In BK and MthK channels, all four assembly interfaces are fixed, forcing the ring to accommodate the change in the angle of the N-lobe cleft through a uniform, four-fold symmetric dilation. In KtrA and TrkA, two of the four assembly interfaces are able to move relative to each other, resulting in a conformational change that is twofold symmetric (Fig. 3f). It may be that the change in the angle of the cleft between the N1-N2-lobes represents a conserved response of RCK domains to ligand binding that introduces a strain on the gating ring, and that differences in the architecture and strength of the intersubunit or assembly interfaces determines how this strain is relieved, resulting in an overall conformational change to the ring that is suitable for the particular associated channel.
Mechanistic implications for channel regulation by TrkA
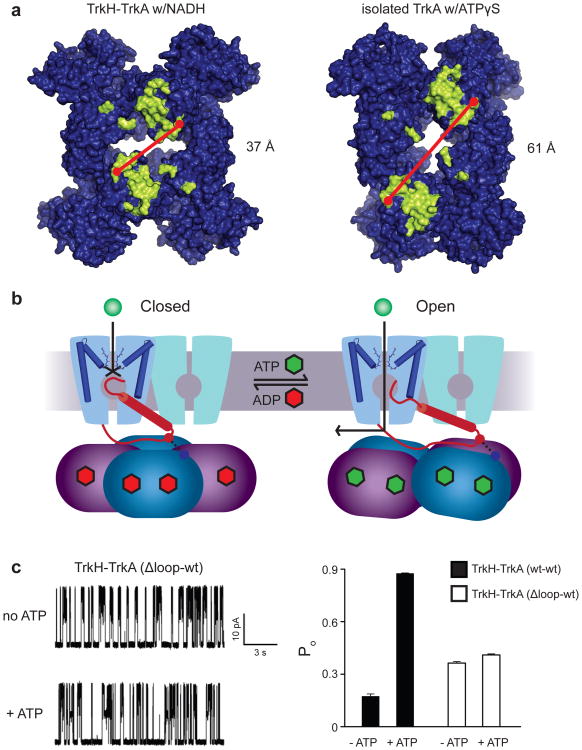
Comparison of the two TrkA conformations suggests a possible mechanism for opening of the channel by nucleotide-induced conformational changes on the gating ring. In the closed state bound to ADP, likely corresponding to the structure of the TrkH-TrkA complex, the ion permeation pathway is blocked by the intramembrane loop connected to helix D3M2b. While the “twisted” conformation of TrkA was obtained in the absence of the channel, possible effects of this conformational change on the channel can be probed by mapping the locations of residues forming the interface in the TrkH-TrkA structure onto the TrkA only structure (Fig. 4a). The surface residues interacting with the channel are located primarily in two patches on the diagonally positioned N2 domains facing the channel, each of which interacts with one of the channel D3M2b helices. These two interface regions are farther apart by ∼24 Å in the isolated TrkA structure (Fig. 4a, Supplementary Video 5). If ATP binding causes the gating ring to adopt a twisted conformation similar to the free TrkA structure, separation of the two interfaces on the tetramer could pull the helix D3M2b away from the center of the ion permeation pathway, leading to movement of the loop out of the pore and opening of the channel (Fig. 4b). As a first test of whether the intramembrane loop is in fact required for the complex's response to ATP, currents were measured on the complex of the WT TrkA and a TrkH mutant with residues 348-354 of the intramembrane loop deleted. The loop deletion mutant had a higher basal open probability than the WT channel, and was largely insensitive to perfusion of ATP (Figure 4c). Although this result is consistent with the hypothesized role of the intramembrane loop in gating of TrkH by TrkA, further structural and functional studies are necessary to reveal the molecular mechanism governing regulation of channel activity.
