Abstract
The spontaneously hypertensive rat (SHR) has been widely used as a model for studies of hypertension and attention deficit/hyperactivity disorder. The inbred Wistar-Kyoto (WKY) rat, derived from the same ancestral outbred Wistar rat as the SHR, are normotensive and have been used as the closest genetic control for the SHR, although the WKY has also been used as a model for depression. Notably, however, substantial behavioral and genetic differences among the WKY substrains, usually from the different vendors and breeders, have been observed. These differences have often been overlooked in prior studies, leading to inconsistent and even contradictory findings. The complicated breeding history of the SHR and WKY rats and the lack of a comprehensive understanding of the genetic background of different commercial substrains make the selection of control rats a daunting task, even for researchers who are mindful of their genetic heterogeneity. In this study, we examined the genetic relationship of 16 commonly used WKY and SHR rat substrains using genome-wide SNP genotyping data. Our results confirmed a large genetic divergence and complex relationships among the SHR and WKY substrains. This understanding, although incomplete without the genome sequence, provides useful guidance in selecting substrains and helps to interpret previous reports when the source of the animals was known. Moreover, we found two closely related, yet distinct WKY substrains that may provide novel opportunities in modeling psychiatric disorders.
Keywords: WKY, SHR, autism, ADHD, inbred rat, animal models, SNP, behavior
the spontaneously hypertensive rat (SHR) is an inbred strain established from outbred Wistar rats selected for high blood pressure (8a). Although the SHR develops hypertension spontaneously at the age of 7–15 wk (36), prior to this age they have been used and validated as a model for attention deficit/hyperactivity disorder (ADHD), demonstrating all three ADHD core symptoms: hyperactivity, impulsivity, and inattentiveness (25, 28). The Wistar-Kyoto (WKY) strain was established from the same parental Wistar stock as the SHR and as a result has been commonly used as a control strain for the SHR (8a). Notably, however, the sources of WKY rats often varies between studies, and significant variability in the comparative results that employed these substrains has been noted, probably because the WKY breeding stock was distributed to different breeders prior to being fully inbred (11, 13, 21). In addition, the littermate pairings that produced the inbred WKY from the Wistar rat were initiated >10 yr after the inbreeding for the SHR was initiated (13), thus increasing the heterogeneity between the WKY and SHR and leading some to question the validity of the WKY as a control strain for SHR (30). Although some genetic substrains of the SHR have also been reported (16), the heterogeneity observed among SHR rats is usually less than that seen among WKY rats, as evidenced by an almost 100% incidence of hypertension in most lines (13).
In studying ADHD phenotypes in SHR rats, our group has previously reported substantial behavioral and genetic differences between two WKY substrains, WKY/NHsd (Harlan Laboratories, UK) and WKY/NCrl (Charles River Laboratories, Germany), as well as large genetic differences between the SHR and both WKY substrains (26). We determined that the three strains differed from each other in ∼30% of their genome sequence, based on single nucleotide polymorphism (SNP) data acquired using the Affymetrix Targeted Genotyping Rat Panel 5 K Arrays. These three strains also differed significantly in ADHD behaviors. The WKY/NCrl rats (Charles River, Germany), although not hyperactive, showed deficits in sustained attention similar to SHR/NCrl rats, whereas the WKY/NHsd rats (Harlan, UK) did not exhibit any hyperactivity, impulsivity, or inattention. Based on these findings, we suggested that WKY/NCrl rats would be a useful model for inattentive ADHD, while the WKY/NHsd rats should continue to serve as an appropriate behavioral control (26). As a direct corollary, seemingly contradictory findings should not be surprising if different substrains of WKY rats were used as controls for the SHR.
In addition to serving as genetic controls for the SHR in study of hypertension and ADHD, WKY rats have been found to exhibit some behavioral characteristics consistent with depression and anxiety (1, 19, 20). Importantly, however, variability in depressive and stress-responsive behaviors has been noted in WKY rats obtained from different vendors, such as Taconic, Harlan, and Charles River Laboratories (21). Nevertheless, despite the report of such differences, rats from these different suppliers are still used equally and often interchangeably. In the last 5 yr, equal numbers of studies used Harlan or Charles River WKY rats as a model for depression (n = 17 each).
