Abstract
Introduction
In randomly assigned studies with EGFR TKI only a minor proportion of patients with NSCLC have genetically profiled biopsies. Guidelines provide evidence to perform EGFR and KRAS mutation analysis in non-squamous NSCLC. We explored tumor biopsy quality offered for mutation testing, different mutations distribution, and outcome with EGFR TKI.
Patient and Methods
Clinical data from 8 regional hospitals were studied for patient and tumor characteristics, treatment and overall survival. Biopsies sent to the central laboratory were evaluated for DNA quality and subsequently analyzed for mutations in exons 18–21 of EGFR and exon 2 of KRAS by bidirectional sequence analysis.
Results
Tumors from 442 subsequent patients were analyzed. For 74 patients (17%) tumors were unsuitable for mutation analysis. Thirty-eight patients (10.9%) had EGFR mutations with 79% known activating mutations. One hundred eight patients (30%) had functional KRAS mutations. The mutation spectrum was comparable to the Cosmic database. Following treatment in the first or second line with EGFR TKI median overall survival for patients with EGFR (n = 14), KRAS (n = 14) mutations and wild type EGFR/KRAS (n = 31) was not reached, 20 and 9 months, respectively.
Conclusion
One out of every 6 tumor samples was inadequate for mutation analysis. Patients with EGFR activating mutations treated with EGFR-TKI have the longest survival.
Introduction
The effect of EGFR tyrosine kinase inhibitors (TKI) in patients with non-small-cell lung cancer (NSCLC) depends on the EGFR mutation status. Therefore, selecting the adequate tumor specimen for mutational analysis is an important issue in making treatment decisions in NSCLC. In previous randomized studies comparing EGFR TKI therapy to regular chemotherapy, the proportion of patients with adequate tumor tissue for analysis ranged from 10 to 38% [1], [2], [3], [4], [5], [6]. Most randomized studies used different EGFR mutation tests that only examined a very limited number of hotspot mutations such as L858R and exon 19 deletions [2], [7], [8], [9], [10]. What happened with less frequent mutations is not always obvious. As EGFR mutations are only present in non-squamous NSCLC [11], accurate histological phenotyping is mandatory in order to make decisions on the type of chemotherapy and for predicting the a priori presence of mutations. The IASLC/ATS/ERS guideline recommends mutational testing in non-squamous NSCLC [12].
In Caucasian patients with non-squamous cell lung carcinoma, the KRAS mutation is most common (20–30% of cases) [13], [14], followed in frequency by mutations in the EGFR gene (10–20% of cases) [13], [15]. Within histological phenotypes, certain features appear to be associated with specific mutations, for example the micropapillary aspect of adenocarcinoma with BRAF V600 mutations [16]. Although it is advantageous for patients with activating EGFR mutations to receive EGFR TKI [2], [3], [8], [17], [18], [19], in patients with other types of genetic aberrations this treatment is not effective. For example, in a study on patients with EML4-ALK translocations a lack of tumor response to EGFR TKI was reported [20]. However, for NSCLC patients with KRAS mutations the evidence is inconclusive. Several studies showed a complete lack of response to treatment with an EGFR TKI [17], [21], [22], one study demonstrated that NSCLC patients with tumors harboring KRAS mutations had a similar outcome to either EGFR TKI or chemotherapy [3]. Tumors with KRAS mutations have been shown to have worse outcome compared to patients with wild type KRAS (WT) both when treated with surgery [23] or with chemotherapy [24].
The aim is to study the distribution of common and rare EGFR and KRAS mutations sent from 8 regional hospitals to the university pathology department. The quality of tumor biopsies sent in for mutational analysis was assessed and mutation status was related to treatment with EGFR TKI outcome.
Methods
Patients
This study concerns all the NSCLC tumor samples from eight regional Dutch hospitals during the period of November 2008 until April 2011 that were tested for mutational status by a central pathology department. Data on gender, smoking status, age at diagnosis, stage at diagnosis, localization of metastases, start date and (different) lines of treatment received were collected. Tumor samples were obtained by either bronchoscopy, transthoracic lung biopsies and/or from pulmonary resections and were sent to the respective pathology department for histological examination. Histology was according to 2004 WHO criteria [25]. Response to treatment was performed according to RECIST criteria [26].
