SUMMARY
Glycine riboswitches regulate gene expression by feedback modulation in response to cooperative binding to glycine. Here, we report on crystal structures of the second glycine-sensing domain from the V. cholerae riboswitch in the ligand-bound and unbound states. This domain adopts a three-helical fold that centers on a three-way junction and accommodates glycine within a bulge-containing binding pocket above the junction. Glycine recognition is facilitated by a pair of bound Mg2+ cations, and governed by specific interactions and shape complementarity with the pocket. A conserved adenine extrudes from the binding pocket and intercalates into the junction implying that glycine binding in the context of the complete riboswitch could impact on gene expression by stabilizing the riboswitch junction and regulatory P1 helix. Analysis of riboswitch interactions in the crystal and footprinting experiments indicate that adjacent glycine-sensing modules of the riboswitch could form specific interdomain interactions, thereby potentially contributing to the cooperative response.
Keywords: glycine riboswitch, crystal structure, RNA, cooperativity
INTRODUCTION
The ten-atom amino acid glycine molecule is one of the smallest organic compounds, which apart from direct use in protein synthesis serves as a precursor for other molecules and plays one of the central roles in cellular carbon flow. In bacteria, glycine is predominantly synthesized from serine by the product of the glyA gene and is cleaved by proteins encoded in the gcv operon. In Escherichia coli, glycine metabolism is tightly controlled at the level of transcription initiation by global regulators, a specific transcriptional factor GcvA, and its glycine-binding partner protein GcvR (Heil et al., 2002). The control of the glyA gene is partially preserved (Dartois et al., 1997; Saxild et al., 2001), while Gcv-like regulators of catabolism are missing in Bacillus subtilis (Abreu-Goodger et al., 2004). It came as a surprise that RNA possesses sufficient structural sophistication to specifically recognize glycine and to employ this recognition for riboswitch-based feedback control of glycine cleavage genes in B. subtilis and other bacteria (Mandal et al., 2004).
Riboswitches have emerged as one of the most widespread and important gene regulatory systems in bacteria with representatives found in archaea and eukaryotes (Nudler and Mironov, 2004; Winkler and Breaker, 2005). Riboswitches are cis-acting structured RNA segments typically located in the 5′-untranslated regions (UTRs) of bacterial mRNAs, where they function as both sensors of cellular metabolites and effectors of the regulatory response (Serganov and Patel, 2007). Glycine riboswitches directly bind glycine present in cells at over-threshold concentrations using evolutionarily conserved sensing domains. Most glycine riboswitches function as rare gene activators that turn on expression of glycine cleavage genes (Mandal et al., 2004). The second frequently observed glycine riboswitch controls carbon metabolism through activation of the malate synthase gene glcB in a marine bacterium (Tripp et al., 2009). Glycine riboswitches were also co-localized with other glycine-associated genes in some bacteria, and along with thiamine pyrophosphate (TPP) and cobalamin riboswitches, constitute one of the most abundant riboswitches (Barrick and Breaker, 2007).
Unexpectedly, the majority of glycine riboswitches are composed of two moderately similar adjacently-positioned sensing domains connected by a short linker and followed by a single expression platform (Mandal et al., 2004). Although each sensing domain is capable of specific binding to a separate glycine molecule, tandem sensors in B. subtilis and Vibrio cholerae gcvT riboswitches displayed a concerted response to glycine, characterized by a sigmoidal ligand binding curve. Such binding is best described in terms of positive cooperativity between riboswitch sensing domains that in turn ensure a strong riboswitch response to small changes in glycine concentration.
Cooperativity was first validated in the interaction of oxygen with hemoglobin and was shown later to contribute to ligand binding to multi-subunit proteins and the assembly of protein complexes. In nucleic acids, cooperativity is involved in the formation of paired regions (Siegfried and Bevilacqua, 2009), RNA folding (Sattin et al., 2008), as well as assembly of RNA- and DNA-protein complexes (Recht and Williamson, 2004). Despite frequent utilization by proteins, biologically relevant cooperative binding of small organic molecules to natural RNAs has not been observed prior to the discovery of glycine riboswitches, although its feasibility was demonstrated by a rationally designed ribozyme that required sequential binding of two effector molecules for its activity (Jose et al., 2001). It should also be noted that some tandem glycine riboswitches do not display strong cooperativity (Tripp et al., 2009).
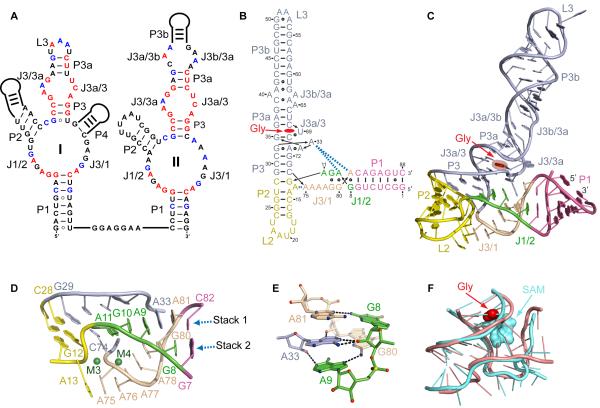
Much effort has already been directed towards the dissection of the ligand binding properties and the molecular mechanism of cooperative binding in glycine-sensing riboswitches (Serganov and Patel, 2009). In-line probing experiments revealed similar regions of glycine-modulated cleavages, despite different junctional architectures of the sensing domains (Figure 1A) (Kwon and Strobel, 2008; Mandal et al., 2004). The majority of cleavage reductions are clustered within the evolutionarily conserved J1/2, J3/1, J3/3a and J3a/3 regions, which constitute the core of the riboswitch fold. Nucleotides from these and adjacent regions were also implicated in glycine binding and/or cooperativity by the nucleotide analog interference mapping (NAIM) approach (Kwon and Strobel, 2008). Specifically, NAIM and mutagenesis studies suggested that the minor groove of helix P1 of sensing domain I and the major groove of helix P3a from both sensing domains appear to participate in cooperative tertiary interactions.
Figure 1. Sequence and Structure of the V. cholerae Riboswitch.
(A) Secondary structure schematics of the glycine riboswitch with tandem sensing domain arrangement. Nucleotides conserved in ≥95 and ≥75 % sequences are in red and blue, respectively.
(B) Crystal structure-based schematic of the VCII RNA fold. The bound glycine is in red. Dashes and circles indicate Watson-Crick and non-canonical base pairs. Key tertiary stacking interactions are shown as blue dashed lines.
(C) Overall crystal structure of VCII RNA in a ribbon representation.
(D) Zoomed-in view of the three-way junction. Green spheres depict Mg2+ cations.
(E) Intercalation of A33 into the junctional region. The RNA is shown in stick representation with color scheme of atoms (nitrogen in blue, oxygen in red, phosphorus in yellow, and carbon in arbitrary colors). Putative hydrogen bonds are shown with black dashed lines.
(F) Superposition of the three-way and four-way junctions of the glycine (light pink) and SAM-I (light blue) riboswitches, respectively. The root mean square deviation (RMSD) is 1.48 Å.
Small-angle X-ray scattering (SAXS) and hydroxyl radical footprinting showed that glycine binding in the presence of Mg2+ cations significantly changed the shape of the tandem riboswitch (Lipfert et al., 2007). After ligand binding, the riboswitch became more compact, thus suggesting that sensing domains might be positioned close to each other in the ligand bound state. However, none of these experiments were able to identify the glycine binding sites and their relation to riboswitch structure, nor could they determine directly whether the pair of sensing domains could interact with each other. Therefore the principles underlying riboswitch cooperativity are currently not understood at the molecular level.
To clarify outstanding issues concerning the glycine riboswitch structure and ligand binding cooperativity, we determined crystal structures of the second sensing domain from the V. cholerae riboswitch in the free and glycine-bound states. These structures coupled to biochemical and biophysical data provide insights into glycine recognition and the mechanism of riboswitch function, and suggest the existence of interdomain interactions that could contribute to cooperative glycine binding in the context of the complete riboswitch.
