Abstract
ADP-ribosylation of proteins regulates protein activities in various processes including transcription control, chromatin organization, organelle assembly, protein degradation, and DNA repair. Modulating the proteins involved in the metabolism of ADP-ribosylation can have therapeutic benefits in various disease states. Protein crystal structures can help understand the biological functions, facilitate detailed analysis of single residues, as well as provide a basis for development of small molecule effectors. Here we review recent advances in our understanding of the structural biology of the writers, readers, and erasers of ADP-ribosylation.
Keywords: ADP-ribosylation, DNA repair, Glycohydrolase, Macro domain, Signaling pathways, Transferase
1. Introduction
Protein ADP-ribosylation, like other covalent posttranslational modifications, is used to modify protein activities in a dynamic fashion. Three major functional classes of proteins are involved in the regulation and turnover of the modification; namely, the writers, readers, and erasers of ADP-ribosylation. In this chapter, we describe the structural aspects of these classes of proteins, and we discuss how X-ray crystallography has helped to further our understanding of the function and regulation of PARP enzymes. In part one of the review, we describe the crystal structures of ADP-ribosyl transferase domains of the cholera toxin-like (ARTC) and the diphtheria toxin-like (ARTD) enzymes, especially the poly(ADP-ribose) polymerases (PARPs), and we compare them to sirtuins. We describe the macro domains, WWE domains, and PBZ domains, which bind one or several metabolites of ADP-ribose and, potentially, modified proteins at the site of ADP-ribose attachment. Finally, we describe the structures of poly(ADP-ribose) glycohydrolases (PARGs) which degrade poly(ADP-ribose) (PAR) chains and of ADP-ribose hydrolases (ARH) which remove mono-ADP-ribosyl moieties from protein side chains. In part two of the review, we describe how recognition of and binding to DNA strand breaks primes the activity of PARP1. Also, we summarize insights into how tankyrases use a specialized ankyrin repeat domain to recognize their targets for modification with poly(ADP-ribose).
2. The writers, readers and erasers of ADP-ribosylation
2.1. Writers: ADP-ribosyl transferases
ADP-ribosyl transferases fall into two major classes based on active site amino acid side chain composition. The cholera toxin like ADP-ribosyl transferases (ARTC) contain an R-S-F-E motif that is involved in NAD+ co-substrate binding and catalysis; whereas the diphtheria toxin like ADP-ribosyl transferases (ARTD), including the PARPs (see Table 1 ), contain an H-Y-Y/F-E motif, Fig. 1 (Otto et al., 2005, Hottiger et al., 2010). In the past decade we have gained structural insights into a significant portion of the ADP-ribosyl transferases, covering diverse enzymes from both of these families. We understand that secondary structures making up the binding pockets differ between the families, whereas anchoring of nicotinamide in the NAD+ binding pocket is conserved. Thus we now have a robust structural basis for interrogating the functions of active site residues. Ultimately, insights from structural biology will be indispensable both for understanding enzyme functions and regulation, and for the development of selective small molecule inhibitors as research tools and as therapeutic agents.
Table 1.
Nomenclature of PARP-family ADP-ribosyl transferases.
| Namea | HUGO name | Alternative names |
|---|---|---|
| ARTD1 | PARP1 | |
| ARTD2 | PARP2 | |
| ARTD3 | PARP3 | |
| ARTD4 | PARP4 | VaultPARP |
| ARTD5 | TNKS | Tankyrase; TNK1; PARP5a |
| ARTD6 | TNKS2 | Tankyrase-2; TNK2; PARP5b |
| ARTD7 | PARP15 | BAL-3 |
| ARTD8 | PARP14 | BAL-2 |
| ARTD9 | PARP9 | BAL-1 |
| ARTD10 | PARP10 | |
| ARTD11 | PARP11 | |
| ARTD12 | PARP12 | |
| ARTD13 | PARP13 | ZAP; ZC3HAV1 |
| ARTD14 | TIPARP | PARP7 |
| ARTD15 | PARP16 | |
| ARTD16 | PARP8 | |
| ARTD17 | PARP6 |
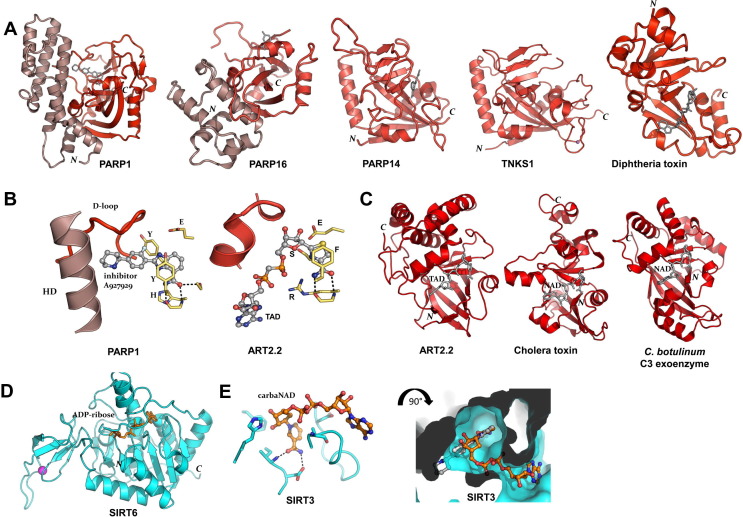
Fig. 1.
Structures of ADP-ribosyl transferases. (A) The ARTD family. Crystal structures of the transferase domains of human PARP1 (PDB: 3L3M), PARP16 (PDB: 4F0D), PARP14 (PDB: 4F1L), TNKS1 (PDB: 2RF5), all with nicotinamide mimicking inhibitors, and diphtheria toxin in complex with NAD+ (PDB: 1TOX). Two central β-sheets, one five stranded anti-parallel and one four stranded mixed β-sheet make up the core of the transferase domain. The β-sheets are surrounded by α-helices on each side and both modules contribute to the NAD+ binding crevice. An additional helical domain in PARP1 and PARP16 are shown in darker shade. N- and C-terminal positions are indicated. (B) Structure surrounding the signature motifs that contribute to the active site. Left, the H-Y-Y/F-E motif exemplified by ARTD member PARP1 in complex with inhibitor A927929 (PDB: 3L3M), with part of the α-helical domain indicated; right, the R-S-F-E motif exemplified by ARTC member rat ART2.2 in complex with NAD+-analog TAD+ (PDB: 1OG1). (C) The ARTC family. Crystal structures of the transferase domains of rat ecto-ART2.2 in complex with the NAD+-analog TAD+ (PDB: 1OG1), cholera toxin A1 in complex with NAD+ (PDB: 2A5F) and Clostridium botulinum C3 exoenzyme in complex with NAD+ (PDB: 2A9K). (D) The sirtuin family: human SIRT6 in complex with ADP-ribose (PDB: 3PKI). (E) Sirtuin NAD+ binding mode illustrated by human SIRT3 in complex with carba-NAD+ (PDB: 4FVT). Note the absence of nicotinamide stacking aromatic sidechains as compared to panel B. Right, the same structure, surface rendered and rotated to view into the nicotinamide pocket. Superimposition of the PARP inhibitor from panel B (in grey) illustrate that sirtuin ligands bind in a conformation different from that of ARTD and ARTC ligands.
2.1.1. The ARTD family
The overall structure of the diphtheria toxin like catalytic domain folds into two central β-sheets; one anti-parallel containing three to five strands, and one four or five stranded mixed β-sheet (Bell and Eisenberg, 1996, Ruf et al., 1996). These elements are surrounded by α-helices on each side and both modules contribute to the NAD+ binding crevice, Fig. 1A. The crystal structure of PARP1/ARTD1 revealed an α-helical domain that packs onto the catalytic domain, burying a large common hydrophobic interface, Fig. 1A (Ruf et al., 1996). Subsequently, structures of PARP2/ARTD2 and PARP3/ARTD3 confirmed the prediction that their catalytic domains are arranged in a very similar fashion (Oliver et al., 2004, Lehtiö et al., 2009). The α-helical domain constrains the NAD+ binding crevice near the opening that is distal to the nicotinamide pocket, Fig. 1B, and likely has regulatory roles (Ruf et al., 1998); but the domains have not been studied in isolation. The crystal structure of PARP16/ARTD15 also reveals an α-helical domain paired with the catalytic domain, but in a different relative position, Fig. 1A (Karlberg et al., 2012). Thus, possibly, some of the uncharacterized ARTD enzymes may also employ currently unrecognized accessory domains for regulation of their activities.
Structures of transferase domains in complex with either NAD+ or a non-hydrolyzable analog have been determined for a few bacterial toxins in the ARTD family, namely diphtheria toxin (Bell and Eisenberg, 1996), Pseudomonas exotoxin A (Jorgensen et al., 2005), and Vibrio cholix toxin (Fieldhouse et al., 2012). Structures of human PARP family ARTD catalytic domains have never been determined in complex with NAD+, but many are available with nicotinamide mimicking ligands, e.g., PARP1 (Kinoshita et al., 2004), PARP2 (Karlberg et al., 2010a), PARP3 (Lehtiö et al., 2009), TNKS1 (Lehtiö et al., 2008), TNKS2 (Karlberg et al., 2010b), PARP10 (PDB: 3HKV), −12 (PDB: 2PQF), and −13 (PDB: 2X5Y), PARP14 (Andersson et al., 2012, Wahlberg et al., 2012), PARP15 (Andersson et al., 2012), and PARP16 (Karlberg et al., 2012). The nicotinamide binding pocket is formed by the PARP signature motif, including a β-strand followed by an α-helix and the donor loop (D-loop) connected to a second β-strand followed by an α-helix.
An important current question concerning PARP activities is how mono-ADP-ribosylation is catalyzed in contrast to ADP-ribose chain elongation. Presence of a glutamate in the active site (the E of the H-Y-Y/F-E motif) has long been seen as a discriminator for PAR chain elongation activity, Fig. 1B. Yet, since bacterial toxins that contain glutamate in the active site only catalyze mono-ADP-ribosylation, it was clear that the mechanism was not fully understood. Later, it was proposed that PARP family transferases with small hydrophobic side chains in place of the glutamate have mono-ADP-ribosyl transferaseADP-ribosyl transferase activity, which was demonstrated for PARP10/ARTD10 and PARP14/ARTD8 (Kleine et al., 2008). Currently, mass spectrometry methods are being developed to reliably evaluate ADP-ribosylated substrates.
2.1.2. The ARTC family
The cholera toxin like ARTC transferases comprise bacterial toxins, with various subgroups (Fieldhouse and Merrill, 2008), and mammalian exo-enzymes (Koch-Nolte et al., 2008). Crystal structures of rat ART2.2 (Mueller-Dieckmann et al., 2002) and 10 different bacterial exotoxins (see http://www.rcsb.org/pdb) have been elucidated. ARTC: NAD+ complex structures have been obtained for rat ART2.2 (Ritter et al., 2003) and several bacterial toxins (Han et al., 1999, Evans et al., 2003, Tsuge et al., 2003, O’Neal et al., 2005, Pautsch et al., 2005, Margarit et al., 2006, Sundriyal et al., 2009, Visschedyk et al., 2012). The overall structure of a typical ARTC is composed of an N-terminal, purely α-helical subdomain and a C-terminal domain of a mixed β-sheet, Fig. 1C. Size and strand composition of the β-sheets varies within the family; a common feature is a core structure of four central β-strands that makes up the base of the nicotinamide pocket of the NAD+ binding crevice. NAD+ binding is accomplished by the side chains of the R-S-F-E motif, and NAD+ is held in a similar configuration as in the ARTD transferases, albeit with different protein side chain involvement, Fig. 1B.
2.1.3. The sirtuins
Sirtuins are most prominent as NAD+-dependent deacetylases of protein acetyl-lysines. Sirtuin assisted protein ADP-ribosylation has also been documented (Haigis et al., 2006, Ahuja et al., 2007, French et al., 2008, Pan et al., 2011). Likely, ADP-ribose-peptidyls are intermediates in the deacetylation reactions, at least for certain sirtuins (Sauve, 2010, Sauve and Youn, 2012). These deacetylases differ substantially from the ARTC and ARTD transferases in both their overall structure, Fig. 1D, and their mode of NAD+ binding, Fig. 1E. Nevertheless, sirtuins might be affected by certain PARP inhibitors. Sirtuins possess a nicotinamide pocket, which is involved in feedback inhibition (Sauve et al., 2005). This pocket is not as prominent as the nicotinamide anchoring pocket of ARTC and ARTD transferases, and it lacks the stacking tyrosine residue that is typical for PARPs and transferase toxins, Fig. 1E, left panel. Consequently, it is important to remember that small nicotinamide-mimicking PARP inhibitors, such as the widely used research tool, 3-aminobenzamide, likely affect sirtuin activities. Larger PARP inhibitors, where affinity and specificity is gained by extending the compound into the ARTD NAD+ pocket, take a different path and are less likely to affect sirtuins.