Sample collection procedure and DNA extraction
From each formalin-fixed and paraffin embedded (FFPE) tumor tissue block that was sent to the pathology department 4 µm sections were cut. After hematoxylin and eosin staining, slides were evaluated by an experienced lung pathologist for the presence of sufficient tumor tissue and estimating the percentage of tumor cells. Samples with clearly less than 50% tumor cells were defined as inadequate for EGFR/KRAS mutation testing. Areas with >50% tumor cells marked by the pathologist on the slide. This area was scraped from the slide using a scalpel and dissolved in TE-4 and 20 mg/ml Proteinase K (Life Technologies, Grand Island, NY, USA). DNA was extracted by incubation overnight at 55°C, followed by heating to 100°C for 5 minutes to inactivate proteinase K and centrifuged at room temperature at 13,000 rpm. The aqueous solution was directly used for PCR analysis or stored at −20°C. DNA concentration was measured on a ND1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). All DNA isolates were set to 10 ng/µl in TE-4 prior to use. For quality control, genomic DNA was amplified in a multiplex PCR containing a control gene primer set resulting in products of 100, 200, 300, 400 and 600 bp according to the BIOMED-2 protocol [27]. Only DNA samples with PCR products of 300 bp and larger were used for mutation analysis. All samples were tested on DNA extracted from two independent slides (duplicates). All standard precautions were taken to avoid contamination of amplification products using separate laboratories for pre- and post-PCR handling. To avoid cross-contamination, a new microtome blade was used each time a new sample was sectioned.
Either direct sequencing or high resolution melting (HRM) with confirmatory direct sequencing was performed according to protocol. Identical mutations in forward and reverse sequencing was required before a positive result is reported. The protocol is detailed in Appendix S1. The primers used for direct sequencing or HRM are described in supplemental table 1.
Table 1. Patient and tumor characteristics from samples sent to central laboratory for mutation analysis.
| N | Percentage | |
| Number of patients | 442 | 100 |
| Number of biopsies | 474 | |
| Histology | ||
| Adenocarcinoma | 353 | 80 |
| SCC | 27 | 6 |
| Large cell undifferentiated | 42 | 9 |
| Adenosquamous | 7 | 1 |
| Carcinoid | 3 | 1 |
| Salivary gland | 2 | 1 |
| NSCLC-NOS | 8 | 2 |
SCC is squamous cell lung carcinoma. NSCLC-NOS is non-small cell lung cancer – not otherwise specified.
Informed Consent and Ethics
When patients first visited the outpatient department, written informed consent for blood and tumor tissue was obtained for mutational analysis. EGFR and KRAS tests were performed as part of routine diagnostic approach and the outcome of these tests was documented in the patient file and communicated with patients. Because this is a retrospective study to collect and analyze clinical patient data, under the Dutch Law for human medical research (WMO), no consent was necessary from the medical ethics committee. Data were coded and not traceable to the individual patient.
Statistics
Descriptive statistics were performed for patient and tumor characteristics. Frequencies of common and rare mutations were tabulated. The frequency of EGFR and KRAS mutations were compared with available data on lung tissue from the Catalogue Of Somatic Mutations In Cancer database, (Cosmic DB; http://www.sanger.ac.uk/genetics/CGP/cosmic/). The relation between the presence or absence of mutations and the occurrence of most common tumor metastases was determined using the two sided Fisher exact test. For this particular analysis the patients with either an EGFR or a KRAS mutation were compared with patients who were scored as being both EGFR and KRAS WT. Overall survival (OS) was calculated from the date of diagnosing stage IV disease until censorship or death. Only patients with available clinical data who had progressed to stage IV disease and subsequently were treated were included for survival analysis. All patients treated with an EGFR TKI irrespective of their mutational status were evaluated for overall survival.
Univariate Cox regression analysis was performed with the covariates age, gender, histology (presence of adenocarcinoma, squamous cell and large cell carcinoma), KRAS and EGFR mutation status, metastatic site (brain, bone, lung) were also analyzed. Variables with p-value less than 0.20 were used for the multivariate analysis.
All statistical analysis was performed using SPSS version 18.0. Nominal P-values less than 0.05 were considered significant.
Results
EGFR and KRAS mutations
From November 2008 until April 2011 474 samples from 442 patients were sent to the central pathology department for mutation analysis. The most common histological classification was adenocarcinoma (80%), 8% of the samples came from histological subtypes not associated with EGFR mutations (Table 1).
