Abstract
Leaf senescence varies greatly among genotypes of cotton (Gossypium hirsutium L), possibly due to the different expression of senescence-related genes. To determine genes involved in leaf senescence, we performed genome-wide transcriptional profiling of the main-stem leaves of an early- (K1) and a late-senescence (K2) cotton line at 110 day after planting (DAP) using the Solexa technology. The profiling analysis indicated that 1132 genes were up-regulated and 455 genes down-regulated in K1 compared with K2 at 110 DAP. The Solexa data were highly consistent with, and thus were validated by those from real-time quantitative PCR (RT-PCR). Most of the genes related to photosynthesis, anabolism of carbohydrates and other biomolecules were down-regulated, but those for catabolism of proteins, nucleic acids, lipids and nutrient recycling were mostly up-regulated in K1 compared with K2. Fifty-one differently expressed hormone-related genes were identified, of which 5 ethylene, 3 brassinosteroid (BR), 5 JA, 18 auxin, 8 GA and 1 ABA related genes were up-regulated in K1 compared with K2, indicating that these hormone-related genes might play crucial roles in early senescence of K1 leaves. Many differently expressed transcription factor (TF) genes were identified and 11 NAC and 8 WRKY TF genes were up-regulated in K1 compared with K2, suggesting that TF genes, especially NAC and WRKY genes were involved in early senescence of K1 leaves. Genotypic variation in leaf senescence was attributed to differently expressed genes, particularly hormone-related and TF genes.
Introduction
Senescence is the age-dependent end of the life span. In plants, it is characterized by the visible yellowing of leaves that accompanies the mobilization of leaf nutrients to the reproductive structures. The yellowing of senescing leaves is correlated with a series of biochemical changes such as loss of chlorophyll contents, degradation of proteins and RNA, and a decline in photosynthetic activity. Because accelerated leaf senescence curtails carbon assimilation, plant growth and yield are reduced [1], [2]. As the final stage of plant development, senescence has a crucial impact on agriculture, especially in crop production where crop yield is enhanced by longer growth periods. As for cotton, senescence may occur too early or too late in the season due to environmental stresses or internal factors [3], [4]. Too early senescence of whole cotton plant is referred to as premature senescence, which has been occurring on an increasing scale since modern transgenic Bacillus thuringiensis Berliner (Bt) cotton (Gossypium hirsutum L.) cultivars were introduced for commercial production [5], [6]. Wright [7], [8] indicated that premature senescence frequently developed during the period of rapid boll filling and this reduced lint yield and fiber quality, thus constituting an important constraint to cotton yield and quality.
During leaf senescence, viability of cells within the leaf is actively maintained until maximum remobilization has occurred [9]. This requires meticulous control of each step of the process, regulated by internal and external signals via a series of interlinking signaling pathways involving gene expression changes and influenced by the balance of hormones and metabolites [10]. Thus, senescence is a very complex process involving the expression of thousands of genes and many signaling pathways [10]–[12]. Elucidation of the relative influences of each pathway and the crosstalk between them is crucial to identify the key regulatory genes that control senescence [10].
Plant hormones play key roles in responses to senescence. Senescence is accelerated by the hormones ethylene, abscisic acid (ABA), and jasmonic acid (JA) that mediate plant responses to biotic and abiotic stresses. Exogenous ethylene enhances visible leaf yellowing and several ethylene biosynthesis genes are up-regulated in senescing leaves [12]–[14]. Ethylene- insensitive mutants such as ethylene-resistant 1 (etr1) and ethylene-insensitive 2 (ein2) display delayed leaf senescence [15], [16]. Similarly, the exogenous application of ABA accelerates leaf senescence [14], [17] and the level of ABA also increases during senescence [18], [19]. In addition, exogenous methyl jasmonic acid has been reported to accelerate leaf senescence. The JA-insensitive mutant, coronatine insensitive 1 (coi1) fails to display JA-dependent senescence [20]. Elevated cytokinin levels accompany delayed senescence, and endogenous cytokinin levels decrease during leaf senescence [19], [21]. Ectopic overproduction of cytokinins was shown to delay leaf senescence in tobacco, petunia, cassava and lettuce [22]–[26]. The induction of the senescence programme is characterized by up-regulation of a set of signature genes that are referred to as Senescence Associated Genes (SAGs) which include genes encoding specific catabolic enzymes and transcription factors. Accordingly, leaf senescence induced by the hormones ABA, JA, and ethylene is characterized by the induction of the expression of some SAGs whereas the delay of senescence by cytokinin is characterized with reduced expression of some SAGs [14], [27].
Auxins play important roles in multiple aspects of plant development and growth, including apical dominance, vascular differentiation, and shoot elongation. However, the role of auxin in regulating senescence is not as clear as that of cytokinins, ABA, and ethylene. Several earlier studies revealed that exogenous application of auxin delayed leaf blade abscission in bean [28], [29]. The exogenous application of auxin represses the transcription of senescence-response gene SAG12 [30], and mutation of the ARF2 and ARF1 can delay senescence and SAG12 expression [31]–[33]. The dominant activation mutant, yuc6-1D and 35S:YUC6 transgenic plants, which have been shown to contain an elevated free IAA level and to display typical high-auxin phenotypes, exhibit a delayed senescence phenotype [34]. On the other hand, auxin can stimulate the biosynthesis of senescence-promoting hormones such as ethylene and ABA [35], [36]. Further, the concentration of free IAA in senescing leaves of Arabidopsis was 2-fold higher than in non-senescing leaves [37], suggesting that auxin might have a senescence-promoting effect or accumulate as a consequence of senescence.
Salicylic acid (SA) plays a key role as a mediator of plant stress responses, including disease and systemic acquired resistance. It was also reported that the SA-signaling pathway was active in the control of developmental senescence [38]. A transcriptome analysis in senescing Arabidopsis leaves from wild-type plants and SA-deficient NahG mutants revealed that many SAGs are dependent on the SA-signaling pathway [11] and many SA related genes were up-regulated during senescence [12]. The roles of GA in leaf senescence are still not clear. van der Graaff et al. [12] reported that GA 2-oxidase 2 (AtGA2OX2) that deactivates GA was up-regulated during leaf senescence, suggesting that at least some GAs are deactivated during Arabidopsis leaf senescence. Greenboim-Wainberg et al. [39] reported that SPY acts as both a repressor of GA responses and a positive regulator of cytokinin signaling and plays a central role in the regulation of GA/cytokinin cross talk during plant senescence. Brassinosteroids appear to promote developmental senescence, as mutants deficient in BR biosynthesis or the BR receptor BRI1 have a retarded senescence progression [40]. Most of the BR related genes were up-regulated during senescence in Arabidopsis [12].
About 100 TFs belonging to the APETALA2, basic-leucine zipper, MYB, NAC (NAM/ATAF1, 2/CUC2), WRKY, zinc finger, and GRAS families are differentially regulated during leaf senescence [11]. Many of the TFs involved in the developmental processes also play diverse roles in hormone signaling, stress responses, and metabolism, suggesting that extensive signaling crosstalk exists between intrinsic aging programs and environmental stress signals [41]–[43]. The NAC TFs constitute one of the largest transcription factor families in plants and Arabidopsis genome contains about 106 NAC members involved in diverse developmental processes and biotic and abiotic stress responses [44], [45]. The NAC TFs also constitute a large fraction of senescence- regulated genes in many plant species [11], [46]–[48]. The Arabidopsis NAP (NAC-LIKE, ACTIVATED BY AP3/PI) gene is transcriptionally regulated during senescing of Arabidopsis leaves [49]. Overexpression of NAP can lead to premature senescence, whereas the NAP-deficient mutant exhibits delayed leaf senescence. Notably, Os NAP and Pv NAP, restore the delayed leaf senescence in the At NAP-deficient mutant, showing that the NAP genes are conserved in diverse plant species [49]. At ORE1 and At ORS1 have been identified as nonredundant positive regulators of senescence, because inhibiting them individually delays leaf senescence [48], [50]. ABA responsive NAC transcription factor VNI2 (VND-INTERACTING2) can integrate ABA- mediated abiotic stress signals into leaf aging by regulating a subset of COR (COLD-REGULATED) and RD (Responsive TO Dehydration) genes [43]. In addition, spatial and temporal expression patterns of the VNI2 gene are correlated with leaf aging and senescence. Accordingly, leaf aging was delayed in transgenic plants overexpressing the VNI2 gene but significantly accelerated in a VNI2-deficient mutant [43]. A hydrogen peroxide induced NAC transcription factor JUB1 gene was also reported to regulate Arabidopsis senescence [51]. Leaf aging was delayed in transgenic plants overexpressing JUB1 gene but significantly accelerated in JUB1 deficient mutant, suggesting that JUB1 is a negative regulator of senescence [51].