In the last decade, with technical advances in microarray genotyping and genome sequencing, we can now use whole genome information to sort out the genetic relationships of different rat substrains, including the WKY and SHR. This knowledge can guide future choices of substrains for research and help improve the interpretation of results from published studies. In 2008, The STAR Consortium published the first whole genome-wide SNP data for 167 distinct inbred rat strains, including 13 SHR and 9 WKY inbred substrains and many other related congenic strains. During the same time, our group, in collaboration with Dr. Terje Sagvolden (1945–2011) from University of Oslo (Oslo, Norway), began to use Affymetrix genotyping arrays and gene expression arrays to examine the genetic differences of SHR and WKY substrains in order to understand variations in their ADHD-like behaviors (5, 22, 23, 26). The observations of the complex and large genetic variations among these strains from these studies and the concerns raised due to their phenotypic heterogeneity prompted us to further analyze their genetic relationship. In this paper, we combine our data and that from the STAR consortium and use statistical methods to identify new genome patterns and relationships that can be associated with behavioral differences between different rat substrains.
MATERIALS AND METHODS
Genome-wide SNP genotyping.
Whole genome-wide SNP genotyping was performed on Affymetrix Targeted Genotyping Rat Panel 1.0 10K arrays for four spontaneously hypertensive rats (SHR/NCrl) and four Wistar/Kyoto (WKY/NCrl) rats from Charles River Germany (Sulzfeld, Germany), three WKY/NCrl rats from Charles River US (Kingston, NY), and four WKY/NHsd rats from Harlan Europe (Blackthorn, Bicester, UK). Two males and two females were used for each of the substrains except the US WKY/NCrl rats (3 females). Genomic DNA was extracted from either tail or brain tissue with the Epicenter MasterPure DNA Purification Kit (Madison, WI). Brain tissue was used for all European-sources strains and was given by Dr. Sagvolden's laboratory at the University of Oslo. Animal procedures were approved by the Norwegian Animal Research Authority, and animals were housed and euthanized at the University of Oslo. Experiments were conducted in accordance with the laws and regulations controlling experimental procedures in live animals in Norway and the European Union. Brain parts were dissected and preserved in RNAlater or AllProtect (Qiagen) and shipped at room temperature to State University of New York Upstate Medical University. Tail tissue for the three US Charles River WKY/NCrl rats was purchased from the US Charles River Laboratories (Kingston, NY).
A total of 4 μg of DNA was prepared for hybridization using the Targeted Genotyping Rat Panel 1.0 10K Array, according to manufacturer protocol. The arrays were washed and stained using the TrueTag_Chip_Wash_R7_450 protocol on an Affymetrix Fluidics Station 450, scanned using an Affymetrix GeneChip Scanner 3000 7G/4C, and analyzed with Affymetrix GeneChip Targeted Genotyping Analysis Software. A total of 10,847 SNP markers were present on the array. After removing SNPs with missing genotype calls for all the animals, we obtained a set of 10,498 SNPs that we used to generate consensus genotypes for each strain.
We also obtained genome-wide SNP genotypes for 14 different strains from the STAR Project (http://www.snp-star.eu), including six SHR substrains, seven WKY substrains, and one Sprague-Dawley (SD) strain (See Table 1 for all the strain information). The SD strain was included as a contrast for the common Wistar origin of all the WKY and SHR substrains. Substrains that had been further bred out for special phenotypes, such as the SHRSP and WKHA rats, were not included. Strains with <2,000 SNP genotypes were also not included because they could not provide fully informative comparisons. The STAR project produced SNP genotypes for ∼21,000 markers. Of these, 9,407 SNPs matched with the SNP markers in the Rat Panel 1.0 10K Array and were used for the cross-strain comparison. Table 1 lists the source of the strains, and in each strain the number of SNPs with the available genotypes (AA, AB, or BB) used in the analysis and the percentage of heterozygous genotypes observed.
Table 1.