Two hundred and twenty one patients (60.1% of all tested patients, 50% of all patients) were EGFR and KRAS WT. Thirty eight patients (10.9% of all tested patients, 8.6% of all patients) had an EGFR mutation (Table 2). In 5 patients, 2 different EGFR mutations coincided in the same tumor tissue resulting in a total of 43 mutations. Thirty of 38 patients with EGFR mutations (79%) were activating EGFR mutations. Only one patient had a T790M mutation in the primary tumor. TTF-1 positive adenocarcinomas showed an EGFR mutation more often than those who were TTF-1 negative (26/150 vs 1/50, Fisher's exact 2-sided test, p = 0.01).
Table 2. Distribution of EGFR mutations in advanced non-squamous cell lung carcinoma.
| Type of EGFR mutation | Sensitivity | Frequency of mutations | Percentage % | Frequency in COSMIC1 |
| p.K708N | Unknown | 1 | 2.3 | ND |
| p.G709_T710>M | Unknown | 1 | 2.3 | ND |
| p.G719 | Sensitive | 3 | 7.0 | 0.008 |
| Exon 19 deletion | Sensitive | 16 | 37.2 | 0.157 |
| p.S768I | Resistant | 1 | 2.3 | <0.5% |
| p.V769M | Resistant | 1 | 2.3 | <0.5% |
| p.D770GY | Unknown | 2 | 4.7 | <0.5% |
| p.D770_N771>SVD | Resistant | 1 | 2.3 | <0.5% |
| p.T790M | Resistant | 1 | 2.3 | 0.011 |
| p.L833F | Unknown | 2 | 4.7 | <0.5% |
| p.A840T | Unknown | 2 | 4.7 | <0.5% |
| p.L858R | sensitive | 11 | 25.6 | 0.145 |
| p.L861R | sensitive | 1 | 2.3 | <0.5% |
| Total | 43 mutations* | 100 |
From the Cosmic data base (retrieved on 05-02-2013) containing 13030 mutations in 48781 samples.
43 mutations were observed in 38 patients, 5 patients had double mutations.
The combination of double EGFR mutations were p.G719C, p.S768I, G719S L861R, G719C D770GY, L833F L858R and T790M L858R.
A total of 110 of patients (30% of all tested patients, 24% of all patients) had a KRAS mutation with G12C (41%) and G12V (18%) being the most frequent mutations and showing a similar distribution as in the Cosmic database (Table 3). We also found 1 (1%) rare KRAS mutation in codon 13, (p.G13Y). In addition, 2 patients had KRAS mutations outside the hotspot (p.V14L and p.L19F), these are non-functional. This means that in a total of 108 patients a functional KRAS mutation was detected in our cohort. The comparison of mutational results in the different subtypes of NSCLC in our population is shown in table 4.
Table 3. Distribution of codon 12/13 KRAS mutations in advanced non-squamous cell lung carcinoma from this study compared with the frequency distribution in the Cosmic database.
| Mutation type | Frequency/no of pts | Percentage | Frequency in Cosmic1 |
| p.G12C | 45 | 41.7 | 40.5 |
| p.G12V | 20 | 18.5 | 19.7 |
| p.G12D | 17 | 15.7 | 16.7 |
| p.G12A | 11 | 10.2 | 6.4 |
| p.G13C | 5 | 4.6 | 2.9 |
| p.G12F | 4 | 3.7 | 0.7 |
| p.G12S | 2 | 1.9 | 4.3 |
| p.G13D | 2 | 1.9 | 2.5 |
| p.G12R | 1 | 0.9 | 2 |
| p.G13Y2 | 1 | 0.9 | ND |
| Total | 108 | 100% | 100% |
ND = Not Described.
1 From the Cosmic data base (retrieved on 05-02-2013) containing 3504 mutations in 21589 samples.
2 This mutation (c37_38GG>TT, p.G13Y) was detected in 2 independent non-synchronous biopsies of the same patient.
Two KRAS mutations (p.V14L (not present at Cosmic) and p.L19F 2/2742 (present at Cosmic retrieved on 05-02-2013) were found outside codon 12/13 (considered as non-functional).
