Abstract
Introduction
Venous thromboembolism may recur in up to 30% of patients with a spontaneous venous thromboembolism after a standard course of anticoagulation. Identification of patients at risk for recurrent venous thromboembolism would facilitate decisions concerning the duration of anticoagulant therapy.
Objectives
In this exploratory study, we investigated whether whole blood gene expression data could distinguish subjects with single venous thromboembolism from subjects with recurrent venous thromboembolism.
Methods
40 adults with venous thromboembolism (23 with single event and 17 with recurrent events) on warfarin were recruited. Individuals with antiphospholipid syndrome or cancer were excluded. Plasma and serum samples were collected for biomarker testing, and PAXgene tubes were used to collect whole blood RNA samples.
Results
D-dimer levels were significantly higher in patients with recurrent venous thromboembolism, but P-selectin and thrombin-antithrombin complex levels were similar in the two groups. Comparison of gene expression data from the two groups provided us with a 50 gene probe model that distinguished these two groups with good receiver operating curve characteristics (AUC 0.75). This model includes genes involved in mRNA splicing and platelet aggregation. Pathway analysis between subjects with single and recurrent venous thromboembolism revealed that the Akt pathway was up-regulated in the recurrent venous thromboembolism group compared to the single venous thromboembolism group.
Conclusions
In this exploratory study, gene expression profiles of whole blood appear to be a useful strategy to distinguish subjects with single venous thromboembolism from those with recurrent venous thromboembolism. Prospective studies with additional patients are needed to validate these results.
Keywords: genomics, risk factors, deep vein thrombosis
Introduction
Venous thromboembolism (VTE) is a major cause of morbidity and mortality. Each year, approximately 350,000 to 600,000 individuals in the United States will develop a VTE and up to 100,000 will die [1]. Recurrent VTE develops within 8 years in as many as 30% of patients after stopping a standard course of anticoagulant therapy [2]. This fact is consistent with a growing body of data suggesting that VTE is a chronic disease with acute exacerbations (manifested as recurrent thromboembolism). Inherited and acquired risk factors contribute to an individual person’s risk for VTE [3], but these factors are frequently inadequate at predicting who will develop an initial VTE or recurrent VTE. Current evidence suggests against evaluating most patients with spontaneous VTE for thrombophilia [4–5]. As a consequence of this increased risk for recurrent VTE, many patients with VTE would benefit from a longer course of anticoagulant therapy [6]. Furthermore, recent studies in subjects with VTE have shown that an extended course of anticoagulation will decrease the risk of recurrence [7–8]. Determining which patients are at highest risk for recurrence is therefore clearly a vital health concern.
D-dimer has been studied extensively for its effectiveness as a biomarker for recurrent VTE. The PROLONG study found that a normal D-dimer level obtained 1 month after anticoagulation therapy was discontinued following treatment for an unprovoked VTE was associated with a low rate of recurrence [9]. In addition, Cosmi et al. found that an elevated D-dimer either while on warfarin or after stopping warfarin for one month was associated with an increased risk for recurrent VTE [10]. Kyrle and colleagues also followed patients with unprovoked VTE after stopping anticoagulation therapy and found that higher levels of P-selectin were associated with an increased risk for recurrent VTE [11].
Peripheral blood gene expression patterns have been successfully used to predict outcomes in a variety of disease contexts including myocardial infarction and systemic lupus erythematosis [12]. Using whole blood gene expression profiles Julia et al. built and validated a model that predicts response to infliximab in patients with rheumatoid arthritis [13]. This approach has also been used to look at patients with primary antiphospholipid syndrome [14], but most patients with VTE will not have this specific syndrome. Consequently, strategies that are more broadly applicable to patients with VTE are needed.
In this exploratory study, we used a heterogeneous group of patients with VTE to investigate whether selected biomarkers, including D-dimer, P-Selectin, and thrombin-antithrombin complex levels, and/or gene expression information could be used distinguish patients with a single VTE event from patients with recurrent VTE.
Materials and methods
Patient Population
Patients were screened and enrolled through the Duke Anticoagulation and Adult Hemostasis and Thrombosis Clinics [15]. Patients over 18 years of age who had one or more prior venous thromboembolic events were recruited for the study. All patients were on warfarin and were at least 4 weeks from their most recent thromboembolic event. Exclusion criteria included: (1) diagnosis of antiphospholipid syndrome; (2) presence of an active cancer (other than minor skin cancer); (3) more than three years since their most recent VTE; and (4) currently on an anticoagulant other than warfarin, or on no anticoagulation. Documentation for all VTE events was reviewed by the investigators to confirm the number of events, thrombus type (i.e. DVT or PE), thrombus location, and other clinical data. Since this was an exploratory study, patients with spontaneous as well as provoked VTE were included. Blood was also collected from 10 healthy controls. This study was approved by the Duke Institutional Review Board and informed consent was obtained from all subjects enrolled.