Figure 4. Proposed mechanism of regulation of TrkH gating by TrkA.
a. Surface representation of the TrkA tetramer from the TrkA-only (top) and TrkH-TrkA structures, viewed from the membrane-facing side. Residues forming the channel-gating ring interface, defined as residues with at least one atom within 4 Å of TrkH in the complex structure, are marked in yellow. b. Diagram illustrating a possible gating mechanism, with the closed channel shown on the left, and the open channel on the right. The D3M2b helix is represented as a red cylinder. For clarity, gating of only one TrkH protomer is shown. c. Left panel, single-channel currents through the TrkH-TrkA (Δ-loop-wt) complex before and after addition of ATP. The holding potential is +50 mV. Right panel, the Popen of TrkH-TrkA (wt-wt) and TrkH-TrkA (Δ-loop-wt) before and after addition of 5mM ATP. The error bars are s.e.m from 3 independent patches.
Discussion
In summary, this study presents the first demonstration that TrkH, and by extrapolation, the TrkG and KtrB family of proteins are ion channels. Additionally, we demonstrate that ion flux through the TrkH-TrkA complex is up-regulated by ATP and down-regulated by ADP. It is likely that in a cell the ratio of ATP to ADP determines the activity of TrkH, thus providing a way for regulating ion conduction in response to the metabolic activity of an organism. However, it is puzzling that a protein that promotes growth of bacteria in low external K+ concentrations only has a modest selectivity between K+ and Na+.
One curious feature of the Na+ and Li+ currents is that the amplitude of level 2 was not twice that of level 1. Similar behavior was observed for the I220C channels modified by MTSET (Supplementary Fig. 1-2 and Supplementary Table 1). One possible interpretation of this result is that under these conditions the conductance through an individual channel is lower when its partner is closed than when both are open. Because the D3M2b helix is connected to the intramembrane loop and also forms significant contacts with the neighboring protomer, it represents a possible avenue for coupling of gating in one channel to conductance in its partner. However, we currently have no explanation for why this behavior would occur more prominently when Na+ or Li+ occupies the selectivity filter.
In addition, we present two different structures of the TrkA gating ring, solved in complex with TrkH or in isolation. From these structures we have proposed a model for the gating of TrkH by TrkA that assigns the conformation in the complex to the closed state of TrkA, and the conformation in isolation to the open state. However, while the “open” structure was obtained bound to an analogue of the channel opener ATP, it should be noted that crystal packing and the absence of the channel likely have significant effect on the gating ring's energy landscape, and these factors rather than the identity of the bound ligand may be responsible for the conformation observed in the crystal structure of the isolated TrkA tetramer. In fact, a recently reported structure of TrkA from Vibrio vulnificus (PDB accession code 4G65) assumes a conformation highly similar to the twisted conformation reported here despite containing no bound nucleotide. Nonetheless, while it may not correspond exactly to the structure in the open complex, the isolated TrkA structure reveals the range of flexibility accessible to the gating ring. Confirmation for the proposed model will require further structural and functional studies of the intact complex.
Full Methods
Cloning and purification of the TrkH-TrkA complex
The TrkH gene was amplified from Vibrio parahaemolyticus genomic DNA and was cloned into the pET31b plasmid (Novagen) with an ampicillin-resistance gene. The Vibrio parahaemolyticus TrkA gene was cloned into a modified pET plasmid (Novagen) with a C-terminal polyhistidine tag and a TEV protease recognition site with a kanamycin-resistance gene. For large-scale purification of the VpTrkH-TrkA complex, BL21(DE3) cells were co-transformed with a 1:1.5 ratio of plasmids carrying VpTrkH and VpTrkA, respectively. The transformants were grown in Luria broth supplemented with 100 mg/l kanamycin and 100 mg/l ampicillin at 37°C and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) after OD600nm reached 1.0. The cells membranes were solubilized with 40 mM n-Decyl-β-D-maltoside (Anatrace) and the TrkA-TrkH complex was purified with TALON Metal Affinity Resin (Clontech Inc.). After removal of the His tag with TEV protease, the TrkH-TrkA was subjected to size exclusion chromatography with a Superdex 200 10/300 GL column (GE Health Sciences) pre-equilibrated in a buffer of 150 mM KCl, 20 mM HEPES, pH 7.5, 5 mM β-mercaptoethanol and 3.5 mM n-Decyl-β-D-maltoside. For crystallization, the complex was concentrated to 10 mg/ml as approximated by ultraviolet absorbance and NADH was added to a final concentration of 4 mM.
