Abstract
Maternal nutrient restriction causes the development of adult onset chronic diseases in the intrauterine growth restricted (IUGR) fetus. Investigations in mice have shown that either protein or calorie restriction during pregnancy leads to glucose intolerance, increased fat mass, and hypercholesterolemia in adult male offspring. Some of these phenotypes are shown to persist in successive generations. The molecular mechanisms underlying IUGR remain unclear. The placenta is a critical organ for mediating changes in the environment and the development of embryos. To shed light on molecular mechanisms that might affect placental responses to differing environments we examined placentas from mice that had been exposed to different diets. We measured gene expression and whole genome DNA methylation in both male and female placentas of mice exposed to either caloric restriction or ad libitum diets. We observed several differentially expressed pathways associated with IUGR phenotypes and, most importantly, a significant decrease in the overall methylation between these groups as well as sex-specific effects that are more pronounced in males. In addition, a set of significantly differentially methylated genes that are enriched for known imprinted genes were identified, suggesting that imprinted loci may be particularly susceptible to diet effects. Lastly, we identified several differentially methylated microRNAs that target genes associated with immunological, metabolic, gastrointestinal, cardiovascular, and neurological chronic diseases, as well as genes responsible for transplacental nutrient transfer and fetal development.
Keywords: intrauterine growth restriction, DNA methylation, placentas, caloric restriction
maternal nutrient restriction of the fetus is known to increase the risk of eventually developing adult-onset chronic diseases. Investigations in mice related to either protein or calorie restriction (CR) have shown that this nutrient deficiency leads to glucose intolerance, increased fat mass, and hypercholesterolemia in adult male offspring (7). In humans, suboptimal intrauterine nutrient environments, such as maternal malnutrition or placental disease that interferes with the fetal growth potential, have been linked to an increased incidence of metabolic and cardiovascular disease (9).
While much is known about the phenotypic consequences of the effects of maternal diet during gestation, the underlying molecular mechanisms that are responsible for these are still poorly understood. However, it is thought that the placenta, which is the critical organ for transporting nutrients from the maternal blood to the embryo, must play a critical role in intrauterine growth restriction (IUGR). In utero placentas are sensitive to the immediate environment, thereby contributing toward programming that affects the health, growth, and survival of the developing fetus (37). Because of its centrality for mediating maternal nutrition effects, several studies have focused on the response of placental gene expression to differing maternal diets. One such study measured placental gene expression changes between high- and low-fat diet-fed mice and identified sexually dimorphic patterns, as well as genes regulating ion balance and chemoreception (39). Similarly, sex-specific expression patterns are also found in mice with maternal overnutrition, although only a limited number of genes were found to be consistently affected (22).
However, gene expression is not the only molecular phenotype that is affected by maternal nutrition. DNA methylation patterns can also respond to environmental perturbations and mark chromatin changes that affect transcription. To address these epigenetic responses, a recent study of infant growth restriction in humans (40) reported that the methylation patterns of ∼27,000 loci in placentas from pregnancies yielding IUGR were found to be significantly different from those of appropriate for gestational age placentas. DNA methylation is also known to play a key role in the regulation of placental imprinted genes. The expression of imprinted genes within the placenta affects the allocation of resources between a mother and her offspring (17, 28, 46). It is thought that maternally imprinted genes tend to limit fetal growth, while paternally imprinted genes tend to favor fetal growth (15). Thus imprinted genes in the placenta may have a direct effect on fetal development by affecting nutrient flow and fetal size (19, 41). Furthermore, as one allele in imprinted genes is already functionally silenced, an epigenetic alteration of the other allele can significantly affect transcription. In a previous study of mice reared on a high-fat diet, several imprinted genes were shown to have sex- and diet-specific changes of expression and DNA methylation (23).
To measure the effect of the intrauterine environment on the placenta, we have previously assessed transplacental nutrient transport in wild-type and null mouse models, specifically the glucose transporter isoform-3 (GLUT3; placenta)-null heterozygous+/− mutation-carrying mice (25). We found that when the glucose transporter Glut3, which is expressed in the placenta, is deleted on one allele it leads to reduced transplacental glucose transport and sexually dimorphic adiposity with insulin resistance in the adult offspring.
Based on this accumulation of information, we hypothesized that maternal CR that results in IUGR would differentially affect placental gene expression and DNA methylation in a sex-specific manner. To test this hypothesis, we employed a murine model of IUGR and measured both changes in gene expression and DNA methylation. This model differs from previous mouse models of maternal diet effects, which primarily focused on overnutrition. Instead we model the effect of CR, which mimics the human IUGR phenotype encountered world-wide of low-birth-weight offspring that are predisposed to metabolic disorders (24, 27, 30).
We identified differentially expressed genes from RNA-Seq data and examined their associated pathways that are affected by late-gestation maternal CR. Subsequently, we asked whether the DNA methylation profiles are also altered by maternal CR. To answer this question, we used reduced representation bisulfite sequencing (RRBS) to examine genome-wide DNA methylation in placentas (42, 52). We observed a decrease in the overall methylation of CR placentas compared with those exposed to normal diets. We performed a genome-wide scan of differentially methylated genes between CR vs. control (CON) groups and identified chromosomal hotspots for methylation changes that are affected by maternal caloric restriction. We also found an enrichment of known imprinted genes among the differentially methylated genes and identified several differentially methylated microRNAs.
MATERIALS AND METHODS
Mouse placenta samples.
C57/BL6 mice were housed in 12:12 h light-dark cycles with ad libitum access to a standard rodent chow diet (Harlan Teklad 7013) and water. At 8 wk of age, male and female mice were mated overnight and the presence of a vaginal plug in the female was designated as gestational day 1. Pregnant females were transferred to individual cages and reared on the same chow diet ad libitum. At gestational day 10, the pregnant mice were arbitrarily divided into two groups; the group that served as the control continued to receive ad libitum chow diet. The second group was subjected to CR by providing 50% (wt) of their daily intake until gestational day 19. This particular time of late gestation was used because the impact of placental function dramatically affects fetal growth at this time. Late gestation is the time period during which placental gene expression significantly affects fetal growth patterns. It has been shown that when glucose transport across the placenta is affected, fetal growth is diminished during late gestation including day 19 (25, 26). At this time, animals were euthanized by 100 mg/kg phenobarbital ip. The placentas were separated from the respective fetuses and collected. After accurate weighing of the placentas in a Mettler AB104 precision balance (0.01 mg sensitivity, see Supplemental Table S8) they were snap-frozen immediately and stored at −80°C until further analyses.1 This study protocol (24) was approved by the Animal Research Committee of the University of California Los Angeles in accordance with the guidelines set by the National Institutes of Health.
RNA-Seq library generation and data processing.
After RNA extraction (see Supplemental Information), total RNA was quantified using Qubit RNA assay, and 1,000 ng were used as starting material for each sample. The library preparation was performed using the Illumina TruSeq RNA Sample Preparation kit and the manufacturer's instructions. Libraries were run using 50-bp single-end reads on the HiSeq 2000 System (Illumina). The reads are mapped with Tophat (55), allowing up to two mismatches, and only unique alignments are kept. The quality of alignments were checked with FastQC. The resulted alignment file were processed through HTSeq program along with annotation file to create gene matrix, as the input for downstream analysis. The differential expression was calculated with DESeq (2) to generate Reads Per Kilobase per Million mapped reads (RPKM) per gene (Supplemental File 1).
Reduced representation bisulfite sequencing.