RESULTS AND DISCUSSION
Riboswitch Structure Conforms to a Three-Way Junction-Based Architecture
We have determined the 2.85 Å crystal structure of domain II (designated as VCII RNA) from the gcvT–type glycine riboswitch identified in the 5′-UTR of the V. cholerae VC1422 gene encoding a putative amino acid transporter (Mandal et al., 2004) (see Experimental Procedures and Table 1). The architecture of the glycine-bound VCII RNA conforms to a three-way junctional fold that comprises three helical stems, P1, P2 and P3/3a/3b, connected by joining regions J1/2 and J3/1 (Figures 1B and 1C), with the glycine binding pocket located above the junction. The most notable feature of the scaffold, not predicted by phylogeny (Figure 1A), is that helix P2, not P1, stacks under P3. This continuous stacking alignment is supported by stacking of G29 from P3 and G12-C28 base pair from P2 (Figure 1D). P1 is positioned at a roughly 55° angle relative to P3 and connects with the P2/P3 stem through J1/2 and J3/1. J1/2 provides a smooth transition between P1 and P2, while J3/1 connects P1 and P3 via a sharp turn that is facilitated by splayed apart nucleotides C74 and A75 (Figure 1D) and stabilized by Mg2+ cations M4 and M3 (Figure 1D and S1A). The junctional region is tied to P2 using a segment of J3/1 and to P3 and J3/3a via J1/2. These contacts contribute to the juxtaposition of the P2/P3 stack and joining segments and help constrain the relative orientation of P1 and P2/P3.
Table 1.
Data Collection and Refinement Statistics, Glycine-Bound VCII RNA
| Crystal | Soaked in 2 mM [Ir(NH3)6]Cl3 | Native Co-crystals |
Native Soaking |
|||
|---|---|---|---|---|---|---|
|
| ||||||
| PDB ID | 3OWZ | 3OWI | 3OWW | |||
| Data Collection | ||||||
|
| ||||||
| Space group | P3221 | P3221 | P3221 | |||
| Cell dimensions a,b,c (Å) | 82.7, 82.7, 149.5 | 83.8, 83.8, 198.7 | 83.5, 83.5, 198.5 | |||
| Peak | Inflection | Remote (L) | Remote (H) | |||
| Wavelength (Å) | 1.1053 | 1.1056 | 1.1256 | 1.0859 | 1.0809 | 1.0750 |
| Resolutiona | 20.00-2.95 (3.00-2.95) |
20.00-2.95 (3.00-2.95) |
20.00-3.00 (3.05-3.00) |
20.00-3.05 (3.10-3.05) |
20.00-2.85 (2.95-2.85) |
20.00-2.80 (2.90-2.80) |
| Rmerge (%)a | 9.0 (38.5) | 7.5 (47.7) | 7.0 (52.5) | 8.4 (42.3) | 5.9 (57.2) | 5.5 (45.1) |
| <I>/σ(I)a | 38.0 (6.1) | 35.0 (4.6) | 33.0 (4.6) | 33.0 (6.0) | 58.2 (5.5) | 41.2 (4.1) |
| Completeness (%)a | 99.9 (100.0) | 99.9 (100.0) | 99.9 (99.8) | 99.9 (99.8) | 98.6 (98.5) | 99.3 (99.4) |
| Unique reflectionsa | 12,921 (620) | 12,945 (632) | 12,318 (608) | 11,724 (548) | 19,420 (1,893) | 20,267 (1,983) |
| Redundancy | 6.9 (7.1) | 6.9 (7.0) | 6.9 (7.0) | 6.9 (7.1) | 14.9 (15.4) | 6.0 (6.1) |
|
| ||||||
| Phasing | ||||||
|
| ||||||
| Number of [Ir(NH3)6] 3+sites | 23 | |||||
| Figure of merit (acentric/centric) | 0.56/0.36 | |||||
|
| ||||||
| Structure Refinement | ||||||
|
| ||||||
| Resolution (Å) | 20.0-2.95 | 20.00-2.85 | 20.00-2.80 | |||
| Working set reflections | 11,610 | 18,400 | 18,182 | |||
| Test set reflections | 1,280 | 996 | 2,065 | |||
| Rfactor / Rfree (%) | 21.0 / 23.1 | 20.8 / 23.6 | 20.4 / 23.6 | |||
|
| ||||||
| Number of Nonhydrogen Atoms | ||||||
|
| ||||||
| RNA | 3,703 | 3,807 | 3,785 | |||
| Glycine | 10 | 10 | 10 | |||
| Cations | 171 | 33 | 33 | |||
|
| ||||||
| Average B-factors (Å2) | ||||||
|
| ||||||
| RNA | 58.5 | 36.8 | 33.6 | |||
| Glycine | 44.5 | 34.1 | 28.5 | |||
| Cations | 37.4 | 74.6 | 54.2 | |||
|
| ||||||
| RMSD values | ||||||
|
| ||||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.005 | |||
| Bond angles (°) | 0.976 | 1.024 | 1.003 | |||
| Estimated coordinate errorb | 0.272 | 0.253 | 0.248 | |||
Values for the highest resolution shell are in parentheses.
Estimated coordinate error based on maximum likelihood was calculated with REFMAC (Murshudov et al., 1997).
The junction adopts an intricate extended fold that combines nucleotides from J1/2, J3/1, P2 and P3 and is based on two stacks formed with participation of cross-strand stacking interactions (Figure 1D). The first base stack joins residues from three regions, J1/2 (A11, G10 and A9), J3/3a (A33), and J3/1 (A81), and continues into P1. The key feature of this alignment is highly conserved A33, that extrudes from J3/3a and intercalates between two junctional segments. The second stacking group begins at A13 in P2, spans J3/1 from A75 to G80, and extends towards G8 in J1/2 and P1. In proximity to P2, the two stacks spreads out and are brought together by three non-canonical purine base pairs G10•G79, A9•G80 and G8•A81 near P1 (Figures 1B and 1D). The junction is reinforced by tertiary pairing interactions that include packing of the A11-G10 segment with the minor groove of P3 (Figures S1B-S1D) and a set of hydrogen bonds formed by intercalated A33 with bases and sugars of J1/2 and J3/1 (Figure 1E). Nucleotides involved in tertiary contacts are evolutionarily conserved while the A75-A76-A77 region of the J3/1 stack is non-conserved and can accommodate an additional helix as predicted in domain I.
Although architectures based on three-way junctions (Lescoute and Westhof, 2006) are a common feature of riboswitches, the VCII fold is distinct from the three-way junctions of other wide-spread riboswitches. Unlike the glycine riboswitch, where helix P2 stacks under P3, in purine (Batey et al., 2004; Serganov et al., 2004) and thiamine pyrophosphate (TPP) riboswitches (Edwards and Ferre-D’Amare, 2006; Serganov et al., 2006; Thore et al., 2006) P2 and P3 are aligned in parallel and anchored to each other through long-range tertiary contacts, while the regulatory P1 helix is collinearly stacked with one of these helices. Unexpectedly, the three-way junction of the glycine sensing domain strikingly resembles the core of the four-way junction in the SAM-I riboswitch (Montange and Batey, 2006) (Figures 1F and S1E). Interestingly, despite placement of the ligands above the central portion of the junction in both the SAM-I and glycine-sensing riboswitches, S-adenosyl methionine directly stabilizes the junction through extensive contacts, while glycine is extruded from the interior of the junction and contributes indirectly to its organization.