Our previous study showed that removal of early fruiting branches delayed leaf senescence but main-stem girdling accelerated leaf senescence in cotton. Leaf cytokinin content was increased in fruit-removal plants but decreased in girdled plants; in contrast, the ABA content was decreased in fruit removal plants and increased in girdled plants [6], [52]. Our previous results also suggested that the cotton line, K1 senescence earlier than K2 due to its lower cytokinin and higher ABA contents [19], [52]. Although studies have indicated possible correlation of leaf senescence with cytokinins and ABA in cotton [6], [19], [52], the expression profiles of senescence-related genes, particularly hormone related and TF genes and their involvement in senescence regulation in cotton have not been documented. In the present study, we compared the difference between the transcriptomes of main-stem leaves obtained from an early senescence (K1) and a late senescence (K2) cotton using the Solexa sequencing system during leaf senescence. By investigating changes in the expression of genes that contribute to senescence, a number of candidate genes related to cotton senescence were identified.
Materials and Methods
Plant Material and Growth Conditions
An early-senescence (K1) and a late-senescence (K2) cotton line with the same genetic background, developed by the Cotton Research Center, Shandong Academy of Agricultural Sciences, Jinan, were used in the experiment. Our previous trial showed that both K1 and K2 grew and developed in a similar time-course and performed similarly in shoot and root weights, shoot: root ratio, and plant biomass. However, leaf senescence based on chlorophyll content and photosynthesis in the late season differed significantly between the two cotton lines [19]. Leaves of K1 do senesce earlier than those of K2 after flowering. So the fourth leaves from the apex of both lines at 110 DAP were selected for gene expression profiling.
Acid-delinted seeds of each transgenic cotton line were sown in plastic pots (50 cm in height and 40 cm in diameter) filled with fertile soil (1.2% organic matter, 500 mg kg−1 total N, 15 mg kg−1 available P, and 120 mg kg−1 available K) on April 20, 2012, and allowed to germinate and grow in a greenhouse (30/20°C). When the first fully expanded true leaf occurred, seedlings were thinned to one per pot and allowed to grow under natural environment conditions. Potted plants were watered to 75% of the field water capacity daily to minimize water stress, and fertilized with 5 g compound fertilizer (25% N, 25% P, and 20% K) per pot at flowering stage. The fourth leaves from the apex on the main-stem of both lines were harvested separately at 65, 80, 95 and 110 DAP, frozen in liquid nitrogen and stored at −80°C for RNA extraction.
RNA Extraction
Total RNA was extracted using the TRIzol reagent (Invitrogen), and mRNA was isolated from total RNA using Dynabeads Oligo (dT) (Invitrogen Dynal), following the manufacturer’s instructions. Dried RNA samples were dissolved in diethylpyrocarbonate- treated H2O, and the concentration determined spectroscopycally. The quality of the RNA was checked on 1.0% denaturing agarose gels. For Solexa sequencing, total RNA from 6 representative individual plants of each line was mixed together.
Solexa Sequencing
At least 20 µg of total RNA was sent to Beijing Genomics Institute for Solexa sequencing (commercial service). Initially, poly (A)-containing mRNA molecules were purified from total RNA using poly (T) oligo-attached magnetic beads. Following the purification step, mRNA was fragmented into small pieces using divalent cations at elevated temperatures. The cleaved RNA fragments were then copied into first-strand cDNA fragment using reverse transcriptase and a high concentration of random hexamer primers. This was followed by a second-strand cDNA synthesis using DNA polymerase I and RNaseH. Finally, the short cDNA fragments were prepared for Solexa sequencing on Illumina HiSeq™ 2000, using the manufacturer’s protocol and reagents of the genomic DNA sequencing sample prep kit. The read lengths were 49 bp for leaf samples of both cotton lines.
The Illumina/Solexa approach involved sequencing of cDNA fragments and counting the number of particular fragments. The terminators were labeled with fluorescent compounds of four different colors to distinguish among the different bases at the given sequence position. The template sequence of each cluster was deduced by reading the color at each successive nucleotide addition step. We obtained 12.5 and 12.2 million reads from Illumina/Solexa sequencing for K1 and K2 cotton lines, and deposited them in Sequence Read Archive (SRA) database of GenBank under the accession no. SRR867611. At present, the full genome sequence for cotton is not available. The overall approach to non-model organism transcriptome analysis (Fig. S1) was to use high-throughput short-read sequences, optimize assembly and mapping parameters using partial data, and then process the total data using these optimized mapping and de novo parameters. Assembled contigs were compared with themselves and with the nominated reference EST using BLAST, leading to the extraction of candidate duplicate genes [53]. The results were visualized at different stages for validation purposes [54].
Gene Annotation
mRNA and EST of cotton (ftp://ftp.ncbi.nih.gov/repository/UniGene/Gossypium_ hirsutum/Ghi.seq.uniq.gz) were used as a reference sequence to align and identify the sequencing reads. To map the reads to the reference, the alignments and the candidate gene identification procedure were conducted using the mapping and assembly with qualities software package [55]. This was done essentially by following the protocols described in the online documentation (http://maq.sourceforge.net) and adopting the default parameter values. Briefly, the reference sequences were converted to binary FASTA format, and each Solexa read data subset (corresponding to one lane on the instrument) was transformed from Solexa FASTQ to Sanger FASTQ format. As recommended, each subset was separately mapped to the reference, and these maps were then merged to form general maps for assembling the consensus sequences (Fig. S1) [53].
Identification of Differentially Expressed Genes and Functional Analysis
To eliminate the influence of different gene length and sequencing level on the calculation, the RPKM method was used for normalization and the result can be directly used for comparing differences in gene expression between samples. The expression of genes was calculated by RPKM (Reads Per kb per Million reads) method [56] according to the formula:
Where C is the number of reads that uniquely aligned to the gene, N is the total number of reads that uniquely aligned to all genes in the specific sample, and L is number of bases of the gene. The P-value corresponding to differential transcript expression in two samples was determined from Audic’s algorithm [57], and FDR (False Discovery Rate) method was applied to determine the threshold of P-values in multiple tests. We used ‘‘FDR ≤0.001 and the absolute value of log2Ratio ≥1” as the threshold to determine the significance of gene expression difference [58].
GO enrichment analysis was performed for functional categorization of differentially expressed transcripts using agriGO software [59] and the P-values corrected by applying the FDR correction to control falsely rejected hypotheses during GO analysis. The pathway analysis was conducted using KEGG (www.genome.jp/kegg/).
Real-time PCR (RT-PCR) Analysis
The expression of some important genes was determined using RT-PCR. The leaves from 6 representative individual plants of each line were harvested and every 2 leaves were combined into one biological replicate and then extracted for total RNA using the TRIzol reagent (Invitrogen). A first-strand cDNA fragment was synthesized from total RNA using Superscript II reverse transcriptase (Invitrogen). Gene-specific primers were designed according to the gene sequences using the Primer Premier 5.0 (Premier Biosoft International, Palo Alto, CA) and then synthesized commercially (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China). The primers are listed in Fig. S2. The amplification of β-actin was used as an internal control to normalize all data. 20 µL samples were run in triplicate on a Bio-red IQ2 Sequence Detection System and Applied Biosystems software using 0.1 µL first-strand cDNAs and SYBR Green PCR Master Mix (Applied Biosystems). Thermal cycling was performed at an initial denaturation step at 95°C for 3 min followed by 40 cycles at 95°C for 10 s, at annealing temperatures of 60°C for 10 s, and at 72°C for 10 s. Relative quantization of gene expression was calculated and normalized to β-actin.
Results
Seedling emergence, squaring, flowering, peak flowering, peak boll-setting, and boll-opening for both K1 and K2 occurred at about 6, 40, 60, 75, 90, and 110 DAP. The growth and development of both lines followed a similar time-course and growth performance was considerably similar. However, leaf senescence based on chlorophyll content and photosynthesis in the late season differed significantly between the two lines (Fig. S3). The chlorophyll content and photosynthesis of K2 were considerably higher than those of K1 at 110 DAP.