Cloning and purification of isolated TrkA
For expression of VpTrkA alone, the VpTrkA gene was cloned into a modified pGEX-5x-1 GST expression vector (GE lifescience) with a TEV protease recognition site replacing the original Factor Xa site. The plasmid was transformed into BL21 (DE3) cells, and the transformants were grown in Luria broth supplemented with 100 mg/l Ampicillin at 37°C. Once the OD600nm reached 1.0, expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 20°C for 15 hours. GST-VpTrkA was purified with Glutathione Sepharose 4B Resin (Amersham Bioscience). After removal of the GST with TEV protease, VpTrkA was subjected to size exclusion chromatography with a Superdex 200 10/300 GL column (GE Health Sciences) pre-equilibrated in a buffer of 150 mM KCl, 20 mM HEPES, pH 7.5, and 5 mM β-mercaptoethanol. The protein was concentrated to 8 mg/ml as approximated by BCA method.
Crystallization and structure solution
TrkH-TrkA crystals were grown by microbatch crystallization under mineral oil where 1.5 μl of the protein solution was mixed with equal volume of crystallization solution containing 37.5% PEG400, 400 mM ammonium sulfate and 100 mM HEPES, pH 6.75. Before harvesting crystals, tantalum bromide clusters (Jena Bioscience) were applied by dissolving approximately 25 μg/μl powder directly into the crystal growth solution and incubating for 10-12 hrs. Before flash-freezing in liquid nitrogen, the crystals were cryoprotected by soaking for 2-5 seconds in a solution obtained by sitting drop vapor diffusion with equal volumes of protein solution and well solution containing 35% PEG400, 400 mM ammonium sulfate, 100 mM Hepes, pH 6.75 and 25 μg/μl Ta6Br12.
Isolated VpTrkA was co-crystallized with ATPγS by vapor diffusion in sitting drops, where 1.5 μl of the protein solution supplemented with 10 mM ATPγS was mixed with an equal volume of crystallization solution. Crystals were obtained in 2 conditions: 10% polyacrlicacid5100, 100 mM Tris, pH 8.0, 20 mM MgCl2, and 5% glycerol; or 30% PEG400, 100 mM Glycine, pH 9.0, and 200 mM MgSO4. Before flash-freezing in liquid nitrogen, the crystals were cryoprotected by increasing the concentration of glycerol (25% v/v) and PEG400 (35% v/v) in the mother liquor.
X-ray diffraction data were collected at the beamlines X29 and X4A and C at the National Synchrotron Light Source, ID-24 and ID-17 at the Advanced Photon Source, 5.0.2 and 8.2.2 at the Advanced Light Source, and A1 and F1 at Chess. The structure of the TrkH-TrkA complex was solved by MRSAD using the Phenix software suite28, with the TrkH structure (3PJZ) as a search model and experimental phases from the eighteen Ta6Br12 clusters in the asymmetric unit. Iterative rounds of manual and automatic structure refinement were carried out with Coot32 and phenix.refine, respectively, and protein geometry was monitored using Molprobity33. The final model contains eighteen tantalum bromide clusters, four NADH molecules, four TrkA molecules and four TrkH molecules. Residues 63-65, and 159-177, located on cytoplasmic loops, are unresolved in the TrkH molecules; residues 162-163 and 178-181 are unresolved in TrkA. The isolated TrkA structure was solved using molecular replacement with the TrkA structure from the complex, and refined by a similar method to the structure of the complex. The final model of the isolated TrkA structure contained four molecules of ATPγS and two TrkA molecules.