Glycine Binding Pocket Is Adjacent to the Junction
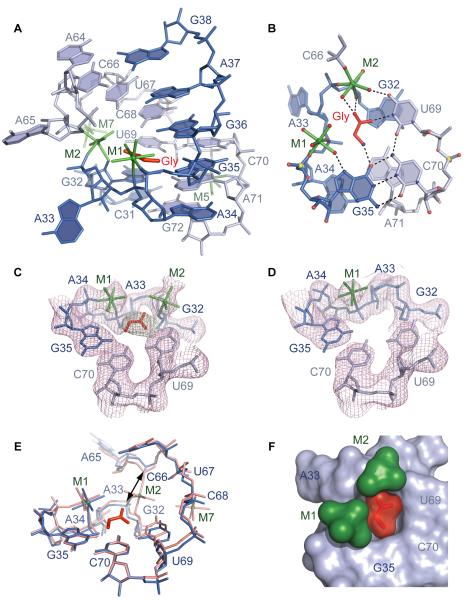
The experimental electron density map assigned to bound glycine was located in the conserved internal loop J3/3a-J3a/3, a segment whose in-line probing cleavage pattern was modulated by bound glycine (Kwon and Strobel, 2008; Mandal et al., 2004) (Figure S2A). The helical structure of adjacent regions is distorted to create a nested pocket within the internal loop for the bound ligand through formation of two stacked major grove A34•A71•G32 and (G35-C70)•U69 base triples (Figures 2A and 2B). The floor of the pocket is built by the first triple centered on A71, which uses its sugar edge for interaction with A34 and N2 amino group for pairing with the Hoogsteen edge of G32 (Figures 2B and S2B). The second triple is positioned above the first triple and together with the backbone of the A33-A34-G35 segment encircles the bound glycine (Figures 2A and 2B). The roof of the pocket is formed by the Watson-Crick G36-C68 base pair (Figures 2A and S2C) that nearly seals the ligand-binding site, and by the backbone of the A65-C66 segment that additionally contributes to formation of the pocket wall. The bases of A33 and A65 are flipped out of the pocket, and while A33 participates in formation of the three-way junction, A65 makes stacking contacts with the second molecule within the asymmetric unit.
Figure 2. Glycine Binding Pocket of VCII RNA and Recognition of Glycine.
(A) Overall view of the glycine-binding pocket. The C31 to G38 and A64 to G72 segments are in blue and light blue, respectively. Hydrated Mg2+ cations are in green with coordination bonds shown in stick representation.
(B) Base triples in the binding pocket and putative hydrogen bonds (dashed lines) contributing to glycine recognition.
(C) Refined 2∣Fo∣-∣Fc∣ electron density map contoured at 1σ level (pink) and superposed with the refined model of the binding pocket of the glycine-bound structure. Green map shows omit ∣Fo∣-∣Fc∣ map (3σ level) calculated prior to the addition of glycine.
(D) The same view as (C) for the binding pocket in the unbound state.
(E) Superposition of the binding pockets of glycine-bound (salmon) and unbound (blue) states. Arrow shows the distance between phosphorus atoms of C66 and A33 in the glycine-bound structure.
(F) Surface view inside of the glycine-bound pocket, with bound glycine in red and a pair of Mg2+ cations in green. The helix above J3/3a-J3a/3 is omitted for clarity.
The glycine binding pocket contains two metals, M1 and M2, assigned to hydrated Mg2+ cations based on the octahedral coordination geometry and the anomalous signal from Mn2+ cations that mimic Mg2+. Both cations are positioned in the vicinity of the backbone of the A33-A34-G35 segment (Figures 2A and 2B). M1 buttresses the compact conformation of the pocket through inner-sphere coordination with the non-bridging phosphate oxygens of A34 and G35 and outer-sphere contacts with A34, G35, and G36. M2 locks up the pocket by direct coordination to the 2′-OH group of G32 and the non-bridging phosphate oxygens of A33 and C66. Water-mediated interactions with bases of U67, C68, and U69 further constrain the dimensions of the pocket. Two more metals, M5 and M7, assigned to hydrated Mg2+ cations, reinforce the RNA structure around the binding pocket (Figure 2A).
To confirm the location of the bound glycine, we determined the 3.05 Å structure of VCII RNA in the unbound state (Table 2). As reported earlier for the lysine and SAM-I riboswitches (Garst et al., 2008; Serganov et al., 2008; Stoddard et al., 2010), the ligand-free VCII RNA structure superpositioned well with the glycine-bound structure (overall r.m.s.d. is 0.96 Å). However, the map lacked the electron density corresponding to glycine and high-occupancy cations M2 and M7 (Figures 2C, 2D and 2E). The subtle shifts of pocket nucleotides on strands held by M2 resulted in a slight opening of the pocket, as indicated by an increase in the distance from ~4.7 to ~6.2 Å between phosphorus atoms of C66 and A33 (Figure 2E). It should be emphasized that the ligand-free structure most likely represents one conformation that crystallizes from an ensemble of conformations present in solution prior to glycine binding. Crystallization of the ligand-free glycine riboswitch was facilitated by high (80 mM) Mg2+ concentration in the crystallization solution and by the choice of a stable RNA construct, which lacked the expression platform and had a long helix P1 that closes the junction.
Table 2.
Data Collection and Refinement Statistics, Glycine-free VCII RNA, Mutants, and Cation Identification
| Crystal | Free | Mutation U69C |
Mutation G35C-C70G |
Soaked in 10 mM MnCl2 |
Soaked in 10 mM Tl-acetate |
Soaked in 10 mM BaCl2 |
|---|---|---|---|---|---|---|
|
| ||||||
| PDB ID | 3OX0 | 3OXB | 3OXD | 3OXE | 3OXM | 3OXJ |
| Data Collection | ||||||
|
| ||||||
| Space group | P3221 | P3221 | P3221 | P3221 | P3221 | P3221 |
| Cell dimensions a,b,c (Å) | 84.3, 84.3, 199.9 | 84.0, 84.0, 197.8 | 84.0, 84.0, 200.0 | 83.4, 83.4, 199.0 | 83.7, 83.7, 200.1 | 84.2, 84.2, 201.4 |
| Wavelength (Å) | 1.0809 | 1.0809 | 1.0750 | 1.5000 | 0.9763 | 1.5000 |
| Resolutiona | 20.00-3.05 (3.16-3.05) |
20.00-2.95 (3.06-2.95) |
20.00-3.00 (3.11-3.00) |
20.00-2.90 (3.00-2.90) |
20.00-2.95 (3.00-2.95) |
30.00-3.20 (3.31-3.20) |
| Rmerge (%)a | 5.2 (48.7) | 6.2 (54.2) | 7.4 (52.4) | 6.3 (56.4) | 6.7 (51.2) | 8.1 (54.3) |
| <I>/σ(I)a | 34.8 (3.9) | 55.5 (4.0) | 41.5 (5.2) | 31.3(3.0) | 47.7 (5.5) | 29.2 (3.1) |
| Completeness (%)a | 99.8 (99.9) | 99.7(99.8) | 99.8 (99.9) | 99.9 (100.0) | 100.0 (100.0) | 99.9 (100.0) |
| Unique reflectionsa | 16,320 (1,600) | 17,666 (1,719) | 16,953 (1,624) | 18,565(1,814) | 17,793 (881) | 14,230 (1,390) |
| Redundancy | 7.1 (7.3) | 16.5 (12.2) | 12.1 (12.5) | 5.3 (5.4) | 11.2 (11.5) | 5.2 (5.4) |
|
| ||||||
| Structure Refinement | ||||||
|
| ||||||
| Resolution (Å) | 20.00-3.05 | 20.00-2.95 | 20.00-3.00 | 20.00-2.90 | 20.00-2.95 | 20.00-.20 |
| Working set reflections | 15,445 | 16,755 | 16,063 | 17,579 | 16,839 | 13,482 |
| Test set reflections | 824 | 899 | 857 | 947 | 904 | 714 |
| Rfactor /Rfree (%)a | 20.7 / 24.0 | 19.8 / 22.1 | 19.6 / 21.4 | 20.1 / 21.9 | 20.0 / 22.5 | 19.4 / 22.4 |
|
| ||||||
| Number of Nonhydrogen Atoms | ||||||
|
| ||||||
| RNA | 3,807 | 3,785 | 3,785 | 3,807 | 3,807 | 3,790 |
| Glycine | 0 | 0 | 0 | 10 | 10 | 10 |
| Cations | 28 | 33 | 34 | 42 | 37 | 45 |
|
| ||||||
| Average B-factors(Å2) | ||||||
|
| ||||||
| RNA | 56.5 | 58.2 | 46.9 | 40.9 | 33.9 | 69.9 |
| Glycine | - | - | - | 40.3 | 35.0 | 66.6 |
| Cations | 65.8 | 79.3 | 69.0 | 84.0 | 73.0 | 123.5 |
|
| ||||||
| RMSD Values | ||||||
|
| ||||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 | 0.005 |
| Bond angles (°) | 1.069 | 1.082 | 1.064 | 1.011 | 1.021 | 1.110 |
| Estimated coordinate errorb | 0.290 | 0.304 | 0.253 | 0.236 | 0.255 | 0.302 |
Values for the highest resolution shell are in parentheses.