2.2. Reader domains
At least four protein modules and motifs recognize and bind ADP-ribose and its metabolites: the macro domains, PAR binding zinc finger (PBZ) domains, WWE domains, and the PAR binding motif (PBM) (Kalisch et al., 2012, Gibson and Kraus, 2012). These fulfill three physiological functions, namely (i) PAR binding and localization of proteins to sites of PARP activity; (ii) binding of mono-ADP-ribosylated substrate proteins; and (iii) sequestration and turnover of ADP-ribose metabolites. PAR binding is mediated by all four modules and motifs; the remaining two functions are indicated for certain macro domain proteins.
PAR binding motifs (PBM) are short amino acid sequences found in proteins related to chromatin regulation, DNA damage response, and RNA processing (Pleschke et al., 2000). Structural information is limited to ligand free solution structures of the PBM containing first BRCT domain of XRCC1 (PDB: 2D8M) and a crystal structure of AIF1 (Mate et al., 2002), although PAR binding by AIF1 has subsequently been characterized using site-directed mutagenesis (Wang et al., 2011).
2.2.1. Macro domains
Originally identified in virus proteins (Gorbalenya et al., 1991) and subsequently found in histone macro H2A protein (Pehrson and Fried, 1992), the ∼120 residue macro domains have now been identified in the genomes from species across all kingdoms of life. The module appears to be utilized to recognize poly(ADP-ribose) and several ADP-ribose metabolites in the processes of DNA repair, gene regulation, signaling events, and control of NAD metabolism (Till and Ladurner, 2009). Macro domains have been shown to bind ADP-ribose as well as PAR chains (Ahel et al., 2009, Gottschalk et al., 2009, Timinszky et al., 2009; and summarized by Kleine and Lüscher (2009), some bind ADP-ribosylated proteins (Forst et al., 2013) and some can bind RNA (Malet et al., 2009). Ten human genes encode macro domains, of which PARP9/ARTD9, PARP14/ARTD8, and PARP15/ARTD7 are unique in containing several macro domains in a series.
The first macro domain crystal structure to be determined was Af1521 from the thermophilic bacterium Archeoglobus fulgidus (Allen et al., 2003). It is a single-domain protein that binds ADP-ribose with high affinity (Karras et al., 2005). Several other macro domain structures have been described (e.g., Kustatscher et al., 2005, Egloff et al., 2006, Tan et al., 2009, Malet et al., 2009). The typical macro domain fold has a compact globular shape with a central 7-stranded mixed β-sheet flanked by five α-helices, packed onto both sides of the sheet, with one helix at the ridge, Fig. 2 . The ADP-ribose binding sites are situated at a crest, with the central phosphates typically buried inside a tunnel, Fig. 2A.
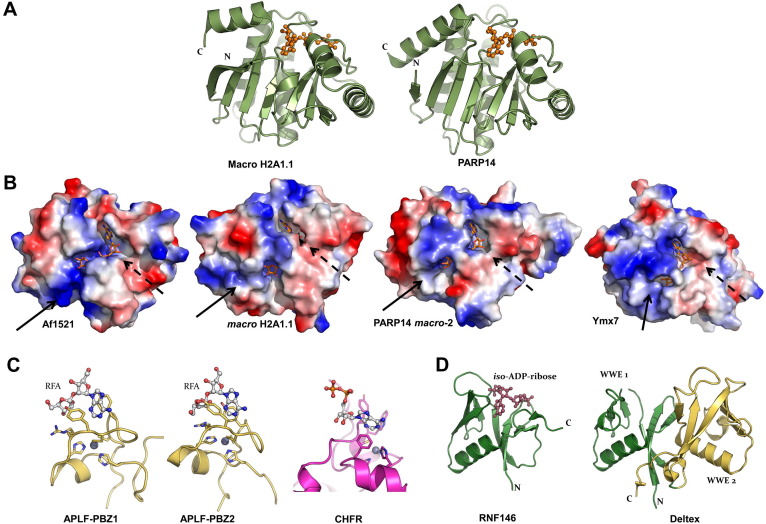
Fig. 2.
Structures of ADP-ribosyl binder domains. (A) Crystal structures of the macro domain of human histone macro H2A1.1 (PDB: 3IID) and of human PARP14 macro domain 2 (PDB: 3Q71), both in complex with ADP-ribose. (B) Charge potential surface rendering of Af1521 (PDB: 2BFQ), histone macro H2A1.1, PARP14 macro domain 2, and yeast Ymx7 (PDB: 1TXZ), all in complex with ADP-ribose. Solid arrows indicate the putative PAR crevice or its corresponding position; dashed arrows indicate the either exposed or buried adenine ribose site. (C) Structures of the two PBZ domains of APLF (PDB: 2KQD, 2KQE) in complex with ribofuranosyladenosine (RFA) and the PBZ domain of CHFR (PDB: 2XOC). (D) Crystal structures of the WWE domain of E3 ubiqutin protein ligase RNF146 in complex with iso-ADP-ribose (PDB: 3V3L) and the tandem WWE-domain of Drosophila melanogaster Deltex protein (PDB: 2A90).
Evidence for recognition of mono-ADP-ribosylated proteins by macro domains has been presented (Dani et al., 2009, Forst et al., 2013, Rosenthal et al., 2013). How do macro domains bind their respective targets, i.e. PAR chains and, potentially, mono-ADP-ribosylated proteins? Crystal structures of PAR binding macro domains show extended surface crevices that are continuous with the sites where ADP-ribose is bound. The A-ribose (adenine ribose, or ribose′) of the co-crystallized ADP-ribose is typically rather buried, (cf. the “ribose cap” of PARG; Section 2.3.1.). By contrast, the proximal N-ribose (nicotinamide ribose, or ribose″) is situated in the continuous crevice that is lined by overall positive charge, suggestive of PAR phosphate group accommodation. This is exemplified by Af1521 and histone macro H2A1, both of which bind PAR chains (Karras et al., 2005), Fig. 2B. The structure of Af1521 suggests binding along PAR chains, as the A-ribose 2′-hydroxyl is exposed; whereas the structure of H2A1 suggests binding the terminal end of a chain, unless a conformational rearrangement could expose the buried A-ribose, Fig. 2B. A putative PAR binding crevice is also found in PARG structures (Section 2.3.1.), for which a model of PAR4 interaction has been constructed (Dunstan et al., 2012). By contrast, the macro domains of PARP14/ARTD8 and PARP15/ARTD7 (Forst et al., 2013) bury the A-ribose, but the N-ribose is notably more exposed compared to PAR binding macro domains, and the N-ribose does not reside near a positively charged crevice, Fig. 2B. However, crystal structures with PAR chain fragments are unavailable.
Whereas most macro domains appear to be binding modules, some have evolved catalytic activities. For example, the human MacroD1 and MacroD2 proteins, the C6orf130 gene product, and several related bacterial proteins remove the acetyl group from O-acetyl-ADP-ribose, the product of sirtuin-mediated protein lysine deacetylation (Peterson et al., 2011, Chen et al., 2011). MacroD1, MacroD2 and C130Orf6 also de-ADP-ribsylate mono-ADP-ribosylated PARP10 substrate GSK3b (Rosenthal et al., 2013). Yeast Ymx7, Fig. 2B, SARS coronavirus non-structural protein-3, and several other viral macro domain proteins have ADP-ribose-1′-phosphatase activity (Kumaran et al., 2005, Saikatendu et al., 2005, Malet et al., 2009, Neuvonen and Ahola, 2009). Crystal structures and results from site-directed mutagenesis are consistent with this activity (Malet et al., 2009, Neuvonen and Ahola, 2009), but there is no consensus catalytic mechanism. Nevertheless, it is clear that the surface loops surrounding the N-ribose are variable, and may have evolved for enzymatic activities. This is also highlighted by the case of PARG: these glycohydrolases that break down PAR chains have a macro domain core, but also contain acidic sidechains in the ADP-ribose binding site, which activate the N-ribose for nucleophilic attack (elaborated in Section 2.3.1. below).
2.2.2. PAR binding zinc finger (PBZ) domains
PAR binding zinc fingers have been identified in only a handful of proteins. They function in DNA break repair, check-point regulation, and PAR metabolism (Ahel et al., 2008). The zinc finger is of C2H2 type, with a consensus [K/R]x 2Cx[F/Y]Gx 2Cxbbx 4Hx 3[F/Y]xH motif. Solution NMR structures of PBZ domains exist for human APLF (aprataxin polynucleotide kinase-like factor) (Eustermann et al., 2010, Li et al., 2010) and Drosophila melanogaster CG1218-PA (Isogai et al., 2010). In addition, a crystal structure of human E3 ubiquitin protein ligase CHFR (checkpoint with forkhead and RING finger domains) (Oberoi et al., 2010) includes a C-terminal PBZ domain. The small domain folds into two short parallel β-strands and a small α-helix with a loop connecting the first strand and the helix, Fig. 2C. The zinc ion is coordinated by two cysteine residues located in the loop, one histidine in the helix, and a second histidine just after the second β-strand. APLF contains two PBZ-domains connected via a short linker; the domains are structurally independent and were both shown to bind ADPR and PAR (Eustermann et al., 2010, Li et al., 2010). A fragment of PAR containing the unique O-glycosidic bond between two ribose units (2′-O-α-D-ribofuranosyladenosine) was successfully used to probe PAR binding in the PBZ domains of APLF (Eustermann et al., 2010). Crystal structures of CHFR also revealed two consecutive binding sites for adenosine, with each site sandwiched between stacking aromatic or arginine sidechains, consistent with PAR chain recognition (Oberoi et al., 2010).
2.2.3. WWE domains
The WWE domain, named after three conserved residues in single letter code, is mainly found in two families of proteins, namely the PARPs and the E3 ubiquitin protein ligases. WWE domains bind PAR and play roles in ubiquitylation mediated proteasomal degradation (Andrabi et al., 2011, Kang et al., 2011, Zhang et al., 2011, Wang et al., 2012). Recently, the crystal structure of the WWE domain from the E3 ubiquitin protein ligase RNF146 (RING finger protein-146/Iduna) was determined in complex with iso-ADP-ribose (Wang et al., 2012). This ligand contains the glycosidic bond between two ribose units that is also found in PAR chains, Fig. 2D. Crystal structures and solution NMR structures of individual WWE-domains have been described for RNF146 (Wang et al., 2012) as well as PARP11/ARTD11 and PARP14/ARTD8 (He et al., 2012), and a tandem WWE domain structure of the Deltex protein from the fruitfly has been reported (Zweifel et al., 2005), Fig. 2D. The domain contains six anti-parallel β-strands that form a half β-barrel structure with an α-helix covering the convex side. The ligand binding site is made up by extended loops on the crest of the domain. In the RNF146:iso-ADP-ribose complex, the adenine moiety forms hydrogen bonds with a conserved glutamine and is sandwiched between two conserved tyrosines. The phosphates face solution and interact with both basic amino acid sidechains and backbone nitrogens (Wang et al., 2012). In the tandem domain structure of the Deltex protein, the two WWE domains pack against each other with approximately 2-fold rotational symmetry, with a small α-helix inserted between them (Zweifel et al., 2005).
2.3. Erasers: ADP-ribose glycohydrolases
In the category of PAR catabolism, two activities are of immediate relevance for ADP-ribose mediated signaling. Breakdown of PAR chains is catalyzed by poly(ADP-ribose) glycohydrolases (PARG) and by ADP-ribosyl hydrolase-3 (ARH3). Removal of ADP-ribose from mono(ADP-ribos)ylated proteins (or the terminal ADP-ribose moiety after PAR chain breakdown) is assisted by at least one of several ARH isoforms in vertebrates. The ARH class of enzymes is closely related to the DRAG bacterial enzymes with ADP-ribose glycohydrolase activity.
2.3.1. Poly(ADP-ribose) glycohydrolase (PARG)
PAR degradation is an essential enzymatic activity, as evidenced by the lethality of PARG knockout in mice (Koh et al., 2004). A canonical PARG enzyme is conserved in bacteria and eukaryotes. The bacterial and fungal enzymes have a catalytic core with a short N-terminal accessory domain extension, whereas protozoan and vertebrate PARG orthologs feature a longer accessory domain, and vertebrate PARG orthologs have an additional regulatory domain, Fig. 3 A. The N-terminal segments confer regulatory activities that are not fully understood, but they are needed for recruitment to sites of DNA damage (Mortusewicz et al., 2011). N-terminally truncated human PARG has significant activity in vitro (Botta and Jacobson, 2010, Dunstan et al., 2012). The catalytic fragment of PARG is essentially a macrodomain (Slade et al., 2011) with an inserted loop region that contains the PARG signature sequence, GGG-X6−8-QEE (Patel et al., 2005), Fig. 3B and C.