Strains included in the analysis
| Strains | SNPs, n | Heterozygous Calls, %AB | Data Source | Animal Origin |
|---|---|---|---|---|
| SHR/NCrl(German) | 9,307 | 0.17 | our study | Sulzfeld, Germany |
| WKY/NCrl(German) | 9,298 | 0.12 | our study | Sulzfeld, Germany |
| WKY/NCrl(US) | 9,214 | 0.21 | our study | Kingston, NY |
| WKY/NHsd(UK) | 9,150 | 0.20 | our study | Blackthorn, Bicester, UK |
| SD/Han(STAR) | 9,364 | 8.61 | STAR | Berlin |
| SHR/Izm (STAR) | 9,388 | 0.03 | STAR | Kyoto |
| SHR/Kyo(STAR) | 9,377 | 0.03 | STAR | Kyoto |
| SHR/molBbb(STAR) | 9,310 | 0.15 | STAR | Berlin |
| SHR/NCrl-1(STAR) | 9,380 | 0.02 | STAR | London |
| SHR/NHsd (STAR) | 9,188 | 2.22 | STAR | Harlan UK |
| SHR/OlaIpcv(STAR) | 9,266 | 0.16 | STAR | Prague, Czech Rep. |
| WKY/Bbb (STAR) | 9,270 | 0.19 | STAR | Berlin |
| WKY/Gla (STAR) | 9,294 | 0.26 | STAR | Glasgow |
| WKY/Izm (STAR) | 9,069 | 0.35 | STAR | Kyoto |
| WKY/NCrl (STAR) | 9,350 | 0.12 | STAR | London |
| WKY/Nmna(STAR) | 9,353 | 0.10 | STAR | Kyoto |
| WKY/Ztm (STAR) | 9,263 | 0.15 | STAR | Hannover |
| WKYO/Kyo (STAR) | 9,355 | 0.05 | STAR | Kyoto |
SNP, single nucleotide polymorphism; SHR, spontaneously hypertensive; WKY, Wistar-Kyoto.
Strain comparisons.
Two different methods were used for the genome-wide comparisons of different strains. We used PLINK (v1.07) to compute a pairwise identity-by-state (IBS) similarity matrix using the genotypes of 9,407 SNPs. We then imported it into STATA 12 for a classical metric multidimensional scaling (MDS) and cluster analyses. We also used principal factors factor analysis (PFFA) with varimax rotation in STATA 12 to extract factors representing the main characteristics of the subgroups of different strains. For the PFFA, only SNP genotypes present for all substrains were used (n = 8,209).
Functional annotation of divergent genomic regions.
The functional consequences of the divergent SNPs were annotated with Ensembl Variant Effect Predictor (VEP), which derives information such as whether they are intergenic, or within the known genes, either coding or noncoding regions (Ref. 14 and Supplementary File).1 The majority of the informative SNPs are not functional or within known genes they likely tag other causal variants within the linkage disequilibrium (LD) region. Thus, we retrieved known genes within the average LD region of the target SNPs using the University of California Santa Cruz Table Browser (9). We focused on regions 665 Kb upstream and downstream to the target SNPs that were divergent between the SHR/NCrl, WKY/NCrl, and WKY/NHsd rats. The 665 Kb region was used as the average size of LD blocks predicted for rats following Saar and colleagues (24). Genes from these regions were imported to Ingenuity Pathway Analysis (IPA; Ingenuity Systems, http://www.ingenuity.com) to determine if the genes in the divergent regions were enriched for canonical biological pathways or lists of candidate genes for specific disease/phenotypes. Table 2 lists the disease/phenotype gene lists, their sources, and the number of genes. Six of the diseases/phenotypes were chosen based on reported phenotypes in WKY and SHR strains: anxiety, depression, ADHD, epilepsy, hypertension, and obesity. We used two sources to derive the disease candidate genes, the Genotator database (34) and the IPA Ingenuity Knowledge Base. To use genes with a higher confidence of association with each disorder, we selected only those genes that were present in both data bases. Autism spectrum disorder (ASD) was also tested because one gene with known genetic mutations in WKY rats, Slc9a9, is a known candidate gene for autism. For ASD, we used the SFARI autism gene database, which was manually curated by ASD experts. Alzheimer's Disease and Parkinson's disease were chosen to serve as negative phenotype controls.
Table 2.