Estimated coordinate error based on maximum likelihood was calculated with REFMAC (Murshudov et al., 1997).
The unbound VCII RNA can be converted to the glycine-bound form by soaking the ligand-free crystals in glycine-containing solution. The structure of the glycine-soaked VCII RNA was practically identical to the glycine-bound structure crystallized under the same conditions, and the 2.8 Å omit ∣Fo∣-∣Fc∣ map (Table 1) revealed three electron density peaks that correspond to glycine and cations M2 and M7 not present in the free structure. This experiment validates the position of the bound glycine and establishes that M2 does not critically contribute to overall scaffold formation. Instead, M2 most likely shields the negative charge of the glycine carboxylate, blocks the entrance to the pocket following glycine binding, and prevents dissociation of glycine by locking up the RNA fold. Thus, the bound glycine is completely buried inside of the pocket formed by the RNA scaffold and bound cations and has virtually no solvent accessible surface area. A striking parallel is observed with the sensing domain of the lysine riboswitch, which also recognizes the carboxylate moiety of the amino acid with the assistance of a positively charged cation (Serganov et al., 2008). However, instead of divalent Mg2+, lysine is directly bound by monovalent K+ cation, that in effect replaces cation M2 of the glycine riboswitch.
Glycine Binds in a Ligand-shaped Pocket Assisted by a Pair of Mg2+ Cations
The experimental density map of bound glycine extends from the Hoogsteen edge of G35 along the Watson-Crick edge of U69 in the direction of Mg2+ cation M2 (Figures 2C, S2D and S2E). The elliptical shape of this map and the omit ∣Fo∣-∣Fc∣ map (Figure 2C), calculated prior to the addition of glycine to the structure, orients glycine termini towards G35 and M2. Analysis of the surface electrostatic potential within the pocket suggests the glycine should be positioned with the negatively charged carboxylate moiety oriented towards the positively charged cation M2, while the positively charged ammonium group should be directed away from cation M1 (Figure 2B). In this orientation, the ammonium group is placed within hydrogen bond distance (2.7 Å) from O6 of G35 (Figure 2B) and close (distances less than 3.5 Å) to N1 of A71 (Figure S2B) and O6 of G36 (Figure S2C). Therefore, the ammonium group has the potential for connecting bases from all three layers of the binding pocket, although rotational flexibility could limit interactions to two layers (Figures S2D and S2E, insets). At the other glycine terminus (Figure 2B), one carboxylate oxygen atom makes a hydrogen bond with the N3 proton of U69, while the second oxygen interacts with 2′-OH group of G32 and also coordinates to the outer sphere of M2. By contrast, positioning of the glycine in an opposite orientation within the pocket would place both glycine termini in an unfavorable electrostatic environment, namely the carboxylate oxygen would face oxygens of G35 and U69 and the ammonium group would be positioned next to the cation M2.
In attempts to validate the orientation of glycine in the pocket, we mutated nucleotides around glycine and analyzed their effects using isothermal titration calorimetry (ITC). We measured a Kd = 3.5 ± 1.5 μM for wild-type VCII RNA with a long P1 helix as compared to a Kd = 24.0 μM determined by in-line probing for domain II flanked by single-stranded regions at the same 20 mM Mg2+ concentration (Mandal et al., 2004). For the first mutant, we replaced a proton donor at N3 of U69 by a proton acceptor following U69C substitution (Figure S2F). This mutation disrupted a single hydrogen bond between U69 and glycine and caused a drop of affinity to a negligible level (Figures 3A and 3B). Next, the G35-C70 base pair was substituted with C-G and A-U base pairs (Figure S2F). These mutations, which disrupt the hydrogen bonds between glycine and G35, also resulted in a total loss in glycine binding. To confirm the integrity of the binding pocket in the mutant VCII RNAs, we crystallized the U69C and G35C/C70G mutants in the presence of glycine (Table 2). The ~3.0 Å resolution structures (Figure S2G) revealed formation of well-organized glycine binding pockets that lack bound glycine and M2 and M7 cations. The G35A/C70U mutant did not crystallize, apparently due to significant perturbations of the RNA structure.
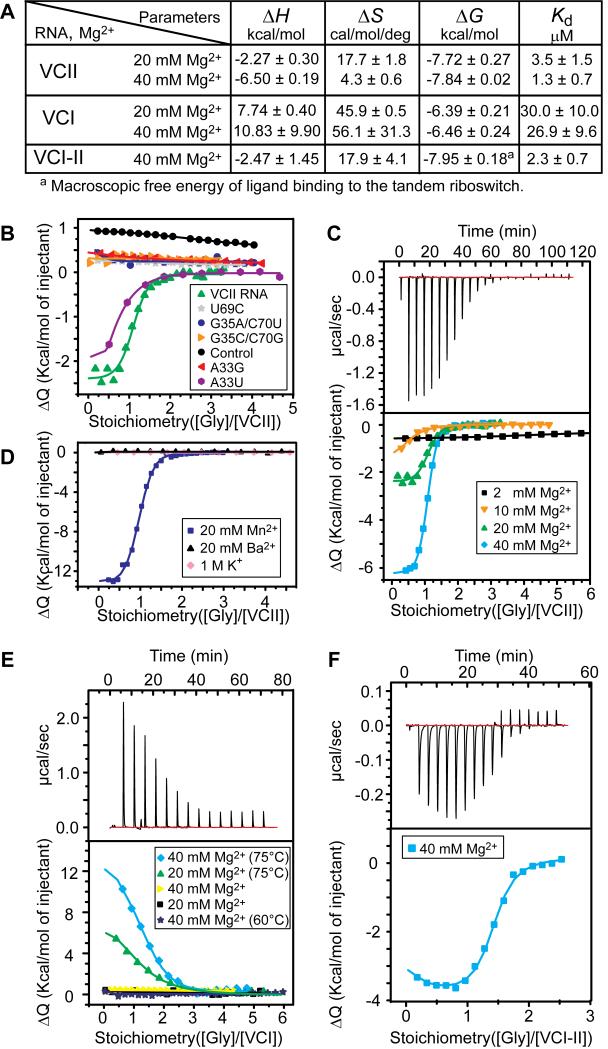
Figure 3. Glycine Binding to RNAs determined by ITC.
(A) Summary of ITC-based parameters for single and tandem RNAs.
(B) Integrated fitted heat plots of glycine binding to VCII RNA and mutants. Control: FMN riboswitch sensing domain.
(C) Effect of magnesium concentration on glycine binding to VCII RNA. Experimental data for 40 mM MgCl2 and integrated plots for all MgCl2 concentrations are shown in top and bottom panels, respectively.
(D) Cation effects on glycine binding to VCII RNA.