Table 4. Distribution of EGFR and KRAS mutations and their wild types in histological NSCLC subtypes of 442 patients.
| EGFR mutation | % | KRAS mutation | % | EGFR/KRAS WT | % | Insufficient material | % | Total | |
| Adenocarcinoma | 33 | 9.3 | 98 | 27.8 | 164 | 46.5 | 58 | 16.4 | 353 |
| Squamous cell carcinoma | 0 | 0 | 2 | 7.4 | 21 | 77.8 | 4 | 14.8 | 27 |
| Adenosquamous | 1 | 14.3 | 1 | 14.3 | 4 | 57.1 | 1 | 14.3 | 7 |
| NSCLC NOS | 3 | 5.4 | 9 | 16.4 | 32 | 58.2 | 11 | 20 | 55 |
| Total | 37* | 8 | 110** | 25 | 221 | 50 | 74 | 17 | 442 |
Not including a patient with dual EGFR/KRAS mutation.
Including 2 patient with a non-functional KRAS mutation and 1 patient with a dual EGFR/KRAS mutation.
Quality of tumor samples for mutation analysis
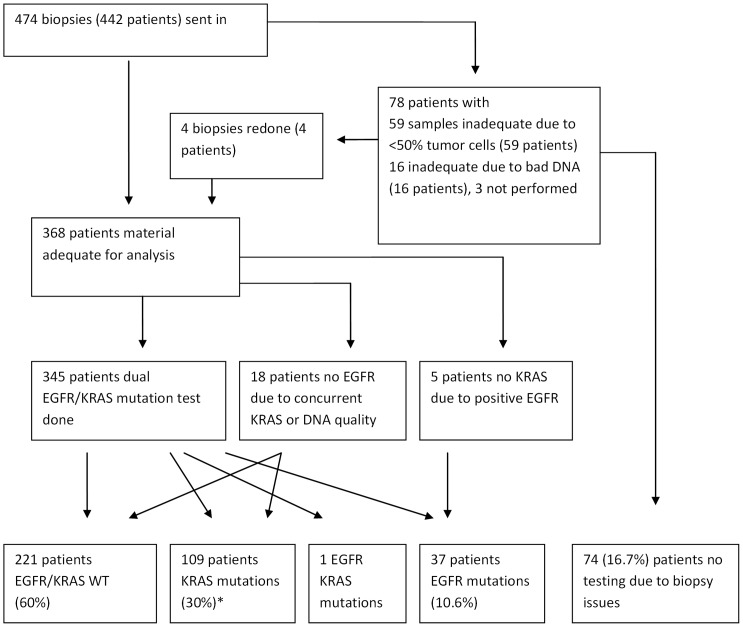
Seventy five tumor samples ((16%) were not adequate for mutation analysis. In 59 samples tissue contained less than 50% tumor cells (mostly because of extensive intermingling inflammation) and in 16 the quality of DNA appeared unsuitable for mutation testing. In 4 of these patients an adequate tissue sample was yielded by re-biopsy. In 3 tumors no further mutation analysis was performed (SCC/carcinoid). This means that from 74(75+3–4) (17%) patients no results were obtained from mutational analysis. In a total of 345 patients the tumor samples were adequate for both EGFR and KRAS analysis. A single KRAS or EGFR mutation analysis was performed in the tumor samples of 18 and 5 patients, respectively (Figure 1).
Figure 1. Flow chart for biopsy specimens sent in and result of mutation analysis.
* 2 KRAS mutations are outside of the hotspot, these are probably non functional.
EGFR and KRAS mutations and metastases distribution
Using the clinical data from 303 patients, we were able to analyze the preference for the known common metastatic regions for the patients with NSCLC with KRAS and EGFR mutational status. Pulmonary nodules (p = 0.01), vertebral (p = 0.03) and other bone metastasis (p = 0.04) were identified to be significantly associated with EGFR mutations. No association was found between EGFR mutations and pleural (p = 0.15), cerebral (p = 1.0), hepatic (p = 0.46) or adrenal (p = 0.37) metastatic localizations. None of these sites were associated with KRAS mutations.