Candidate genes for disease phenotypes
| Disease List | Genes, n | Source |
|---|---|---|
| ADHD | 28 | IPA and Genotator |
| Anxiety | 43 | IPA and Genotator |
| Depression | 60 | IPA and Genotator |
| Obesity | 98 | IPA and Genotator |
| Hypertension | 183 | IPA and Genotator |
| Parkinson's disease | 325 | IPA |
| Autism spectrum disorder | 328 | SFARI |
| Alzheimer's disease | 679 | IPA |
| Epilepsy | 129 | IPA and Genotator |
ADHD, attention deficit/hyperactivity disorder; IPA, Ingenuity Pathway Analysis.
For both the pathway and candidate disease gene analyses, IPA calculates the probability that associations between a given collection of genes and a pathway/or gene list are due to simple chance. For each pathway and disease/phenotype list, a Fisher's exact test is computed by classifying all genes in the genome into a two-by-two table with rows defined by the presence/absence of the gene in the divergence list and columns by presence or absence of the gene's product in the canonical pathway or disease/phenotype list. The P value for each pathway/list is then calculated by considering 1) the number of genes that participate in that pathway, or list and 2) the total number of genes that are known to be associated with that pathway or list in Ingenuity's knowledge base. The more genes involved in a given pathway, the more likely the association is not due to chance, and the more significant the P value. Similarly, the larger the total number of genes known to be associated with the process, the greater the likelihood that an association is due to random chance, and the P value accordingly becomes less significant. Pathways with empirical P value 0.05 or stronger were subjected to multiple-testing correction via the Bonferroni method. The fold enrichment was calculated as the ratio of the percentage of disease candidate genes in each test gene list vs. the percentage of total disease genes in the genome background. Therefore a ratio of 1 indicates no enrichment for the disease genes.
Sequencing to verify genetic divergence.
We used PCR and sequencing to verify previously identified genetic variation in two genes, Slc9a9 (37) and Slc6a3 (15), in the four strains of interest, SHR/NCrl, WKY/NHsd, and the two WKY/NCrl substrains (from the US and Germany vendors). Primers that amplify the region that harbors these genetic variations were designed using the Primer3 program (http://frodo.wi.mit.edu/). PCR products were visualized on a 1.5% agarose gel, purified, and sequenced using GeneWiz services (http://Genewiz.com). Sequencing results were aligned using Sequencher 5.0 (GeneCodes) to show sequence variations. Primers flanking the Slc6a3 exon 4 genomic sequences are: 5′-TGACTGACCCTTGGGGATAC-3′ and 5′-GGAGAAAACCTCATGGCAAG-3′; primers flanking the Slc6a3 cDNA sequences are: 5′-CGATGCCCATGCCAGCAACT-3′ and 5′-AGACAGCAGCCCAGGCAGAAGA-3′; primers flanking the Slc9a9 exon 16 genomic sequences are: 5′-AATTGGATTTGGTTTGACGTG-3′ and 5′-CCAGATGGGGTGCAGAGTAG-3′.
RESULTS
Genetic divergence of WKY and SHR rat strains.
A total of 9,407 SNPs had genotypes available from both the STAR rat substrains and the rat substrains we genotyped using the Affymetrix Targeted Genotyping Rat Panel 1.0 10K (US WKY/NCrl and WKY/NHsd rats, as well as SHR/NCrl and German WKY/NCrl rats). These 9,407 SNPs were used in our strain comparison analysis. The number of available genotypes and the percentage of SNPs with heterozygous genotypes for each strain are summarized in Table 1. Notably, the outbred SD rats had 8.6% heterozygous SNPs, and SHR/NHsd rats had 2.2%. All other stains showed very low frequencies of heterozygous genotypes, consistent with a high degree of inbreeding, with the highest observed frequency only 0.35%.