Genomic DNA from our mice placentas was extracted for making RRBS libraries following the standard RRBS protocol (42). The genome was digested with the MspI enzyme, a methylation-insensitive restriction enzyme. Fragments from 100–200 bases were selected as these are enriched for CpG-rich regions, such as CpG islands, promoter regions, and enhancer elements. In total we selected 500K fragments for sequencing. These MspI-digested samples were ligated with Illumina adaptors, and size selected, denatured, and treated with sodium bisulfite to reveal their methylation status. These libraries were sequenced with Solexa sequencing technology (Illumina Hiseq 2000 sequencers). The reads were aligned to the reference genome (mouse mm9) using the modified bisulfite aligner, BS Seeker, to keep track of the fragment that each alignment was uniquely mapped to. To generate genome-wide DNA methylation profiles, we calculated the methylation level for each covered cytosine on the genome. As bisulfite treatment converted unmethylated cytosines (Cs) to thymines (Ts), we estimated the methylation level at each cytosine by #C/(#C + #T), where #C is the number of methylated reads and #T is the number of unmethylated reads. The methylation level per cytosine serves as an estimate of the percentage of cells that are methylated at this cytiosine. In this study we included only cytosines that are covered by at least four reads for the analysis. The resulting methylation profile per sample covered ∼1.4 M CpG sites. These methylation profiles and the processed data sets can be accessed at http://genomes.mcdb.ucla.edu/IUGR/.
Identifying differentially methylated regions and the associated genes.
We first searched for differentially methylated regions (DMR) that show significant differential methylation. Genes that are close to these DMR are considered differentially methylated.
For each CG site we calculated a t-score from the t-test of mean difference between the two groups of comparison and then select sites with |t-score| ≥1.5 (approximately top 10%) as markers of differential methylation. If two markers are within 80 bp (in our data median distance = 74 bp) then the region between them is deemed a candidate DMR. For each candidate DMR we then calculate a z-score of the average t-score from all CG sites within the region, as a measure of the differential methylation within this candidate DMR. When the |z-score| is greater than a threshold and the mean methylation levels in the two groups differ by at least 15%, this region is considered as differentially methylated (DMR). The selection of z-score threshold is based the false discovery rate (FDR) estimated as described below. In our analysis, hypermethylation in CR group, or in female group, has positive z-scores. Finally, if the genes overlap with any of these DMRs or if their transcription start sites are within 5 Kbp of the DMR, these genes are deemed differentially methylated. In total, we identified 297 genes that are differentially methylated between CR and CON, and 527 between male and female.
The DMR of CR vs. CON with sex effect was based on the comparison of two t-tests (male comparison: male CR vs. male CON, and female comparison: female CR vs. female CON). The t-score here is estimated by (t-score from female) − (t-score from male), and the same approach is used for calculating Δ m.
Estimating FDR.
To assess the FDR for our DMRs, we constructed 10 simulated methylomes, with the same read coverage per site as the real samples. For each CG site in each simulated sample, we then simulated the reads (C if methylated or T if unmethylated) based on the average methylation level (Pm) from all real samples at this CG site. The number of methylated reads (Cs) at a site of coverage n is a random sample from the binomial distribution B(n,Pm). We repeated our simulation of reads throughout the genome for all 10 samples. The resulting samples have the sample average methylation levels as the real sample, since the reads were simulated from the binomial distribution with the same average methylation levels as in the real samples, so the differences in methylation patterns across genes, repeats, promoters, etc. are preserved. The simulated data also have the same coverage as the real samples so the statistical power is not affected. The simulated methylomes should have no difference in methylation levels between the two comparison groups (i.e., no DMR), since we use the same methylation frequency to select them. Any DMR (and the DMR-associated genes) identified from these simulated samples are thus considered false positives. Finally, for each comparison (e.g., CR vs. CON) we repeated the whole procedure to detect the DMR on simulated samples. The resulting FDR are <5% in all comparisons (See Supplemental Table S9 for FDR).
To identify hotspots of differential methylation (i.e., genomic regions that are significantly clustered with differentially methylated genes) we tested the enrichment of differentially methylated genes within nonoverlapping windows of 3 Mb in the genome using the hypergeometric distribution. The P value cutoffs are selected such that the FDR [estimated by R module p.adjust using Benjamin's method (5)] is <5% (P ≤ 0.002 for the comparison between CR vs. CON, P ≤ 0.0007 for diet response with sex effect, P ≤ 0.003 for sex comparisons).
RESULTS
To profile transcription and DNA methylation in mouse placentas, we employed our established model of maternal CR in mice and compared the expression and methylation state of CR placentas to those of normal CON placentas (see materials and methods and Supplemental Information for the details of our mouse models). In total we collected 10 placentas for genome-wide DNA methylation profiling (RRBS) and another 10 placentas for expression analysis (RNA-Seq). All placentas arose from separate pregnancies. Equal numbers of samples were collected in each group, i.e., five from CR mice, and five placentas from CON (see Supplemental Information for sex determination of mice and Supplemental Table S1A/S1B for the number of samples in different diet and sex groups.). The placentas were collected after 10 days of CR, and therefore we hypothesize that the changes in expression and methylation between CR and CON are associated with this altered environment. From previous data we know that CR mice tend to develop increased fat mass and glucose intolerance compared with CON mice (30).
The RNA and genomic DNA from placenta samples were extracted for RNA-Seq and reduced representation bisulfite library (RRBS) library preparation, followed by massively parallel sequencing with Illumina HiSeq 2000 sequencers. The resulting RNA-Seq reads were mapped to mouse reference genome (mm9) using Tophat (55), and the differential expression was calculated using DEseq (2) (see Supplemental File 1 for table of expression level per gene). The RRBS reads were mapped using BS Seeker (12) to generate DNA methylation profiles at single base resolution with ∼50× coverage per strand (see materials and methods for the RNA-Seq library generation and data processing, RRBS protocols, mapping, and the estimation of methylation levels per cytosine). We compared gene expression and DNA methylation profiles between CR and CON. The differentially expressed genes are defined as those with log 2 ratio >0.2. For differential methylation, the statistical significance is controlled by FDR at <5%, which was estimated by generating synthetic reads from an empirically guided null model in which CR and control samples have the same methylation rates (See materials and methods for the details of computing FDR).
Maternal CR resulted in differentially expressed pathways.
We began by identifying differential expression between CR and CON (Fig. 1A), in which we observed sets of genes that are differentially regulated due to maternal diet. The comparison between sex (Supplemental Fig. S1) shows fewer changes, suggesting that the effect due to maternal diet is stronger than that of sex. Alpha fetoprotein is a protein produced by the liver that changes during pregnancy with growth restriction and other fetal congenital malformations such as neural tube defects, omphalocele, and liver defects (49). It is also a clinical marker for IUGR. We found that in our mouse model that this was the most significantly differentially expressed gene based on counts (RPKM), with the affected group showing higher levels than CON, validating our mouse model by reproducing human clinical data. However, this gene also had strong sex-specific effect within females but not in males; females CR showed higher levels of expression and methylation than female CON.