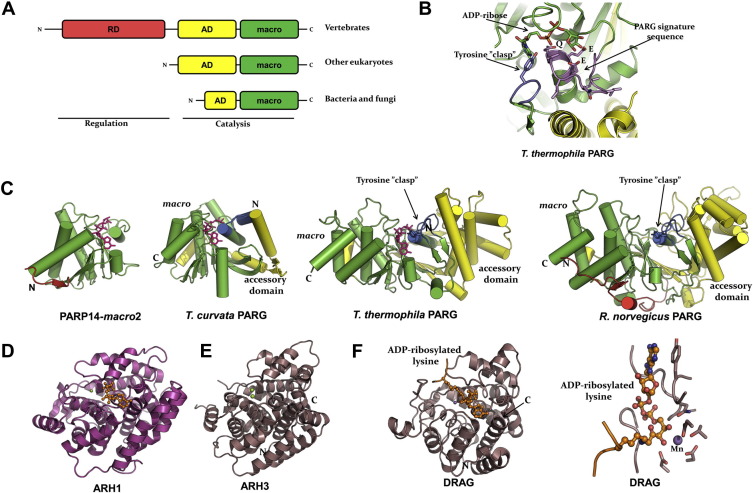
Fig. 3.
Structures of ADP-ribosyl erasers. (A) Schematic domain arrangement in PARG orthologs. RD, regulary domain; AD, accessory domain; macro, macro homology domain. (B) Closeup of the ADP-ribose binding site including the PARG signature motif in a eukaryotic PARG (T. thermophila; PDB: 4EPP). Green and yellow indicate domains as in panel A. (C) Crystal structures of PARP14 macro domain 2 (PDB: 3Q71), a bacterial PARG (PDB: 3SIJ), and the T. thermophila and rat orthologs (PDB: 3UEK). Colors indicate domains as in panel A. (D) Structure of human ARH1 (PDB: 3HFW). (E) Structure of human ARH3 (PDB: 2FOZ). (F) Structure of the dinitrogenase reductase activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribosyllysine (PDB: 2WOD), and right, closeup view of ADP-ribosyllysine trapped in the active site of DRAG.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this book.)
The crystal structures of a bacterial and of Tetrahymena PARG orthologs (Slade et al., 2011, Dunstan et al., 2012) explain PAR binding and exo-glycohydrolase activities: the ligand ADP-ribose resides in a surface crevice with the A-ribose blocked by a “ribose cap”, marking the position of the terminal moiety of a PAR chain. Two glutamate sidechains near the N-ribose are important for catalysis; one of them is ideally positioned to activate the leaving group on N-1 ADP-ribose and a water molecule for attack on the ribose, resulting in O-glycosydic bond cleavage. The rat and human PARG catalytic regions (core and the N-terminal accessory domains) are very similar; they adopt a bean-shaped structure with a deep central cleft containing the conserved PARG signature motif and a “tyrosine clasp”, Fig. 3B and C, which contributes strongly to PARG catalytic efficiency and inhibitor binding (Koh et al., 2003, Kim et al., 2012; PDB: 4A0D). The ribose cap that shields the A-ribose 2′-hydroxyl in bacterial PARG is absent in the rat, human, and Tetrahymena PARG structures, explaining their activities as both exo- and endoglycohydrolases (Dunstan et al., 2012). PARG orthologs cannot remove the last remaining ADP-ribose moiety on their substrate proteins (Slade et al., 2011, Dunstan et al., 2012).
2.3.2. ADP-ribosyl hydrolases (ARH)
Mammals express a family of three ADP-ribosyl hydrolases (ARH1-3) (Koch-Nolte et al., 2008). ARH1 removes ADP-ribose from ADP-ribosylated arginine residues, ARH3 predominantly hydrolyses O-acetyl-ADP-ribose and PAR in vitro, whilst the activity of ARH2 remains elusive (Moss et al., 1997, Mueller-Dieckmann et al., 2006, Oka et al., 2006). ARH3 has also been observed to hydrolyze O-acetyl-ADP-ribose (Ono et al., 2006).
The crystal structures of human ARH1 (Kernstock et al., 2009; PDB: 3HFW), Fig. 3D, and mouse and human ARH3 (Mueller-Dieckmann et al., 2006, Mueller-Dieckmann et al., 2008), Fig. 3E, show globular, purely α-helical protein folds composed of four helical bundle subdomains. In ARH3, the active site is situated in the top center of the molecule, with major sidechain contributions from two of the subdomains. The active site contains a bi-magnesium center, and each metal ion has octahedral coordination. A glutamic acid or one of two aspartic acid sidechains are believed to be involved in catalysis by activating a water molecule for nucleophilic attack on the scissile bond (Mueller-Dieckmann et al., 2006). The ARH1 structure was solved as a complex with ADP and one magnesium ion. The positions of the three ARH3 sidechains proposed to be involved in catalysis are conserved in ARH1; however the active site is shielded by a triad of hydrophobic sidechains (F26, A98, W266) emerging from extended surface loops. Thus it appears that a protein substrate binding event might be needed for access to the catalytic site.
Our understanding of ARH activities might make a leap forward once substrate proteins have been verified. Since ARH2 activity has not been documented, it will be important to investigate whether it works on a limited range of substrates only. If so, a next question will be whether ARH1 catalyzes the removal of ADP-ribose from a variety of substrates and sidechains, or whether there are other, as yet unidentified ADP-ribose glycohydrolases. Since ARH3 is mitochondrial (Mueller-Dieckmann et al., 2006, Niere et al., 2012), it will be interesting to see whether it can remove also the last remaining ADP-ribose moiety of its modified substrate.
2.3.3. Bacterial ADP-ribosyl hydrolases (DRAG)
ADP-ribosylation contributes to regulation of dinitrogenase reductase in nitrogen fixing bacteria (Nordlund and Ludden, 2004). ADP-ribosylation of an arginine residue by dinitrogenase reductase ADP-ribosyl transferase (DRAT) inactivates the enzyme. The reverse reaction, removal of ADP-ribose and re-activation of the enzyme, is catalyzed by dinitrogenase reductase activating glycohydrolase (DRAG).
Crystal structures of DRAG from two different bacteria, Rhodospirillum rubrum and Azospirillum brasilense (Berthold et al., 2009, Li et al., 2009), show extensive structural homology with the ARH enzymes. The overall structures are compact and purely α-helical, and the active sites are located in a deep cleft surrounded by loops, Fig. 3F. The active site center contains a bi-nuclear metal site, occupied by two magnesium or two manganese atoms coordinated by eight sidechain contacts. While the highest activity is observed with manganese (Berthold et al., 2009), mammalian ARH proteins prefer magnesium (Mueller-Dieckmann et al., 2006). Interestingly, Berthold et al. presented an ADP-ribosyllysine complex that was formed when an active-site mutant was crystallized in presence of large excess of ADP-ribose. A lysine residue from a neighboring DRAG molecule had been ADP-ribosylated and trapped in the active site. The ribosyllysine was observed in an open conformation possibly mimicking the Michaelis complex, Fig. 3F. Based on available structural data, a detailed reaction mechanism of mono-ADP-ribosyl hydrolysis was suggested (Berthold et al., 2009).
3. Regulation of poly(ADP-ribose) formation and target selection for posttranslational modification
There is currently a gap in our overall understanding of how the poly(ADP-ribosyl)ation activity of PARP enzymes is regulated, and how the targets for modification are selected. The following sections describe recent structural studies of PARP1/ARTD1 and tankyrases (ARTD5 and ARTD6) that have made key contributions to our understanding of specific regulation mechanisms. A better understanding of the regulation mechanisms of PARP1 and tankyrases will advance our knowledge of poly(ADP-ribose)-mediate cellular signaling events, and will enhance our ability to specifically inhibit these enzymes for therapeutic purposes.
3.1. PARP1: a multi-domain, multi-function enzyme
PARP1/ARTD1 is the founding and most studied member of the PARP superfamily. PARP1 is responsible for the majority of the poly(ADP-ribosyl)ation activity in the cell and is involved in various cellular processes including DNA repair, transcription, chromatin regulation, and cell death signaling (reviewed by D’Amours et al., 1999, Krishnakumar and Kraus, 2010). PARP1 modifies various cellular substrates, but its major target is PARP1 itself, an activity termed automodification. Using NAD+ as a substrate, PARP1 is capable of producing long and branched poly(ADP-ribose) modifications. In response to cellular DNA damage, PARP1 binds avidly to DNA breaks, which stimulates its catalytic activity up to 500-fold and subsequently allows the recruitment of the DNA repair machinery and chromatin remodeling factors to the site of damage (reviewed in (D’Amours et al., 1999, De Vos et al., 2012)). PARP1 has emerged as a promising therapeutic target for the treatment of cancer due to its role in responding to DNA damage and through its role in transcriptional regulation (Fong et al., 2009, Brenner et al., 2011, Schiewer et al., 2012).
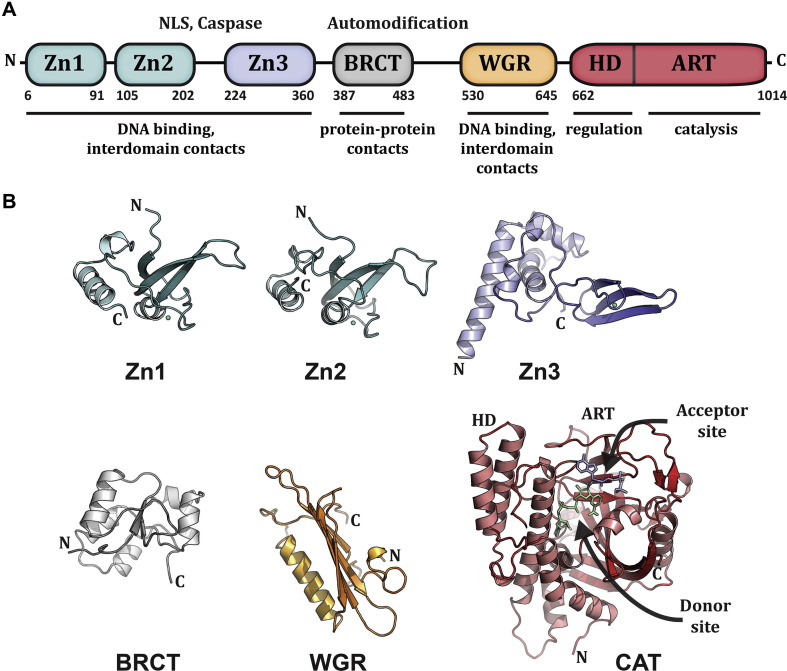
PARP1/ARTD1 is 116 kDa with a modular architecture of six independent domains, Fig. 4 A. The N-terminus of the protein is composed of two zinc fingers, Zn1 and Zn2, which are involved in binding to DNA breaks. The Zn1 and Zn2 domains are specialized zinc fingers that recognize DNA structures rather than specific sequences (D’Silva et al., 1999, Pion et al., 2003). The Zn3 domain has a structure distinct from Zn1 and Zn2 and is involved in PARP1 interdomain communication and PARP1 ability to compact chromatin (Langelier et al., 2008, Langelier et al., 2010). The automodification domain (AD) contains a BRCT motif and several of the residues that are targeted for automodification (Altmeyer et al., 2009, Tao et al., 2009). The WGR is a domain for which the function has only recently begun to emerge (Langelier et al., 2012). WGR is named for a strongly conserved patch of residues, Trp-Gly-Arg (WGR in single-letter code), and it is essential for PARP1 DNA-dependent activity (Altmeyer et al., 2009). The catalytic domain of PARP contains two subdomains: the helical subdomain (HD) and the ADP-ribosyl transferase subdomain (ART). The HD is composed of six α-helices (αA to αF) with connecting linkers. The ART is of the ARTD class described in a previous section, and contains the conserved residues involved in NAD+ binding and catalysis. The Zn1, Zn3, WGR and CAT domains are strictly required for PARP1 DNA-dependent activity, while the Zn2 and BRCT domains are not essential (Langelier et al., 2008, Tao et al., 2008, Altmeyer et al., 2009, Langelier et al., 2011, Langelier et al., 2012). The Zn2 domain does contribute to PARP1 activation by single-strand breaks, but has no apparent role in double-strand break activation of PARP1 (Ikejima et al., 1990, Altmeyer et al., 2009, Langelier et al., 2011, Langelier et al., 2012).
Fig. 4.
PARP1 modular domain structure. (A). Schematic representation of human PARP1 domains. The amino acid numbering for human domain boundaries are noted. Homologous DNA-binding zinc finger domains, Zn1 and Zn2, are located at the N-terminus of PARP1. These domains are followed by a linker region containing a bipartite nuclear localization signal (NLS) and a caspase three cleavage site (Caspase). This regulatory region is followed by a third zinc finger domain (Zn3), which has a distinct structure and function from that of Zn1 and Zn2. A BRCA C-terminus (BRCT) fold is located within the region of PARP1 that is primarily targeted for automodification. The C-terminal end of PARP1 contains a WGR domain, named after a conserved Trp-Gly-Arg sequence, and the catalytic domain, which is composed of an α-helical subdomain (HD) and an ADP-ribosyl transferase subdomain (ART). The known functions for the individual domains are noted beneath the schematic. (B). Crystal and/or NMR structures have been determined for each of the PARP1 domains in the absence of DNA: the NMR structures for the homologous Zn1 and Zn2 domains [PDB: 2DMJ and 2CS2; see also reference (Eustermann et al., 2011)], the NMR structure of the Zn3 domain is shown [PDB: 2JVN; see also (Langelier et al., 2008) for crystal structure determination], the NMR structure of the BRCT fold [PDB: 2COK; see also (Loeffler et al., 2011), the NMR structure of the WGR domain (PDB: 2CR9), and the crystal structure of the catalytic domain (PDB: 1A26) (Ruf et al., 1996)]. Catalytic subdomains are labeled (HD and ART). The crystal structure was determined in the presence of an NAD+ analog that has defined the proposed “acceptor site” for poly(ADP-ribose) formation. The NAD+ binding site is modeled in this figure based on the structure of an ADP-ribosylating toxin structure (Bell and Eisenberg 1996). Reprinted from (Langelier and Pascal 2013) with permission from Elsevier.