(E) Glycine binding to VCI RNA. Values in parentheses depict RNA refolding temperature. Representative raw data at 20 mM MgCl2 and 75 °C refolding temperature are shown in top panel.
(F) Glycine binding to VCI-II RNA.
The glycine-bound structure of VCII RNA readily explains the detrimental effects of functional group substitutions on glycine binding reported previously in NAIM experiments (Kwon and Strobel, 2008). For instance, the replacement of G32 by deoxyribonucleotide eliminates the hydrogen bond with the carboxylate of glycine and the direct coordination bond with M2, while methylation of N6 of A34 or N7 of G32 prevents formation of the A34•A71•G32 triple (Figure S3A, top inset).
Shape Complementarity and Heteroatom Recognition Contribute to Glycine Selectivity
Glycine riboswitches selectively recognize glycine and efficiently discriminate against similar compounds. Like purine, lysine and other riboswitches (Montange and Batey, 2008), the sensing domain of the glycine riboswitch forms a ligand-shaped pocket, which snugly fits bound glycine. The surface views (Figures 2F and S2H) illustrate that the binding pocket (volume of ~72 Å3) of VCII RNA is almost completely filled by the bound glycine (volume ~68 Å3). Consistent with published data (Kwon and Strobel, 2008; Mandal et al., 2004), the ligand-shaped pocket cannot accommodate methyl groups attached either to the amino group of glycine, as in sarcosine, or to the Cα atom, as in alanine. At the carboxyl end, both oxygens are critical for interactions with the riboswitch. Replacement of a single oxygen atom by amine or hydroxylamine decreased binding affinity in in-line probing assays (Kwon and Strobel, 2008; Mandal et al., 2004) and abolished ligand recognition in ITC experiments (Figure S4A). In agreement with our biochemical data, the crystal structure of VCII RNA obtained in the presence of hydroxamate lacked both the ligand and M2 cation (Figure S4B). Modifications of the carboxyl moiety that preserved both oxygens, such as methyl, ethyl and tert-butyl esters, were reported to be tolerated with progressively reduced binding affinity (Kwon and Strobel, 2008; Mandal et al., 2004) (Figure S4C). Though the methyl ester could theoretically be accommodated in a slightly widened pocket, the larger ethyl and tert-butyl moieties would require more dramatic adjustments, such as the loss of M2. Unfortunately, electron density maps calculated from the crystals grown with glycine esters, did not reveal the density for additional groups (Figure S4D). Moreover, the pocket volumes, ranging from ~70 to 81 Å3, are incompatible with large glycine analogs. Possibly, these crystals contained either glycine as a contaminant of the original ester samples or alternately reflected the release of glycine as a product of ester decomposition during crystallization (Figure S4E).
Mg2+ Cations Participate in Both Ligand Recognition and RNA Folding
The function of many cellular RNAs, including glycine riboswitches (Lipfert et al., 2007; Lipfert et al., 2010), depends critically on the presence of Mg2+ cations. The crystal structure of VCII RNA revealed fifteen Mg2+ cations distributed along P1, P2, J1/2 and J3/1, where they participate in the formation of the intrinsic junctional fold. Five more Mg2+ cations are bound around and within the glycine binding pocket. Remarkably, six out of eight NAIM sites (Kwon and Strobel, 2008), located in the vicinity of the glycine binding pocket, can affect coordination of Mg2+ cations (Figure S3A). Our ITC experiments showed that glycine binding to VCII RNA decreased ~13 (Kd = 45.9 ± 6.6 μM) and ~300 fold (Kd greater than 1 mM), when Mg2+ concentration was reduced from 20 mM to 10 mM and 2 mM, respectively (Figures 3A and 3C). Increase of Mg2+ concentration from 20 to 40 mM did not significantly affect binding affinity, although it resulted in a reduction in favorable entropy and a substantial increase in favorable enthalpy (Figure 3A). Nevertheless, ligand binding to VCII is enthalpy driven at both 20 and 40 mM Mg2+ (Figures 3A and 3C). Notably, Mg2+ could not be replaced by 1 M K+ (Figure 3D), as has been reported for the flavin mononucleotide (FMN) riboswitch (Serganov et al., 2009), suggesting that monovalent cations cannot fulfill the function of divalent Mg2+. In agreement with the structures and ITC results, Tl+ soaks identified 5 anomalous peaks (Figures S5B) mostly coincident with divalent cation sites, further confirming that VCII RNA does not require specific monovalent cations for glycine binding.
Large divalent cations, such as Ba2+, abolished glycine binding, while smaller Mn2+ cations supported ligand recognition (Figure 3D). These data are in line with earlier SAXS measurements that revealed the critical role of Mg2+ and similar cations in ligand-dependent conformational changes of the tandem glycine riboswitch (Lipfert et al., 2007; Lipfert et al., 2010). Both Mn2+ and Ba2+ cations were located in the structure based on their anomalous signals. Mn2+ cations were found in seventeen sites occupied by Mg2+ cations in the native structure, including M1 and M2 (Figures S5A and S5C). M1 was substituted by Mn2+ to a smaller extent, suggesting that this partially hydrated cation is tightly bound to RNA in the glycine-bound structure (Figures 2C and S2I). Ba2+ cations replaced only several Mg2+ cations (Figure S5D); M1 and M2 were not substituted. Therefore, the VCII RNA is more restrictive to the nature of divalent cations than earlier studied sensing domains of TPP (Edwards and Ferre-D’Amare, 2006) or FMN (Serganov et al., 2009) riboswitches.
Interestingly, while binding to VCII RNA is exothermic and enthalpy driven, binding to the isolated domain I of V. cholerae riboswitch (VCI RNA) is ~10-fold weaker, endothermic, entropy driven and requires high re-folding temperatures (Figures 3A and 3E). On the other hand, glycine binding to the RNA that contains both sensing domains of the natural riboswitch (VCI-II) is highly dependent on Mg2+ concentration (data not shown) and is characterized by Kd = 2.3 ± 0.7 μM, a value comparable to VCII RNA binding affinity (Figures 3A and 3F).
Contribution of Junctional Folding to Riboswitch Mechanism
In the majority of riboswitches, modulation of helix P1 formation is the key conformational rearrangement affecting gene expression. In the presence of glycine, helix P1 of the glycine riboswitch sequesters the regulatory sequence in the 3′-region of the sensing domain so that the downstream region folds into an anti-terminator hairpin and allows the RNA polymerase to complete gene transcription (Mandal et al., 2004). Unlike other riboswitches, gene expression control in the glycine riboswitch could rely heavily on a single conserved nucleotide A33, which is looped out of the ligand-binding pocket (Figure 1E) and organizes the junctional region through continuous stacking interactions and non-canonical base-pairing that facilitates formation of P1 (Figures 1D). It is conceivable that glycine binding facilitates extrusion of A33 from the pocket and synergizes with the A33-dependent stabilization of the three-way junction. Although A33 is not flexible in the glycine-free VCII structure, solution binding assays of A33 mutants are in line with this suggestion. Disruption of the junction is the likely cause of glycine binding loss in the A33G mutant (Figure 3B), in which the NH2 group of the introduced guanine clashes with sugar atoms of G80. On the other hand, substitution of A33 with the small uridine residue, found in 1% of glycine riboswitches, preserved glycine binding albeit to smaller extent (Kd = 16.2 ± 5.2 μM ). In addition, less than a half of the A33U RNA was found capable of glycine binding (n = 0.35-0.55), most likely because of difficulties in junction formation due to the loss of two hydrogen bonds with the G8 sugar. Therefore, without intercalation of A33, it is likely that the junction and P1 would not be stably folded and the regulatory sequence would engage in base pairing with the downstream region, thereby forming a transcription terminator that aborts gene transcription. The formation of the stable junction and P1 is also precluded by the tested earlier G8C mutation (Mandal et al., 2004) that disrupts G8•A81 base pair formation (Figure 1B), G10C mutation that prevents interactions of G10 with G79 and nucleotides of helix P3 (Figures 1B and S1D), and by six functional group substitutions identified in the junctional core by NAIM experiments (Kwon and Strobel, 2008) (Figure S3A, bottom inset).