Solexa Sequencing and Assessment
We generated 12.5 and 12.2 million reads from one lane of Illumina/Solexa sequencing for K1 and K2 lines. Prior to mapping these sequencing reads to the reference mRNA and expressed sequence tag (EST), only adaptor reads, containing N reads and low-quality reads were filtered. After filtration, 12.4 and 12.1 million usable reads for K1 and K2 were obtained. Since the full genome sequence for cotton is not available, mRNA and EST contigs of cotton (ftp://ftp.ncbi.nih.gov/repository/UniGene/Gossypium_hirsutum/Ghi.seq.uniq.gz) were used as reference sequences to align and identify the sequencing reads. This allowed for the mapping of 44.01 and 44.19% of the K1 and K2 reads that passed our filters, representing about 5.45 and 5.36 million reads (Table 1).
Table 1. Total number of sequencing reads obtained from each sample.
| Early-senescence cotton line | Late-senescence cotton line | |||
| Map to gene | Read number | Percentage | Read number | Percentage |
| Total good reads | 12392329 | 100% | 12135428 | 100.00% |
| Total mapped reads | 5454055 | 44.01% | 5362278 | 44.19% |
| unique match | 5167284 | 41.70% | 5091444 | 41.96% |
| multi-position match | 286771 | 2.31% | 270834 | 2.23% |
| Total Unmapped Reads | 6938274 | 55.99% | 6773150 | 55.81% |
Identification and Functional Classification of Differentially Expressed Genes
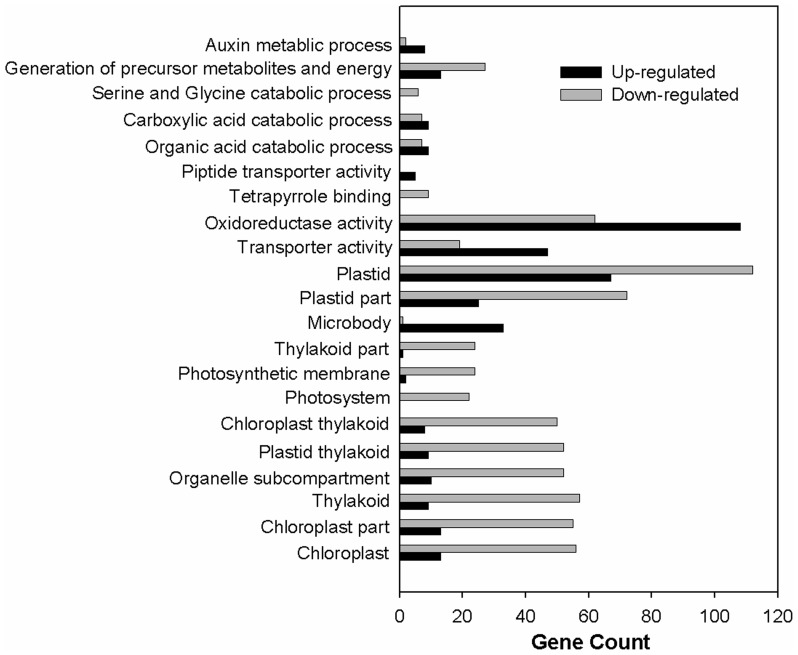
Putative differentially expressed genes were finally selected depending on the expression profiles and whether: a) the average fold change between K1 and K2 genes was more than or equal to twofold, and b) the FDR was less than 0.001. Accordingly, 1132 genes were identified as having been up-regulated in K1 compared with K2. Meanwhile, the expression of 455 genes was decreased by more than twofold in K1 compared with K2. Gene ontology (GO) analysis was performed by mapping each differentially expressed gene into the records of the GO database (http://www.geneontology.org/). The GO annotation of these genes is presented in Fig. 1. The main functional groups of up-regulated genes were related to auxin metabolic process, carboxylic acid and organic acid catabolic process, oxidoreductase activity, transporter activity and microbody; the main functional groups of down-regulated genes were related to serine and glycine catabolic process, generation of precursor metabolites and energy, tetrapyrrole binding, plastid and plastid part, thylakoid and thylakoid part, photosystem and photosynthetic membrane, organelle subcompartment and chloroplast and chloroplast part (Fig. 1).
Figure 1. GO analysis of differentially expressed genes obtained from Solexa sequencing.
The abscissa of the bar plot represents the gene count within each GO category. All processes listed had enrichment p values <0.05.
To understand the functions of the differentially expressed genes, we mapped all the genes to terms in KEGG database and, compared this with the whole transcriptome background, with a view to searching for genes that were significantly enriched (Table 2). Among the differentially expressed genes with KEGG pathway annotation, 250 genes were attributed to the first significantly enriched metabolic pathway. There were 152 up- and 98 down-regulated genes in the metabolic pathway. Notably, the specific enrichment of metabolic pathway genes was observed to be involved in valine, leucine, isoleucine, lysine and tryptophan metabolism and degradation, alpha-linolenic acid metabolism, fatty acid metabolism, nitrogen metabolism, glutathione metabolism, glycerophospholipid metabolism, ascorbate and aldarate metabolism; more of the differently expressed genes were up-regulated in K1 than K2. Photosynthesis, photosynthesis-antenna proteins and carbon fixation in photosynthetic organisms pathways were also enriched in KEGG pathway analysis, with most of the differently expressed genes been down-regulated in K1 compared with K2. Plant-pathogen interaction pathway was another enriched pathway with most of the differentially expressed genes been significantly up-regulated in K1 compared with K2.
Table 2. KEGG pathway annotation of differentially expressed genes obtained from Solexa sequencing.
| Pathways | DEGs with pathway annotation (783) | All genes with pathway annotation (9335) | Up-regulated gene (K1/K2) | Down-regulated gene (K1/K2) |
| Metabolic pathways | 250 | 2227 | 152 | 98 |
| Alpha-linolenic acid metabolism | 22 | 72 | 14 | 8 |
| Fatty acid metabolism | 21 | 74 | 18 | 3 |
| Biosynthesis of secondary metabolites | 133 | 1141 | 87 | 46 |
| Photosynthesis | 17 | 78 | 0 | 17 |
| Biosynthesis of unsaturated fatty acids | 14 | 59 | 10 | 4 |
| Photosynthesis-antenna proteins | 7 | 19 | 0 | 7 |
| Peroxisome | 23 | 134 | 21 | 2 |
| Carbon fixation in photosynthetic organisms | 18 | 98 | 3 | 15 |
| Plant-pathogen interaction | 68 | 568 | 56 | 12 |
| Valine, leucine and isoleucine degradation | 11 | 51 | 10 | 1 |
| Glyoxylate and dicarboxylate metabolism | 9 | 39 | 6 | 3 |
| Limonene and pinene degradation | 14 | 81 | 11 | 3 |
| Flavonoid biosynthesis | 19 | 124 | 11 | 8 |
| Indole alkaloid biosynthesis | 5 | 16 | 3 | 2 |
| Glycosphingolipid biosynthesis globo series | 3 | 6 | 3 | 0 |
| Steroid biosynthesis | 8 | 38 | 2 | 6 |
| Phenylpropanoid biosynthesis | 25 | 189 | 15 | 10 |
| Nitrogen metabolism | 10 | 56 | 7 | 3 |
| Lysine degradation | 6 | 26 | 6 | 0 |
| Glutathione metabolism | 14 | 91 | 11 | 3 |
| Vitamin B6 metabolism | 5 | 20 | 3 | 2 |
| Glycolysis/Gluconeogenesis | 20 | 150 | 10 | 10 |
| Glycerophospholipid metabolism | 14 | 95 | 10 | 4 |
| Tryptophan metabolism | 7 | 36 | 7 | 0 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 14 | 96 | 9 | 5 |
| Ascorbate and aldarate metabolism | 12 | 84 | 9 | 3 |
All pathways listed had enrichment p values <0.05.
Hormone Related Genes and Transcription Factor Genes
There were 51 differently expressed hormone-related genes, of which 39 were up-regulated and 12 down-regulated in K1 compared with K2 (Table 3). Isopentenyltransferase (IPT, Gene ID: DW494123) which is involved in cytokinin biosynthesis was down-regulated in K1 compared with K2. JA is critically involved in senescence. There are 6 differentially expressed JA-related genes, 5 of which were up-regulated in K1 compared with K2. Four of the 5 up-regulated genes were JA biosynthesis genes. Two differentially expressed SA related genes were both down-regulated in K1 compared with K2. In contrast to SA related genes, the 5 ethylene and 3 BR related genes were all up-regulated in K1 compared with K2. Eighteen of the 20 differently expressed auxin-related genes were up-regulated in K1 compared with K2. As for the 11 differently expressed GA-related genes, 8 genes were up-regulated and 3 down-regulated in K1 compared with K2. Furthermore, 2 GA biosynthesis genes, gibberellin 20-oxidase (gene ID: DQ122188) and Gibberellin 3-hydroxylase (gen ID: ES800765) were up-regulated but the GA degradation gene, GA 2-oxidase (gene ID: DW485236) was down-regulated in K1 compared with K2 (Table 3). Unexpectedly, only one ABA-related gene was up-regulated while 3 were down-regulated in K1 compared with K2. Moreover, the expression level of ABA biosynthesis gene NCED (9-cis-epoxycarotenoid dioxygenase, gene ID: EX168449) in K2 was 44 folds higher than that in K1.