3.1.1. PARP1: DNA damage detection
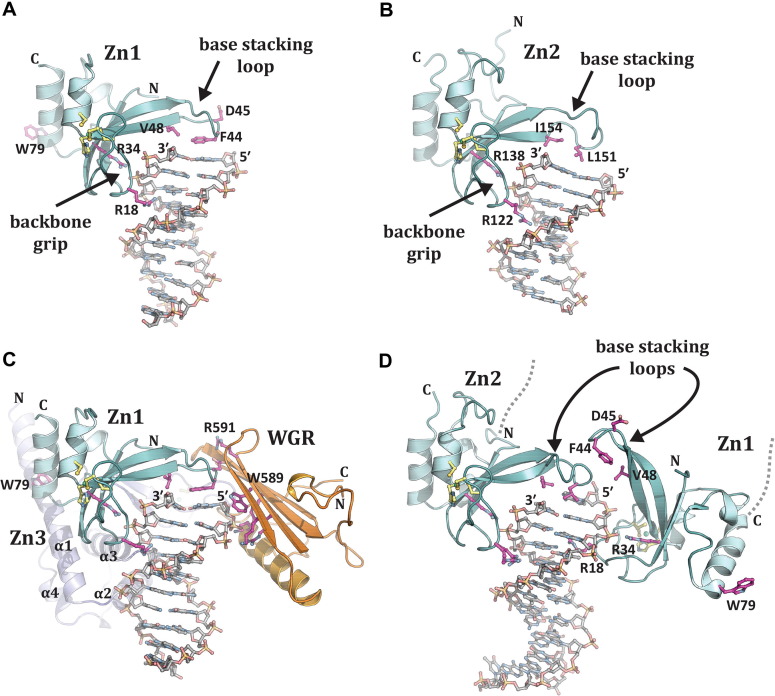
NMR and/or crystal structures have been reported for all of the individual domains of PARP1/ARTD1 (Ruf et al., 1996, Langelier et al., 2008, Tao et al., 2008, Eustermann et al., 2011, Loeffler et al., 2011; PDB: 2D8M), Fig. 4B; however, until recently there was no comprehension of how PARP1 detects DNA breaks and how the multiple domains of PARP1 coordinate DNA binding and catalytic activity. The first crystal structures of the individual Zn1 and Zn2 domains bound to DNA double-strand breaks have shed light on how these domains interact with DNA damage (Langelier et al., 2011). The Zn1 and Zn2 domains, which are structurally homologous, bind to DNA ends in a very similar fashion, Fig. 5 A and B. Zn1 and Zn2 engage the DNA break in a sequence-independent manner using two conserved structural features named (i) the backbone grip and (ii) the base stacking loop (Langelier et al., 2011). The backbone grip contacts the sugar phosphate backbone near the 3′ end of the break, engaging roughly three nucleotides. The backbone grip does not interact directly with the break per se, thus leaving the 3′ terminus exposed. Instead, the backbone grip interacts with a continuous part of the DNA duplex using Zn1 residues 15–22, and Zn2 residues 119–126. A key contact is also observed between a conserved arginine (R34 in Zn1; R138 in Zn2) and the phosphate group of the penultimate nucleotide at the 3′ end. In addition to contacting the sugar phosphate backbone, the phosphate backbone grip of Zn1 and Zn2 also inserts a conserved arginine residue into the minor groove of the DNA (R18 in Zn1; R122 in Zn2).
Fig. 5.
Structures of PARP1 domains bound to DNA damage. (A) Crystal structure of human Zn1 domain in complex with a DNA double strand break (PDB: 3OD8). (B) Crystal structure of human Zn2 domain in complex with a DNA double strand break (PDB: 3ODC). Together, the structures in panels A and B illustrate the features of damage DNA that are recognized by PARP1 zinc finger domains: a continuous phosphate backbone engaged primarily by conserved Arg (R) residues on the backbone grip, and exposed nucleotide bases engaged by hydrophobic residues on the base stacking loop. Key Zn1 residues that are involved in communicating with other PARP1 domains are shown (D45 and W79). (C) The Zn1, Zn3, and WGR domains collectively assemble on a DNA double strand break, with each domain forming specific protein-DNA, and protein–protein contacts with adjacent domains. The Zn1 domain has the same orientation as shown in panel A, and the same key residues are shown as sticks. WGR residues W589 and R591 are essential to PARP1 DNA-dependent activity and form protein-DNA and protein–protein contacts, respectively. Zn1 residue W79 is critical for PARP1 DNA-dependent activity and is located at the interface with Zn3. (D) Crystal structure of the Zn1–Zn2 domains of PARP1 bound to a DNA double strand break with a single 5′ nucleotide overhang. In this complex, the Zn2 domain forms contacts with the DNA similar to that seen for the isolated domain in panel B (the same conserved residues are shown). The Zn1 domain binds to DNA with a opposite polarity to that seen in panels A and C, with Arg18 inserting into the major groove rather than the minor groove, and the base stacking loop forming protein–protein contacts rather than protein-DNA contacts. Reprinted from (Langelier and Pascal 2013) with permission from Elsevier.
The base stacking loop extends the DNA contacts made by the backbone grip toward the 5′ terminus of the break, thus covering the end of the DNA duplex, Fig. 5A and B. Two hydrophobic residues (F44, V48 in Zn1; L151, I154 in Zn2) stack against the nucleotide bases exposed at the end of the duplex. Therefore, the base stacking loop and the backbone grip mediate interactions with the DNA that are sequence-independent and related to the DNA structure i.e. exposed base pairs and a continuous DNA backbone. These structural features are found in a variety of DNA breaks and abnormal DNA structures such as hairpins and cruciforms that are substrates bound by PARP1 (Lonskaya et al., 2005). Unlike the backbone grip, the base stacking loop exhibits some important differences between Zn1 and Zn2. Indeed, the Zn2 base stacking loop has a two-residue insert and lacks some key residues present in Zn1, which are crucial for interdomain contacts that lead to DNA-dependent activation, as discussed later (Langelier et al., 2011, Langelier et al., 2012). This explains why Zn2 cannot perform Zn1 function in PARP1 DNA-dependent mechanism of activation. Zn2, which exhibits a higher affinity than Zn1 for DNA ends (Eustermann et al., 2011, Langelier et al., 2011) is likely to function at the level of break recognition, but not at the level of activation. Mutational analysis has confirmed the importance of residues of the backbone grip and the base stacking loop on Zn1 and Zn2 abilities to bind to DNA (Langelier et al., 2011).
NMR structures of the individual unbound Zn1 and Zn2 domains have shown a conformation similar to the ones observed in the Zn1/DNA and Zn2/DNA crystal structures (Eustermann et al., 2011). The base stacking loop exhibits multiple conformations in the NMR structures (Eustermann et al., 2011; PDB: 2D8M), suggesting high mobility for this region. Alignment of the Zn1 NMR structure with the Zn1/DNA structure reveals a significant shift in the position of the base stacking loop upon DNA binding (Langelier et al., 2011). This repositioning and stabilization of the base stacking loop on the DNA end is necessary to allow its interaction with other PARP1 domains during DNA-dependent activation, as discussed below. Thus the base stacking loop likely serves to trigger PARP1 activation in response to DNA damage.
The crystal structure of multiple domains of human PARP1 in the presence of DNA revealed that the Zn3 and WGR domains cooperate with the Zn1 domain to collectively assemble on DNA damage (Langelier et al., 2012), Fig. 5C. The structure has thus provided roles for the Zn3 and WGR domains in DNA damage detection, and insights into how the multiple regulatory domains of PARP1 function together. In this structure, the Zn1 domain binds to the DNA break in a manner that is identical to that observed in the Zn1/DNA crystal structure (Langelier et al., 2011), Fig. 5A and C. The Zn3 domain extends the contacts made by Zn1 with the DNA by sitting next to Zn1 on the 3′ terminus of the break and contacting the continuous phosphate backbone and spanning the minor groove, Fig. 5C. The WGR domain binds to the 5′ terminus of the DNA, adjacent to the Zn1 base stacking loop. Collectively, the Zn1, Zn3, and WGR domains form a continuous surface that traverses the DNA double-strand break, Fig. 5C.
A recent structure of the tandem Zn1–Zn2 domains was determined in complex with DNA damage represented by a double-strand break with a single 5′ nucleotide overhang (Ali et al., 2012). In this structure, Zn2 engages DNA as seen in the individual Zn2–DNA complex, Fig. 5 (compare Zn2 in panels B and D), with the base stacking loop engaging the nucleotide bases at the end of the DNA duplex and conserved arginine residues bracing the phosphate backbone. Unexpectedly, Zn1 binds to DNA with an opposite polarity to that seen in other PARP1 zinc finger structures, Fig. 5 (compare Zn1 in panel A and D). The backbone grip uses the same conserved R34 residue to bind to the phosphate backbone, but the entire domain is flipped such that R18 is directed into the DNA major groove rather than the minor groove, Fig. 5D. Furthermore, the base stacking loop of Zn1 does not engage DNA, but rather forms contacts with the base stacking loop of Zn2, Fig. 5D. It was not possible to model the linker connecting the two zinc fingers; however, the termini of the two zinc fingers engaged on one DNA end are not close enough in the structure to be physically linked. A model was therefore proposed in which the two zinc fingers originate from different polypeptides, thus forming a dimer, and suggesting that the observed interaction between Zn1 and Zn2 is responsible for assembling two PARP1 molecules onto DNA. Further experimentation is necessary to establish whether the observed conformation of Zn1 and Zn2 is consistent with the activation of PARP1.
The stoichiometry of PARP1 binding to DNA breaks remains controversial. Several structural studies indicate that PARP1 binds to DNA as a monomer, including an NMR study of Zn1–Zn2 interaction with DNA (Eustermann et al., 2011), crystal structures of individual Zn1 and Zn2 domains bound to DNA (Langelier et al., 2011), a SAXS analysis of a Zn1-Zn2-Zn3-BRCT complex with DNA (Lilyestrom et al., 2010), and a sedimentation analysis of full-length PARP1 in the presence of DNA (Langelier et al., 2012). However, a biochemical study of PARP1 binding to various DNA structures indicated a dimer interaction with certain DNA (Pion et al., 2003), and a dimer structure has been inferred from the crystal structure of Zn1–Zn2 bound to a DNA double strand break (Ali et al., 2012). A confounding issue is that through biochemical analysis the individual domains of PARP1 are able to act quite efficiently when provided in trans. Although this has been interpreted as evidence of dimerization, it could also be the inherent nature of the modular domain architecture of PARP1, which might allow some plasticity in how the multiple domains are contributed to the assembly of PARP1.