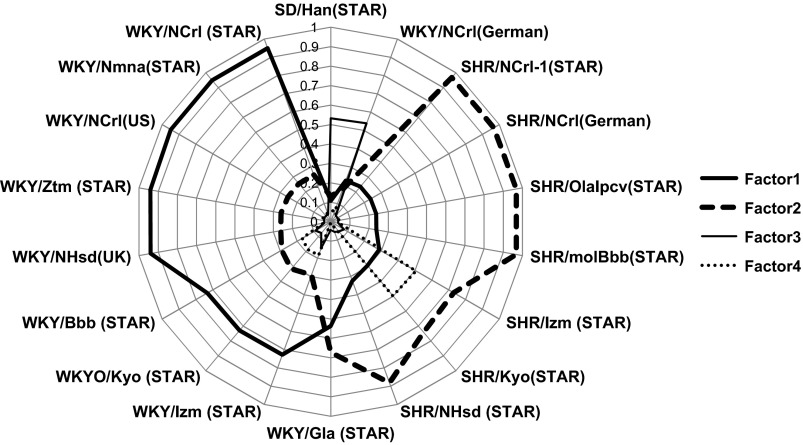
The results of the PFFA yielded four factors that accounted for 100% of the genetic variation among the 16 strains (Table 3). Figure 1 plots the rotated factor loading scores. The first two factors each explained ∼45% of the total variance across all the strains and clearly distinguished the two main strain types in our data set: SHR and WKY. Thus these two factors each represent the bulk of genetic variation for the SHR and the WKY strains. In contrast, the German WKY/NCrl strain did not load heavily on either factor 1 or 2 but instead loaded equally with the SD strain on factor 3, suggesting that the German WKY/NCrl strain is the most genetically divergent inbred strain.
Table 3.
PFFA main factors
| Factor | Eigenvalue | Variance | Proportion | Cumulative |
|---|---|---|---|---|
| Factor 1 | 11.03 | 6.92 | 0.46 | 0.46 |
| Factor 2 | 2.91 | 6.70 | 0.45 | 0.92 |
| Factor 3 | 0.58 | 0.64 | 0.04 | 0.96 |
| Factor 4 | 0.37 | 0.62 | 0.04 | 1.00 |
PFFA, principal factors factor analysis.
Fig. 1.
Rotated factor loadings for each strain on each of the four main factors extracted by principal factors factor analysis (PFFA) are plotted on a radar plot. Most of the Wistar-Kyoto (WKY) strains loaded high on factor 1, and all spontaneously hypertensive (SHR) strains loaded high on factor 2. The WKY/NCrl German strain and Sprague-Dawley (SD) strain loaded moderately on factor 3.
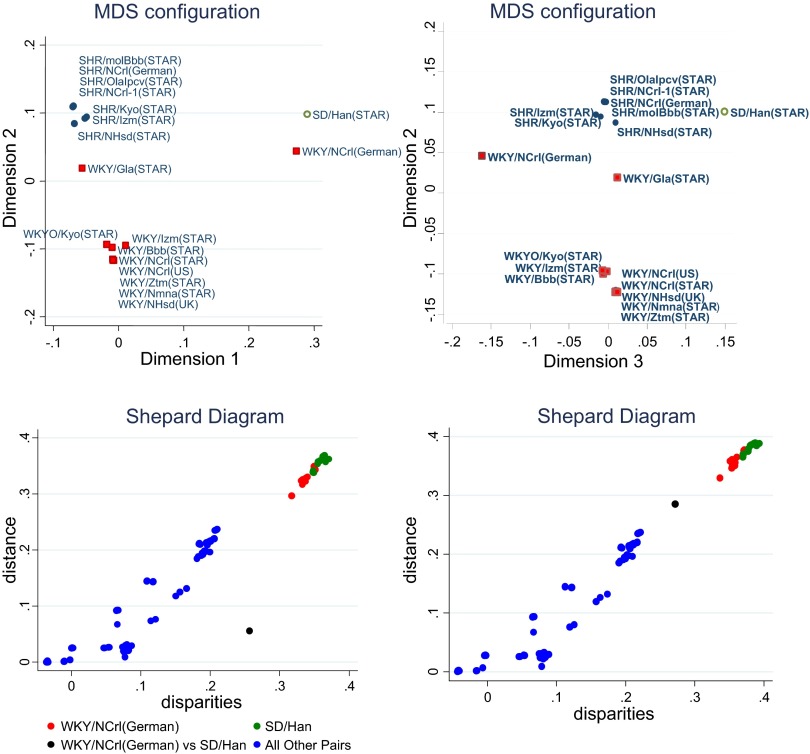
MDS analysis of the IBS dissimilarity matrix retained two main dimensions, which accounted for 41 and 39% of the distance observed, respectively. Strains were plotted in these two dimensions to visualize their genetic distance (Fig. 2, top left). Consistent with the PFFA, strains were grouped into two main clusters: an SHR cluster and a WKY cluster. Many SHR and WKY substrains were located in close proximity in the configuration plots. Dimension 2 represented the main SHR and WKY differences, with the WKY/Gla substrain intermediate between the SHR and WKY clusters. The German WKY/NCrl strain deviated far from the SHR and WKY clusters as did the outbred SD strain. The Shepard diagram (Fig. 2, bottom left) shows that the genetic distances in a two-dimensional model provide a good fit to the actual genetic disparities with one exception, which is shown as the black dot representing the German WKY/NCrl comparison with the SD strain. This indicated that the actual genetic disparity between German WKY/NCrl and the SD strains