D-dimer, P-Selectin, Thrombin-Antithrombin Complexes, and Antiphospholipid Antibody Testing
D-dimers were measured using the Vidas D-dimer exclusion (DD2) assay from bioMerieux (Durham, NC). P-Selectin was measured using the humans P-Selectin/CD62P ELISA kit from R&D systems (Minneapolis, MN) and thrombin-antithrombin (TAT) complexes were determined using the Enzygnost TAT micro ELISA kit from Siemens (Washington, DC). The absence of antiphospholipid and anti-β2-glycoprotein 1 (β2GP1) antibodies in all VTE subjects and healthy controls was confirmed using the Asserachrom APA IgG,M, Asserachrom anti-β2GP1 IgG, and Asserachrom anti-β2GP1 IgM ELISA kits from Diagnostica Stago, Inc. (Parsippany, NJ).
RNA Isolation and Microarray Hybridization
Blood was collected in PAXgene RNA tubes, and RNA was isolated using the Paxgene Blood RNA kit (Qiagen, Valencia, CA). The quality was checked using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). RNA was submitted to Expression Analysis, Inc (Durham, NC) for β-globin reduction and one round of in-vitro transcription. Gene expression data was obtained using Affymetrix HU133A 2.0 Arrays (Affymetrix, Santa Clara, CA). All steps involved in RNA processing, probe preparation, microarray hybridization, and data processing used MIAME (Minimal Information About a Microarray Experiment) compliant guidelines [16].
Data Processing
A comprehensive quality control process was performed on all arrays using Bioconductor [17] extension packages (affyPLM, affyQAReport) for the R statistical environment [18]. The affyQAReport package provides a method for creating and viewing QA reports on an experiment run on Affymetrix CEL files to ensure the inclusion of quality array data prior to the normalization process. affyPLM extends and improves the functionality of the base affymetrix package to implement methods for fitting probe-level models (PLM) and provides PLM modeling based quality assessment tools. In addition to evaluating the standard quality control measures provided by Affymetrix, a series of graphical illustrations (e.g. boxplots of perfect match intensities, density histograms, raw and corrected chip images, MA plots, median absolute deviation plots, and multidimensional scaling plots) were produced and evaluated [19–20]. Also, residual plots from PLM were used to identify blemishes, and artifacts on each chip. The expressions from the arrays were pre-processed using Robust Multichip Averaging (RMA) pre-processing algorithm using PLM summarization (RMAExpress 1.0 release, http://rmaexpress.bmbolstad.com)[21]. The resulting summary expressions were log (base 2) transformed. Some of the pathways analyzed by the score signature program (see below) required MAS5 pre-processed data, which was done using Affymetrix Expression Console Version 1.0.
The samples were submitted for microarray analysis in 2 batches. Batch 1 consisted of 21 samples from VTE subjects (single VTE, 12 samples; recurrent VTE, 9 samples), and batch 2 consisted of 19 samples from VTE subjects (single VTE, 11 samples; recurrent VTE, 8 samples) and 10 samples from healthy controls. Potential batch effects within our VTE patient expression data for the single versus recurrent comparison were adjusted using ComBat [22]. The microarray data files were submitted to the gene expression omnibus (http://www.ncbi.nlm.nih.gov/geo/, accession number GSE19151).
Statistical Analysis
Descriptive statistics of the characteristics of study subjects including means, standard deviations, frequencies, and proportions were performed with SAS Enterprise Guide 4.0 (SAS, Cary, NC). For the descriptive analyses, parametric tests (t-test) were used when data met normality assumptions and non-parametric tests (Mann-Whitney-Wilcoxon) were used when normality assumptions were not met. All p-values are 2 tailed with p < 0.05 considered significant. Bayesian binary probit regression was performed in MATLAB (The Math Works Inc. Natick, MA) using metagenes, defined as linear combinations of genes identified to be associated with a specific phenotype [23]. A metagene classifier consisting of the top 50 probe sets, ranked according to the absolute value of the two-sample t test using a pooled standard error estimate, was developed for each comparison. To guard against over-fitting, leave-one-out cross validation was performed. Each sample was left out of the data set one at a time, the top features were reselected and the model was retrained using the remaining samples. The phenotype of the “left-out” sample was then predicted and the certainty of the classification was calculated based on the model. For model assessment, the Receiver Operating Characteristic (ROC) curves and the corresponding area under the curve (AUC) were generated for each model using Prism 5 (GraphPad software Inc. La Jolla, CA). Mean classification probabilities with 25th and 75th percentiles were calculated for each group using Prism 5. Data for D-dimer, P-Selectin, and TAT were analyzed using the Mann-Whitney-Wilcoxon test in Prism 5.