Table 3. Differently expressed hormone related genes identified using Solexa sequencing in cotton.
| Hormone | Gene ID | log2 Ratio(K1/K2) | P-value | Blast nr (annotation) |
| Cytokinin | DW494123 | −1.61 | <0.01 | Gossypium hirsutum Isopentenyltransferase [Gossypium hirsutum] |
| JA | DT466567 | 3.09 | <0.01 | JA and ethylene-dependent systemic resistance, zinc finger protein [Ricinus communis] |
| DW482657 | 1.66 | <0.01 | JA biosynthetic process, AMP dependent CoA ligase [Ricinus communis] | |
| ES813117 | 1.59 | <0.01 | JA biosynthetic process, 4-coumarate-coa ligase [Populus trichocarpa] | |
| ES850570 | 1.12 | <0.01 | JA biosynthetic process, AMP dependent CoA ligase [Ricinus communis] | |
| HM462002 | 1.06 | <0.01 | JA biosynthetic process, 0/3-hydroxyacyl-CoA dehyrogenase [Ricinus communis] | |
| DT548121 | −1.16 | <0.01 | JA biosynthetic process, AMP dependent CoA ligase [Ricinus communis] | |
| SA | EX165509 | −1.63 | <0.01 | Response to salicylic acid stimulus, fiber protein Fb31 [Gossypium barbadense] |
| ES828698 | −1.54 | <0.01 | Salicylic acid-binding protein 2 [Nicotiana tabacum] | |
| Ethylene | DT466567 | 3.09 | <0.01 | JA and ethylene-dependent systemic resistance, zinc finger protein [Ricinus communis] |
| DW503700 | 1.64 | <0.01 | Ethylene-responsive transcription factor 2 | |
| DW504358 | 1.45 | <0.01 | Response to ethylene stimulus, unknown [Glycine max] | |
| EX172462 | 1.18 | <0.01 | Response to ethylene stimulus, Methylthioribose kinase [Ricinus communis] | |
| ES804001 | 1.07 | <0.01 | Ethylene response factor 11 [Actinidia deliciosa] | |
| Auxin | DT466567 | 3.09 | <0.01 | Auxin mediated signaling pathway, zinc finger protein [Ricinus communis] |
| DW476189 | 2.15 | <0.01 | Auxin metabolic process, mitochondrial substrate carrier family protein [Arabidopsis] | |
| DW488782 | 1.95 | <0.01 | Response to auxin stimulus, LRR-repeat protein [Ricinus communis] | |
| EX166666 | 1.82 | <0.01 | Auxin metabolic process, peroxisomal membrane protein pmp34, [Ricinus communis] | |
| AI731637 | 1.73 | <0.01 | Auxin-regulated protein, SAUR family protein [Populus trichocarpa] | |
| DW482657 | 1.66 | <0.01 | Auxin metabolic process, AMP dependent CoA ligase [Ricinus communis] | |
| ES813117 | 1.59 | <0.01 | Auxin metabolic process, 4-coumarate-coa ligase [Populus trichocarpa] | |
| CO498303 | 1.59 | <0.01 | Auxin-responsive family protein [Arabidopsis lyrata subsp. lyrata] | |
| DT552463 | 1.55 | <0.01 | Auxin-induced in root cultures protein 12 precursor [Ricinus communis] | |
| ES816485 | 1.36 | <0.01 | Auxin metabolic process, dehydrogenase/reductase family protein [Glycine max] | |
| DW497941 | 1.30 | <0.01 | Auxin-responsive protein IAA1 [Ricinus communis] | |
| ES852227 | 1.20 | <0.01 | Auxin transport, aminopeptidase [Arabidopsis thaliana] | |
| DW500898 | 1.15 | <0.01 | Auxin metabolic process, anthranilate synthase, beta subunit, ASB1 [Populus trichocarpa] | |
| DT568077 | 1.15 | <0.01 | Response to auxin stimulus, carnitine racemase [Ricinus communis] | |
| DW499567 | 1.15 | <0.01 | Auxin : hydrogen symporter [Ricinus communis] | |
| ES808260 | 1.15 | <0.01 | Auxin-regulated protein [Populus tremula x Populus tremuloides] | |
| ES850570 | 1.12 | <0.01 | Auxin metabolic process, AMP dependent CoA ligase [Ricinus communis] | |
| EF467065 | 1.10 | <0.01 | Gossypium hirsutum auxin response factor 3 (ARF3) | |
| DT548121 | −1.16 | <0.01 | Auxin metabolic process, AMP dependent CoA ligase [Ricinus communis] | |
| ES837803 | −1.03 | <0.01 | Auxin metabolic process, nodulin-like protein [Arabidopsis thaliana] | |
| GA | ES812534 | 2.47 | <0.01 | Gibberellin 3-beta-dioxygenase [Ricinus communis] |
| ES800765 | 2.24 | <0.01 | Gibberellin 3-hydroxylase 1 [Gossypium hirsutum] | |
| DQ829776 | 1.98 | <0.01 | Gossypium hirsutum gibberellic acid receptor | |
| DT551192 | 1.47 | <0.01 | Gibberellin receptor GID1 [Ricinus communis] | |
| EF607794 | 1.32 | <0.01 | Gossypium hirsutum gibberellic acid receptor-b | |
| DQ122188 | 1.24 | 0.02 | Gossypium hirsutum gibberellin 20-oxidase | |
| ES826231 | 1.06 | <0.01 | Gibberellin 3-beta-dioxygenase [Ricinus communis] | |
| FJ790128 | 1.01 | 0.02 | Gibberellic acid mediated signaling pathway, GID1-4 [Gossypium hirsutum] | |
| DW485236 | −3.48 | <0.01 | Gibberellin 2-oxidase [Populus trichocarpa] | |
| FJ384629 | −1.72 | <0.01 | Gibberellin mediated signaling pathway, tonoplast intrinsic protein [Gossypium hirsutum] | |
| ES807419 | −1.29 | <0.01 | Gibberellin mediated signaling pathway, unnamed protein product [Vitis vinifera] | |
| BR | DT466203 | 4.44 | <0.01 | Brassinosteroid insensitive 1-associated receptor kinase 1 [Ricinus communis] |
| ES794445 | 2.55 | <0.01 | Brassinosteroid-6-oxidase [Vitis vinifera] | |
| ES811041 | 1.52 | <0.01 | Brassinosteroid insensitive 1-associated receptor kinase 1 precursor, [Ricinus communis] | |
| ABA | EX170109 | 1.07 | <0.01 | Abscisic acid responsive elements-binding protein 2 [Populus trichocarpa] |
| EX168449 | −5.48 | <0.01 | 9-cis-epoxycarotenoid dioxygenase [Ricinus communis] | |
| ES836088 | −2.21 | 0.02 | Response to abscisic acid stimulus, unnamed protein product [Vitis vinifera] | |
| EX165509 | −1.63 | <0.01 | Response to abscisic acid stimulus, fiber protein Fb31 [Gossypium barbadense] |
It was also noted that 48 TFs including 11 NAC domain proteins were differentially expressed during leaf senescence (Table 4, 5). Eight WRKY and 3 R2R3-myb TFs along with 5 of the 6 differentially expressed ERF TFs were up-regulated in K1 compared with K2. Interestingly, the 11 differentially expressed NAC domain proteins were all up-regulated in K1 compared with K2, suggesting that NAC domain proteins may have important roles in cotton leaf senescence (Table 5).