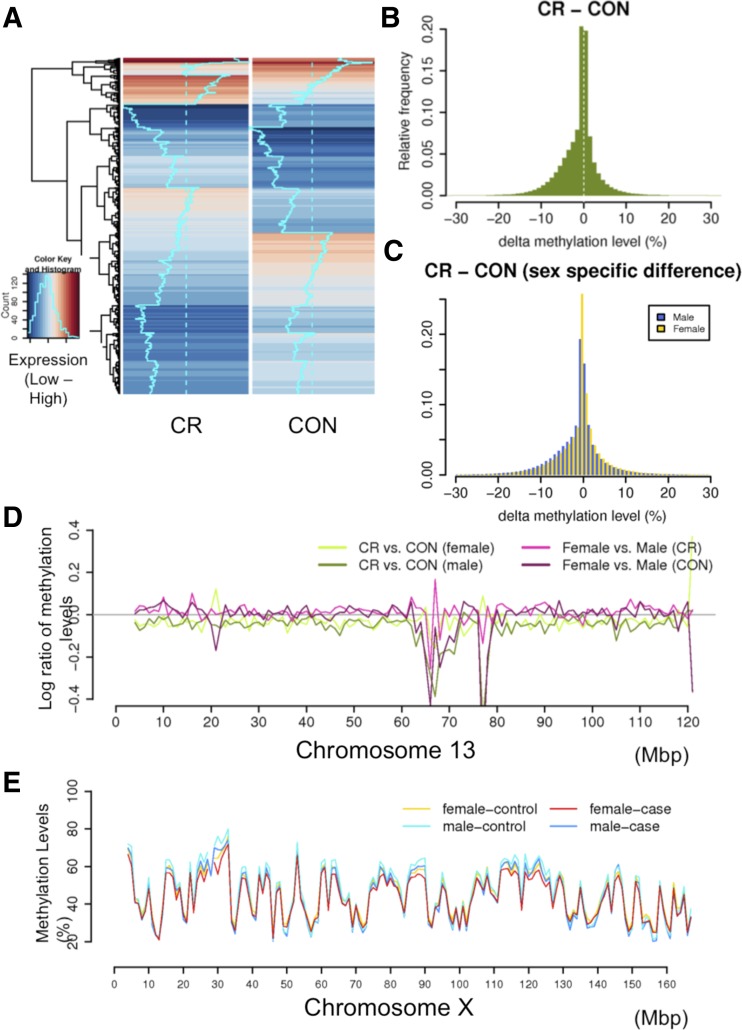
Fig. 1.
Comparisons of gene expression and DNA methylation levels in mouse placenta samples. A: heat map of gene expression levels in caloric restricted (CR) and wild-type (CON) groups; B: average methylation levels in CR vs. CON; C: histogram of Δ methylation levels per CG site between CR and CON. Log2 ratios of methylation levels between sample groups in genome-wide view (D) and in chromosome 13 (E).
We next searched for gene groups that are enriched in differentially expressed genes and found several associated with maternal CR (see Table 1 for selected pathways). Among these were pregnancy-specific glycoproteins and lipid-handling proteins. We found that 13 of the 16 pregnancy-specific glycoproteins sequenced were downregulated in CR samples (P <0.0001). Pregnancy-specific glycoproteins are postulated to have an immunomodulatory role in protecting the fetal allograft, as well as a role in promoting maternal angiogenesis to support the fetus. Lower levels of pregnancy-specific glycoproteins have been associated with IUGR, fetal hypoxia, and increased risk for abortion, thus providing a link to the CR-associated IUGR phenotype (58, 59). Among the 21 apolipoproteins sequenced, eight were upregulated in the CR condition, with the remaining 13 expressed equally in both conditions (enrichment test: P value < 0.0001). Similarly, two of the five fatty acid-binding proteins (FABPs) were upregulated in the CR condition, with the remaining three equally expressed (P = 0.03). This upregulation likely represents an attempt to increase the nutritional uptake of the CR fetus. Indeed, placental expression of some of these apolipoproteins and one or both of the FABPs has been linked to mother-fetus lipid transfer (38, 48). This compensatory response may represent an attempt to maintain the nutrient equipoise and thus represent one mechanism for the increased incidence of adult disease in malnourished fetuses. The FABPs and several of the apolipoproteins involved have been implicated with cardiovascular and metabolic disease in adults, and elevated fetal blood levels of one, ApoB, have already been associated with IUGR and adult atherosclerosis (11, 21, 45).
Table 1.
Differentially expressed pathways
| Pathway | Total Genes in Pathway | Upregulated in CR | Downregulated in CR | P of Enrichment Test | Note |
|---|---|---|---|---|---|
| Pregnancy-specific glycoproteins | 16 | Psg16 Psg17 | 5.46E-16 | elevated expression in males (P = 4.48E-22) | |
| Psg18 Psg19 | facilitate maternal immune tolerance of fetus | ||||
| Psg20 Psg21 | lower levels associated with IUGR, fetal hypoxia, and increased abortion risk | ||||
| Psg22 Psg23 | |||||
| Psg25 Psg26 | |||||
| Psg27 Psg28 | |||||
| Psg-ps1 | |||||
| Apolipoproteins | 21 | Apoa2 Apoc1 | 1.45E-6 | mostly upregulated in males vs. females | |
| Apoc2 Apom | implicated with cardiovascular and metabolic disease in adults, and elevated fetal blood levels of one, ApoB, have already been associated with IUGR and adult atherosclerosis | ||||
| Apoe Apob | |||||
| Apoa4 Apoa1 | |||||
| Fatty acid binding proteins | 7 | Fabp4 Fabp5 | 0.03 | maternal-fetal lipid exchange | |
| cardiovascular and metabolic disease | |||||
| Cathepsins and Granzymes | 27 | Ctss Ctsh | Cts3 Ctsm | 1.90E-8 | granzymes have higher expression in males. (P = 2.82E-8) |
| Gzmf Gzmd | potential role in trophoblast invasion | ||||
| Ctsk Gzmg | |||||
| Gzmc Gzme |
P value calculated from the hyergeometric test of enrichment. CR, caloric restriction; IUGR, intrauterine growth restriction.
DNA methylation changes and sex differences.
We next compared the global DNA methylation levels across our samples (see Supplemental Table S2A/B for individual samples). The average methylation levels are calculated at 1,195,334 cytosines for which we have methylation data across all samples. We observed that the maternal CR group is less methylated than the CON mice (P = 0.018, t-test, Fig. 1A, Supplemental Table S2C). The average Δ methylation level is ∼2%. The histogram of Δ methylation levels also shows more sites are less methylated in CR than in CON (Fig. 1B). These differences were observed across the genome where particular regions appear to be more differentially methylated (Fig. 1C). Furthermore, we found that there is a more significant level of demethylation in male than female mice following CR (P < 2e-16, Kolmogorov-Smirnov test, see Supplemental Fig. S1 for histograms of Δ methylation levels by sex). The methylation difference due to maternal diet in males is 67% more than that in females.
We next investigated whether differential methylation is associated with specific genomic features, such as coding genes, exons, CpG islands, and repetitive sequences, and observed no distinct patterns between these, suggesting that the change of methylation is generally nonspecific (Supplemental Fig. S2). These methylation patterns in mouse placentas are similar to those in mouse embryos, except the overall methylation level is lower (18). However, we do observe that IUGR dramatically affects the methylation of specific loci. For example, a detailed view of chromosome 13 (Fig. 1D) shows that certain megabase-sized regions are hypermethylated in CR compared with CON, with sex-specific differences. The genes within this region are found to be associated with genetic disorders, skeletal and muscular disorders, and developmental disorders (Supplemental Table S3). Lastly, we compared male and female placentas independent of diet, to identify general sex-specific methylation differences in placentas (see Supplemental Information).
Differentially methylated genes are clustered across the genome.