We performed a limited permutation analysis to further evaluate the sampling distribution of the AUC under the noise model (i.e., none of the probe sets are associated with the phenotype of interest). This was accomplished by shuffling the phenotype labels (single versus recurrent VTE) using a script written for the R statistical environment while holding the expression matrix fixed and then repeating the entire leave-one-out cross validation analysis described above. The resulting ROC and AUC from this could be considered permutation replicates of the corresponding observed ROC and AUC.
Pathways Analysis
GATHER (http://gather.genome.duke.edu)[24] and Gene decks (http://www.genecards.org//index.php?path=/GeneDecks) [25] were used to help understand the gene ontology enrichment, biological annotation and shared descriptors of the 50 gene probes used in the models. We also used an alternative strategy to assess patterns of gene expression in patients with VTE, using expression signatures of pathway activation [26–28]. Gatza et al developed and validated pathway-specific signatures for the following pathways: BCAT, E2F1, EGFR, HER2, INFα, INFγ, PI3K, PR, SRC, AKT, ER, MYC, P53, P63, RAS, and STAT3 using either cells infected with viral vectors expressing one of the above proteins or cell lines of known phenotype [26]. These signatures have been shown to identify tumor samples from patients with cancer that share patterns of pathway activity and exhibit similar clinical and biological properties [26]. We used the binary regression algorithm score signatures implemented in GenePattern to determine if these pathway activities were differentially expressed in patients with single or recurrent VTE.
Results
Clinical Characteristics of Subjects
Clinical characteristics of the subjects with VTE are shown in Table 1. The recurrent VTE group had a significantly longer time between their most recent thrombotic event and enrollment compared to the single event group (P=0.012), and the subjects in the recurrent group (mean age=55.7 years) were slightly older than the subjects in the single event group (mean age = 43.8 years). None of the study subjects had antibodies to phospholipids or β2 GP1 (data not shown).
Table 1.
Characteristics of VTE Subjects
| Single Events (n=23) |
Recurrent Events (n=17) |
|
|---|---|---|
| Female, n (%) | 13 (57%) | 11 (65%) |
| Caucasian, n (%) | 17 (74%) | 11 (65%) |
| Mean age, years (SD) | 43.8 (16)† | 55.7 (17)† |
| Mean BMI,# (SD) | 33.8 (6.8) | 35.3 (7.0) |
| PE, n (%) | 10 (43%) | 8 (47%) |
| Median years from most recent thrombosis, (25th, 75th percentile) | 0.7 (0.4, 1.3)† | 1.8 (0.8, 2.3)† |
| Provoked*, n (%) | 10/23 (43%) | 7/14 (50%) |
| Statin use, n (%) | 3 (13%) | 2 (12%) |
| Aspirin use, n (%) | 2 (9%) | 2 (12%) |
| Hypertension | 7 (30%) | 11 (64%) |
| Diabetes | 1(4%) | 1(6%) |
P<0.05
BMI data was not available for 6 subjects in the single event and 4 subjects in the recurrent event group
Provoked VTE refers to events occurring in patients with acquired risk factors for thrombosis, such as in the post-operative state, prolonged immobilization, during pregnancy, etc. Clinical data on whether the thrombosis was provoked or not was unavailable for 3 subjects in the recurrent group
D-dimer, P-Selectin, and TAT Levels
The recurrent event group had significantly higher D-dimer levels (P= 0.01) as compared to the single event group, but this was primarily due to three individuals (Figure S1 in Supplementary Material). The median (interquartile range for the single event and recurrent events groups are 164 ng/ml (113, 282) and 290 ng/ml (238, 598) respectively. Five of 17 subjects in the recurrent group (29%) had elevated D-dimer levels (>0.500 ng/ml) as compared to 2 of 23 in the single group (9%). The P-Selectin and TAT levels of the single VTE event and recurrent VTE event groups were not significantly different (data not shown).