Table 4. Differently expressed transcription factor genes identified using Solexa sequencing in cotton (K1/K2).
| Transcription factor | Gene ID | log2 Ratio(K1/K2) | P-value | Blast nr (annotation) |
| ERF | DW233991 | 3.92 | <0.01 | ERF transcription factor 4 [Vitis pseudoreticulata] |
| DT468163 | 2.78 | 0.02 | AP2/ERF domain-containing transcription factor [Populus trichocarpa] | |
| DW227971 | 2.60 | <0.01 | AP2/ERF domain-containing transcription factor [Populus trichocarpa] | |
| DW503700 | 1.64 | <0.01 | Ethylene-responsive transcription factor 2 | |
| DT554469 | 1.13 | <0.01 | AP2/ERF domain-containing transcription factor [Populus trichocarpa] | |
| DW236983 | −1.90 | <0.01 | AP2/ERF domain-containing transcription factor [Populus trichocarpa] | |
| WRKY | DT463609 | 2.39 | <0.01 | WRKY transcription factor 16 [Populus tomentosa x P. bolleana)] |
| DT562541 | 2.04 | <0.01 | WRKY transcription factor 33 [Populus tomentosa x P. bolleana)] | |
| DT468618 | 1.90 | <0.01 | WRKY transcription factor [Ricinus communis] | |
| DT544987 | 1.85 | <0.01 | WRKY transcription factor [Ricinus communis] | |
| DW240842 | 1.37 | <0.01 | WRKY transcription factor [Ricinus communis] | |
| ES817646 | 1.36 | <0.01 | WRKY transcription factor [Ricinus communis] | |
| EX168442 | 1.35 | <0.01 | WRKY transcription factor 1 [Populus tomentosa x P. bolleana)] | |
| ES839303 | 1.23 | <0.01 | WRKY Transcription factor 1 [Gossypium arboreum] | |
| R2R3-myb | ES808205 | 2.20 | <0.01 | R2R3-myb transcription factor [Ricinus communis] |
| ES820794 | 1.24 | <0.01 | R2R3-myb transcription factor [Ricinus communis] | |
| ES821169 | 1.20 | <0.01 | R2R3-Myb transcription factor [Citrus sinensis] | |
| Putative TF | ES842342 | 2.30 | 0.04 | Transcription factor, putative [Ricinus communis] |
| EX165732 | 1.43 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| ES844147 | 1.11 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| ES829573 | 1.04 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| DW500579 | −1.62 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| DW503784 | −1.09 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| DT463358 | −1.06 | <0.01 | Transcription factor, putative [Ricinus communis] | |
| Heat stress | EX166632 | 1.62 | <0.01 | Heat stress transcription factor [Carica papaya] |
| ES800773 | 1.08 | <0.01 | Heat stress transcription factor [Carica papaya] | |
| BZIP | DR454207 | 1.60 | <0.01 | BZIP domain class transcription factor [Malus x domestica] |
| GRAS | DT051524 | 1.41 | <0.01 | GRAS family transcription factor [Populus trichocarpa] |
| GATA | ES803228 | 1.36 | <0.01 | GATA transcription factor [Ricinus communis] |
| TCP | ES817176 | 1.33 | <0.01 | TCP domain class transcription factor [Malus x domestica] |
| TGA | ES848366 | 1.25 | <0.01 | Transcription factor TGA7 [Ricinus communis] |
| CCAAT-binding | ES841782 | 1.10 | <0.01 | CVAAT-binding transcription factor [Ricinus communis] |
| ES822081 | 1.09 | 0.06 | CVAAT -binding transcription factor subunit A [Ricinus communis] | |
| MYB | DW481287 | −1.81 | <0.01 | MYB transcription factor MYB127 [Glycine max] |
| MYBR | DW238551 | −1.14 | <0.01 | MYBR domain class transcription factor [Malus x domestica] |
| BIM | DW508557 | −1.49 | <0.01 | Transcription factor BIM1 [Ricinus communis] |
| ARF | ES834779 | −1.33 | <0.01 | ARF domain class transcription factor [Malus x domestica] |
Table 5. Differently expressed NAC genes identified using Solexa sequencing in cotton.
| Gene ID | log2 Ratio (K1/K2) | P-value | Blast nr (annotation) |
| DW517699 | 3.07 | <0.01 | NAC domain protein NAC6 [Gossypium hirsutum] |
| DR458413 | 2.75 | <0.01 | NAC domain-containing protein 21/22 [Ricinus communis] |
| CA992724 | 2.21 | <0.01 | NAC domain-containing protein [Ricinus communis] |
| EU706340 | 2.20 | <0.01 | Gossypium hirsutum NAC domain protein NAC6 |
| EU706342 | 2.06 | <0.01 | Gossypium hirsutum NAC domain protein NAC4 |
| CA992692 | 1.84 | <0.01 | NAC domain-containing protein [Ricinus communis] |
| ES806311 | 1.62 | <0.01 | NAC domain-containing protein [Ricinus communis] |
| EU372996 | 1.45 | <0.01 | NAC domain-containing protein [Ricinus communis] |
| EX166759 | 1.21 | <0.01 | NAC domain protein [Citrus trifoliata] |
| EU706343 | 1.68 | <0.01 | Gossypium hirsutum NAC domain protein NAC5 |
| ES839947 | 1.09 | <0.01 | NAC transcription factor [Vitis pseudoreticulata] |
Confirmation of Solexa Expression Patterns by RT-PCR Analysis
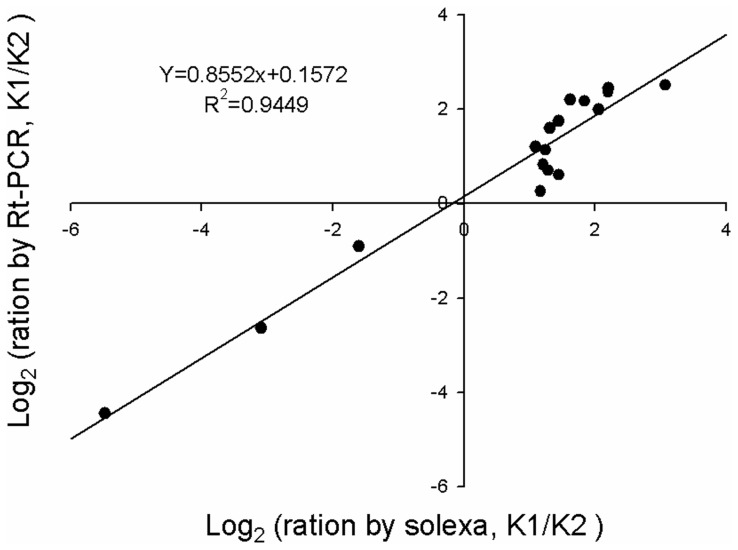
To test the reliability of Solexa sequencing further, RT-PCR analysis was performed with specific primers for a subset of 18 genes, which have been identified by Solexa sequencing in which 14 genes were up-regulated and 4 genes down-regulated. The results showed that 17 of the 18 genes had the same expression profiles as the original Solexa sequencing. This indicates that the original Solexa pattern was validated in 94.4% of the cases. This was not the case for the other gene presumably because of the mutations within the primer sites or because the RNA used for Solexa sequencing and qRT-PCR were extracted from different plants. The expression patterns of the 17 genes were highly consistent with the Solexa sequencing ratios, with a relative R2 of 0.9449 (Fig. 2).
Figure 2. Comparison of the expression ratios of some selected genes using Solexa sequencing and qRT-PCR.