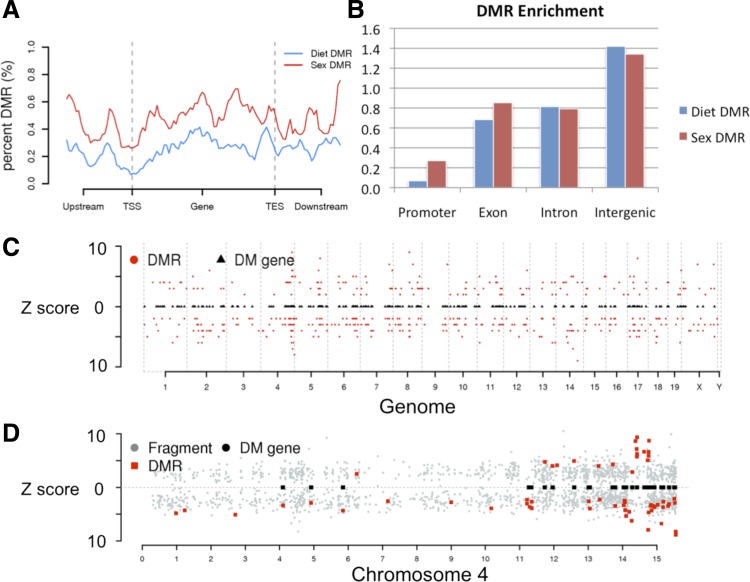
Given the observed global changes of methylation profiles in CR, we searched for hotspots of differential methylation that are affected by maternal diet. RRBS fragments are enriched for CG-rich regions and CpG islands and hence are also enriched for cis-regulatory sites. Thus changes in their methylation could either cause or be associated with changes in transcriptional regulation. As expected, we find that the distribution of methylation levels in CR fragments tends to be slightly lower than CON fragments in both sexes (P < 2.2e-16, Kolmogorov-Smirnov test, see Supplemental Fig. S3). Within these testable fragments we identified 477 DMR (FDR <5%) of significance between CR and CON (for the detailed statistical procedure see materials and methods). These DMRs are depleted from promoter regions (Fig. 2A). Instead, intergenic regions are enriched with DMRs (Fig. 2B), which agrees with a recent finding that distal regulatory regions, where transcriptional regulatory enhancers are often located, show altered methylation status (53). Our DMRs are proximal to 297 genes. Of these, 131 genes are hypermethylated in CR samples, and 168 are hypomethylated (Supplemental Table S4, see Supplemental File 2 for list of DMR and genes). Although the differentially methylated genes are generally spread across the genome, we did find some clusters within chromosomes (Supplemental File 3). Figure 2, C and D, shows the distribution of DMRs in the genome and in chromosome 4 where some of these clusters reside.
Fig. 2.
Distribution of differentially methylated regions (DMR) and genes. A: metagene plot of percent DMR of reduced representation bisulfite sequencing (RRBS) fragments; B: fold enrichment of DMRs in promoters, exons, introns, and intergenic regions. Distribution of DMRs and the associated genes in a genome-wide view (C) and in chromosome 4 (D). Circles are all candidate regions with the z-scores in y-axis, squares are the differentially methylated genes associated to DMR (red circles).
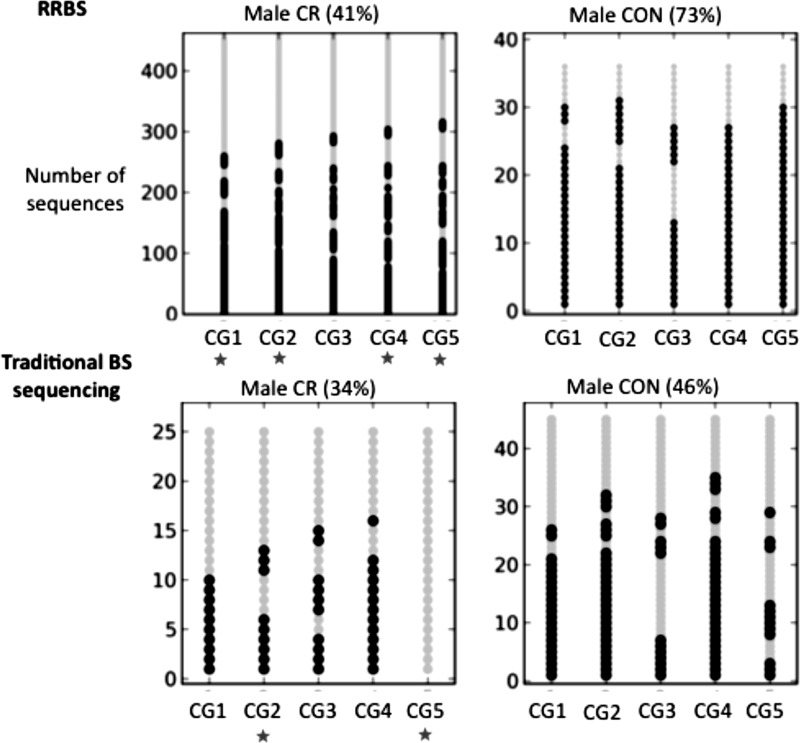
As a validation of the RRBS-predicted DMRs, we performed traditional bisulfite sequencing (see Ref. 10 for method and Supplemental Table S5 for primers) on three DMRs that are flagged to be differentially methylated between male CR and male CON (Fig. 3 and Supplemental Fig. S4). Using this approach we were able to validate two of the three loci that showed significant changes in the validation data. Figure 3 shows one of the DMRs with a significant change in methylation level in both RRBS and the traditional bisulfite sequencing (32 and 13%, respectively). In this locus we find several sites that have significant differential methylation between CR and CON, as shown by the red stars. Despite the fact that one of the loci was not significantly differentially methylated in the validation data, all three DMRs were consistent with respect to the direction of methylation differences.
Fig. 3.
DMR validation using traditional bisulfite sequencing. Bubble plots show the RRBS sequences aligned to a DMR (chr14:99765624–99765704) (top) and the traditional bisulfite sequencing data generated from the same locus (bottom). Each row shows the methylation status of the 5 CpG sites within this region (black and gray circles stand for methylated and unmethylated cytosines, respectively). The average methylation levels are shown in parentheses. *P < 0.05 for a binomial test of differential methylation for that site between CR and CON placentas.
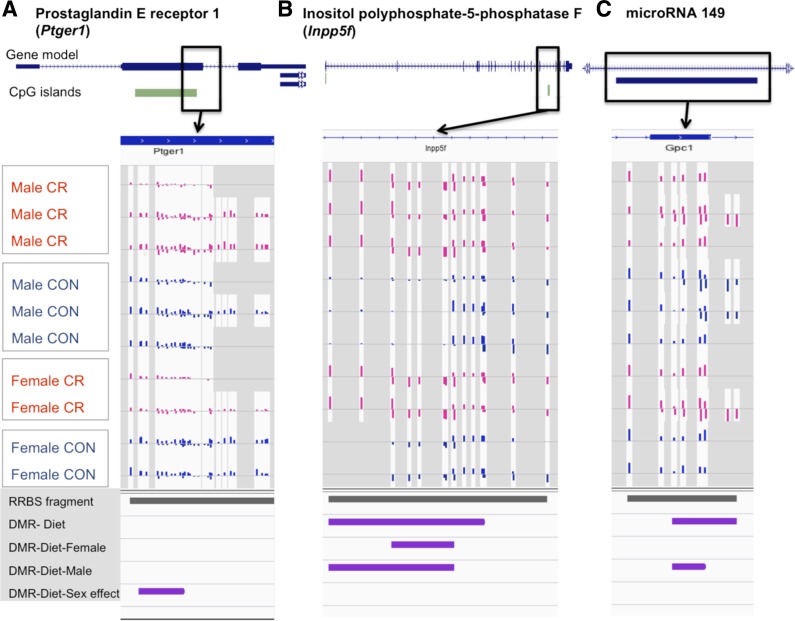
To investigate if imprinted genes in placentas are more susceptible to methylation changes in response to diet, we compared our list of differentially methylated genes, between CR and CON and between sexes, with 113 known imprinted genes (Supplemental File 4) (43). We found that differentially methylated genes are enriched among the 81 known imprinted genes that are covered by our data; nine (Igf2, Inpp5f, Dlk1, Gnas, Usp29, Wt1, Kcnk9, Grb10, Cdkn1c) show differential methylation in CR with or without sex preference, and seven between sexes (Nnat, Mest, Blcap, Peg13, Snrpn, Grb10, Gnas) (P < 0.09, using the hypergeometric test of enrichment). This enrichment of imprinted genes suggests that the methylation of these genes may be more sensitive to changes in the environment.