Genomic Comparison of Patients with Single VTE and Recurrent VTE
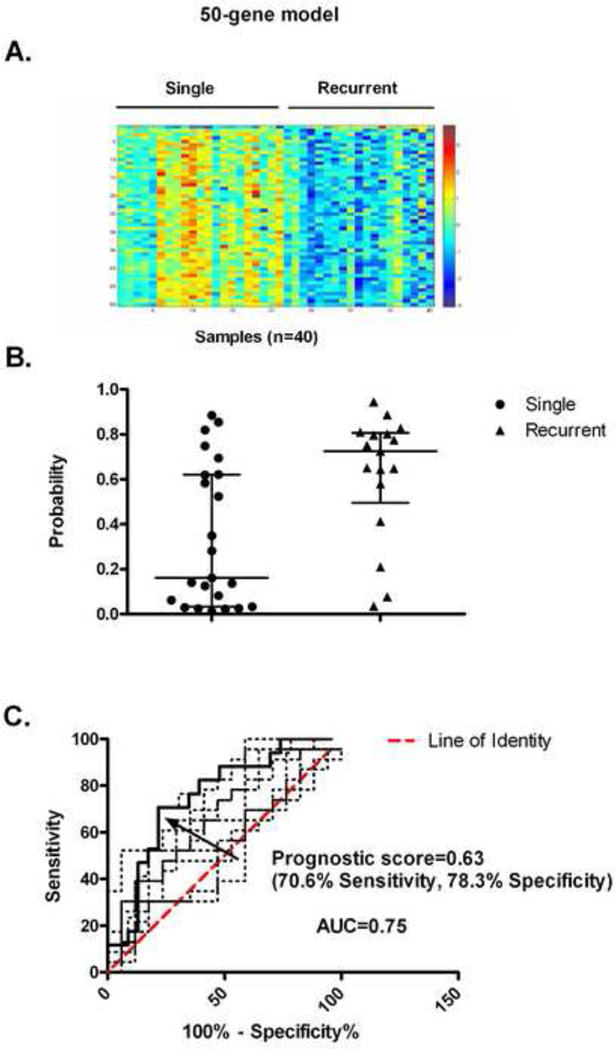
Using binary regression, we developed a 50 gene probe model that distinguished subjects with a single VTE event from subjects with recurrent VTE. The heat map for this model revealed that the expression of these 50 genes was generally increased in the single VTE group relative to the recurrent VTE group (Fig 1A). After performing a leave-one-out cross validation, we found that there was a significant difference in the average probability scores of the 2 groups. For the single VTE event group the median (25th percentile, 75th percentile) score was 0.16 (0.033, 0.62) while for the recurrent event group the median (25th percentile, 75th percentile) score was 0.72 (0.49, 0.81) (figure 1B). The ROC curve is shown in Figure 1C with an AUC of 0.75 and a 95% confidence interval of 0.60 to 0.90. The optimal probability score cut point was 0.63 which corresponds to a sensitivity of 70.6% and specificity of 78.3% and a likelihood ratio of 3.25. The ROC curves for 10 random permutations where the phenotype labels were randomly assigned to single or recurrent VTE are shown in Figure 1C. Eight of the 10 random permutations had AUC’s less than the model (0.75). A complete list of genes from this model can be found in Table S1 (Supplementary Material). When various characteristics of these 50 genes were analyzed by Gather [24] , the only significant enrichment was in the gene ontology terms of RNA spliceosome assembly and mRNA splice site selection. However, there is no significant enrichment in chromosomal locations, transfection factor binding sites or KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. Several of the genes used in the model are potentially related to thrombotic risk. Two genes, IGF1R and PPARD, are involved in platelet aggregation [29–30], ten genes are involved in immune and inflammatory responses (e.g, gene ITPR2, LTB), and eight genes have been associated with oncogenesis (e.g, gene PHF20, UBE2D2).
Figure 1.
Development of the gene model to distinguish subjects with a single VTE from those with recurrent VTE. (A) The heat map representing the expression of the 50 genes probes used to develop the metagenes model is shown with blue and red representing extremes of expression with visually apparent differences in gene expression. Samples from single VTE subjects are on the left (n=23), and samples from subjects with recurrent VTE are on the right (n=17). (B) Leave-one-out cross validation: a comparison of the individual and median (with interquartile range) estimated classification probability of recurrent VTE predicted by the 50 gene model is shown. (C) Leave-one-out cross validation: the ROC curve identifying the probability score of 0.63 as the optimal cut-point to be used to classify samples is shown (black solid line). The area under the curve (AUC) is 0.75. The ROC curves for the 10 random permutations are shown as dotted lines.