Survival analysis
In univariate analysis from the clinical data, large cell histology (HR 1.8, 95% CI., 1.2–2.8, p<0.01) and spinal bone metastasis (HR 1.5, 95% CI., 1.0–2.2, p = 0.05) were associated with a worse survival while EGFR mutation (HR 0.4, 95% CI., 0.2–0.7, p<0.01) was associated with a better survival. In a multivariate model, histology (large cell carcinoma, HR 2.2, 95% CI., 1.4–3.4, p<0.01), spinal bone metastasis (HR 1.7, 95% CI., 1.2–2.6, p<0.01), and mutational status (EGFR mutation, HR 0.3, 95% CI., 0.1–0.6 p<0.01) were significantly associated with survival. (Table 5).
Table 5. Univariate and multivariate hazards ratios for overall survival in 248 patients with metastatic non-small cell lung cancer.
| Univariate | Multivariate | |||||
| Variables | HR | 95% CI | P | HR | 95% CI | P |
| Histology | ||||||
| Adeno | 1 | 1 | ||||
| Squamous | 1.2 | 0.7–2.1 | 0.41 | 1.2 | 0.7–2.1 | 0.48 |
| Large Cell | 1.8 | 1.2–2.8 | <0.01 | 2.2 | 1.4–3.4 | <0.01 |
| Mutation result | ||||||
| EGFR/ KRAS WT | 1 | 1 | ||||
| EGFR mutation | 0.4 | 0.2–0.7 | <0.01 | 0.3 | 0.1–0.6 | <0.01 |
| KRAS mutation | 1.1 | 0.7–1.5 | 0.70 | 1.1 | 0.8–1.8 | 0.34 |
| No test performed | 1.2 | 0.8–1.9 | 0.33 | 1.4 | 0.8–2.1 | 0.31 |
| Metastasis * | ||||||
| Spinal bone | 1.5 | 1.0–2.2 | 0.05 | 1.7 | 1.2–2.6 | <0.01 |
| Brain | 0.9 | 0.6–1.4 | 0.67 | |||
| Lung | 1.1 | 0.8–1.5 | 0.64 | |||
HR>1 means a shorter survival.
denotes presence of metastasis at specific site.
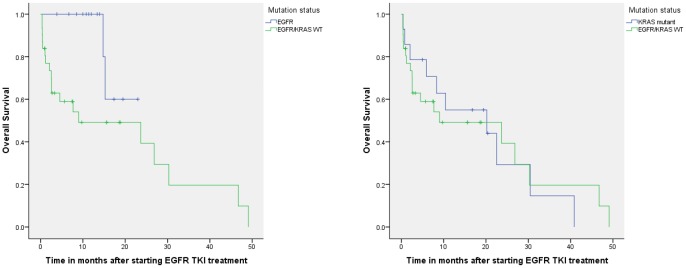
When selecting patients who received EGFR TKI treatment in the first or second line, the median overall survival after start of this treatment was not reached in patients with EGFR mutation (n = 14), 20 months (95% CI., 0–46, n = 14) for patients with KRAS mutation, and 9 months (95% CI., 0–28, n = 31) for patients with EGFR/KRAS WT. (Figure 2A and 2B).
Figure 2. A: Overall survival in patients with non-small cell lung cancer treated with EGFR-TKI in the first and second line with or without an EGFR mutation. The median overall survival for patients with EGFR mutations (n = 14) was not reached, in patients with EGFR/KRAS WT it was 9 months (95% CI., 0–28 months, n = 31). 2B: Overall survival in patients with non-small cell lung cancer treated with EGFR-TKI in the first and second line with or without KRAS mutation. The median overall survival for patients with KRAS mutations was 20 months (95% CI., 0–46, n = 14), in patients with EGFR/KRAS WT it was 9 months (95% CI., 0–28 months, n = 31).
Rare EGFR and KRAS mutations and response to treatment
Mutations that were not previously described in COSMIC DB are described in table 6. Treatment with an EGFR TKI in patients with these rare EGFR mutations did not result in clinical benefit except in one patient who also had an additional activating EGFR mutation.