For example, Inpp5f is an imprinted gene that regulates cardiac hypertrophic responsiveness. It has been reported to be paternally expressed and maternally methylated (13). We identified a CpG island within the gene that was significantly hypermethylated in CR (see Fig. 4B). Another imprinted gene Igf2 serves as a fetal growth factor associated with fetal growth (14). In a study of Igf2P0 (placenta-specific knockout) mouse, a decrease in system A amino acid transport across the placenta was reported with fetal growth restriction (50). In our data we observed altered methylation patterns in exon 3 of Igf2 corresponding to a hypermethylated state in CR samples. These findings are consistent with previous reports in IUGR that imprinted genes in placentas are susceptible to epigenetic changes (46).
Fig. 4.
Screenshots of DNA methylation tracks at ptger1 (A), inpp5f (B), and microRNA 149 (C). Gene annotation tracks are at top, followed by 10 tracks of methylation levels from CR and CON samples, and the DMR tracks at bottom. The methylation levels at each measured CG site are represented as bars whose length represents methylation levels from 0 to 100%.
Differential methylation is associated with genes related to cardiovascular, metabolic, and neurological diseases.
We performed a functional analysis of the differentially methylated genes using the DAVID Bioinformatics Resources 6.7 (29) and the Ingenuity Pathway Analysis tool (29a). We found that differentially methylated genes in CR are enriched for functional categories such as transcriptional regulation, learning, cytoplasmic vesicle, cell morphogenesis involved in neuron differentiation, lipid binding, regulation of neuron apoptosis, behavior, and fatty acid metabolic process (Supplemental Table S6). In the network analysis, we found that the major networks are enriched with functions in embryonic development, nervous system development, and cardiovascular system development and function (Table 2). The associated disorders include cardiovascular disease (e.g., heart disease, vascular disease), neurological disease (e.g., bipolar, Parkinson's, Alzheimer's diseases), and metabolic disorders (e.g., Crohn's disease, noninsulin-dependent diabetes mellitus). Although there may not be direct links between the differential methylation we observe and these diseases, it is proposed that alterations to placental pathways could potentially be similar to those found within the fetus and thus link IUGR to the above late onset disorders (40). Supplemental Figure S5 (network view) shows a major network including 21 differentially methylated genes that are enriched with functions in cardiovascular system development and function, cellular movement and embryonic development.
Table 2.
Differentially methylated genes and enriched gene networks
| Comparison | DMR | Differentially Methylated Genes | Top Major Networks [significance score = −log10 (P value)] |
|---|---|---|---|
| CR vs. CON | nervous system development and function, embryonic development, organ development (41) | ||
| cardiovascular system development and function, cellular movement, embryonic development (36) | |||
| 477 | 297 | cellular assembly and organization, cellular function and maintenance, cellular movement (32) | |
| cell-to-cell signaling and interaction, developmental disorder, endocrine system disorders (31) | |||
| cardiovascular system development and function, tissue development, protein synthesis (27) | |||
| CR vs. CON (with sex effect) | cellular development, nervous system development and function, visual system development and function (49) | ||
| cellular function and maintenance, auditory and vestibular system development and function, organ morphology (41) | |||
| 1,141 | 667 | cell signaling, nucleic acid metabolism, small molecule biochemistry (41) | |
| cancer, endocrine system disorders, gastrointestinal disease (37) | |||
| tissue morphology, connective tissue development and function, embryonic development (37) | |||
| CR vs. CON (female) | connective tissue development and function, connective tissue disorders, dermatological diseases and conditions (43) | ||
| cell signaling, molecular transport, vitamin and mineral metabholism. (41) | |||
| 881 | 459 | cellular assembly and organization, cellular compromise, embryonic development (38) | |
| cell signaling, connective tissue disorders, dental disease (32) | |||
| cellular assembly and organization, cellular compromise, carbohydrate metabolism (28) | |||
| CR vs. CON (male) | 892 | 477 | connective tissue development and function, embryonic development, organ development (43) |
| endocrine system development and function, molecular transport, small molecule biochemistry (36) | |||
| digestive system development and function, embryonic development, endocrine system development and function (36) | |||
| cell morphology, hematological system development and function, cell-to-cell signaling and interaction (31) | |||
| cellular function and maintenance, molecular transport, cell-to-cell signaling and interaction (29) | |||
| Female vs. male | 855 (413 from autosome, 114 from chrX) | 572 | cellular assembly and organization, cellular function and maintenance, cellular compromise (45) |
| embryonic development, organismal development, skeletal and muscular system development and function (45) | |||
| developmental disorder, skeletal and muscular disorders, hereditary disorder (42) | |||
| cell cycle, organismal development, auditory disease (33) | |||
| cell death and survival, cellular function and maintenance, cell cycle (31) |
DMR, differentially methylated region; CON, control group.
Sex-specific effects on maternal CR.
We identified 667 differentially methylated genes (FDR ≤ 4.03%) that are significantly differentially methylated between CR and CON in a sex-specific manner (see materials and methods). Of these genes, 380 genes are hypermethylated in male CR vs. male CON as opposed to the female CR vs. female CON, and 309 genes show inversely hypermethylation in female CR vs. female CON (Supplemental Table S4, see Supplemental File 2 for list of DMR and genes). This large number of genes is distributed throughout the genome (Supplemental Fig. S6), suggesting the changes of methylation in response to maternal diet have a strong sex-specific component. In the functional analysis, we found that these differentially methylated genes are enriched with functions in embryonic morphogenesis, metabolic processes (Supplemental Table S6), and with the networks of cellular development, nucleic acid metabolism, and auditory and vestibular system development (Table 2). Specifically, the genes that are hypermethylated in female CR are enriched in the network of lipid metabolism, nerve system development and function, and developmental disorders, whereas those hypermethylated in male CR are enriched in networks of cell and organ morphology. Placentas are functional during a critical developmental window in which both the gametes and the sex organs are determined. The functional categories we identified contain key genes that are possibly under epigenetic control within the fetus and affect these processes.
Prostaglandin receptor and glucose transporter.