Contacts between RNAs in Crystallographic Asymmetric Unit
The asymmetric unit contains two similarly-folded VCII RNA molecules which pack against each other and exhibit good surface complementarity and a surprisingly extensive buried surface area of 1262 Å2 per molecule (Figures 4A and 4B). This area is larger than the 900 Å2 value characteristic of potential biologically relevant contact areas in protein dimers (Ponstingl et al., 2000). Although crystal contacts do not necessarily correspond to physiological interactions (Bahadur et al., 2004), there are examples when such correspondence has been proven for proteins (Shi et al., 1997). The same analysis cannot be performed on RNA since only a few structures of small biological RNA dimers have been determined.
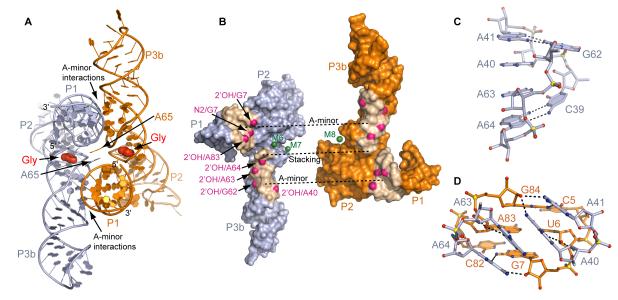
Figure 4. Intermolecular Interactions in the Asymmetric Unit of the VCII RNA structure.
(A) Two VCII RNA molecules (light blue and orange) in the asymmetric unit. Contact areas are indicated.
(B) Split-up view of the interacting VCII RNAs in surface representation. Green spheres indicate Mg2+ cations located in the riboswitch interface. Nucleotide analog interference sites (Kwon and Strobel, 2008) which cannot be explained by the structure of the individual VCII RNA are shown in magenta.
(C) Zoomed-in view of the J3b/3a-J3a/3b region.
(D) A-minor interactions between J3b/3a-J3a/3b region and P1.
The contacting surface of VCII molecules involves three RNA regions and three Mg2+ cations, M6, M7 and M8. The first contact features π-stacking interaction between A65 residues from each VCII RNA that are looped out of the glycine binding pocket (Figure 4A). Practically all glycine riboswitches have a residue at this position, with adenine observed in ~56 % of cases. The other two contact regions are symmetrical and formed between P1 of one molecule and internal loop J3a/3b-J3b/3a of the second molecule. To make these interactions, J3a/3b-J3b/3a adopts a twisted helical conformation constrained by the non-canonical A41•G62 and A64•C39 base pairs at each end, with four adenines, A40, A41, A63 and A64, involved in cross-strand stacking interactions (Figure 4C). The stacked adenines are inserted into the minor groove of P1 where they comprise four A-minor interactions (Nissen et al., 2001): two type I, A64•(G7-C82) and A41•(G84-C5), and two type II, A63•(U6-A83) and A40•(U6-A83), triples (Figures 4D and S6A). The consecutive adenine bases are inclined with respect to the P1 base pairs to form double A-minor interactions (Gilbert et al., 2008; Klein and Ferre-D’Amare, 2006; Serganov et al., 2006), such that A40 forms tertiary contacts with both the A83-U6 base pair and adjacent G84. The folding of J3a/3b-J3b/3a apparently depends on glycine binding as suggested by in-line probing of the individual VCII RNA (Mandal et al., 2004). Therefore, it is conceivable that glycine binding could induce conformational changes required for allosteric communication between tandem domains in the context of the complete riboswitch.
Interdomain Interactions in Engineered Tandem Riboswitch in Solution
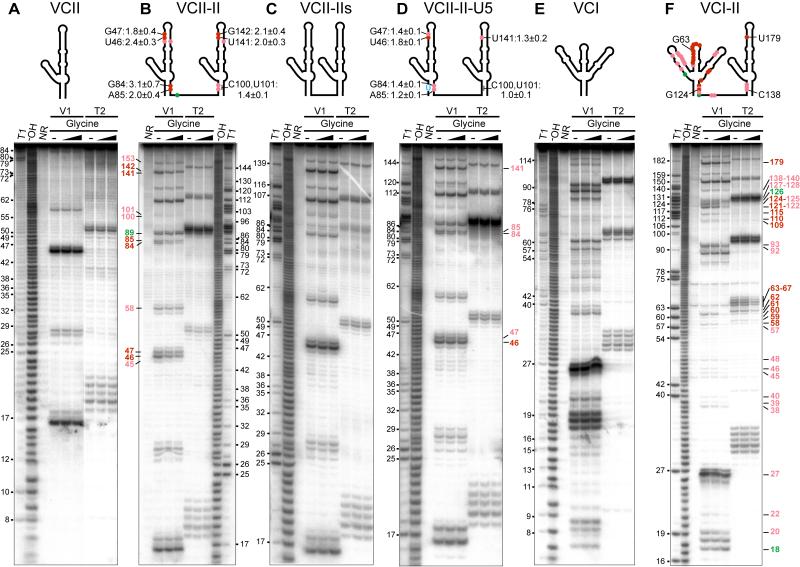
To test if interdomain interactions are not solely the property of the crystal lattice but could also take place in solution, we conducted nuclease footprinting experiments with the individual VCII RNA and its tandem VCII-II variant, composed of a pair of VCII RNAs connected by a 7-nt linker. The linker can easily span the ~35 Å distance between the 5′- and 3′-ends of the packed VCII RNA molecules observed in the crystallographic asymmetric unit (Figure 4A). Addition of 200-fold excess of glycine to the VCII RNA did not alter V1 and T2 nuclease cleavages, suggesting that the individual VCII RNA, stabilized by long P1 helix, did not significantly affect its conformation upon glycine binding, at least in the context of tracking by large nucleases (Figure 5A). It should be noted that other probing techniques that are less dependent on the local structural context, such as in-line probing, detected changes in the individual domain II upon glycine binding (Mandal et al., 2004). Since VCII RNA did not stably dimerize in gel-shift experiments at similar concentrations (not shown), the nuclease cleavage patterns of VCII RNA most likely correspond to a monomeric conformation. The digestion patterns of individual VCII and tandem VCII-II RNAs in the absence of glycine are similar (Figures 5A, 5B, S7A, S7B), suggesting that VCII-II RNA does not form stable interdomain contacts prior to glycine binding. Addition of glycine to VCII-II RNA reduced many V1 cleavages in P1 (nts 84-85, 100-101) and P3b (nts 45-47, 58, 141, 142, 153) (Figures 5B and schematic representation above panel, S7B, S7C). These regions are adjacent to the intermolecular contacts in the crystallographic dimer of the VCII RNA, and hence one can infer that the cleavage reductions are most likely associated with the formation of interdomain interactions in the tandem VCII-II RNA in the glycine-bound state. To reinforce our conclusion, we conducted footprinting on mutated tandem VCII-II RNAs, containing a short 2-nt linker (designated VCII-IIs RNA), that cannot connect the ends of the crystallographic dimer in the VCII structure. Glycine did not modulate nuclease cleavages in this RNA (Figures 5C, S7D), and as anticipated, prevented formation of interdomain interactions.
Figure 5. Ribonuclease V1 and T2 Probing of Interdomain Interactions.
(A-F) Projections of weak and strong nuclease cleavage reductions (light and dark red) and enhancement (green) on the secondary RNA structures are shown on top and the corresponding gels are on the bottom. T1 and −OH designates RNase T1 and alkaline ladders, respectively. NR, no reaction. Cleavage reduction values (mean ± standard deviation from at least 3 gels) in the presence of 200-fold excess of glycine are shown on the secondary RNA structures at the top of panels B and D. Note that the 5′ and 3′ regions of RNAs have not been analyzed.