Table 6. Rare EGFR and KRAS mutations and tumor response to EGFR TKI.
| Mutations | N | Response | Response | Published response to |
| to chemotherapy | to EGFR TKI | EGFR TKI | ||
| EGFR mutations | ||||
| p.K708N | 1 | PD | PD (E) | [47]PR with |
| gefitinib with | ||||
| p.K708M | ||||
| p.V769M | 1 | PR | PD (E) | [33], [48]; |
| No treatment | ||||
| information | ||||
| p. D770GY | 1 | PD | PR (E) | [36], [37] |
| with a secondary | No treatment | |||
| p.G719C mutation | information | |||
| p.D770GY; | 1 | PR | PD (G) | [36], [37] |
| without secondary | No treatment | |||
| mutation | information | |||
| p.L833F | 1 | PR | PD (G) | ND |
| (dual KRAS mutation) | ||||
| p.A840T | 2 | PR/PR | PD (E)/– | ND |
| KRAS mutations | ||||
| p.G13Y | 1 | PD | – | ND |
| p.V14L | 1 | PR | – | ND |
PR is partial response, PD is progressive disease, – = no EGFR TKI treatment; (E) = erlotinib, (G) = gefitinib, ND = Not described.
Discussion
EGFR is a cell surface protein that leads to activation of proliferation and invasion via different signal transduction pathways [28]. KRAS is a downstream target of EGFR. Activating or sensitizing mutations cause a constitutive activation of the tyrosine kinase domain of the EGFR protein, by destabilizing the autoinhibiting conformation [29]. EGFR TKI such as gefitinib have increased binding abilities for these mutant proteins. The ratios of this increased binding ability is up to 100 fold compared to wild-type EGFR protein [29].
The two most common sensitizing EGFR mutations to EGFR TKI, in frame deletions of exon 19 and the L858R mutation, [19], [30], [31], [32], [33] represented over half of all EGFR mutation patients. Other sensitizing aberrations were found in three patients having a G719X mutation and in another patient a L861R mutation [33], [34], [35]. We observed 5 rare or previously undescribed mutations (Table 2) and have characterized their response to TKI treatment (Table 6). Of specific note is the p.D770GY mutation, which was found in two patients, with different response. The first of these patients had a combination of p.D770GY and a p.G719C mutation while the second had only a p.D770GY mutation. The first patient responded to EGFR TKI and remains disease free after 15 months while the patient without the secondary mutation had progressive disease diagnosed at 4 weeks. Previously 2 cases of this mutation were described without information on tumor response [36], [37]. Our data suggest that the p.G770GY mutation does not provide benefit for EGFR TKI treatment. Furthermore, we demonstrated that also patients with one of the other 4 rare EGFR mutations (p.K708N, p.G709_T710>M, p.L833F and p.A840T) had no benefit from EGFR-TKI.
Small tumor samples mainly from bronchoscopic or transthoracic core biopsies may be a problem for adequate mutation testing. We identified causes why mutational analysis at our lab was not possible in 17% of patients. This was either due to insufficient number of tumor cells (12%) or due to insufficient DNA quality (4%) highlighting the need for adequate tumor tissue selection for mutational analysis. Retrospective studies in which long- term archived paraffin embedded tissue was used to determine EGFR status showed a low proportion of adequate tumor tissue available [1], [2], [3], [4], [5], [6]. One way to obtain more tumor cells is by repeated biopsies or cryobiopsies [38]. New technological developments are far more sensitive than previously, allowing fewer tumor cells both qualitatively (%) and quantitatively (absolute number) required for detecting mutations. However, regarding tumor heterogeneity, this increased sensitivity harbors an increased risk of sampling errors and detection of minor clones that may be less relevant for therapy. A study showed that about two thirds of all somatic mutations seemed not to be detectable across every tumor region [39].
EGFR mutations occurred most often in TTF-1 positive adenocarcinoma. Two recent studies showed this cell lineage association [40], [41]. Functionally, TTF-1 induced ROR-1 is necessary to sustain the EGFR signaling pathway in lung adenocarcinoma cell lines [42].
We identified the preference of EGFR mutant tumors to spread to intrapulmonary and to both the vertebra and other bone localizations. This contrasts with a study by Doebele et al, who observed only a preference for hepatic metastatic spread in EGFR mutant tumors [43].In contrast, we observed the typical miliary pattern of tumors with EGFR exon 19 deletion as described previously [44]. Our results for KRAS mutant tumors (71 patients) were as described previously by Doebele et al (49 patients) [43].