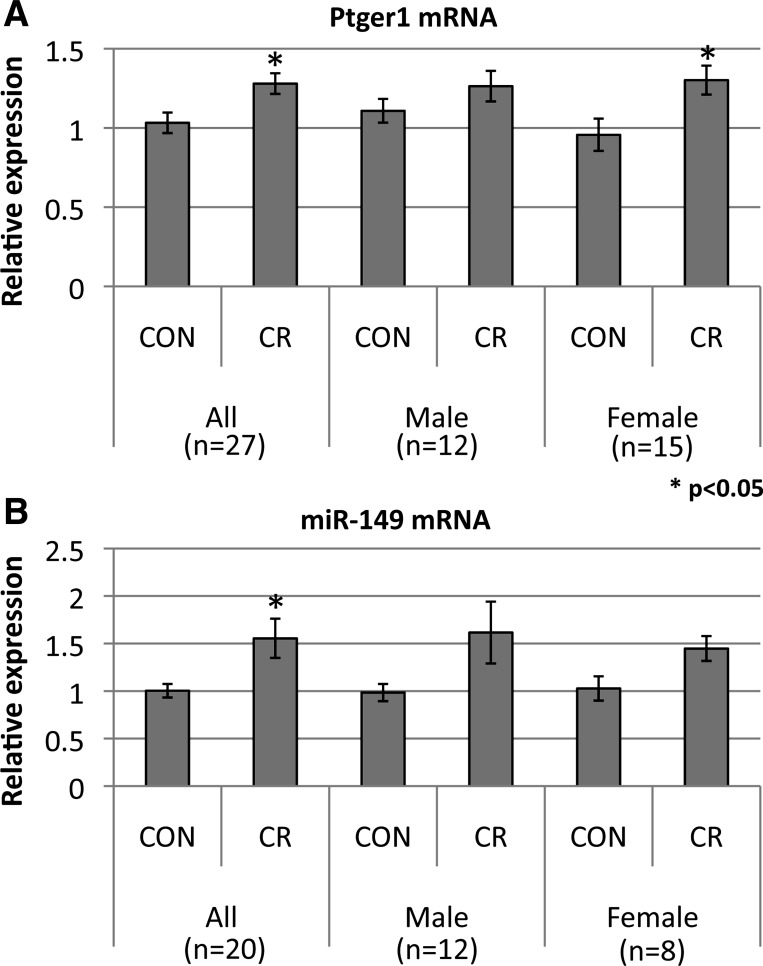
To further examine the relationship between DNA methylation and transcription in the differentially methylated genes, we performed qRT-PCR (see Supplemental Information for RNA preparation and qRT-PCR) on the prostaglandin E receptor 1 (ptger1), which we observed to be less methylated in CR at a CpG island in the second exon (Fig. 4A). Interestingly, the change of methylation is significantly greater in females than in males. An increased expression level is observed in CR (P = 0.012, t-test, 26 biological replicates, see Fig. 5A), suggesting that the change of methylation at ptger1 is associated with its transcription. However, only the female CR group shows a significant change in expression, which is consistent with the female-specific methylation change. Ptger1 is a receptor of prostaglandin E2 (PGE2), which is a vasodilator that acts to lower blood pressure and also acts to induce labor. Ptger1 with its function of regulating blood pressure has also been associated with diabetes, preeclampsia, and premature birth; all three conditions can be associated with IUGR and have long-term fetal programming implications. Ptger1 is also known to mediate hypertension resulting in end organ damage (4).
Fig. 5.
Expression level of ptger1 and miR-149 between CR and CON groups in all, male, and female samples. The size of biological replicates (n) for each comparison is shown in parentheses.
A second gene of interest is Glut3. We have previously shown that maternal CR in mice led to a decrease in placental Glut3 protein expression along with a functional decrease in trans-placental glucose transport (24, 26). Furthermore, using candidate gene-specific methylation-sensitive PCR we detected hypermethylation of this gene in placentas exposed to CR (A. Ganguly, SU Devaskar, unpublished data). The CpGs within a stretch that extended from −805 to 922 bp 5′ to the transcription start site (TSS) were specifically hypermethylated. This hypermethylation was associated with a decrease in placental Glut3 expression. In our present investigation, we explored this further using our genome-wide data and confirmed that the Glut3 gene, situated on the negative strand of chromosome 6, is hypermethylated in CR vs. CON (Supplemental Fig. S7). Specifically +717 to +1,040 bp, 5′ to the TSS was hypermethylated (P = 0.008). Thus in the case of a gene that is critically important for transplacental glucose transport and embryonic survival (26), DNA methylation in CR was associated with changes in gene expression and its ultimate function.
Differentially methylated microRNAs target metabolic genes.
MicroRNAs affect posttranscriptional regulation of genes, typically regulating the transcription and translation of many target genes. MicroRNAs have also been shown to play a role in epigenetic inheritance in mice (57). We found within our list of differentially methylated genes 19 microRNAs (Supplemental Table S7). miR-149 is only 65 bp in length and is encoded in one hypermethyated fragment in CR vs. CON (Fig. 4C). RT-qPCR (see Supplemental Information for qRT-PCR of miR-149) shows that this gene is differentially expressed in CR vs. CON in placentas (P = 0.026, t-test, 20 biological replicates, see Fig. 5B), suggesting that the difference of methylation may be associated with its expression. Gene ontology analysis of its target genes finds that these are enriched for functions associated with embryonic development and associated with cardiovascular and metabolic diseases. One of its target genes is the system L amino acid transporter isoform 2 (LAT2), which in females is hypermethylated in CR vs. CON but not in males. The expression of placental LAT2, however, shows no expression change in response to maternal CR but a decline in protein abundance, supporting the notion that in this case the microRNA regulates protein translation (A. Ganguly, SU Devaskar, unpublished data). The decrease in LAT2 mediates diminished transplacental leucine transport in CR. Thus in CR, hypermethylation of the miR-149 gene body leads to increased miR-149 gene expression, which is associated with decreased LAT2 protein concentrations.
Association between changes of DNA methylation and gene expression.
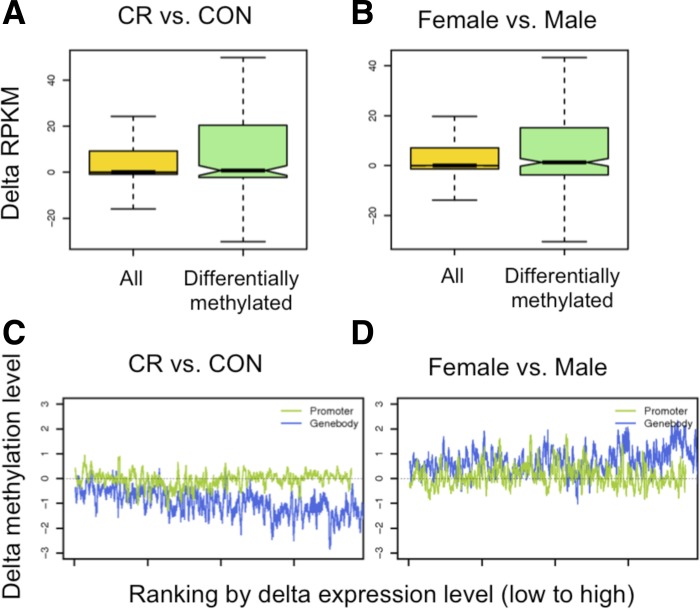
The global correlation between the changes of DNA methylation and gene expression is weak (Pearson correlation <5%). However, we found that differentially methylated genes had a greater variation in gene expression (measured via RPKM) compared with all genes (Fig. 6, A and B; Supplemental Fig. S10), suggesting that changes in DNA methylation tend to increase the variability of gene expression. To further study the effect of methylation changes in transcription changes, we plot methylation levels in promoters and gene bodies ranked by their changes in gene expression levels (Fig. 6, C and D; Supplemental Fig. S11). The result shows that in our data gene bodies have a stronger association with expression than promoters. We find that genes whose expression increases in CR vs. CON tend to lose methylation within the gene body but are unaffected in their promoters. This suggests a subtle coupling between the rate of transcription and methylation, suggesting that the latter is dynamically regulated by the transcriptional machinery.
Fig. 6.
Change of methylation vs. change of expression. Boxplots of changes of expression levels in differentially methylated genes between CR and CON (A) and between female and male (B). Changes of methylation levels in promoters and genes ranked by the change in gene expression level between CR and CON (C) and between female and male (D).