Pathway Signature Analysis
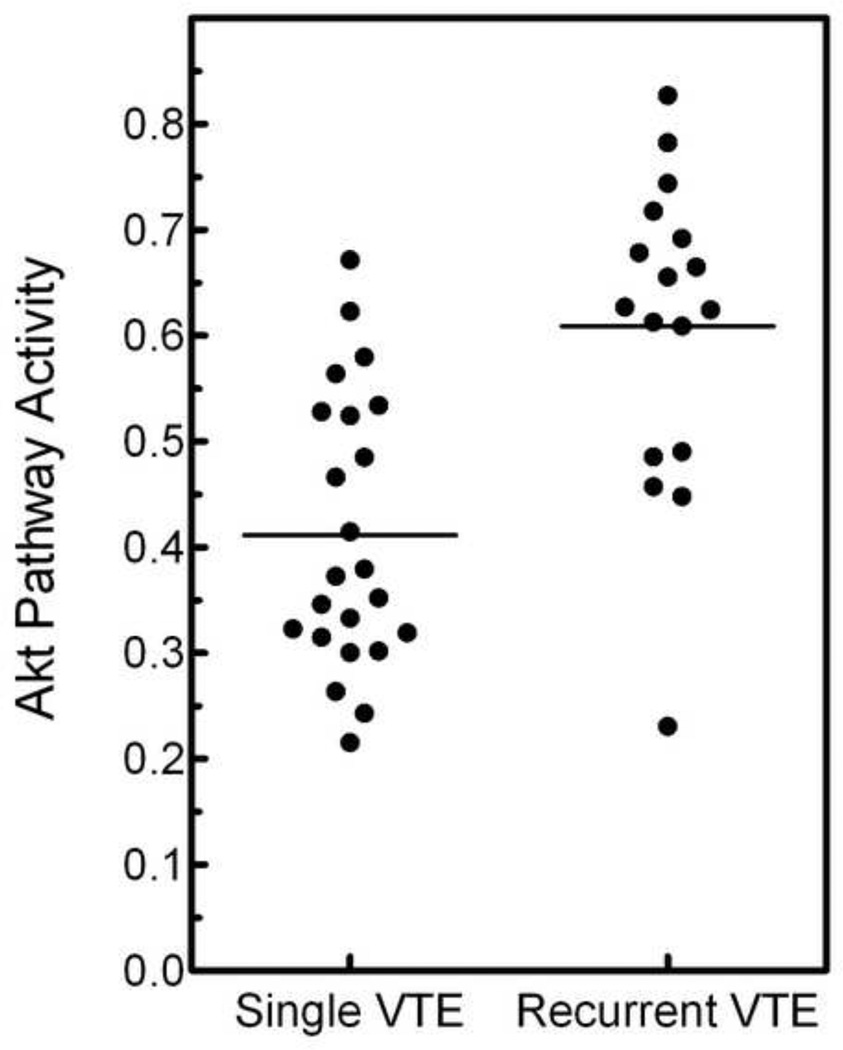
As an alternative strategy to review the data, we investigated whether expression signatures associated with pathway-specific activation were differentially expressed in patients with single versus recurrent VTE. A binary regression algorithm was used to predict the probability of pathway activity for 16 pathways. The average activity for each pathway in the single and recurrent groups is listed in Table 2. Three pathway-specific signatures, AKT, E2F1 and P53, are differentially expressed in the single and recurrent groups (p < 0.05), but the results for the Akt pathway are clearly the most significant (p = 0.0001). The predicted Akt pathway activity of each VTE subject is shown in figure 2 with an average predicted Akt pathway activity for the single VTE group of 0.411, compared to an average predicted Akt pathway activity for the recurrent VTE group of 0.609. The AKT signature contains 250 gene probes [26] and has one gene probe (Golga8A) in common with the single versus recurrent VTE comparison. The entire Akt signature can be accessed at: http://data.genome.duke.edu/breast_subgroups.
Table 2.
Pathway Signature Analysis
| Pathway | Single VTE (average pathway activity) |
Recurrent VTE (average pathway activity) |
P value |
|---|---|---|---|
| BCAT | 0.441 | 0.509 | 0.356 |
| E2F1 | 0.416 | 0.658 | 0.010 |
| EGFR | 0.492 | 0.506 | 0.843 |
| HER2 | 0.582 | 0.419 | 0.109 |
| IFNα | 0.385 | 0.514 | 0.252 |
| IFNγ | 0.447 | 0.512 | 0.520 |
| PI3K | 0.469 | 0.549 | 0.108 |
| PR | 0.522 | 0.479 | 0.436 |
| SRC | 0.566 | 0.456 | 0.210 |
| AKT | 0.411 | 0.609 | 1.09×10−4 |
| ER | 0.513 | 0.460 | 0.362 |
| MYC | 0.542 | 0.448 | 0.081 |
| P53 | 0.552 | 0.453 | 0.036 |
| P63 | 0.480 | 0.545 | 0.249 |
| RAS | 0.445 | 0.528 | 0.086 |
| STAT3 | 0.506 | 0.458 | 0.346 |
Figure 2.