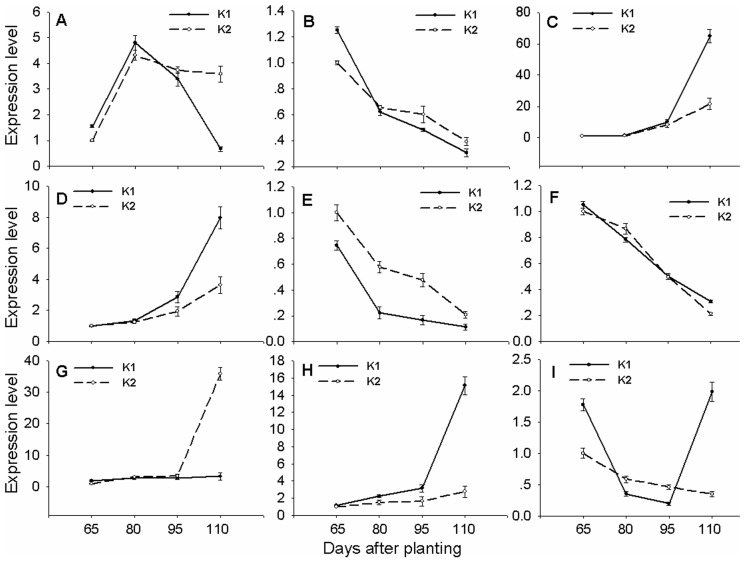
To test if the differentially expressed genes were developmental stage-dependent, the expression patterns of 9 senescence-related genes were analyzed by RT-PCR at 65, 80, 95 and 110 DAP. For both K1 and K2, the expression level of LHCB (Chlorophyll binding protein, gene ID: EX169337) was increased gradually before 80 DAP but decreased after 80 DAP during leaf senescence and the LHCB expression level of K1 was distinctly lower than K2 at 110 DAP (Fig. 3A). The expression level of RBCL (large subunit of Rubisco, ribulose-1, 5-bisphosphate carboxylase/oxygenase; gene ID: ES820978) decreased as K1 and K2 plants started to senesce from 65 to 110 DAP and the RBCL expression level of K1 was lower than K2 at 80 and 110 DAP (Fig. 3B). The expression level of SOD (superoxide dismutase, Gene ID: ES824305) and ATG (autophagy, gene ID: CO493577) in K1 and K2 plants increased during senescence and SOD and ATG expression levels of K1 were distinctly higher than K2 at 95 and 110 DAP (Fig. 3C, D ). The IPT (isopentenyltransferase, gene ID: DW494123) expression level decreased as K1 and K2 started senescence and the level of K1 was lower than that of K2 from 65 to 110 DAP (Fig. 3E). The GhNCED2 (gene ID: HM014161) expression level also decreased as K1 and K2 started to senesce but the level of K1 was higher than that of K2 at 110 DAP (Fig. 3F). However, the NCED1 (gene ID: EX168449) expression level increased as K1 and K2 started to senesce but the level of K1 was lower than that of K2 at 110 DAP (Fig. 3G). The expression level of NAC (NAC domain protein, gene ID: CA992724) in K1 and K2 increased during senescence but that in K1 was higher than that in K2 from 65 to 110 DAP (Fig. 3H). On the contrary, the expression level of another NAC domain protein gene GhNAC6 (gene ID: Dw517699) in K2 gradually decreased during senescence and that in K1 also gradually decreased before 95 DAP. The GhNAC6 expression level was distinctly increased after 95 DAP, therefore, the level in K1 was higher than that in K2 at 110 DAP (Fig. 3I).
Figure 3. Senescence-dependent changes in gene expression determined by quantitative RT-PCR in the main-stem leaves of K1 and K2 cotton lines at 65, 80, 95 and 110 DAP.
A, Chlorophyll binding protein (LHCB; gene ID: EX169337). B, Large subunit of Rubisco, ribulose- 1,5-bisphosphate carboxylase/oxygenase (RBCL; gene ID: ES820978). C, Superoxide dismutase (SOD; gene ID: ES824305). D, Autophagy (ATG; gene ID: CO493577). E, Isopentenyltransferase (IPT; gene ID: DW494123). F, 9-cis-epoxycarotenoid dioxygenase (GhNCED2; gene ID: HM014161); G, 9-cis-epoxycarotenoid dioxygenase (NCED1; gene ID: EX168449). H, NAC domain protein (NAC; gene ID: CA992724). I, NAC domain protein (GhNAC6; gene ID: Dw517699). Expression ratios are presented relative to K2 values at 65 DAP. Data are means of three biological replicates ± SE.
Discussion
Early leaf senescence has become one of the important factors limiting cotton production in recent years [8], [60]. Understanding its causes and mechanisms would help reduce yield loss due to premature senescence. Leaf senescence varies greatly among cotton genotypes. Our previous study suggested that K1 aged earlier than K2 because of more cytokinins but less ABA in leaves of K1 than K2 [19]. In this study, the genome-wide changes in gene expression between K1 and K2 at 110 DAP were investigated by Illumina/Solexa sequencing method. We found that K1 differed significantly from K2 in the expression of senescence-related genes during leaf senescence. About 1132 genes were up-regulated and 455 genes down-regulated in K1 compared with K2.
Leaf senescence begins with the transition from anabolism of carbohydrates and other biomolecules to catabolism of proteins, nucleic acids, and lipids, and culminates in cell death. The onset and progression of leaf senescence is accompanied by expression of a large number of genes [11], [12]. Down-regulated genes are significantly enriched for genes linked to the plastid and thylakoid, and with functions in metabolic processes, particularly photosynthesis and carbohydrate and amino acid metabolism [10]. Genes involved in chloroplast activity such as photosystem (PS) I and II, carbon fixation, chlorophyll (tetrapyrrole) biosynthesis, and amino acid metabolism are also down-regulated during senescence [10]. In our experiment, 1132 genes were up-regulated and 455 genes down-regulated in K1 compared with K2. Down-regulated genes were related to serine and glycine catabolism, tetrapyrrole binding, plastid and thylakoid, photosystem and photosynthetic membrane, organelle subcompartment, chloroplast and chloroplast part, carbon fixation and steroid biosynthesis (Fig. 1 and table 2). The senescence-repressed gene, LHCB1 has been used to monitor the progress of senescence in otsB-expressing plants and the LHCB1 expression level in otsB-expressing plants was higher than the wild type plants [61]. In the present study, the LHCB expression level in K1 was lower than in K2 at 95 and 110 DAP, which might be an attribute to earlier senescence in K1 than K2 (Fig. 3A). The levels of RBCL mRNA exhibited a gradual decrease which started before the onset of visible senescence in two different senescent types of maize and a decline in RBCL could lead to degradation of Rubisco, thereby decreasing the photosynthetic rate during leaf senescence [62], [63]. Therefore, in the present study the decreased chlorophyll content and photosynthesis in K1 might be attributed to reduced expression of LHCB and RBCL at 110 DAP (Fig. 3A, B and Fig. S3).
During senescence different pathways contribute to degradation of proteins and other macromolecules, of which autophagy (ATG) is an important pathway. The ATG plays a key role in the senescence process, and accelerated senescence has been observed in a number of autophagy-defective mutants [64]–[66]. Knockout or RNAi mutants of AtATG4a/b, AtATG5, AtATG7, AtATG9, and AtATG18a exhibit accelerated senescence and hypersensitivity to nutrient starvation, indicating that autophagy is not only essential for the senescence program but also plays an important role in nutrient remobilization [64]–[68]. A key role of senescence in plant tissues is the ordered degradation of macromolecules and mobilization of the products, during which supposedly transporters (TPs) are critically involved. There are many Arabidopsis TPs which are up-regulated during senescence [11], [12], [46], [69], [70]. The preponderance of up-regulated TPs corresponds well with the substrates (amino acids, inorganic phosphorus, sugars, purines, pyrimidines and metal ions), which are known to be transported from senescent leaves to sink organs [71], [72]. The up-regulation of amino acid and oligopeptide TPs correlates with the increase in protein degradation, and the breakdown products were exported to the sink organs by TPs during senescence [9]. The expression levels of protein degradation, Peroxisome, ATG and TP related genes in K1 were higher than those in K2, indicating that proteins and other macromolecules degrade and then export the nutrients to sink organs much earlier in K1 than in K2 (Fig. 1, Fig. 3D and Table 2).
Cytokinin is involved in leaf senescence, because its levels are reduced in senescing leaves [6], [19] and exogenous application of cytokinin or endogenous overexpression of IPT gene can delay senescence [22], [73]. In this study, the expression level of IPT in both K1 and K2 were gradually decreased, but he IPT level in K1 was distinctly lower than K2 from 65 to 110 DAP (Fig. 3E and table 3). The results well matched our previous observation that the cytokinin contents in both K1 and K2 were gradually decreased, but the cytokinin contents in K1 were distinctly lower than in K2 from 65 to 110 DAP. This implied that the decreased cytokinin contents in K1 and K2 may be caused by decreased IPT expression level [19]. Thus it was suggested that enhanced leaf senescence and reduced cytokinin level of K1 was attributed to decreased IPT expression in K1 relative to K2. On the contrary, ABA is considered a promoter of senescence because ABA treatment can induce leaf senescence and many genes related to ABA synthesis, metabolism and signaling are up-regulated during senescence [10]–[12], [14], [74], [75]. However, not all ABA-response genes were up-regulated during senescence. Therefore, the estimate of the impact of ABA as a regulator during senescence is not straightforward [12]. There are many NCED gene members and different NCED genes have different temporal- and tissue-specific expression patterns [76]–[78]. The expression of the CsNCED1 gene was consistent with the accumulation of ABA but the CsNCED2 gene showed a different and tissue-specific expression during fruit maturation [77]. In our study, 3 of the 4 differently expressed ABA-related genes were down-regulated in K1 compared with K2, though K1 senescence earlier than K2 (Table 3). One NCED (EX168449) was gradually increased but another GhNCED (HM014161) was gradually decreased from 65 to 110 DAP during senescence and the expression level of NCED (EX168449) in K1 was considerably lower than in K2. However, the GhNCED (HM014161) expression level in K1 was distinctly higher than K2 at 110 DAP (Fig. 3F, G). The expression of NCED genes was not consistent with our previous observation that the ABA level in K1 and K2 were gradually increased during senescence and K1 was higher than K2 in ABA level [19]. Such inconsistency might be due to the fact that the 2 NCED genes were not exactly the senescence-related genes or ABA was not a straightforward senescence regulator in K1 and K2 cotton plants. Further study on the exact roles of NCED genes may help better explain such inconsistency.