DISCUSSION
In this study, we generated placental genome-wide DNA methylation and transcriptional profiles in CR and CON pregnant mice. We identified differentially regulated pathways associated with IUGR, including a significant change in alpha-fetoprotein as well as pregnancy-specific glycoproteins. We also observed a mild decrease of global methylation levels in CR. We find that CR samples consistently have lower methylation levels throughout the genome. This result is consistent with previous reports from specific loci in rats that showed that prenatal nutritional constraints result in hypomethylation (33, 34) and that the reduced expression of DNMT1 is likely involved in this impaired methylation. Furthermore, these studies examine the connection between hypomethylation and a wide range of developmental and metabolic processes (6, 32).
We identified 297 genes that are significantly differentially methylated due to maternal CR. These genes are clustered within chromosomes, supporting the notion that while maternal diet affects global DNA methylation levels of the placenta, certain regions appear to be more susceptible than others. These placental mechanisms that are mediated by epigenetic adaptation may promote fetal survival at the expense of achieving optimal energy balance and growth. While surviving the adverse in utero environment, some of these perturbations in gene methylation may predispose the offspring for adult-type chronic diseases. Thus, specific genes that are epigenetically regulated could serve as placental biomarkers for predicting the potential of developing the disease phenotype in the adult, which could be clinically important for diagnosing and predicting the outcome of low-birth-weight babies.
We also found that known imprinted genes are significantly enriched in our lists of differentially methylated genes between CR and CON and between sexes. This suggests that imprinted genes in placentas are particularly sensitive to environmental changes. This observation supports previous findings that these genes are often critical for establishing the growth and size of the developing fetus and are under different adaptive pressures depending on whether they are maternally or paternally imprinted.
Ptger1 was found to be one of the most significant sex-specific differentially methylated genes in CR vs. CON. This raises a possibility that dysregulation of DNA methylation of the Ptger1 gene in females may underlie the mechanism of preterm birth in maternal undernutrition. It has been clinically and experimentally shown that preterm birth rate increases in the cohort conceived during famine and animals fertilized under CR (8, 20, 35, 36, 51, 54). Ptger1 mediates the effect of prostaglandin E2 (PGE2), an uterotonic agent that is clinically used for prevention of postpartum hemorrhage as well as induction of abortion. Upregulation of Ptger1 in the female CR group in our study increases the effect of PGE2, which may lead to the contraction of the uterus and preterm delivery. These functional consequences need to be further investigated in our murine model of maternal CR in the future.
An unexpected result in our data is the enrichment of differentially methylated genes that are associated with cardiovascular disease. This observation is consistent with the previous publications by us and others demonstrating that extraembryonic tissues display latent cardiogenic potential: a population of yolk sac cells show contractile phenotype in ex vivo culture (44) or by genetic manipulation (56), and placental cells may contribute to the maternal heart during peripartum cardiomyopathy (31). Thus, our current data might suggest that this latent cardiogenic potential is sensitive to the nutrition status.
Fetal malnutrition is linked to the risk of adult cardiovascular diseases including coronary heart disease (47). This may be due to the placental malfunction, as reconstitution of the placenta restores heart development in an experimental setting (3). Alternatively, our data raise the possibility that the methylation status of the cardiovascular genes in the placenta is sensitive to the nutritional state in other tissues including the heart. The direct impact of malnutrition on DNA methylation of cardiovascular tissues remains to be elucidated in the future.
MicroRNAs have been implicated in epigenetic regulation by posttranscriptionally altering transcripts (16). Our finding of differential methylation of certain placental microRNAs by maternal CR is novel, in particular the observation that the hypermethylation of the miR-149 gene body is associated with enhanced miR-149 gene expression. While LAT2 was known to be a target of this microRNA, previous investigations by us have demonstrated that maternal CR led to no change in placental LAT2 mRNA. However, measurement of the LAT2 protein revealed that it has reduced concentrations in response to CR (24). Thus it appears that maternal CR may affect initiation of LAT2 protein translation via activation of miR-149 expression. LAT2 is a system L amino acid transporter that mediates transplacental branched chain amino acid transfer (e.g., leucine, isoleucine). In the IUGR fetus, there is a perceptible decrease in circulating branched chain amino acids, perhaps related to a diminution of transplacental transfer. Combining the information on placental Glut3 and LAT2 in CR, it appears that while Glut3 DNA hypermethylation transcriptionally affects its gene expression, hypermethylation of miR-149 posttranscriptionally may affect the protein translation of LAT2. Both of these genes mediate transplacental nutrient transport and are critically important in fueling fetal energy metabolism and growth. In CR, diminished concentrations of both these proteins results in reduced materno-fetal glucose and leucine transport, thereby contributing to diminished fetal growth and its associated consequences (24).
Our study investigates altered transcription and methylation in placentas. We identified the enriched functions of differentially expressed or methylated genes as possible phenotypes. Our results strongly support the notion that the expression and methylation state of the placenta is sensitive to the intrauterine environment, and it is likely that these changes have profound effects on fetal development. Measuring genes in the placenta is important to identify potential markers of IUGR as well as suggesting new biochemical mechanisms that could affect fetal development.
GRANTS
This work was supported by National Institutes of Health Grants HD-46979, HD-33997, HD-25024 and HD-41320 to S. U. Devaskar, and R01 GM095656-01A1 to M. Pellegrini. S. Feng is Special Fellow of the Leukemia & Lymphoma Society.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: P.-Y.C., D.A., and A.J. analyzed data; P.-Y.C., D.A., S.E.J., A.N., S.U.D., and M.P. interpreted results of experiments; P.-Y.C. prepared figures; P.-Y.C. drafted manuscript; P.-Y.C., S.U.D., and M.P. edited and revised manuscript; P.-Y.C., S.U.D., and M.P. approved final version of manuscript; A.G., L.R., L.D.O., M.M., S.F., and S.E.J. performed experiments; S.U.D. and M.P. conception and design of research.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Weihong Yan for the data visualization on genome browser.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 2. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 11: R106, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell 4: 585–595, 1999 [DOI] [PubMed] [Google Scholar]
- 4. Bartlett CS, Boyd KL, Harris RC, Zent R, Breyer RM. EP1 disruption attenuates end-organ damage in a mouse model of hypertension. Hypertension 60: 1184–1191, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300, 1995 [Google Scholar]
- 6. Bertram C, Trowern AR, Copin N, Jackson AA, Whorwood CB. The maternal diet during pregnancy programs altered expression of the glucocorticoid receptor and type 2 11beta-hydroxysteroid dehydrogenase: potential molecular mechanisms underlying the programming of hypertension in utero. Endocrinology 142: 2841–2853, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Bhasin KK, van Nas A, Martin LJ, Davis RC, Devaskar SU, Lusis AJ. Maternal low-protein diet or hypercholesterolemia reduces circulating essential amino acids and leads to intrauterine growth restriction. Diabetes 58: 559–566, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bloomfield FH, Oliver MH, Hawkins P, Campbell M, Phillips DJ, Gluckman PD, Challis JR, Harding JE. A periconceptional nutritional origin for noninfectious preterm birth. Science 300: 606, 2003 [DOI] [PubMed] [Google Scholar]