Akt pathway activities for the single and recurrent VTE subjects. The mean and standard error for each is shown.
Discussion
Current methods for identifying patients at high risk for recurrent VTE are limited. D-dimer is the most studied biomarker in this area, and a recent systematic review found that the risk for recurrent VTE in patients with an elevated D-dimer level is 2.2 times that in patients with a normal D-dimer level [31]. Consistent with these results, we found that D-dimer levels were higher in the study subjects with recurrent VTE compared to those individuals with a single VTE. However, only five of 17 subjects with recurrent VTE actually had an elevated D-dimer level, suggesting that using D-dimer levels alone in patients on warfarin therapy would not be sufficient to identify patients at risk for recurrent VTE.
We used two strategies to investigate whether data from whole blood gene expression analyses could distinguish patients with a single VTE from patients with recurrent VTE. First, using binary regression we developed a gene expression model with good ability to distinguish these two groups of patients (ROC curve AUC = 0.75). Leave-one-out cross validation and a limited random permutation test (Figures 1B and 1C) supported that this model was able to distinguish these two groups. In addition, several genes from the single versus recurrent VTE gene expression model are involved in either platelet function or structure, immune or inflammatory responses, or cancer, adding biological plausibility in support of this model. For example, the most differentially expressed gene was insulin-like growth factor receptor 1, which is found in the alpha granules of platelets.
Our second strategy involved using pathway-specific activation signatures applied to the gene expression data from our patients with single and recurrent VTE. This revealed that the Akt pathway is up-regulated in patients with recurrent VTE compared to individuals with single events. Studies with human and mouse platelets have shown that Akt plays a role in modulating platelet responses and has an important role in hemostasis. Chen and colleagues, using Akt-1-deficient mice, found that Akt-1 activity was required for optimal platelet integrin αIIbβ3 activation, thrombin and collagen induced platelet aggregation and normal tail bleeding times [32]. Using human platelets, Resendiz et al found that activated Akt regulated platelet function by modulating secretion and integrin αIIbβ3 activation [33]. Our finding that the Akt pathway is over expressed in the recurrent VTE group is potentially consistent with a prothrombotic role of this pathway [34].
The D-dimer, binary regression analysis of gene expression profiles and the pathway specific activation analysis all support the hypothesis that these markers would be useful in analyzing patients with VTE. Similar to the ROC analysis for the binary regression analyses (Figure 1C), ROC analyses for the D-dimer and Akt pathway demonstrated that all three methods were comparable in their ability to identify patients with recurrent thrombosis (Supplemental Figures S2 and S3, respectively).
Although these data support the hypothesis that whole blood gene expression analysis and D-dimer would be useful in the analysis of patients with VTE, several limitations of this study merit discussion. First, the patient population was a heterogeneous mixture of patients with provoked and non-provoked VTE, although the relative distributions of patients with provoked and non-provoked VTE were similar in the two groups (Table 1). Second, the two groups of patients also differed in the duration of time since their last VTE as well as duration of time on warfarin therapy. Third, some patients with a single VTE would likely sustain a recurrent event if anticoagulant therapy was stopped, resulting in reclassification of any affected individual patients. Even with this possibility, our current phenotype assignment still accurately represents the majority of individuals with single and recurrent VTE phenotypes seen in a typical clinical practice. Additional cohorts are needed to confirm the accuracy of the signatures obtained in these initial comparisons.
Despite these limitations, our results support the hypothesis that whole blood gene expression analyses can distinguish patients with single VTE from patients with recurrent VTE. A larger prospective study with an independent validation set needs to be performed to confirm these results. Enrolling patients after a first unprovoked VTE and following them for several years would provide a more robust dataset.
Supplementary Material
Figure S1. D-dimer levels in subjects with single vs. recurrent VTE. The horizontal bar indicates the median. The mean result from 9 normal donors was 306 ng/mL (median, 276 ng/mL).
Figure S2. ROC Curve for D-dimer levels in subjects waith single vs. recurrent VTE. The ROC curve is shown as a solid line and the line of identity is shown as a dashed line. The AUC is 0.74
Figure S3. ROC Curve for Akt pathway activities for the single and recurrent VTE subjects. The ROC curve is shown as a solid line and the line of identity is shown as a dashed line. The AUC is 0.84.