is larger than the distance shown in the two-dimensional plot (Fig. 2, top left). Among all the two-by-two comparisons, that fit well along the diagonal line of the Shepard diagram, we also see that the difference between the SD strain and all other strains, except the German WKY/NCrl rats, are the largest, whereas the difference between German WKY/NCrl rats with all other rats, except the SD rats, are the second largest. The differences between the German WKY/NCrl and SD strains are fully represented in dimension 3 (Fig. 2, top right), which explained an additional 8.6% of the total genetic distances. When dimension 3 is retained in the MDS configuration, the Shepard diagram showed an improved fit of estimated and the actual genetic distances (Fig. 2, bottom right).
Fig. 2.
Multidimensional scaling analysis (MDS) of the identity-by-state (IBS) similarity matrix. Top left: each strain is plotted in the 2 main dimensions of the MDS configuration. The distance between each dot represents the genetic distance of any given 2 strains. Notice the 2 main clusters: an SHR cluster (●) and a WKY cluster (■). However, the WKY/Gla substrain was intermediate between the SHR and WKY clusters. The German WKY/NCrl strain deviated far from the SHR and WKY clusters as did the outbred SD strain. Top right: MDS configuration with the inclusion of the 3rd dimension. The graph plots each strain in dimensions 2 and 3, showing the differences between the German WKY/NCrl and the SD rats in dimension 3. Bottom left: Shepard diagram for the 2-dimensional configuration. The y-axis is the calculated distance between any 2 strains in the MDS solutions. The x-axis is the actual genetic disparities between the 2. Most of the dots fall in the diagonal line, meaning that the genetic distances in the MDS solutions provides a good prediction to the actual genetic disparities. One exception is shown as the black dot which represents the German WKY/NCrl comparison with the SD on the 2-dimensional MDS plot. Among all the 2×2 comparisons, the differences between the SD strain and all other strains, except the German WKY/NCrl rats, are the largest (green dots), whereas the differences between German WKY/NCrl rats with all other rats, except the SD rats, are the 2nd largest (red dots). All other 2×2 strain comparisons showed less distance and are plotted in blue. Bottom right: Shepard diagram for the 3-dimensional configuration, showing an improved fit for the German WKY/NCrl and SD rats comparisons.