To test putative A-minor interactions in VCII-II RNA more directly, the C5-G84 base pair was mutated to a U•G pair in P1 of the first domain (designated VCII-II-U5 RNA). This mutation changes the base alignment in the A41•(G84-C5) A-minor triple and should prevent the formation of stable tertiary interactions between P1 of the first domain and J3a/3b-J3b/3a of the second domain. In the presence of glycine, the reduction of average V1 cleavage (~1.3) in this contacting area for the VCII-II-U5 mutant is 1.8 fold smaller than the corresponding footprint in unaltered RNA (~2.3) (Figures 5D, S7E). Although the contact between P1 of the second domain and J3a/3b-J3b/3a of the first domain was not mutated, the V1 footprint in this contacting area was also reduced (~1.4), but to a lesser extent (~1.4 fold) relative to unaltered RNA (~1.9). These results are indicative of synergistic formation of two A-minor mediated tertiary interdomain contacts in the glycine-bound VCII-II RNA. The binding affinities of the mutated tandem RNAs were preserved and their apparent Kd values, 3.9 ± 0.03 μM for VCII-IIs mutant, and 7.4 ± 2.3 μM for VCII-II-U5 mutant, were in range of Kd values of 7.0 ± 1.6 μM for VCII-II, and 3.5 ± 1.5 μM for VCII. Our footprinting and glycine binding data are consistent with the engineered tandem VCII-II RNA forming interdomain interactions in solution, and we suggest that these interactions could reflect the intermolecular contacts observed for the crystallographic VCII dimer.
Interdomain Interactions in Natural Tandem Riboswitch in Solution
To test for interdomain contacts in the natural tandem riboswitch in solution, we performed nuclease footprinting with tandem VCI-II and individual VCI RNAs. As in VCII RNA, the cleavage of VCI RNA was not affected by glycine (Figure 5E), while VCI-II RNA showed modulated cleavage patterns (Figure 5F). Most footprints observed in VCII-II RNA were also found in VCI-II RNA, with the exception of V1 protections in helix P3b of domain I in VCII-II RNA (Figure 5B). In domain I of VCI-II RNA, the P3b/L3 hairpin was reduced to the apical loop L3, which was strongly protected against nuclease T2 cleavage (Figure 5F). These data suggest that both tandem natural VCI-II RNA and engineered VCII-II RNAs likely experience similar conformation changes upon glycine binding in solution and corresponding regions of both RNAs might form analogous interdomain interactions. In addition to protections common to both tandem RNAs, VCI-II RNA displayed strong cleavage reductions in J3/3a, and helices P3a, P2-P2a and P4-P4a (Figure 5F). These footprints indicate that VCI-II RNA forms a compact structure that prevents access of nucleases to these regions (Figures S7F and S7G).
Correlation of Interdomain Interactions with NAIM Data on Natural Tandem Riboswitch
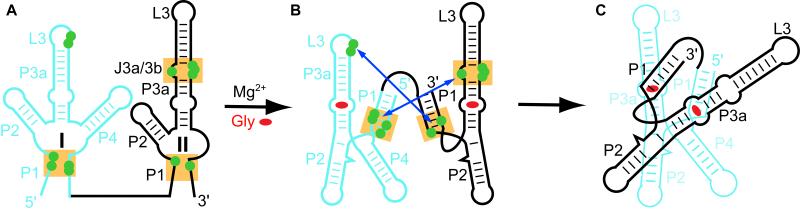
The NAIM data on VCI-II RNA (Kwon and Strobel, 2008) include several interference sites within domain II which cannot be explained by the structure of VCII RNA alone, but could be rationalized if these sites interfere with interdomain interactions. Projection of these NAIM sites on the VCII structure shows that they are located at the interface of the riboswitch dimer in the asymmetric unit (Figure 4B), where they should interfere with the formation of intermolecular A-minor interactions (Figure S3B). Since VCI and VCII RNAs resemble each other, some of these interactions could be involved in formation of biologically relevant interdomain interactions in the natural tandem riboswitch and could be responsible for riboswitch cooperativity. Indeed, the C5U mutation that disrupted the interdomain A-minor interactions in VCII-II RNA in solution (Figure 5) also abrogated cooperative binding in the natural tandem VCI-II RNA (Kwon and Strobel, 2008). Therefore, combined structural and NAIM data indicate that in the natural tandem VCI-II RNA the J3b/3a-J3a/3b region of domain II may interact with the upper part of P1 of domain I (Figures 6A, 6B and S6B), which resembles the corresponding region in domain II. Although in domain I the A-rich apical loop L3 replaces the A-rich internal loop J3b/3a-J3a/3b of domain II, it is possible that the L3 loop participates in tertiary A-minor interactions with the top part of P1 from domain II (Figures 6A and 6B), as suggested by the NAIM sites at positions 67-68, 140 and 216 (Figure S7G numbering) (Kwon and Strobel, 2008).
Figure 6. Likely Interdomain Interaction in V. cholerae Glycine Riboswitch.
(A) Projection of intermolecular interactions found in the VCII crystallographic dimer and NAIM sites (Mandal and Breaker, 2004) on the secondary structure of natural tandem riboswitch. Domain I is shown in cyan and domain II in black. Green circles indicate NAIM sites which cannot be explained by the structure of the individual VCII RNA. Orange shading shows areas that correspond to the regions involved in interdomain interactions in the crystallographic dimer of VCII RNA.
(B)-(C) Proposed interdomain interactions in the tandem natural riboswitch.
The tertiary interaction scheme (Figure 6), based on the crystal contacts observed for the VCII RNA dimer, also allows prediction of interdomain contacts in the cooperative Fusobacterium nucleatum and B. subtilis glycine riboswitches (Figures S6C-S6E), with the interdomain contact areas proposed by us in agreement with the NAIM data by Kwon and Strobel (2008) on the F. nucleatum glycine riboswitch. The regions of interdomain contacts, proposed by us, also coincide with the areas of glycine-induced reductions of hydroxyl radical cleavage in the tandem V. cholerae riboswitch (Lipfert et al., 2007). Note that tandem RNAs, such as VCII-II, do not necessarily have to display a strong cooperativity since interdomain interactions may not improve glycine binding affinity if both domains are capable of strong and identical glycine binding.
Although crystallization facilitates formation of the intermolecular RNA dimer in the absence of glycine in the crystal lattice, the solution nuclease footprinting data indicated that stable interdomain interactions can be observed in the presence of glycine and when domains are connected by a linker. Lack of stable interdomain interactions prior to glycine binding suggests that glycine likely interacts with domains of the tandem riboswitch sequentially. It is conceivable that glycine binds initially to domain I, which is transcribed first. On the other hand, domain II may initially interact with the ligand, since this domain binds glycine tighter than domain I, according to our ITC (Figures 3C and 3E) and published mutagenesis (Mandal et al., 2004) data. In addition, domain I had to be re-folded at 75 °C to bind glycine efficiently (Figure 3E) and, therefore, it may require assistance from domain II to adopt a conformation competent for glycine binding in the tandem riboswitch. Since other factors, such as transcriptional pausing, are likely involved in the riboswitch folding, the order of glycine binding to domains of the tandem riboswitch cannot be deduced at this time.
In conclusion, the VCII RNA crystal structures provide a comprehensive picture of glycine recognition by RNA, governed by electrostatics, shape complementarity, and specific direct and Mg2+-mediated interactions. Although the exact mechanism of the cooperative response remains an open question for future studies, our structural and footprinting data combined with the results of in-line probing (Kwon and Strobel, 2008; Mandal et al., 2004), NAIM (Kwon and Strobel, 2008; Mandal et al., 2004) and SAXS (Lipfert et al., 2007) experiments, have identified specific interdomain interactions between tandem sensing domains and are supportive of their participation in the cooperative binding of glycine by the tandem glycine riboswitch.