In our population the outcome of patients with a KRAS mutation responded similarly to KRAS WT both with respect to chemotherapy and to EGFR TKI. Previously it was demonstrated that patients with KRAS wild type have a better outcome than patients with KRAS mutations when treated with an EGFR TKI [22]. Other studies showed the presence of KRAS mutations in lung cancer to be indicative of worse outcome regardless of the treatment they received [45], [46]. In the TITAN study, there was some evidence for a higher risk of death in KRAS mutant tumor patients treated with erlotinib compared to chemotherapy but there was no elevated risk of tumor progression [4]. In our study, we did not pool the EGFR mutation positive patients with the EGFR/KRAS WT when comparing these patients with KRAS mutant patients. As patients with EGFR mutations tend to have better outcomes then EGFR WT patients, this could explain our results.
In conclusion, we found in 10.9% and 30% of all the tested patients an EGFR or KRAS mutation, respectively. We also identified 5 novel or rare EGFR mutations and 2 novel KRAS mutations in our population. Seventeen percent of patients had inadequate tumor tissue to perform mutation analysis, mostly due to insufficient tumor volume and/or percentage. There was no difference in overall survival after starting EGFR-TKI in patients with KRAS mutation and EGFR/ KRAS WT.
Supporting Information
(DOC)
Acknowledgments
We are grateful to Klaas Kooistra, Erik Nijhuis, Anke van de Berg for their contribution to EGFR mutation analysis and to Roel Soesbeek for helping with the gathering of data.
Funding Statement
This work was partly funded by the CTMM Air Force consortium (http://www.ctmm.nl). CTMM pays for GSMAK's salary and had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. No other external funding sources for this study.
References
- 1. Maruyama R, Nishiwaki Y, Tamura T, Yamamoto N, Tsuboi M, et al. (2008) Phase III study, V-15-32, of gefitinib versus docetaxel in previously treated Japanese patients with non-small-cell lung cancer. J Clin Oncol 26: 4244–4252. [DOI] [PubMed] [Google Scholar]
- 2. Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, et al. (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 3. Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, et al. (2010) Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol 28: 744–752. [DOI] [PubMed] [Google Scholar]
- 4. Ciuleanu T, Stelmakh L, Cicenas S, Miliauskas S, Grigorescu AC, et al. (2012) Efficacy and safety of erlotinib versus chemotherapy in second-line treatment of patients with advanced, non-small-cell lung cancer with poor prognosis (TITAN): a randomised multicentre, open-label, phase 3 study. Lancet Oncol 13: 300–308. [DOI] [PubMed] [Google Scholar]
- 5. Kim ES, Hirsh V, Mok T, Socinski MA, Gervais R, et al. (2008) Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet 372: 1809–1818. [DOI] [PubMed] [Google Scholar]
- 6. Garassino MC, Marsoni S, Floriani I (2011) Testing epidermal growth factor receptor mutations in patients with non-small-cell lung cancer to choose chemotherapy: the other side of the coin. J Clin Oncol 29: 3835–3837. [DOI] [PubMed] [Google Scholar]
- 7. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, et al. (2012) Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 13: 239–246. [DOI] [PubMed] [Google Scholar]
- 8. Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, et al. (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 11: 121–128. [DOI] [PubMed] [Google Scholar]
- 9. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, et al. (2010) Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362: 2380–2388. [DOI] [PubMed] [Google Scholar]
- 10. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, et al. (2011) Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 12: 735–742. [DOI] [PubMed] [Google Scholar]
- 11. Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, et al. (2005) EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 23: 857–865. [DOI] [PubMed] [Google Scholar]
- 12. Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, et al. (2011) International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6: 244–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blons H, Cote JF, Le Corre D, Riquet M, Fabre-Guilevin E, et al. (2006) Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol 30: 1309–1315. [DOI] [PubMed] [Google Scholar]
- 14. Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, et al. (2008) Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 14: 5731–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reinmuth N, Jauch A, Xu EC, Muley T, Granzow M, et al. (2008) Correlation of EGFR mutations with chromosomal alterations and expression of EGFR, ErbB3 and VEGF in tumor samples of lung adenocarcinoma patients. Lung Cancer 62: 193–201. [DOI] [PubMed] [Google Scholar]
- 16. Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, et al. (2011) Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 29: 3574–3579. [DOI] [PubMed] [Google Scholar]
- 17. Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, et al. (2005) Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900–5909. [DOI] [PubMed] [Google Scholar]
- 18. Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, et al. (2008) Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol 26: 2745–2753. [DOI] [PubMed] [Google Scholar]
- 19. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, et al. (2009) Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 361: 958–967. [DOI] [PubMed] [Google Scholar]
- 20. Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, et al. (2009) Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27: 4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, et al. (2005) KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, et al. (2009) Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia 11: 1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Slebos RJ, Kibbelaar RE, Dalesio O, Kooistra A, Stam J, et al. (1990) K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med 323: 561–565. [DOI] [PubMed] [Google Scholar]
- 24. Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, et al. (2011) Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol 29: 4113–4120. [DOI] [PubMed] [Google Scholar]
- 25.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CCE (2004) World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Lung, Pleura, Thymus and Heart. Lyon: IARC Press.