3.1.2. PARP1: coupling DNA damage detection to poly(ADP-ribosyl)ation activity
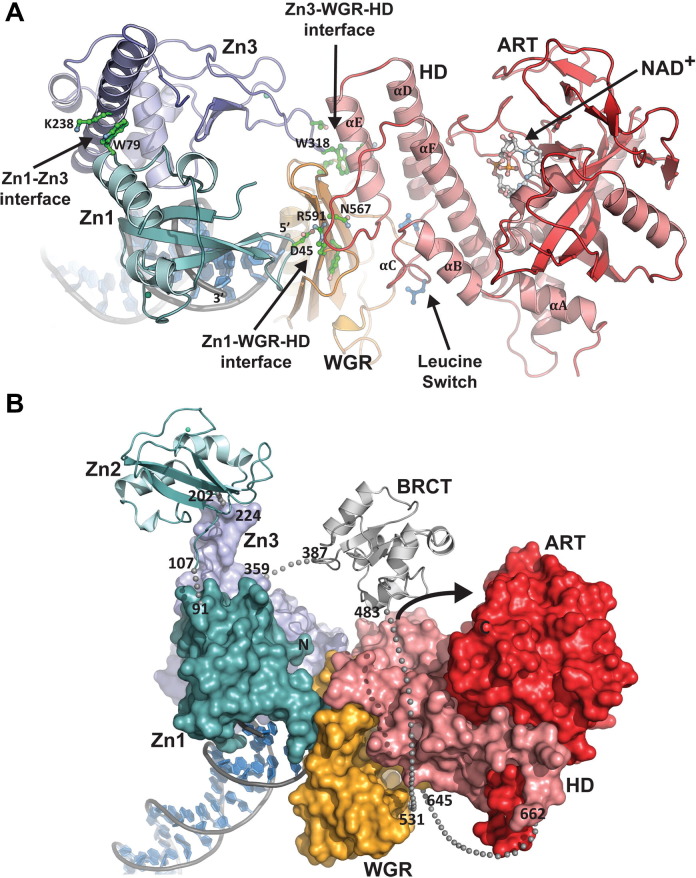
The crystal structure of the essential domains of PARP1/ARTD1 (Zn1, Zn3, WGR-CAT) bound to DNA has illustrated how the domains of PARP1 assemble on DNA and communicate with each other, and how the multidomain assembly results in structural alterations that regulate PARP1 catalytic activity (Langelier et al., 2012), Fig. 6 A. The PARP1/DNA crystal structure was obtained using individual fragments of the Zn1, Zn3, and WGR-CAT domains and a blunt-ended DNA as a model for a double-strand break. The Zn2 and BRCT domains, which are not required for PARP1 activation by DNA double-stranded breaks, were not included in the structure. The assembly of PARP1 regulatory domains on DNA forms a network of interdomain contacts that ultimately lead to activation of the CAT. The Zn1 domain contacts WGR using the face of the base stacking loop not involved in binding to DNA, Fig. 6A. WGR in turn interacts with the HD providing a bridge between the DNA damage interface and the CAT. Using a different surface, Zn1 forms a domain interface with Zn3. The main contacts of the Zn1-Zn3 interface are formed by residues on the first α-helix of Zn3 and a turn preceding the C-terminal α-helix of Zn1. The Zn3 domain also forms an interface with the WGR and the HD using an extended loop in the zinc ribbon motif. In particular, residue W318 located at the tip of the extended loop is wedged into a pocket formed by the WGR and HD, interacting with WGR residue K633 and HD residue R735, Fig. 6A. Importantly, mutational analysis performed in full-length PARP1 has confirmed the importance of the interdomain contacts observed in the PARP1/DNA structure on DNA-dependent activity (Langelier et al., 2012).
Fig. 6.
Crystal structure of essential PARP1 domains in complex with DNA damage. (A) Human PARP1 domains Zn1, Zn3, and WGR-CAT were crystallized in complex with a DNA double-strand break (PDB: 4DQY) (Langelier et al., 2012). The PARP1/DNA complex illustrates how DNA damage detection is coupled to structural transitions in the catalytic domain that elevate poly(ADP-ribosyl)ation activity (Langelier et al., 2012). Three interdomain contact regions form upon PARP1 interaction with DNA. The Zn1-WGR-HD interface is the most direct coupling of DNA damage recognition to the catalytic domain, with Zn1 contacting both DNA damage and WGR, and the WGR contacting the HD. The Zn3-WGR-HD interface forms where the zinc ribbon motif of Zn3 contacts conserved WGR and HD residues, with W318 as a central component of the interaction. The Zn1-Zn3 interface forms where these two domains rest together on DNA. Mutations that target these interfaces have a severe impact on DNA damage-dependent PARP1 activity (Langelier et al., 2010, Langelier et al., 2011, Langelier et al., 2012), indicating that the structure has captured interdomain contacts that are relevant to PARP1 regulation. Collectively, the interdomain communication with the HD displaces conserved Leu residues from the hydrophobic interior of the HD, leading to destabilization of the CAT that correlates with an elevation in PARP1 catalytic activity. (B) A model for the approximate positioning of the Zn2 and BRCT domains within the PARP1/DNA complex. Zn1, Zn3, and WGR-CAT are shown as surfaces, labeled, and colored as in Fig. 4A. The Zn2 and BRCT domains are drawn in schematic representation. Their positioning is based on the relative location of the termini of adjacent domains in the structure. The numbering and location of linker residues are shown. The arrow indicates the location of the PARP1 automodification region near the catalytic active site. Reprinted from (Langelier and Pascal 2013) with permission from Elsevier.
The PARP1/DNA structure has revealed that the regulatory domains of PARP1 only contact the catalytic domain through the HD. The ART domain, which is responsible for catalysis, is not involved in any contacts with the other domains or with DNA, suggesting that the HD plays an important role in connecting DNA break recognition to catalytic activation. Comparison of the CAT structure in the PARP1/DNA complex to the isolated CAT structure has revealed that the HD is distorted, whereas the ART domain maintains the same overall structure. The most notable change is that conserved residues L698 and L701 are displaced from the HD hydrophobic interior. Thus, the DNA damage-induced organization of the PARP1 regulatory domains forms an extended interface with HD that imposes a disruption of the HD hydrophobic core, suggesting that this alteration to HD structure underlies the activation mechanism of PARP1 in response to DNA damage. Mutagenesis of the residues facing the interior of the HD hydrophobic core results in an increase in PARP1 activity in the absence of DNA (Langelier et al., 2012), thus supporting that distortion of HD structure is the underlying mechanism that elevates PARP1 activity. The HD mutants that increase PARP1 activity decrease the thermal stability of the CAT. The decrease in thermal stability is also observed in full-length PARP1 upon binding to a DNA break, and this destabilizing effect requires the CAT domain and residues located at PARP1 interdomain interfaces (Langelier et al., 2012). Together with the crystal structure, these biochemical results suggest that the assembly of PARP1 regulatory domains on DNA ends establishes a network of interdomain contacts that lead to a disruption of the HD hydrophobic core, and subsequently to a decrease in thermal stability.
A decrease in thermal stability suggests an increase in the flexibility and dynamics of PARP1 upon binding to DNA damage. The PARP1 DNA damage-dependent poly(ADP-ribosyl)ation reaction is a multi-step process involving attachment of ADP-ribose to the target protein, repeated cycles of NAD+ binding and hydrolysis, and displacement of the ADP-ribose from the donor site to the acceptor site. An increase in protein flexibility and dynamics of the ART could increase the rate of the catalytic reaction by affecting the turnover of one or more of these steps. PARP1 affinity for substrate NAD+ does not change upon binding to DNA breaks, but rather PARP1 exhibits an increase in the rate of the poly(ADP-ribose) synthesis (Langelier et al., 2010, Clark et al., 2012); therefore it is not expected that the changes in HD structure and CAT dynamics affect the binding of substrate NAD+. Crystal structure analysis is not ideally suited to assess the fluctuations in structural dynamics that are proposed to enhance poly(ADP-ribose) synthesis; therefore further experimentation with other techniques, such as NMR, will likely be required to fully understand the changes in ART that increase the rate of PAR synthesis. Moreover, there are likely to be other contributions to the dramatic increase in PARP-1 activity in response to DNA damage. Upon binding to DNA breaks, the regulatory domains of PARP1 collapse onto each other to form a more compact structure (Langelier et al., 2012). Positioning of the Zn2 and BRCT domains on the PARP1/DNA crystal structure suggests a model of full-length PARP1, Fig. 6B. In this model the automodification domain (AD), containing the residues targeted for modification, is located in close proximity to the catalytic site. This increase in catalytic domain access to substrate could contribute to elevation in the rate of poly(ADP-ribose) synthesis, due to the proximity of the catalytic and automodification domains. Furthermore, the positioning of the AD can explain why PARP1 has a strong preference to modify itself over other target proteins.
The complex of PARP1/ARTD1 domains bound to DNA illustrates a damage-dependent intramolecular mechanism for rapidly elevating the low basal level of PARP1 activity through perturbations to the HD. PARP1 activity is also regulated through other signals besides DNA damage, such as posttranslational modifications and interaction with nucleosomes. It will be interesting to investigate whether these other signals also act through perturbations to the HD structure. PARP2/ARTD2 and PARP3/ARTD3 catalytic domains also contain an HD, but their overall domain organization is quite different from PARP1. Thus, it will also be interesting to see how the HD of PARP2 and PARP3 will be regulated, and whether they ultimately use destabilization of the HD as a mechanism of activation.
3.2. Overview of the biology of tankyrases (ARTD5 and ARTD6)
Tankyrase (TRF1-interacting ankyrin-related ADP-ribose polymerase) was first identified in association with human telomeres, where it acts as a regulator of telomeric DNA through its interaction with and modification of telomere repeat-binding factor 1 (TRF1) (Smith et al., 1998, Smith and de Lange, 2000). In addition to this nuclear function, it was soon appreciated that the majority of tankyrase is located in the cytoplasm of cells, in particular as a peripheral membrane protein localized to the Golgi complex and found in association with transport vesicles (Chi and Lodish, 2000). Over the past few years, there has been a growing appreciation of the large number of proteins that are targeted for modification by tankyrase, and greater insights into how tankyrase activity contributes to specific cellular processes. A number of studies have indicated that tankyrase-mediated poly(ADP-ribosyl)ation activity acts in concert with ubiquitin ligase activity to regulate protein turnover and thus affect the stability and lifetime of key components in signaling pathways (Huang et al., 2009, Levaot et al., 2011, Callow et al., 2012, Wang et al., 2012). A notable example of a protein targeted for tankyrase modification and subsequent ubiquitination is the Axin1 component of the β-catenin destruction complex, which is integral to Wnt signaling. Tankyrase involvement in cellular glucose homeostasis, mitotic progression, and signaling pathways disregulated in cancer have made the protein of broad scientific interest to basic scientists, with strong implications for use as a therapeutic target.
3.2.1. Tankyrase domain organization
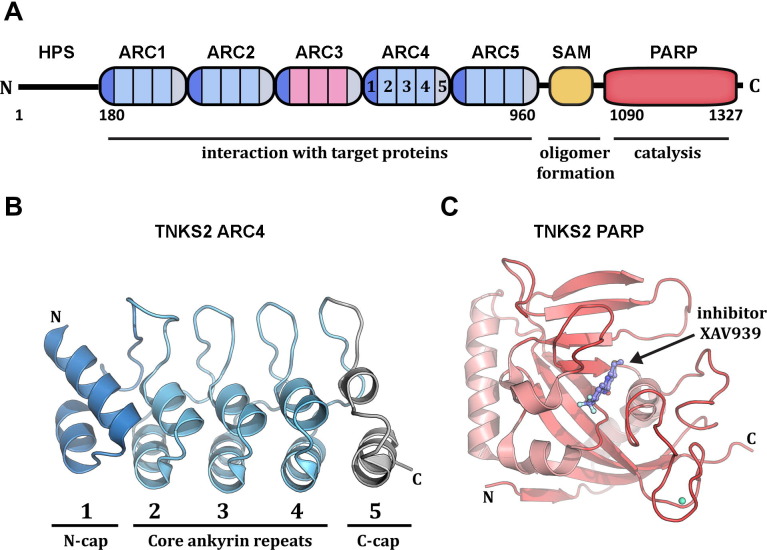
There are two tankyrase genes in humans, TNKS1 (ARTD5) and TNKS2 (ARTD6). Overall, the tankyrases share a similar domain organization, Fig. 7 A. The ART domain that creates PAR is located at the extreme C-terminus. Unlike PARPs-1, -2, and -3, tankyrases do not have an α-helical regulatory subdomain, Fig. 1A. The majority of the tankyrase polypeptide is composed of a series of structural modules known as ankyrin repeats. In general, an individual ankyrin repeat has a loop-helix-loop-helix-loop structure, Fig. 7B, and the ankyrin repeat structure contacts adjacent repeats to form continuous structures like that found in the human protein ankyrin. A sterile-alpha motif (SAM) domain separates the N-terminal ankyrin repeat region and the C-terminal catalytic region of the tankyrases. SAM domains are found in a variety of proteins with different functions; therefore the presence of a SAM domain does not indicate a function unambiguously. However, many SAM domains are known to multimerize, and the SAM domain of tankyrase 1 has been suggested to mediate multimerization of tankyrase molecules (De Rycker et al., 2003, De Rycker and Price, 2004). Although there are structures for a number of SAM domains from different proteins, there are currently no structures for the SAM domains of tankyrases. The most significant difference between tankyrase 1 (ARTD5) and tankyrase 2 (ARTD6) is an extended N-terminal region of tankyrase 1, approximately 150 amino acids, which has low sequence complexity and thus is not likely to have a defined globular structure. The N-terminus of tankyrase 1 has been termed the HPS region due to repeats of histidine, proline, and serine. The HPS is likely to play a regulatory function.
Fig. 7.
Tankyrase domain structure. (A) Tankyrase 1 and 2 have a similar domain construction. The C-terminus contains a SAM domain implicated in tankyrase oligomerization and the PARP catalytic domain that synthesizes poly(ADP-ribose). The N-terminus is primarily composed of an ankyrin repeat region that is segmented into five ankyrin repeat clusters (ARC1 through ARC5), each containing five ankyrin modules (numbered 1–5 in ARC4). The ARCs interact with proteins targeted for modification by tankyrase, with the exception of ARC3 that lacks key conserved residues involved in target binding. The extreme N-terminus of tankyrase 1 contains runs of His, Pro, and Ser and is thus termed the HPS domain. HPS likely serves a regulatory function. Tankyrase 2 does not have an HPS domain. The residue numbering of domain boundaries is shown for tankyrase 1. (B) The crystal structure of ARC4 from tankyrase 2 illustrates the organization and specialization of the five ankyrin repeat modules that form each ARC (Guettler et al., 2011). Three typical ankyrin repeats form the core of the ARC (ankyrin modules 2–4). Modified ankyrin repeats initiate the N-terminus of the ARC (module 1, N-cap) and terminate the C-terminus of the ARC (module 5), thus capping each end of the ARC and preventing continuous stacking of ankyrin repeats. (C) X-ray structure of the catalytic domain of tankyrase 2 bound to inhibitor XAV939 (Karlberg et al., 2010b).