- 9. Burton GJ, Jauniaux E, Charnock-Jones DS. The influence of the intrauterine environment on human placental development. Int J Dev Biol 54: 303–312, 2010 [DOI] [PubMed] [Google Scholar]
- 10. Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99, Suppl 4: 16491–16498, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan DC, Barrett HP, Watts GF. Dyslipidemia in visceral obesity: mechanisms, implications, and therapy. Am J Cardiovasc Drugs 4: 227–246, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 11: 203, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi JD, Underkoffler LA, Wood AJ, Collins JN, Williams PT, Golden JA, Schuster EF, Loomes KM, Oakey RJ. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol 25: 5514–5522, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Constancia M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, Fowden A. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci USA 102: 19219–19224, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature 432: 53–57, 2004 [DOI] [PubMed] [Google Scholar]
- 16. Devaskar SU, Raychaudhuri S. Epigenetics–a science of heritable biological adaptation. Pediatr Res 61: 1R–4R, 2007 [DOI] [PubMed] [Google Scholar]
- 17. El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, Walter J, Horsthemke B. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 27: 341–344, 2001 [DOI] [PubMed] [Google Scholar]
- 18. Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res 65, Suppl 3: 50–58, 2006 [DOI] [PubMed] [Google Scholar]
- 20. Fowden AL, Silver M. The effect of the nutritional state on uterine prostaglandin F metabolite concentrations in the pregnant ewe during late gestation. Q J Exp Physiol 68: 337–349, 1983 [DOI] [PubMed] [Google Scholar]
- 21. Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7: 489–503, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gabory A, Ferry L, Fajardy I, Jouneau L, Gothie JD, Vige A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS One 7: e47986, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vige A, Breton C, Reusens B, Remacle C, Vieau D, Ekstrom TJ, Jais JP, Junien C. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One 5: e14398, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ganguly A, Collis L, Devaskar SU. Placental glucose and amino acid transport in calorie-restricted wild-type and Glut3 null heterozygous mice. Endocrinology 153: 3995–4007, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ganguly A, Devaskar SU. Glucose transporter isoform-3-null heterozygous mutation causes sexually dimorphic adiposity with insulin resistance. Am J Physiol Endocrinol Metab 294: E1144–E1151, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Ganguly A, McKnight RA, Raychaudhuri S, Shin BC, Ma Z, Moley K, Devaskar SU. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am J Physiol Endocrinol Metab 292: E1241–E1255, 2007 [DOI] [PubMed] [Google Scholar]
- 27. Garg M, Thamotharan M, Pan G, Lee PW, Devaskar SU. Early exposure of the pregestational intrauterine and postnatal growth-restricted female offspring to a peroxisome proliferator-activated receptor-γ agonist. Am J Physiol Endocrinol Metab 298: E489–E498, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haig D, Westoby M. Parent-specific gene-expression and the triploid endosperm. Am Nat 134: 147–155, 1989 [Google Scholar]
- 29. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 29a. Ingenuity Systems Ingenuity Pathway Analysis software. Redwood City, CA: Ingenuity Systems [Google Scholar]
- 30. Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, Patti ME. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 58: 460–468, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kara RJ, Bolli P, Karakikes I, Matsunaga I, Tripodi J, Tanweer O, Altman P, Shachter NS, Nakano A, Najfeld V, Chaudhry HW. Fetal cells traffic to injured maternal myocardium and undergo cardiac differentiation. Circ Res 110: 82–93, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135: 1382–1386, 2005 [DOI] [PubMed] [Google Scholar]
- 33. Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr 100: 278–282, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. Br J Nutr 97: 1064–1073, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lumey LH, Ravelli AC, Wiessing LG, Koppe JG, Treffers PE, Stein ZA. The Dutch famine birth cohort study: design, validation of exposure, and selected characteristics of subjects after 43 years follow-up. Paediatr Perinat Epidemiol 7: 354–367, 1993 [DOI] [PubMed] [Google Scholar]
- 36. Lumey LH, Van Poppel FW. The Dutch famine of 1944–45: mortality and morbidity in past and present generations. Soc Hist Med 7: 229–246, 1994 [DOI] [PubMed] [Google Scholar]
- 37. Maccani MA, Marsit CJ. Epigenetics in the Placenta. Am J Reprod Immunol 62: 78–89, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Madsen EM, Lindegaard ML, Andersen CB, Damm P, Nielsen LB. Human placenta secretes apolipoprotein B-100-containing lipoproteins. J Biol Chem 279: 55271–55276, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci USA 107: 5557–5562, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marsit CJ, Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics 6: 920–927, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27: 540–549, 2006 [DOI] [PubMed] [Google Scholar]
- 42. Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 33: 5868–5877, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res 29: 275–276, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murakami Y, Hirata H, Miyamoto Y, Nagahashi A, Sawa Y, Jakt M, Asahara T, Kawamata S. Isolation of cardiac cells from E8.5 yolk sac by ALCAM (CD166) expression. Mech Dev 124: 830–839, 2007 [DOI] [PubMed] [Google Scholar]
- 45. Radunovic N, Kuczynski E, Rosen T, Dukanac J, Petkovic S, Lockwood CJ. Plasma apolipoprotein A-I and B concentrations in growth-retarded fetuses: a link between low birth weight and adult atherosclerosis. J Clin Endocrinol Metab 85: 85–88, 2000 [DOI] [PubMed] [Google Scholar]
- 46. Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2: 21–32, 2001 [DOI] [PubMed] [Google Scholar]
- 47. Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Schroeder-Tanka JM, van Montfrans GA, Michels RP, Bleker OP. Coronary heart disease after prenatal exposure to the Dutch famine, 1944–45. Heart 84: 595–598, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scifres CM, Chen B, Nelson DM, Sadovsky Y. Fatty acid binding protein 4 regulates intracellular lipid accumulation in human trophoblasts. J Clin Endocrinol Metab 96: E1083–E1091, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sebire NJ, Spencer K, Noble PL, Hughes K, Nicolaides KH. Maternal serum alpha-fetoprotein in fetal neural tube and abdominal wall defects at 10 to 14 weeks of gestation. Br J Obstet Gynaecol 104: 849–851, 1997 [DOI] [PubMed] [Google Scholar]
- 50. Sferruzzi-Perri AN, Vaughan OR, Coan PM, Suciu MC, Darbyshire R, Constancia M, Burton GJ, Fowden AL. Placental-specific Igf2 deficiency alters developmental adaptations to undernutrition in mice. Endocrinology 152: 3202–3212, 2011 [DOI] [PubMed] [Google Scholar]
- 51. Silver M, Fowden AL. Uterine prostaglandin F metabolite production in relation to glucose availability in late pregnancy and a possible influence of diet on time of delivery in the mare. J Reprod Fertil Suppl 32: 511–519, 1982 [PubMed] [Google Scholar]
- 52. Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods 48: 226–232, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495, 2011 [DOI] [PubMed] [Google Scholar]
- 54. Susser M, Stein Z. Timing in prenatal nutrition: a reprise of the Dutch Famine Study. Nutr Rev 52: 84–94, 1994 [DOI] [PubMed] [Google Scholar]
- 55. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, Zhou J, Li X, Pellegrini M, Chen JN, Orkin SH, Kurdistani SK, Evans SM, Nakano A, Mikkola HK. Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 150: 590–605, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell 14: 962–969, 2008 [DOI] [PubMed] [Google Scholar]
- 58. Wessells J, Wessner D, Parsells R, White K, Finkenzeller D, Zimmermann W, Dveksler G. Pregnancy specific glycoprotein 18 induces IL-10 expression in murine macrophages. Eur J Immunol 30: 1830–1840, 2000 [DOI] [PubMed] [Google Scholar]
- 59. Wynne F, Ball M, McLellan AS, Dockery P, Zimmermann W, Moore T. Mouse pregnancy-specific glycoproteins: tissue-specific expression and evidence of association with maternal vasculature. Reproduction 131: 721–732, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.