- 26. Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, et al. (2000) New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92: 205–216. [DOI] [PubMed] [Google Scholar]
- 27. van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, et al. (2003) Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- 28. Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, et al. (2007) Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 11: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500. [DOI] [PubMed] [Google Scholar]
- 31. Wu JY, Yu CJ, Yang CH, Wu SG, Chiu YH, et al. (2008) First- or second-line therapy with gefitinib produces equal survival in non-small cell lung cancer. Am J Respir Crit Care Med 178: 847–853. [DOI] [PubMed] [Google Scholar]
- 32. Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, et al. (2006) Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 12: 3908–3914. [DOI] [PubMed] [Google Scholar]
- 33. Janne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, et al. (2006) A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res 12: 751–758. [DOI] [PubMed] [Google Scholar]
- 34. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 35. Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, et al. (2011) Effectiveness of tyrosine kinase inhibitors on "uncommon" epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 17: 3812–3821. [DOI] [PubMed] [Google Scholar]
- 36. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, et al. (2004) Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 64: 8919–8923. [DOI] [PubMed] [Google Scholar]
- 37. Sequist LV, Joshi VA, Janne PA, Muzikansky A, Fidias P, et al. (2007) Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing. Oncologist 12: 90–98. [DOI] [PubMed] [Google Scholar]
- 38. Hetzel J, Eberhardt R, Herth FJ, Petermann C, Reichle G, et al. (2012) Cryobiopsy increases the diagnostic yield of endobronchial biopsy: a multicentre trial. Eur Respir J 39: 685–690. [DOI] [PubMed] [Google Scholar]
- 39. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, et al. (2006) Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 24: 1679–1688. [DOI] [PubMed] [Google Scholar]
- 41. Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T (2005) EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol 29: 633–639. [DOI] [PubMed] [Google Scholar]
- 42. Yamaguchi T, Yanagisawa K, Sugiyama R, Hosono Y, Shimada Y, et al. (2012) NKX2-1/TITF1/TTF-1-Induced ROR1 Is Required to Sustain EGFR Survival Signaling in Lung Adenocarcinoma. Cancer Cell 21: 348–361. [DOI] [PubMed] [Google Scholar]
- 43. Doebele RC, Lu X, Sumey C, Maxson DA, Weickhardt AJ, et al. (2012) Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 118: 4502–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laack E, Simon R, Regier M, Andritzky B, Tennstedt P, et al. (2011) Miliary never-smoking adenocarcinoma of the lung: strong association with epidermal growth factor receptor exon 19 deletion. J Thorac Oncol 6: 199–202. [DOI] [PubMed] [Google Scholar]
- 45. Graziano SL, Gamble GP, Newman NB, Abbott LZ, Rooney M, et al. (1999) Prognostic significance of K-ras codon 12 mutations in patients with resected stage I and II non-small-cell lung cancer. J Clin Oncol 17: 668–675. [DOI] [PubMed] [Google Scholar]
- 46.Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, et al. (2011) Prospective Molecular Marker Analyses of EGFR and KRAS From a Randomized, Placebo-Controlled Study of Erlotinib Maintenance Therapy in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. [DOI] [PubMed]
- 47. Xu JM, Han Y, Duan HQ, Gao EM, Zhang Y, et al. (2009) EGFR mutations and HER2/3 protein expression and clinical outcome in Chinese advanced non-small cell lung cancer patients treated with gefitinib. J Cancer Res Clin Oncol 135: 771–782. [DOI] [PubMed] [Google Scholar]
- 48. Huang SF, Liu HP, Li LH, Ku YC, Fu YN, et al. (2004) High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 10: 8195–8203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)