X-ray structures of the isolated ART domain of tankyrases 1 and 2 have been determined (Lehtiö et al., 2008, Karlberg et al., 2010b, Gunaydin et al., 2012, Kirby et al., 2012, Narwal et al., 2012, Wahlberg et al., 2012; see tankyrase 2 catalytic domain, Fig. 7C). The catalytic regions have the same fold as other ADP-ribosyl transferases, with two central beta sheets surrounded by alpha helices and connecting loops. The first structure of a tankyrase catalytic domain identified an unexpected zinc binding region that appears to play a structural role, Fig. 7C (Lehtiö et al., 2008). Several structures of tankyrase catalytic domains in complex with inhibitors have highlighted similarities between the NAD+ binding sites of PARP1 and tankyrase, where most inhibitors occupy the region of the active site that interacts with the nicotinamide moiety of NAD+. However, more recent structures have accented the active site differences that might be exploited for the development of more specific inhibitors. In particular, the inhibitor XAV939 in complex with tankyrase 2 indicates that drugs can exploit structural features specific to tankyrase that are not present in PARP1 due to the regulatory helical domain (Karlberg et al., 2010b).
3.2.2. Tankyrase target selection
Bioinformatic and biochemical analysis of the tankyrase ankyrin repeat region suggested that the many ankyrin repeat modules were organized into five segments, in which each segment was composed of ankyrin repeat clusters (ARCs), Fig. 7A (Sbodio et al., 2002, Seimiya and Smith, 2002). Several of these ARCs could be produced individually, or in combinations, that were capable of binding to tankyrase partner proteins, thus indicating a potential redundancy of function for the five ARCs (Seimiya and Smith, 2002, Seimiya et al., 2004). Furthermore, as the list of tankyrase binding partners increased, a consensus binding site emerged as R-X-X-P-D-G (where X denotes any amino acid) (Sbodio et al., 2002). Despite this information, the precise boundaries and number of ankyrin repeats per ARC were not entirely clear, and the basis for ARC:peptide interactions were unknown, due to a lack of structural information. Recent crystal structures of ARCs from tankyrase 1 and 2 have clarified some of the uncertainties in the organization of the ankyrin repeat region, and have provided the first insights into the mechanism of tankyrase selection of target proteins (Guettler et al., 2011, Morrone et al., 2012).
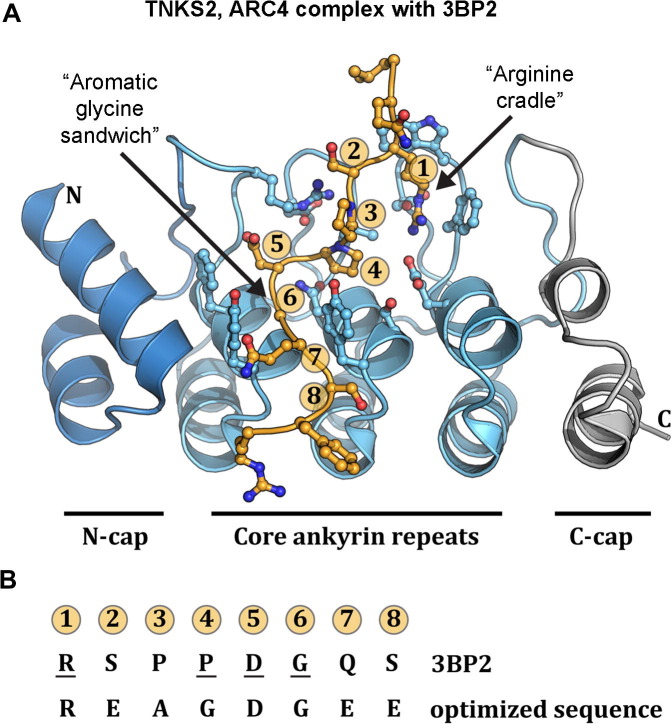
The overall construction of an individual ARC was revealed by the structure of ARC4 from human tankyrase 2 (Guettler et al., 2011), Fig. 7B. The individual ARC is composed of three core ankyrin repeats, which are flanked on the N-terminus and C-terminus by modified ankyrin repeats that cap the core repeats and thus prevent a continuous stacking of all 25 ankyrin modules of tankyrase, Fig. 7B. The N-cap and C-cap ankyrin modules are conserved within each ARC, indicating that each ARC will have a similar arrangement of ankyrin modules. The molecular basis for a single ARC:peptide interaction was defined for the ARC4 of TNKS2 in complex with six different target peptides (Guettler et al., 2011). The three core ankyrin repeats form the peptide binding site, Fig. 8 . Notable features of the peptide binding surface of the ARC are the “Arginine cradle,” in which the conserved Arg at position 1 of the peptide is engaged through hydrophobic and ionic interactions, and the “Aromatic glycine sandwich,” in which the conserved glycine at position 6 of the peptide passes between two tyrosine residues, Fig. 8A. Although each of the five ARCs are expected to have the same organization of ankyrin repeat modules observed in the ARC4 structure (i.e. N-cap, core, C-cap), the peptide binding interface of ARC3 lacks key residues that form the peptide binding interface, consistent with the observation that ARC3 does not interact with partner proteins.
Fig. 8.
Structural basis for ARC recognition of target peptides. (A) The X-ray structure of ARC4 of tankyrase 2 in complex with a peptide derived from the target 3BP2 (Guettler et al., 2011), an adaptor protein in signaling pathways that depend on tankyrase activity for protein turnover. The core ankyrin repeats form the peptide interaction surface. An “arginine cradle” forms around the conserved Arg residue at position 1 of the consensus peptide sequence. Two Tyr residues form an “aromatic glycine sandwich” that surrounds the conserved Gly residue at position 6 of the consensus peptide sequence. (B) The amino acid sequence for 3BP2 is shown for each of the eight positions. Underlined residues are mutated in individuals with cherubism (Levaot et al., 2011), resulting in a loss of interaction with and modification by tankyrase. An optimized sequence was determined through an extensive analysis of amino acid substitutions at each of the eight positions.
Collectively, the ARC4:peptide structures further define the consensus sequence and extend the binding region to eight positions. A systematic evaluation of amino acid substitutions at each of the eight positions defined a peptide with high binding affinity, R-E-A-G-D-G-E-E. However, analysis of the crystal structures of ARC4:peptide complexes demonstrates that certain targets that contain unfavorable amino acids in strong consensus positions are compensated through additional contacts at position 8. For example, the binding partner MCL1 has an isoleucine at position 5, which is detrimental to the peptide contacts at this position, but this weakened contact is compensated through a glutamic acid residue at position 8, which increases binding affinity. This target-specific modulation of the ARC:peptide interaction suggests that the ARC binding sites and their associated binding affinities with specific targets can be fine-tuned.
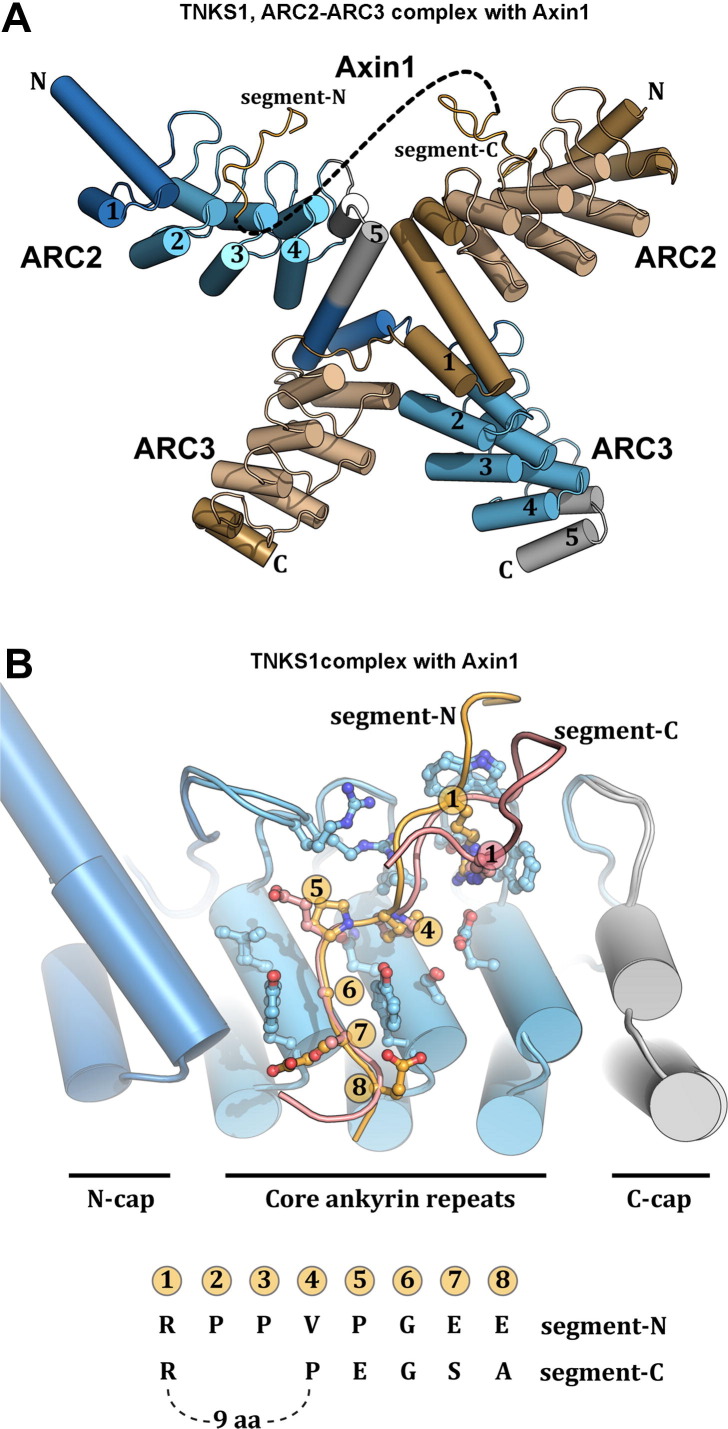
The X-ray structure ARC2–ARC3 of tankyrase 1 was determined in complex with the tankyrase-binding fragment of Axin1 and has provided new insights into the nature of ARC:peptide interactions, and a potential mechanism for organizing the multiple ARCs of tankyrase (Morrone et al., 2012). The ARC2–ARC3:Axin1 structure and associated binding studies revealed that Axin1 has two segments that interact with tankyrase, Fig. 9 A, B, and C. Segment-N conforms to the known consensus peptide and binds in the same manner seen in the ARC4:peptide complexes. Segment-C exhibits an interesting variation on the ARC:peptide interaction, with the primary interactions occurring at positions 1 and 4–7, Fig. 9B and C. Several residues are inserted between the arginine residue that binds to the “arginine cradle” and the Pro residue at position 4, Fig. 9B and C. The residues that contribute to the “arginine cradle” adapt to accommodate the different positioning of the conserved arginine, thus indicating some flexibility to the peptide-binding surface of the ARC, and suggesting that other target protein peptides might interface with tankyrase in new and unexpected ways.
Fig. 9.
Variations in ARC recognition of target peptides and organization of multiple ARCs. (A) The X-ray structure of the ARC2–ARC3 fragment of tankyrase 1 was determined in complex with the tankyrase binding region of Axin1, a key component of the Wnt signaling complex (Morrone et al., 2012). Two regions of Axin1, segment-N and segment-C, each interact with an ARC2 of tankyrase, thus forming a 1:2 complex of Axin1:tankyrase. An ARC2–ARC3 homodimer is formed where the ARC3N-cap of one monomer “crosses over” to stack against the core ankyrin modules of ARC3 in the second monomer. (B) The segment-N and segment-C interactions with ARC2 exhibit variations in how the polypeptides engage the target protein docking site. (C) Comparison of the amino acid sequences for segment-N and segment-C of Axin1. Segment-C has an insertion of 9 amino acids between position 1 and position 4, but otherwise has the same key features of the segment-N interaction with the ARC2, such as the arginine at position 1 and the glycine at position 6.