Acknowledgments
The authors thank: Ara Metjian, MD, and Richard Becker, MD for reviewing the manuscript; William Barry, PhD and Joseph Lucas, PhD for statistical help analyzing microarray data; Jeffrey Chang, PhD and Joseph Nevins, PhD for assistance with pathways analysis in GenePattern, Sheila Lambert-Adams and Christine Mette for enrolling patients; Andrew Hertzberg, Sharon Hall, Mary Pound and Keith Klemp for performing the D-dimer and antiphospholipid and anti-β2GP1 assays; Gwyn Cutsforth, PhD from Diagnostica Stago for providing the anti-β2GP1 ELISA kits; and Linda Karolak from bioMerieux for providing the D-dimer kits. This work was supported by the Centers for Disease Control and Prevention, UO1-DD000014 (TLO), the National Institutes of Health, T32-HL007538 (GS), R21DK080994 (JTC) and U54-HL077878 (TLO) and the Duke IGSP Scholars Program (TLO) and a grant from LabCorp (TLO, JTC).
Abbreviations
- VTE
venous thromboembolism
- DVT
deep vein thromboembolism
- PE
pulmonary embolism
- β2GP1
β2-glycoprotein
- MIAME
Minimal Information About a Microarray Experiment
- RMA
Robust Multichip Averaging
- ROC
Receiver Operating Characteristic
- AUC
area under the curve
- TAT
thrombin-antithrombin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
None of the authors have any conflicts of interest to declare.
References
- 1.Beckman MG, Hooper WC, Critchley SE, Ortel TL. Venous thromboembolism: a public health concern. Am J Prev Med. 2010;38:S495–S501. doi: 10.1016/j.amepre.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Heit JA, Mohr DN, Silverstein MD, Petterson TM, O'Fallon WM, Melton LJ., III Predictors of Recurrence After Deep Vein Thrombosis and Pulmonary Embolism: A Population-Based Cohort Study. Arch Intern Med. 2000;160:761–768. doi: 10.1001/archinte.160.6.761. [DOI] [PubMed] [Google Scholar]
- 3.Lopez JA, Kearon C, Lee AY. Deep venous thrombosis. Hematology Am Soc Hematol Educ Program. 2004:439–456. doi: 10.1182/asheducation-2004.1.439. [DOI] [PubMed] [Google Scholar]
- 4.Bauer KA. The thrombophilias: well-defined risk factors with uncertain therapeutic implications. Ann Intern Med. 2001;135:367–373. doi: 10.7326/0003-4819-135-5-200109040-00013. [DOI] [PubMed] [Google Scholar]
- 5.Lindhoff-Last E, Luxembourg B. Evidence-based indications for thrombophilia screening. Vasa. 2008;37:19–30. doi: 10.1024/0301-1526.37.1.19. [DOI] [PubMed] [Google Scholar]
- 6.Schafer AI. Venous thrombosis as a chronic disease. N Engl J Med. 1999;340:955–956. doi: 10.1056/NEJM199903253401209. [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM, Goldhaber SZ, Danielson E, Rosenberg Y, Eby CS, Deitcher SR, et al. Long-Term, Low-Intensity Warfarin Therapy for the Prevention of Recurrent Venous Thromboembolism. N Engl J Med. 2003;348:1425–1434. doi: 10.1056/NEJMoa035029. [DOI] [PubMed] [Google Scholar]
- 8.Kearon C, Ginsberg JS, Kovacs MJ, Anderson DR, Wells P, Julian JA, et al. Comparison of low-intensity warfarin therapy with conventional-intensity warfarin therapy for long-term prevention of recurrent venous thromboembolism. N Engl J Med. 2003;349:631–639. doi: 10.1056/NEJMoa035422. [DOI] [PubMed] [Google Scholar]
- 9.Palareti G, Cosmi B, Legnani C, Tosetto A, Brusi C, Iorio A, et al. d-Dimer Testing to Determine the Duration of Anticoagulation Therapy. New England Journal of Medicine. 2006;355:1780–1789. doi: 10.1056/NEJMoa054444. [DOI] [PubMed] [Google Scholar]
- 10.Cosmi B, Legnani C, Cini M, Guazzaloca G, Palareti G. D-dimer and residual vein obstruction as risk factors for recurrence during and after anticoagulation withdrawal in patients with a first episode of provoked deep-vein thrombosis. Thromb Haemost. 2011;105:837–845. doi: 10.1160/TH10-08-0559. [DOI] [PubMed] [Google Scholar]
- 11.Kyrle PA, Hron G, Eichinger S, Wagner O. Circulating P-selectin and the risk of recurrent venous thromboembolism. Thromb Haemost. 2007;97:880–883. [PubMed] [Google Scholar]