Electrophysiology
The VpTrkH and VpTrkA containing vectors used in expressing the TrkH-TrkA complex were used. All mutations were made with the QuikChange kit (Agilent Technologies) and verified by sequencing through the entire coding regions. Giant spheroplasts were prepared from E. coli following a protocol described previously29-31, with minor modifications. In brief, a single colony co-transformed with TrkH and TrkA was incubated in 5 ml of modified LB medium (0.5% NaCl, 1% tryptone and 0.5% yeast extract) by shaking at 250 rpm, 37 °C. When the OD600 reached 0.5, the culture was diluted by 10 fold into 5 ml fresh modified LB with the addition of 60 μg/ml cephalexin. The elongated filaments were then harvested by centrifuging 2 ml of the culture at 3,000 g for 1 min. The supernatant was removed carefully and 0.5 ml 0.8 M sucrose was added to resuspend the pellet. The following solutions were added sequentially: 30 μl of 1 M Tris-HCl (pH 8.0), 24 μl of 0.5 mg/ml lysosome, 6 μl of 5 mg/ml DNase I, and 6 μl of 0.125 M EDTA-NaOH (pH 8.0). The mixture was incubated for 7.5 min at room temperature and 100 μl of the stop solution (0.7 M sucrose, 20 mM MgCl2, and 10 mM Tris-HCl, pH 8.0) was added to stop the digestion. The spheroplasts were aliquoted and stored at -20 °C. The frozen spheroplasts were thawed on ice and used in electrophysiology.
Patch clamp currents were recorded on inside-out patches pulled from the spheroplasts placed on clean glass coverslips. Electrodes were drawn from patch glass (TW100F-4, World Precision Instruments) and polished (MP-803, Narishige Co.) to a resistance of 7–10 MΩ. Both of bath solution and the pipette solution contained 200 mM KCl, 0.5 M sucrose, and 10 mM HEPES at pH 7.4. All solutions of perfused nucleotides were prepared from sodium salts. No pressure was applied to the membrane after the GΩ seal formation was achieved, which excluded the endogenous mechanosensitive channel activities31. The analog signals were amplified by an Axon-200B patch clamp amplifier (Molecular Devices Inc), and filtered at 1 kHz using the built-in Bessel filter, digitized at 100 μs by Digidata 1322a (Molecular Devices Inc), and recorded to a computer hard disk. The data was analyzed with pClampfit9 software.
Supplementary Material
Acknowledgments
Diffraction data for this study were measured at beamlines X4A, X4C, X25, and X29 of the National Synchrotron Light Source, ID-17 and ID-24 at the Advanced Photon Source, 5.0.2 and 8.2.2 at the Advanced Light Source, and A1 and F1 at the Cornell High Energy Synchrotron Source. MZ thanks Drs Evert Bakker, Ian Booth, Reinhard Krämer, and Clifford Slayman for discussions and insights. This work was supported by the US National Institutes of Health (DK088057, GM098878, and HL086392 to M.Z, and a subcontract (sub 5808 to M.Z.) from the PSI: Biology grant U54GM095315 to Dr. Wayne Hendrickson), the American Heart Association (12EIA8850017 to M.Z. and 0826067D to Y.P.), and the Irma T. Hirschl Trust Award to MZ. MZ was a Pew Scholar in Biomedical Sciences.
Footnotes
Author Contributions YC, YP, HH, EJL, XJ and MZ designed the experiments. YC and HH expressed, purified and crystallized the proteins, and collected and processed the diffraction data. HH, XJ, and EJL solved and refined the structures. YP prepared spheroplasts and performed patch clamp recordings. BK provided reagents and performed the initial cloning of vpTrkH. All authors participated in data analysis. YP, EJL, and MZ wrote the manuscript with inputs from all authors.
Supplementary Information accompanies the paper.