The structure of the ARC2–ARC3 fragment of tankyrase 1 provides insights into the potential arrangement and architecture of the five consecutive ARCs of tankyrase. Consistent with sequence analysis, the structure shows that there is no linker region between adjacent ARCs, with the terminating helix of one C-cap continuous with the initiating helix of the N-cap on the next ARC, Fig. 9A. This continuous structure indicates that the ARCs will form a segmented structure with consecutive ARCs firmly associated, rather than a “beads-on-a-string” arrangement with the individual ARCs connected by flexible linkers. An exception could be the linker connecting ARC3 to ARC4, which is slightly longer and might provide flexibility within the ankyrin repeat region of tankyrase (Morrone et al., 2012). Interestingly, the ARC2–ARC3 fragment forms a domain swapped homodimer in the crystal with the overall shape of the letter X, Fig. 9A. In the homodimer, the ARC3 N-cap of one monomer “crosses over” to stack against the core ankyrin modules of ARC3 in the second monomer. Solution studies indicate that ARC2–ARC3 is largely monomeric on its own, but in the presence of Axin1 there is greater population of dimeric ARC2–ARC3, suggesting that tankyrase interaction with target proteins containing two binding sites might alter the oligomeric state of tankyrase. However, the dimerizing effect has not been tested in the context of full-length TNKS1, where there will be four binding sites (ARCs 1, 2, 4, and 5), in contrast to the ARC2–ARC3 fragment with only one high affinity binding site (ARC2).
3.2.3. Future directions for understanding tankyrase from a structural perspective
There are several key unresolved questions that will need to be addressed to further build on our understanding of tankyrase structural biology. Although several SAM domain structures have been described, there are no specific insights into the SAM structure of tankyrase. SAM domains serve a variety of disparate functions in different proteins, thus the actual functional role for tankyrase cannot be simply inferred. Work with a maltose-binding protein (MBP)-SAM fusion indicates that tankyrase SAM domains form polymers (De Rycker and Price, 2004). However, the functional relevance of this polymerization is not clear. This gap in understanding is in part due to the lack of structural information that could define the interface used for oligomerization, and therefore suggest mutations that could test the potential function of oligomerization. Despite the significant progress in understanding the ankyrin repeat region, it will be important to determine the overall conformation of the complete ankyrin repeat region. Moreover, what is the overall structural organization and arrangement of tankyrase domains? Do the ankyrin repeats “steer” target proteins toward the catalytic domain? Given the size and complexity of tankyrase, these are challenging questions that are certain to provide interesting answers. Furthermore, the answers to these questions will provide a better understanding of tankyrase interaction with partner proteins that will potentially allow the development of a new class of tankyrase-specific inhibitors.
4. Conclusions
Structural analysis continues to advance our understanding of the writers, readers, and erasers that carry out the biology of ADP-ribose-mediated cell signaling events. As shown in this review, structural studies have helped to define the functions and catalytic activities of these proteins, and they have formed a molecular framework for understanding catalytic mechanism and regulation. Despite the significant progress highlighted here, there are many remaining structural questions that are key to fully understanding the biology of ADP-ribosylation; thus we can look forward to many exciting new structures and insights. The growing collection of a variety of PARP catalytic domain structures provides insights into the selectivity of current clinical PARP inhibitors, but they also suggest new avenues for the development of greater specificity and selective enzyme inhibition. Such inhibitors, with chemical probe qualities, will be of immense value as research tool. Continued progress in this field might also yield a second generation of PARP inhibitors for the treatment of cancer and inflammation, as well as for anti-infectives that are both safer and less prone to resistance mechanisms than current drugs.
Acknowledgments
Work in the Pascal laboratory is supported by the American Cancer Society and the NIH. Work in the Schüler laboratory is supported by the Swedish Foundation for Strategic Research, the Swedish Research Council, the Swedish Cancer Society, and the Structural Genomics Consortium.
Biographies
T. Karlberg, PhD, is a research associate in the Department of Medical Biochemistry and Biophysics at Karolinska Institutet in Stockholm, Sweden. He studied structural biology at the Structural Genomics Consortium as a postdoctoral fellow, and received his PhD from the University of Lund. His current research focuses on the structural biology of ADP-ribose transferases and structure based design of PARP inhibitors.
M. F. Langelier, Ph.D., is a research associate in the Department of Biochemistry and Molecular Biology at Thomas Jefferson University, Philadelphia, USA. She studied transcriptional regulation mechanisms as a postdoctoral fellow at Harvard Medical School and as a Ph.D. student at the University of Montreal. Her current research focuses on structural and biochemical analysis of ADP-ribose transferases.
J. M. Pascal, Ph.D., is an Associate Professor of Biochemistry and Molecular Biology and a member of the Kimmel Cancer Center at Thomas Jefferson University, Philadelphia, USA. He studied the structural biology of DNA replication and repair as a postdoctoral fellow at Harvard Medical School, and earned his Ph.D. at the University of Texas, Austin. His research currently focuses on proteins involved in maintaining genome stability.
H. Schüler, Ph.D., is an independent group leader in the Department of Medical Biochemistry and Biophysics at Karolinska Institutet in Stockholm, Sweden. He headed a team for the Structural Genomics Consortium at Karolinska Institutet that determined crystal structures of ATPases, RNA helicases, and ADP-ribose transferases. He studied protein chaperones as a postdoctoral fellow at New York.University Medical Center, and earned his Ph.D. degree at Stockholm University. Currently his research is focused on the structural biology of proteins involved in ADP-ribosylation as well as the generation of selective PARP inhibitors.
Contributor Information
John M. Pascal, Email: john.pascal@kimmelcancercenter.org.
Herwig Schüler, Email: herwig.schuler@ki.se.
References
- Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P., Owen-Hughes T., Boulton S.J. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ahuja N., Schwer B., Carobbio S., Waltregny D., North B.J., Castronovo V., Maechler P., Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J. Biol. Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Ali A.A., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Buckle A.M., Cordell S.C., Lowe J., Bycroft M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- Altmeyer M., Messner S., Hassa P.O., Fey M., Hottiger M.O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson C.D., Karlberg T., Ekblad T., Lindgren A.E., Thorsell A.G., Spjut S., Uciechowska U., Niemiec M.S., Wittung-Stafshede P., Weigelt J., Elofsson M., Schüler H., Linusson A. Discovery of ligands for ADP-ribosyltransferases via docking-based virtual screening. J. Med. Chem. 2012;55:7706–7718. doi: 10.1021/jm300746d. [DOI] [PubMed] [Google Scholar]
- Andrabi S.A., Kang H.C., Haince J.F., Lee Y.I., Zhang J., Chi Z., West A.B., Koehler R.C., Poirier G.G., Dawson T.M., Dawson V.L. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011;17:692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.E., Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- Berthold C.L., Wang H., Nordlund S., Hogbom M. Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG. Proc. Natl. Acad. Sci. USA. 2009;106:14247–14252. doi: 10.1073/pnas.0905906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta D., Jacobson M.K. Identification of a regulatory segment of poly(ADP-ribose) glycohydrolase. Biochemistry. 2010;49:7674–7682. doi: 10.1021/bi100973m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner J.C., Ateeq B., Li Y., Yocum A.K., Cao Q., Asangani I.A., Patel S., Wang X., Liang H., Yu J., Palanisamy N., Siddiqui J., Yan W., Cao X., Mehra R., Sabolch A., Basrur V., Lonigro R.J., Yang J., Tomlins S.A., Maher C.A., Elenitoba-Johnson K.S., Hussain M., Navone N.M., Pienta K.J., Varambally S., Feng F.Y., Chinnaiyan A.M. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow M.G., Tran H., Phu L., Lau T., Lee J., Sandoval W.N., Liu P.S., Bheddah S., Tao J., Lill J.R., Hongo J.A., Davis D., Kirkpatrick D.S., Polakis P., Costa M. Ubiquitin ligase RNF146 regulates tankyrase and axin to promote Wnt signaling. PLoS One. 2012;6(7):e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Vollmar M., Rossi M.N., Phillips C., Kraehenbuehl R., Slade D., Mehrotra P.V., von Delft F., Crosthwaite S.K., Gileadi O., Denu J.M., Ahel I. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N.W., Lodish H.F. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 2000;275:38437–38444. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- Clark N.J., Kramer M., Muthurajan U.M., Luger K. Alternative modes of binding of poly(adp-ribose) polymerase 1 to free DNA and nucleosomes. J. Biol. Chem. 2012;287:32430–32439. doi: 10.1074/jbc.M112.397067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D., Desnoyers S., D’Silva I., Poirier G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- D’Silva I., Pelletier J.D., Lagueux J., D’Amours D., Chaudhry M.A., Weinfeld M., Lees-Miller S.P., Poirier G.G. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim. Biophys. Acta. 1999;1430:119–126. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- Dani N., Stilla A., Marchegiani A., Tamburro A., Till S., Ladurner A.G., Corda D., Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycker M., Price C.M. Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol. Cell. Biol. 2004;24:9802–9812. doi: 10.1128/MCB.24.22.9802-9812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rycker M., Venkatesan R.N., Wei C., Price C.M. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M., Schreiber V., Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Dunstan M.S., Barkauskaite E., Lafite P., Knezevic C.E., Brassington A., Ahel M., Hergenrother P.J., Leys D., Ahel I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012;3:878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- Egloff M.P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C., Ziebuhr J., Ahola T., Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S., Brockmann C., Mehrotra P.V., Yang J.C., Loakes D., West S.C., Ahel I., Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat. Struct. Mol. Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S., Videler H., Yang J.C., Cole P.T., Gruszka D., Veprintsev D., Neuhaus D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011;407:149–170. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans H.R., Sutton J.M., Holloway D.E., Ayriss J., Shone C.C., Acharya K.R. The crystal structure of C3stau2 from Staphylococcus aureus and its complex with NAD. J. Biol. Chem. 2003;278:45924–45930. doi: 10.1074/jbc.M307719200. [DOI] [PubMed] [Google Scholar]
- Fieldhouse R.J., Merrill A.R. Needle in the haystack: structure-based toxin discovery. Trends Biochem. Sci. 2008;33:546–556. doi: 10.1016/j.tibs.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Fieldhouse R.J., Jorgensen R., Lugo M.R., Merrill A.R. The 1.8 A cholix toxin crystal structure in complex with NAD+ and evidence for a new kinetic model. J. Biol. Chem. 2012;287:21176–21188. doi: 10.1074/jbc.M111.337311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., Ashworth A., Carmichael J., Kaye S.B., Schellens J.H., de Bono J.S. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Forst A.H., Karlberg T., Herzog N., Thorsell A.G., Gross A., Feijs K.L., Verheugd P., Kursula P., Nijmeijer B., Kremmer E., Kleine H., Ladurner A., Schüler H., Lüscher B. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure. 2013;21:264–275. doi: 10.1016/j.str.2012.12.019. [DOI] [PubMed] [Google Scholar]
- French J.B., Cen Y., Sauve A.A. Plasmodium falciparum Sir2 is an NAD+ -dependent deacetylase and an acetyllysine-dependent and acetyllysine-independent NAD+ glycohydrolase. Biochemistry. 2008;47:10227–10239. doi: 10.1021/bi800767t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin E.V., Lai M.M. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W., Conaway R.C. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettler S., LaRose J., Petsalaki E., Gish G., Scotter A., Pawson T., Rottapel R., Sicheri F. Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Gunaydin H., Gu Y., Huang X. Novel binding mode of a potent and selective tankyrase inhibitor. PLoS One. 2012;7(3):e33740. doi: 10.1371/journal.pone.0033740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M.C., Mostoslavsky R., Haigis K.M., Fahie K., Christodoulou D.C., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Karow M., Blander G., Wolberger C., Prolla T.A., Weindruch R., Alt F.W., Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Han S., Craig J.A., Putnam C.D., Carozzi N.B., Tainer J.A. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat. Struct. Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- He F., Tsuda K., Takahashi M., Kuwasako K., Terada T., Shirouzu M., Watanabe S., Kigawa T., Kobayashi N., Guntert P., Yokoyama S., Muto Y. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586:3858–3864. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Hottiger M.O., Hassa P.O., Lüscher B., Schüler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C.J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M.W., Lengauer C., Finan P.M., Tallarico J.A., Bouwmeester T., Porter J.A., Bauer A., Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Ikejima M., Noguchi S., Yamashita R., Ogura T., Sugimura T., Gill D.M., Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Isogai S., Kanno S., Ariyoshi M., Tochio H., Ito Y., Yasui A., Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- Jorgensen R., Merrill A.R., Yates S.P., Marquez V.E., Schwan A.L., Boesen T., Andersen G.R. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- Kalisch T., Ame J.C., Dantzer F., Schreiber V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 2012;37:381–390. doi: 10.1016/j.tibs.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.C., Lee Y.I., Shin J.H., Andrabi S.A., Chi Z., Gagne J.P., Lee Y., Ko H.S., Lee B.D., Poirier G.G., Dawson V.L., Dawson T.M. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA. 2011;108:14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg T., Hammarstrom M., Schütz P., Svensson L., Schüler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- Karlberg T., Markova N., Johansson I., Hammarström M., Schütz P., Weigelt J., Schüler H. Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J. Med. Chem. 2010;53:5352–5355. doi: 10.1021/jm100249w. [DOI] [PubMed] [Google Scholar]