Although auxin effects on leaf abscission were first reported more than 50 years ago, its involvement in leaf senescence is much less understood than that of ET, JA, or cytokinin. In senescing leaves of Arabidopsis, the indole-3-acetic acid (IAA) concentration was 2-fold higher than in non-senescing leaves [37]. However, auxin treatment leads to a transient decrease in SAG12 expression [30]. Arabidopsis plants over-expressing the auxin biosynthesis gene YUCCA6, such as the yuc6-1D activation mutant and 35S:YUC6 transgenic plants, have been shown to contain an elevated level of free IAA and reduced transcript abundances of SAG12, NAC1, and ORE1 during leaf senescence compared with the wild type. It was suggested that auxin delayed senescence by directly or indirectly regulating the expression of senescence-associated genes [34]. In contrast to K2, 18 of the 20 differentially expressed auxin related genes were up-regulated in K1, suggesting that auxin might play an important role in leaf senescence of cotton (Table 3). The exact roles of auxin and the differentially expressed gene effects on cotton senescence need further study.
It is still not well determined if GA activity is involved in leaf senescence. GA can partially inhibit the cytokinin effect from seedling development to senescence through SPINDLY protein [39]. No GA biosynthesis gene but GA-inducible GA 2-oxidase 2 (AtGA2OX2) that deactivates GA was up-regulated during leaf senescence, suggesting that at least some GAs are deactivated during leaf senescence [12]. On the contrary, the GA biosynthesis genes, gibberellin 20-oxidase (gene ID: DQ122188) and Gibberellin 3-hydroxylase (gen ID: ES800765) were up-regulated and GA 2-oxidase (gene ID: DW485236) down-regulated in K1 compared with K2, suggesting that GA content in K1 was higher than K2 (Table 3). In contrast to K2, the early senescence cotton line K1 had higher GA but lower cytokinin content, suggesting a regulatory pathway similar to Arabidopsis that GA inhibits cytokinin effect through SPINDLY-like protein. The plant hormones JA, ethylene and BR are well known as regulators of leaf senescence. Exogenous application of JA and ethylene can enhance senescence [13], [14], [20]. Many JA, ethylene and BR related genes were up-regulated during senescence in Arabidopsis [11], [12]. In this study, 4 JA biosynthesis genes were up-regulated in K1 compared with K2, indicating that JA plays an important role in enhanced leaf senescence of K1 (Table 3). All the differently expressed ethylene and BR related genes were up-regulated in K1 compared with K2, indicating that JA, ethylene and BR are possibly involved in enhancing leaf senescence in K1 (Table 3).
Many TFs are differentially regulated and the NAC and WRKY families are particularly rich in senescence-regulated TFs in many plant species during leaf senescence [10], [11], [46], [70], [79]–[81], suggesting that they are involved in leaf senescence. Forty-eight differentially regulated TFs were found in our experiment (Table 4, 5). Among the TFs, NAC and WRKY that rank the first and second in TF families might play important roles in leaf senescence of cotton, because both were up-regulated during leaf senescence. Although more than 20 of the 106 known NAC genes in Arabidopsis exhibit senescence-dependent expression, the distinct regulatory function with respect to senescence has only been reported for some members so far [43], [48]–[51]. NAC (At NAP, At ORE1 and AtORS1) have been identified as nonredundant positive regulators of senescence, because inhibiting them individually delays leaf senescence [48]–[50]. A dual-function NAC gene VNI2 was recently reported to integrate ABA signaling with leaf senescence and to negatively regulate xylem vessel formation [43]. A hydrogen peroxide induced NAC transcription factor JUB1 gene was also reported to regulate Arabidopsis senescence [51]. Leaf aging was delayed in transgenic plants overexpressing the VNI2 or JUB1 gene but significantly accelerated in a VNI2 or JUB1 deficient mutant, suggesting that VNI2 and JUB1 are negative regulators of senescence [43], [51]. There were 11 up-regulated NAC TFs in our study. The expression level of NAC (gene ID: CA992724) in K1 and K2 was increased during senescence and that in K1 was higher than in K2 from 65 to 110 DAP (Fig. 3H). However, the expression level of GhNAC6 (gene ID: Dw517699) in K1 and K2 gradually decreased before 95 DAP, although the level in K1 was distinctly increased after 95 DAP and that in K1 was higher than K2 at 110 DAP (Fig. 3I). The different expression patterns of both NAC genes in K1 and K2 during senescence suggest that different NAC TFs have different functions in leaf senescence of cotton. Further research on their exact functions in leaf senescence related candidate NAC genes is necessary.
Taken together, our results showed that K1 senescence earlier than K2. This is because most of the genes related to anabolism of carbohydrates and other biomolecules were down-regulated but those for catabolism of proteins, nucleic acids, lipids and nutrient recycle etc. were up-regulated in K1 compared with K2. The transition from anabolism to catabolism occurred earlier in K1 than K2, which might be regulated by up- or down-regulated hormone related and TF genes such as down-regulated cytokinin biosynthesis and GA degradation genes, up-regulated JA and GA biosynthesis genes and up-regulated NAC and WRKY genes in K1. Further studies on hormone related genes and NAC TFs will improve our understanding of the regulatory function of hormone and NAC TFs on leaf senescence.
Supporting Information
Overview of the short-read based transcriptome analysis approach for non-model organisms. Mapping is an option only if a suitable reference genome is available; otherwise FASTA (or FASTQ) sequences must be used [54].
(TIF)
Primers used for RT-PCR analyses.
(TIF)
Chlorophyll (Chl) content and net photosynthetic (Pn) rate of the fourth leaf from the apex on the main-stem at 65–110 d after planting. Values are means ±SD (n = 4). Initial flowering, peak flowering, peak boll-setting, and initial boll-opening occurred at 65, 80, 95, and 110 d after planting.