References
- 1.Corratge-Faillie C, et al. Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cellular and molecular life sciences: CMLS. 2010;67:2511–2532. doi: 10.1007/s00018-010-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y, et al. Crystal structure of a potassium ion transporter, TrkH. Nature. 2011;471:336–340. doi: 10.1038/nature09731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 4.Hanelt I, et al. Membrane region M2C2 in subunit KtrB of the K+ uptake system KtrAB from Vibrio alginolyticus forms a flexible gate controlling K+ flux: an electron paramagnetic resonance study. The Journal of biological chemistry. 2010;285:28210–28219. doi: 10.1074/jbc.M110.139311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanelt I, et al. Gain of function mutations in membrane region M2C2 of KtrB open a gate controlling K+ transport by the KtrAB system from Vibrio alginolyticus. The Journal of biological chemistry. 2010;285:10318–10327. doi: 10.1074/jbc.M109.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossemeyer D, et al. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. The Journal of biological chemistry. 1989;264:16403–16410. [PubMed] [Google Scholar]
- 7.Nakamura T, Yuda R, Unemoto T, Bakker EP. KtrAB, a new type of bacterial K+-uptake system from Vibrio alginolyticus. Journal of bacteriology. 1998;180:3491–3494. doi: 10.1128/jb.180.13.3491-3494.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlosser A, Hamann A, Bossemeyer D, Schneider E, Bakker EP. NAD+ binding to the Escherichia coli K+-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Molecular microbiology. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 9.Kong C, et al. Distinct gating mechanisms revealed by the structures of a multi-ligand gated K+ channel. eLife. 2012;1:e00184. doi: 10.7554/eLife.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y, Pico A, Cadene M, Chait BT, MacKinnon R. Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel. Neuron. 2001;29:593–601. doi: 10.1016/s0896-6273(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 12.Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 13.Roosild TP, et al. KTN (RCK) domains regulate K+ channels and transporters by controlling the dimer-hinge conformation. Structure. 2009;17:893–903. doi: 10.1016/j.str.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albright RA, Ibar JL, Kim CU, Gruner SM, Morais-Cabral JH. The RCK domain of the KtrAB K+ transporter: multiple conformations of an octameric ring. Cell. 2006;126:1147–1159. doi: 10.1016/j.cell.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, Yang Y, Ye S, Jiang Y. Structure of the gating ring from the human large-conductance Ca2+ gated K+ channel. Nature. 2010;466:393–397. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution. Science. 2010;329:182–186. doi: 10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buehner M, Ford GC, Moras D, Olsen KW, Rossman MG. D-glyceraldehyde-3-phosphate dehydrogenase: three-dimensional structure and evolutionary significance. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3052–3054. doi: 10.1073/pnas.70.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roosild TP, et al. Mechanism of ligand-gated potassium efflux in bacterial pathogens. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19784–19789. doi: 10.1073/pnas.1012716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye S, Li Y, Chen L, Jiang Y. Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels. Cell. 2006;126:1161–1173. doi: 10.1016/j.cell.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Tholema N, et al. All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. The Journal of biological chemistry. 2005;280:41146–41154. doi: 10.1074/jbc.M507647200. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda N, et al. Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. The Journal of biological chemistry. 2004;279:54952–54962. doi: 10.1074/jbc.M407268200. [DOI] [PubMed] [Google Scholar]
- 22.Bakker EP, Mangerich WE. The effects of weak acids on potassium uptake by Escherichia coli K-12 inhibition by low cytoplasmic pH. Biochimica et biophysica acta. 1983;730:379–386. doi: 10.1016/0005-2736(83)90355-3. [DOI] [PubMed] [Google Scholar]
- 23.Yuan P, Leonetti MD, Hsiung Y, MacKinnon R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature. 2012;481:94–97. doi: 10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 25.Rhoads DB, Epstein W. Energy coupling to net K+ transport in Escherichia coli K-12. The Journal of biological chemistry. 1977;252:1394–1401. [PubMed] [Google Scholar]
- 26.Stewart LM, Bakker EP, Booth IR. Energy coupling to K+ uptake via the Trk system in Escherichia coli: the role of ATP. Journal of general microbiology. 1985;131:77–85. doi: 10.1099/00221287-131-1-77. [DOI] [PubMed] [Google Scholar]
- 27.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. Journal of molecular biology. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long WS, Slayman CL, Low KB. Production of giant cells of Escherichia coli. Journal of bacteriology. 1978;133:995–1007. doi: 10.1128/jb.133.2.995-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felle H, Porter JS, Slayman CL, Kaback HR. Quantitative measurements of membrane potential in Escherichia coli. Biochemistry. 1980;19:3585–3590. doi: 10.1021/bi00556a026. [DOI] [PubMed] [Google Scholar]
- 31.Kuo MM, Saimi Y, Kung C, Choe S. Patch clamp and phenotypic analyses of a prokaryotic cyclic nucleotide-gated K+ channel using Escherichia coli as a host. The Journal of biological chemistry. 2007;282:24294–24301. doi: 10.1074/jbc.M703618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.