- Karlberg T., Thorsell A.G., Kallas A., Schüler H. Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain. J. Biol. Chem. 2012;287:24077–24081. doi: 10.1074/jbc.M112.379289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G.I., Kustatscher G., Buhecha H.R., Allen M.D., Pugieux C., Sait F., Bycroft M., Ladurner A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernstock S., Koch-Nolte F., Mueller-Dieckmann J., Weiss M.S., Mueller-Dieckmann C. Cloning, expression, purification and crystallization as well as X-ray fluorescence and preliminary X-ray diffraction analyses of human ADP-ribosylhydrolase 1. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2009;65:529–532. doi: 10.1107/S1744309109014067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.K., Kiefer J.R., Ho C.M., Stegeman R.A., Classen S., Tainer J.A., Ellenberger T. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat. Struct. Mol. Biol. 2012;19:653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Nakanishi I., Warizaya M., Iwashita A., Kido Y., Hattori K., Fujii T. Inhibitor-induced structural change of the active site of human poly(ADP-ribose) polymerase. FEBS Lett. 2004;556:43–46. doi: 10.1016/s0014-5793(03)01362-0. [DOI] [PubMed] [Google Scholar]
- Kirby C.A., Cheung A., Fazal A., Shultz M.D., Stams T. Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2012;68:115–118. doi: 10.1107/S1744309111051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H., Lüscher B. Learning how to read ADP-ribosylation. Cell. 2009;139(1):17–19. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kleine H., Poreba E., Lesniewicz K., Hassa P.O., Hottiger M.O., Litchfield D.W., Shilton B.H., Lüscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F., Kernstock S., Mueller-Dieckmann C., Weiss M.S., Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front. Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- Koh D.W., Lawler A.M., Poitras M.F., Sasaki M., Wattler S., Nehls M.C., Stoger T., Poirier G.G., Dawson V.L., Dawson T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh D.W., Patel C.N., Ramsinghani S., Slama J.T., Oliveira M.A., Jacobson M.K. Identification of an inhibitor binding site of poly(ADP-ribose) glycohydrolase. Biochemistry. 2003;42:4855–4863. doi: 10.1021/bi0272048. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran D., Eswaramoorthy S., Studier F.W., Swaminathan S. Structure and mechanism of ADP-ribose-1″-monophosphatase (Appr-1″-pase), a ubiquitous cellular processing enzyme. Protein Sci. 2005;14:719–726. doi: 10.1110/ps.041132005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustatscher G., Hothorn M., Pugieux C., Scheffzek K., Ladurner A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12(7):624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Langelier M.F., Servent K.M., Rogers E.E., Pascal J.M. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- Langelier M.F., Ruhl D.D., Planck J.L., Kraus W.L., Pascal J.M. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010;285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M.F., Planck J.L., Roy S., Pascal J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M.F., Planck J.L., Roy S., Pascal J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011;286:10690–10701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M.F., Pascal J.M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtiö L., Collins R., van den Berg S., Johansson A., Dahlgren L.G., Hammarström M., Helleday T., Holmberg-Schiavone L., Karlberg T., Weigelt J. Zinc binding catalytic domain of human tankyrase 1. J. Mol. Biol. 2008;379:136–145. doi: 10.1016/j.jmb.2008.03.058. [DOI] [PubMed] [Google Scholar]
- Lehtiö L., Jemth A.S., Collins R., Loseva O., Johansson A., Markova N., Hammarstrom M., Flores A., Holmberg-Schiavone L., Weigelt J., Helleday T., Schüler H., Karlberg T. Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3. J. Med. Chem. 2009;52:3108–3111. doi: 10.1021/jm900052j. [DOI] [PubMed] [Google Scholar]
- Levaot N., Voytyuk O., Dimitriou I., Sircoulomb F., Chandrakumar A., Deckert M., Krzyzanowski P.M., Scotter A., Gu S., Janmohamed S., Cong F., Simoncic P.D., Ueki Y., La Rose J., Rottapel R. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Huergo L.F., Gasperina A., Pedrosa F.O., Merrick M., Winkler F.K. Crystal structure of dinitrogenase reductase-activating glycohydrolase (DraG) reveals conservation in the ADP-ribosylhydrolase fold and specific features in the ADP-ribose-binding pocket. J. Mol. Biol. 2009;390:737–746. doi: 10.1016/j.jmb.2009.05.031. [DOI] [PubMed] [Google Scholar]
- Li G.Y., McCulloch R.D., Fenton A.L., Cheung M., Meng L., Ikura M., Koch C.A. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. USA. 2010;107:9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilyestrom W., Van der Woerd M.J., Clark N., Luger K. Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J. Mol. Biol. 2010;395:983–994. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler P.A., Cuneo M.J., Mueller G.A., DeRose E.F., Gabel S.A., London R.E. Structural studies of the PARP-1 BRCT domain. BMC Struct. Biol. 2011;11:37. doi: 10.1186/1472-6807-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonskaya I., Potaman V.N., Shlyakhtenko L.S., Oussatcheva E.A., Lyubchenko Y.L., Soldatenkov V.A. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J. Biol. Chem. 2005;280:17076–17083. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- Malet H., Coutard B., Jamal S., Dutartre H., Papageorgiou N., Neuvonen M., Ahola T., Forrester N., Gould E.A., Lafitte D., Ferron F., Lescar J., Gorbalenya A.E., de Lamballerie X., Canard B. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarit S.M., Davidson W., Frego L., Stebbins C.E. A steric antagonism of actin polymerization by a salmonella virulence protein. Structure. 2006;14:1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Mate M.J., Ortiz-Lombardia M., Boitel B., Haouz A., Tello D., Susin S.A., Penninger J., Kroemer G., Alzari P.M. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat. Struct. Biol. 2002;9:442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- Morrone S., Cheng Z., Moon R.T., Cong F., Xu W. Crystal structure of a tankyrase–axin complex and its implications for axin turnover and tankyrase substrate recruitment. Proc. Natl. Acad. Sci. USA. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O., Fouquerel E., Ame J.C., Leonhardt H., Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Zolkiewska A., Okazaki I. ADP-ribosylarginine hydrolases and ADP-ribosyltransferases. Partners in ADP-ribosylation cycles. Adv. Exp. Med. Biol. 1997;419:25–33. doi: 10.1007/978-1-4419-8632-0_3. [DOI] [PubMed] [Google Scholar]
- Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J.P., Haag F., Weiss M.S., Koch-Nolte F. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. USA. 2006;103:15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C., Kernstock S., Mueller-Dieckmann J., Weiss M.S., Koch-Nolte F. Structure of mouse ADP-ribosylhydrolase 3 (mARH3) Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 2008;640:156–162. doi: 10.1107/S1744309108001413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Dieckmann C., Ritter H., Haag F., Koch-Nolte F., Schulz G.E. Structure of the ecto-ADP-ribosyl transferase ART2.2 from rat. J. Mol. Biol. 2002;322:687–696. doi: 10.1016/s0022-2836(02)00818-5. [DOI] [PubMed] [Google Scholar]
- Narwal M., Venkannagari H., Lehtio L. Structural basis of selective inhibition of human tankyrases. J. Med. Chem. 2012;55:1360–1367. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere M., Mashimo M., Agledal L., Dölle C., Kasamatsu A., Kato J., Moss J., Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J. Biol. Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund, S., Ludden, P.W., (2004). Genetics and regulation of nitrogen fixation in free-living bacteria. In: Klipp, W., Masephol, B., Gallon, J.R., Newton, W.E. (Eds.), Kluwer Academic Publisher, The Netherlands, pp. 175–196.
- O’Neal C.J., Jobling M.G., Holmes R.K., Hol W.G. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- Oberoi J., Richards M.W., Crumpler S., Brown N., Blagg J., Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR) J. Biol. Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S., Kato J., Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- Oliver A.W., Ame J.C., Roe S.M., Good V., de Murcia G., Pearl L.H. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Kasamatsu A., Oka S., Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA. 2006;103:16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Reche P.A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomics. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan P.W., Feldman J.L., Devries M.K., Dong A., Edwards A.M., Denu J.M. Structure and biochemical functions of SIRT6. J. Biol. Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C.N., Koh D.W., Jacobson M.K., Oliveira M.A. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautsch A., Vogelsgesang M., Trankle J., Herrmann C., Aktories K. Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme. EMBO J. 2005;24:3670–3680. doi: 10.1038/sj.emboj.7600813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson J.R., Fried V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Peterson F.C., Chen D., Lytle B.L., Rossi M.N., Ahel I., Denu J.M., Volkman B.F. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 2011;286:35955–35965. doi: 10.1074/jbc.M111.276238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion E., Bombarda E., Stiegler P., Ullmann G.M., Mely Y., de Murcia G., Gerard D. Poly(ADP-ribose) polymerase-1 dimerizes at a 5′ recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42:12409–12417. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- Pleschke J.M., Kleczkowska H.E., Strohm M., Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Ritter H., Koch-Nolte F., Marquez V.E., Schulz G.E. Substrate binding and catalysis of ecto-ADP-ribosyltransferase 2.2 from rat. Biochemistry. 2003;42:10155–10162. doi: 10.1021/bi034625w. [DOI] [PubMed] [Google Scholar]
- Rosenthal, F., Feijs, K.L.H., Frugier, E., Bonalli, M., Forst, A.H., Imhof, R., Winkler, H.C., Fischer, D., Caflisch, A., Hassa, P.O., Lüscher, B., Hottiger, M.O., 2013. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol., in press. http://dx.doi.org/10.1038/nsmb.2521. [DOI] [PubMed]
- Ruf A., de Murcia G., Schulz G.E. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- Ruf A., Mennissier de Murcia J., de Murcia G., Schulz G.E. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc. Natl. Acad. Sci. USA. 1996;93:7481–7485. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve A.A. Sirtuin chemical mechanisms. Biochim. Biophys. Acta. 2010;1804:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve A.A., Moir R.D., Schramm V.L., Willis I.M. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol. Cell. 2005;17(4):595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Sauve A.A., Youn D.Y. Sirtuins: NAD+ -dependent deacetylase mechanism and regulation. Curr. Opin. Chem. Biol. 2012;16:535–543. doi: 10.1016/j.cbpa.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Sbodio J.I., Lodish H.F., Chi N.W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase) Biochem. J. 2002;361:451–459. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., McCue P., Gomella L.G., Raj G.V., Dicker A.P., Brody J.R., Pascal J.M., Centenera M.M., Butler L.M., Tilley W.D., Feng F.Y., Knudsen K.E. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya H., Muramatsu Y., Smith S., Tsuruo T. Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol. Cell. Biol. 2004;24:1944–1955. doi: 10.1128/MCB.24.5.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya H., Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182) J. Biol. Chem. 2002;277:14116–14126. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Giriat I., Schmitt A., de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Smith S., de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- Sundriyal A., Roberts A.K., Shone C.C., Acharya K.R. Structural basis for substrate recognition in the enzymatic component of ADP-ribosyltransferase toxin CDTa from Clostridium difficile. J. Biol. Chem. 2009;284:28713–28719. doi: 10.1074/jbc.M109.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Vonrhein C., Smart O.S., Bricogne G., Bollati M., Kusov Y., Hansen G., Mesters J.R., Schmidt C.L., Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5(5):e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Gao P., Hoffman D.W., Liu H.W. Domain C of human poly(ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry. 2008;47:5804–5813. doi: 10.1021/bi800018a. [DOI] [PubMed] [Google Scholar]
- Tao Z., Gao P., Liu H.W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- Till S., Ladurner A.G. Sensing NAD metabolites through macro domains. Front. Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K., Hottiger M.O., Ladurner A.G. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Tsuge H., Nagahama M., Nishimura H., Hisatsune J., Sakaguchi Y., Itogawa Y., Katunuma N., Sakurai J. Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J. Mol. Biol. 2003;325:471–483. doi: 10.1016/s0022-2836(02)01247-0. [DOI] [PubMed] [Google Scholar]
- Visschedyk D., Rochon A., Tempel W., Dimov S., Park H.W., Merrill A.R. Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus. J. Biol. Chem. 2012;287:41089–41102. doi: 10.1074/jbc.M112.412809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E., Karlberg T., Kouznetsova E., Markova N., Macchiarulo A., Thorsell A.G., Pol E., Frostell A., Ekblad T., Oncu D., Kull B., Robertson G.M., Pellicciari R., Schüler H., Weigelt J. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Kim, N.S., Haince, J.F., Kang, H.C., David, K.K., Andrabi, S.A., Poirier, G.G., Dawson, V.L., Dawson, T.M., (2011). Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal 4, ra20. [DOI] [PMC free article] [PubMed]
- Wang Z., Michaud G.A., Cheng Z., Zhang Y., Hinds T.R., Fan E., Cong F., Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu S., Mickanin C., Feng Y., Charlat O., Michaud G.A., Schirle M., Shi X., Hild M., Bauer A., Myer V.E., Finan P.M., Porter J.A., Huang S.M., Cong F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell. Biol. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- Zweifel M.E., Leahy D.J., Barrick D. Structure and notch receptor binding of the tandem WWE domain of Deltex. Structure. 2005;13:1599–1611. doi: 10.1016/j.str.2005.07.015. [DOI] [PubMed] [Google Scholar]