EXPERIMENTAL PROCEDURES
RNA Preparation, Crystallization and Data Collection
RNAs encoding the V. cholerae riboswitch sequences were transcribed in vitro and purified by denaturing polyacrylamide gel electrophoresis (PAGE) followed by anion-exchange chromatography and ethanol precipitation. Complexes of glycine riboswitch with glycine were prepared by mixing 0.5 mM RNA with 2 mM glycine in a buffer containing 100 mM potassium acetate, pH 6.8, and 4 mM MgCl2. Crystals were grown at 20 °C for one week using hanging-drop vapor diffusion method after mixing the complex at an equimolar ratio with the reservoir solution containing 0.05 M Na-cacodylate, pH 5.1, 0.2 M KCl, 8 % (w/v) PEG8000 and 80 mM magnesium acetate. Crystallization of the mutants and the riboswitch in the unbound state were carried out in a similar manner. For data collection, crystals were quickly transferred into a cryoprotectant solution containing 0.05 M Mes-NaOH buffer, pH 5.5, 150 mM magnesium acetate, 25 % MPD and 25 % ethanol, and flash-frozen in liquid nitrogen. For heavy atom and cation soaking, crystals were transferred into the cryoprotectant solution supplemented with 2 mM [Ir(NH3)6]Cl3, 10 mM thallium acetate, 10 mM BaCl2 or 10 mM MnCl2 and soaked for 8, 8, 3.5, and 4 h, respectively. Data were collected at 100 K at the X29a beamline of the National Synchrotron Light Source (NSLS, Brookhaven) and processed with HKL2000 (HKL Research). Crystals with glycine analogs were obtained by co-crystallization or by soaking glycine-unbound crystals in the cryoprotectant solution supplemented with 2 mM analogues for 6-8 h.
Structure Determination, Refinement and Analysis
The structure of glycine-bound VCII RNA was determined by multi-wavelength anomalous dispersion (MAD) method using 2.95 Å iridium hexamine data and autoSharp (Table 1). Crystals were improved by the replacement of the L3 loop with a GAAA tetraloop. The RNA model was built in Coot (Emsley and Cowtan, 2004), and refined in Phenix (Adams et al., 2002) and Refmac (Murshudov et al., 1997) using a 2.85 Å native data set. Glycine was added to the model at a later stage based on the experimental and refined maps, electrostatic analysis and mutational data. Cations and their coordinated waters were added based on the coordination geometry, coordination distances, omit and anomalous maps. All other riboswitch structures were refined using native riboswitch structure as a starting model (Table 2). The contact surface area and the solvent accessibility were calculated by PISA webserver (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver). The volumes of the binding pocket and ligand were calculated using VOIDOO package (Uppsala Software Factory) and the molinspiration server (http://www.molinspiration.com/). Figures were prepared with PyMOL (http://www.pymol.org).
Nuclease Probing and Footprinting
For footprinting experiments, the riboswitch RNAs were radioactively labelled at the 5′ end. 20 μl samples of the radiolabeled RNA (100,000 c.p.m.) with a final RNA concentration of 0.5×10−7 M were preheated at 75 °C for 1-3 min in 50 mM HEPES-KOH, pH 7.5, 100 mM KCl and 20 mM MgCl2 in the presence, if necessary, of 20- and 200-fold excess of glycine, incubated at room temperature for 1 min, chilled on ice, and additionally incubated at 37 °C for 15 min. Cleavage reactions were performed with 0.13 U RNase V1 (Pierce) or 0.05 U RNase T2 (Sigma) at 37 °C for 10 min. Reactions were quenched by the addition of 80 μl cold buffer, immediately extracted with phenol-chloroform, and precipitated by ethanol. RNA pellets were dissolved and analyzed by PAGE. Nuclease effects were quantified after normalization and background subtraction using ImageGauge (Fuji).
Isothermal Titration Calorimetry Binding Assays
All experiments were performed at least in duplicate on Microcal calorimeter ITC200 at 35 °C. Prior to titration, 0.1-0.3 mM RNA samples were dialyzed overnight at 4 °C against experimental buffer containing 50 mM HEPES-KOH, pH 7.5, 100 mM potassium acetate and 0-40 mM MgCl2 or other divalent cations. If necessary, RNAs were refolded by heating at 60-75 °C for 3-10 min and cooling on ice. For measurements, the ligands, dissolved in the dialysis buffer at ~1-3 mM concentration, were typically titrated into RNA in the sample cell (V=207 μL) by 26 serial injections of 1.5 μL each with a 3 μL sec−1 rate, 180 s intervals between injections, and a reference power of 6 μcal sec−1. c values were between 15 and 25. The thermograms were integrated and analyzed using Origin 7.0 software (Microcal, Inc) and SEDPHAT (Houtman et al., 2007) as described in Extended Experimental Procedures. All values are mean ± standard deviation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the personnel of beamline X29 at NSLS for assistance with data collection. We thank O. Ouerfelli (MSKCC, New York) for the synthesis of iridium hexamine. D.J.P. was supported by funds from the National Institutes of Health.
Footnotes
ACCESSION NUMBERS The atomic coordinates and structure factors were deposited in the Protein Data Bank (IDs 3OWZ, 3OWI, 3OWW, 3OX0, 3OXB, 3OXD, 3OXE, 3OXM, 3OXJ) (Tables 1 and 2).
SUPPLEMENTAL DATA Supplemental data include seven Supplemental Figures, Extended Experimental Procedures, and Supplemental References can be found with this article online at
Author Contributions: L.H. grew the crystals, determined the structures and performed ITC binding assays. A.S. conducted footprinting experiments. D.J.P. supervised the project. All authors were involved in the planning of experiments, interpretation of the results and writing of the manuscript.
REFERENCES
- Abreu-Goodger C, Ontiveros-Palacios N, Ciria R, Merino E. Conserved regulatory motifs in bacteria: riboswitches and beyond. Trends Genet. 2004;20:475–479. doi: 10.1016/j.tig.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. Journal of molecular biology. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome biology. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Dartois V, Liu J, Hoch JA. Alterations in the flow of one-carbon units affect KinB-dependent sporulation in Bacillus subtilis. Molecular microbiology. 1997;25:39–51. doi: 10.1046/j.1365-2958.1997.4491805.x. [DOI] [PubMed] [Google Scholar]
- Edwards TE, Ferre-D’Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- Heil G, Stauffer LT, Stauffer GV. Glycine binds the transcriptional accessory protein GcvR to disrupt a GcvA/GcvR interaction and allow GcvA-mediated activation of the Escherichia coli gcvTHP operon. Microbiology (Reading, England) 2002;148:2203–2214. doi: 10.1099/00221287-148-7-2203. [DOI] [PubMed] [Google Scholar]
- Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16:30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Soukup GA, Breaker RR. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 2001;29:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. Journal of molecular biology. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Sim AY, Herschlag D, Doniach S. Dissecting electrostatic screening, specific ion binding, and ligand binding in an energetic model for glycine riboswitch folding. RNA. 2010 doi: 10.1261/rna.1985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature structural & molecular biology. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annual review of biophysics. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends in biochemical sciences. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. Journal of molecular biology. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. Journal of the American Chemical Society. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxild HH, Brunstedt K, Nielsen KI, Jarmer H, Nygaard P. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J Bacteriol. 2001;183:6175–6183. doi: 10.1128/JB.183.21.6175-6183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Bba-Gene Regul Mech. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry & biology. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massague J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Bevilacqua PC. Thinking inside the box: designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding. Methods in enzymology. 2009;455:365–393. doi: 10.1016/S0076-6879(08)04213-4. [DOI] [PubMed] [Google Scholar]
- Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol. 2009;11:230–238. doi: 10.1111/j.1462-2920.2008.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annual review of microbiology. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.