(TIF)
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. 30971720), the earmarked fund for China Agricultural Research System (CARS-18-21), the Natural Science Foundation of Shandong Province (grant no. ZR2012CM027), and the Seed Improvement Funds (Project No. 2009LZ005-05) from Shandong Province. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gay AP, Thomas H (1995) Leaf development in Lolium temulentum: photosynthesis in relation to growth and senescence. New Phytol 130: 159–168. [Google Scholar]
- 2. Thomas H, Howarth CJ (2000) Five ways to stay green. J Exp Bot 51: 329–337. [DOI] [PubMed] [Google Scholar]
- 3. Guin G (1985) Abscisic acid and cutout in cotton. Plant Physiol 77: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong HZ, Li Wj, Eneji AE, Zhang DM (2012) Nitrogen rate and plant density effects on yield and late-season leaf senescence of cotton raised on a saline field. Field Crop Res 126: 137–144. [Google Scholar]
- 5. Dong HZ, Li WJ, Tang W, Zhang DM, Li ZH (2006) Yield quality and leaf senescence of cotton grown at varying planting dates and plant densities in the Yellow River Valley of China. Field Crop Res 98: 106–115. [Google Scholar]
- 6. Dai JL, Dong HZ (2011) Stem girdling influences concentrations of endogenous cytokinins and abscisic acid in relation to leaf senescence in cotton. Acta Physiol Plant 33: 1697–1705. [Google Scholar]
- 7. Wright PR (1999) Premature senescence of cotton (Gossypium hirsutum L.): predominantly a potassium disorder caused by an imbalance of source and sink. Plant Soil 211: 231–239. [Google Scholar]
- 8. Wright PR (1998) Research into early senescence syndrome in cotton. Better Crop International 12: 14–16. [Google Scholar]
- 9. Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937. [DOI] [PubMed] [Google Scholar]
- 10. Breeze M, Harrison E, McHattie S, Hughes L, Hickman R, et al. (2011) High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, et al. (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585. [DOI] [PubMed] [Google Scholar]
- 12. Van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, et al. (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141: 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grbić V, Bleecker AB (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8: 595–602. [Google Scholar]
- 14. Weaver L, Gan S, Quirino B, Amasino R (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469. [DOI] [PubMed] [Google Scholar]
- 15. Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana . Science 241: 1086–1089. [DOI] [PubMed] [Google Scholar]
- 16. Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, et al. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- 17. Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473. [Google Scholar]
- 18. Gepstein S, Thimann KV (1980) Changes in the abscisic acid content of oat leaves during senescence. Proc Natl Acad Sci USA 77: 2050–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong HZ, Niu YH, Li WJ, Zhang DM (2008) Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. J Exp Bot 59: 1295–1304. [DOI] [PubMed] [Google Scholar]
- 20. He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noodén LD, Singh S, Letham DS (1990) Correlation of xylem sap cytokinin levels with monocarpic senescence in soybean. Plant Physiol 93: 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988. [DOI] [PubMed] [Google Scholar]
- 23. McCabe MS, Garratt LC, Schepers F, Jordi WJ, Stoopen GM, et al. (2001) Effects of PSAG12-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol 127: 505–516. [PMC free article] [PubMed] [Google Scholar]
- 24. Chang H, Jones ML, Banowetz GM, Clark DG (2003) Overproduction of cytokinins in petunia flowers transformed with PSAG12-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol 132: 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, et al. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci USA 104: 19631–19636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang P, Wang WQ, Zhang GL, Kaminek M, Dobrev P, et al. (2010) Senescence- inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J Integr Plant Biol 52: 653–669. [DOI] [PubMed] [Google Scholar]
- 27. Gepstein S, Sabehi G, Carp M, Hajouj T, Nesher MFO, et al. (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642. [DOI] [PubMed] [Google Scholar]
- 28. Shoji K, Addicott FT, Swets WA (1951) Auxin in relation to leaf blade abscission. Plant Physiol 26: 189–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sacher JA (1957) Relationship between auxin and membrane-integrity in tissue senescence and abscission. Science 125: 1199–1200. [DOI] [PubMed] [Google Scholar]
- 30. Noh Y, Amasino R (1999) Identification of a promoter region responsible for the senescence-specific expression of. SAG12. Plant Mol Biol 41: 181–194. [DOI] [PubMed] [Google Scholar]
- 31. Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, et al. (2005) AUXIN RESPONSE FACTOR1and AUXIN RESPONSE FACTOR2 regulates senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574. [DOI] [PubMed] [Google Scholar]
- 32. Okushima Y, Mitina I, Quach HL, Theologis A (2005) AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J 43: 29–46. [DOI] [PubMed] [Google Scholar]
- 33. Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, et al. (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim JI, Murphy AS, Baek D, Lee SW, Yun DJ, et al. (2011) YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J Exp Bot 62: 3981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansen H, Grossmann K (2000) Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol 124: 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vandenbussche F, Smalle J, Le J, Saibo NJ, De Paepe A, et al. (2003) The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiol 131: 1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quirino B, Normanly J, Amasino R (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence including pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278. [DOI] [PubMed] [Google Scholar]
- 38. Morris K, MacKerness SA, Page T, John CF, Murphy AM, et al. (2000) Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J 23: 677–685. [DOI] [PubMed] [Google Scholar]
- 39. Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, et al. (2005) Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clouse SD, Sasse JM (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451. [DOI] [PubMed] [Google Scholar]
- 41. Lim PO, Nam HG (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol 67: 49–83. [DOI] [PubMed] [Google Scholar]
- 42. Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136. [DOI] [PubMed] [Google Scholar]
- 43. Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC Transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, et al. (2000) Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- 45. Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87. [DOI] [PubMed] [Google Scholar]
- 46. Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell and Environ 27: 521–549. [Google Scholar]
- 47. Yoon HK, Kim SG, Kim SY, Park CM (2008) Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol Cells 25: 438–445. [PubMed] [Google Scholar]
- 48. Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, et al. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264. [DOI] [PubMed] [Google Scholar]
- 49. Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612. [DOI] [PubMed] [Google Scholar]
- 50. Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, et al. (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, et al. (2012) JUNGBRUNNEN1, a Reactive Oxygen Species–Responsive NAC Transcription Factor, Regulates Longevity in Arabidopsis . Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dong HZ, Niu YH, Kong XQ, Luo Z (2009) Effects of early-fruit removal on endogenous cytokinins and abscisic acid in relation to leaf senescence in cotton. Plant Growth Regul 59: 93–101. [Google Scholar]
- 53. Wu T, Qin ZW, Zhou XY, Feng Z, Du YL (2010) Transcriptome profile analysis of floral sex determination in cucumber. J Plant Physiol 167: 905–913. [DOI] [PubMed] [Google Scholar]
- 54. Collins LJ, Biggs PJ, Voelckel C, Joly S (2008) An approach to transcriptome analysis of non-model organisms using short-read sequences. Genome Informatics 21: 3–14. [PubMed] [Google Scholar]
- 55. Li H, Ruan J, Durbin R (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 57. Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995. [DOI] [PubMed] [Google Scholar]
- 58. Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188. [Google Scholar]
- 59. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57: 391–399. [DOI] [PubMed] [Google Scholar]
- 61. Wingler A, Delatte TL, ÓHara LE, Primavesi LF, Jhurreea D, et al. (2012) Trehalose 6-Phosphate Is Required for the Onset of Leaf Senescence Associated with High Carbon Availability. Plant Physiol 158: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. He P, Osaki M, Takebe M, Shinano T (2002) Changes of photosynthetic characteristics in relation to leaf senescence in two maize hybrids with different senescent appearance. Photosynthetica 40: 547–552. [Google Scholar]
- 63. He P, Osaki M, Takebe M, Shinano T, Wasaki J (2005) Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J Exp Bot 56: 1117–1128. [DOI] [PubMed] [Google Scholar]
- 64. Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12 activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114. [DOI] [PubMed] [Google Scholar]
- 65. Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, et al. (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, et al. (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546. [DOI] [PubMed] [Google Scholar]
- 69. Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, et al. (2003) The molecular analysis of leaf senescence: a genomics approach. Plant Biotechnol J 1: 3–22. [DOI] [PubMed] [Google Scholar]
- 70. Lin JF, Wu SH (2004) Molecular events in senescing Arabidopsis leaves. Plant J 39: 612–628. [DOI] [PubMed] [Google Scholar]
- 71. Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323. [Google Scholar]
- 72. Soudry E, Ulitzur S, Gepstein S (2005) Accumulation and remobilization of amino acids during senescence of detached and attached leaves: in planta analysis of tryptophan levels by recombinant luminescent bacteria. J Exp Bot 56: 695–702. [DOI] [PubMed] [Google Scholar]
- 73. Smart CM, Scofield SR, Bevan MW, Dyer TA (1991) Delayed leaf senescence in tobacco plants transformed with tmr, a gene for cytokinin production in Agrobacterium. Plant Cell 3: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Oh SA, Lee SY, Chung IK, Lee CH, Nam HG (1996) A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol Biol 30: 739–754. [DOI] [PubMed] [Google Scholar]
- 75. He Y, Tang W, Swain JD, Green AL, Jack TP, et al. (2001) Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol 126: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tan BC, Joseph LM, Deng WT, Liu L, Cline K, et al. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56. [DOI] [PubMed] [Google Scholar]
- 77. Rodrigo MJ, Alquezar B, Zacarías L (2006) Cloning and characterization of two 9-cis- epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J Exp Bot 57: 633–643. [DOI] [PubMed] [Google Scholar]
- 78. Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, et al. (2012) Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J 70: 501–512. [DOI] [PubMed] [Google Scholar]
- 79. Andersson A, Keskitalo J, Sjödin A, Bhalerao R, Sterky F, et al. (2004) A transcriptional timetable of autumn senescence. Genome Biol 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gregersen PL, Holm PB (2007) Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 5: 192–206. [DOI] [PubMed] [Google Scholar]
- 81.Balazadeh S, Riaňo-Pachón DM, Mueller-Roeber B (2008) Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol (Stuttgart) suppl 1: 63–75. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the short-read based transcriptome analysis approach for non-model organisms. Mapping is an option only if a suitable reference genome is available; otherwise FASTA (or FASTQ) sequences must be used [54].
(TIF)
Primers used for RT-PCR analyses.
(TIF)
Chlorophyll (Chl) content and net photosynthetic (Pn) rate of the fourth leaf from the apex on the main-stem at 65–110 d after planting. Values are means ±SD (n = 4). Initial flowering, peak flowering, peak boll-setting, and initial boll-opening occurred at 65, 80, 95, and 110 d after planting.
(TIF)