- 12.Aziz H, Zaas A, Ginsburg GS. Peripheral blood gene expression profiling for cardiovascular disease assessment. Genomic Med. 2007;1:105–112. doi: 10.1007/s11568-008-9017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julia A, Erra A, Palacio C, Tomas C, Sans X, Barcelo P, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4:e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernales I, Fullaondo A, Marin-Vidalled MJ, Ucar E, Martinez-Taboada V, Lopez-Hoyos M, et al. Innate immune response gene expression profiles characterize primary antiphospholipid syndrome. Genes Immun. 2008;9:38–46. doi: 10.1038/sj.gene.6364443. [DOI] [PubMed] [Google Scholar]
- 15.Dowling NF, Beckman MG, Manco-Johnson M, Hassell K, Philipp CS, Michaels LA, et al. The US Thrombosis and Hemostasis Centers pilot sites program. J Thromb Thrombolysis. 2007;23:1–7. doi: 10.1007/s11239-006-9002-y. [DOI] [PubMed] [Google Scholar]
- 16.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team. R: A Language and Environment for Statistical Computing, 2009 http:www.R-project.org. [Google Scholar]
- 19.Gentleman R, Carey VJ, Huber W, Irizarry R, Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. XIX ed. Springer; 2005. [Google Scholar]
- 20.Hahne F, Huber W, Gentleman R, Falcon S. Bioconductor Case Studies. Springer; 2008. [Google Scholar]
- 21.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 22.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 23.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 25.Stelzer G, Inger A, Olender T, Iny-Stein T, Dalah I, Harel A, et al. GeneDecks: paralog hunting and gene-set distillation with GeneCards annotation. OMICS. 2009;13:477–487. doi: 10.1089/omi.2009.0069. [DOI] [PubMed] [Google Scholar]
- 26.Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci U S A. 2010;107:6994–6999. doi: 10.1073/pnas.0912708107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, et al. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- 28.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 29.Hers I. Insulin-like growth factor-1 potentiates platelet activation via the IRS/PI3Kalpha pathway. Blood. 2007;110:4243–4252. doi: 10.1182/blood-2006-10-050633. [DOI] [PubMed] [Google Scholar]
- 30.Ali FY, Davidson SJ, Moraes LA, Traves SL, Paul-Clark M, Bishop-Bailey D, et al. Role of nuclear receptor signaling in platelets: antithrombotic effects of PPARbeta. Faseb J. 2006;20:326–328. doi: 10.1096/fj.05-4395fje. [DOI] [PubMed] [Google Scholar]
- 31.Verhovsek M, Douketis JD, Yi Q, Shrivastava S, Tait RC, Baglin T, et al. Systematic review: D-dimer to predict recurrent disease after stopping anticoagulant therapy for unprovoked venous thromboembolism. Ann Intern Med. 2008;149:481–490. doi: 10.7326/0003-4819-149-7-200810070-00008. W94. [DOI] [PubMed] [Google Scholar]
- 32.Chen J, De S, Damron DS, Chen WS, Hay N, Byzova TV. Impaired platelet responses to thrombin and collagen in AKT 1-deficient mice. Blood. 2004;104:1703–1710. doi: 10.1182/blood-2003-10-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resendiz JC, Kroll MH, Lassila R. Protease-activated receptor-induced Akt activation--regulation and possible function. J Thromb Haemost. 2007;5:2484–2493. doi: 10.1111/j.1538-7836.2007.02769.x. [DOI] [PubMed] [Google Scholar]
- 34.Luyendyk JP, Schabbauer GA, Tencati M, Holscher T, Pawlinski R, Mackman N. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol. 2008;180:4218–4226. doi: 10.4049/jimmunol.180.6.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. D-dimer levels in subjects with single vs. recurrent VTE. The horizontal bar indicates the median. The mean result from 9 normal donors was 306 ng/mL (median, 276 ng/mL).
Figure S2. ROC Curve for D-dimer levels in subjects waith single vs. recurrent VTE. The ROC curve is shown as a solid line and the line of identity is shown as a dashed line. The AUC is 0.74
Figure S3. ROC Curve for Akt pathway activities for the single and recurrent VTE subjects. The ROC curve is shown as a solid line and the line of identity is shown as a dashed line. The AUC is 0.84.