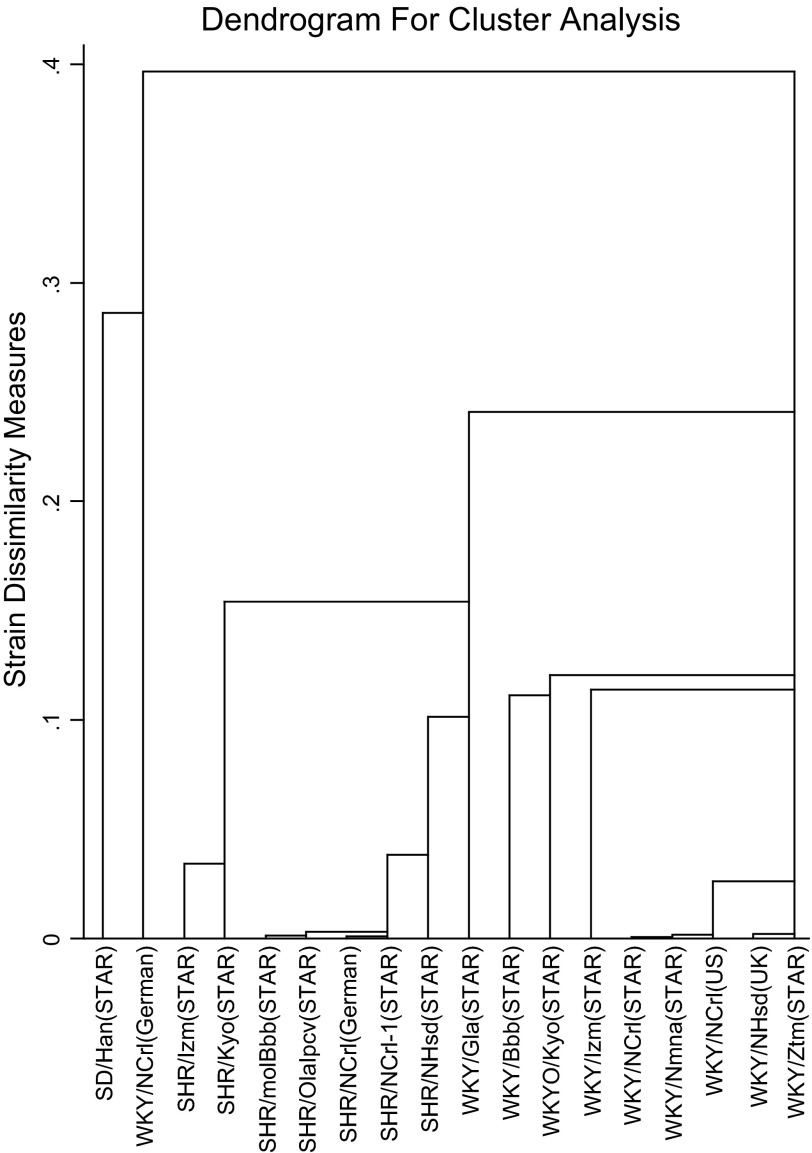
Altogether, these analyses showed that there are two main genetic clusters in the rat strains that we examined: the SHR and the WKY, in addition to one distantly isolated WKY/NCrl German strain. We also observed one small cluster within the SHR substrains, which included the SHR/MolBBB, SHR/Olapcv, and the two SHR/NCrl strains (from UK and Germany, see Table 1). The genetic variation among these strains is <0.5%, which most likely represents residual heterozygosity from a shared common ancestor. Among the WKY strains, there were also two smaller clusters (Fig. 3). One WKY cluster comprises two WKY/NCrl rats from two difference sources (US and UK) and the WKY/Nmna strain. The other WKY cluster comprises the WKY/NHsd strain and the WKY/Ztm strain. Within each of the two clusters, the genetic differences were <0.4%. However, between these two clusters, the genetic differences were ∼2.6%. Table 4 provides a two-by-two comparison of the strains within these three clusters.
Fig. 3.
Hierarchical cluster analysis of the IBS similarity matrix. The genetic differences between the different strains are visualized by a dendrogram. Height of the each branch represents the distance between any 2 pairs. This diagram reveals 1 SHR and 2 WKY clusters, within which the smallest genetic difference was observed.
Table 4.
Two-by-two comparison of the representative SHR and WKY strains in the three clusters
| Strain Comparison | Identical SNPs | Total SNPs Present in Both Strains | % of SNPs Different |
|---|---|---|---|
| SHR/molBbb (STAR) vs. SHR/OlaIpcv (STAR) | 9,157 | 9,177 | 0.22 |
| SHR/NCrl (German) vs. SHR/NCrl-1 (STAR) | 9,265 | 9,283 | 0.19 |
| SHR/molBbb (STAR) vs. SHR/NCrl (German) | 9,179 | 9,218 | 0.42 |
| SHR/molBbb (STAR) vs. SHR/NCrl-1 (STAR) | 9,262 | 9,287 | 0.27 |
| SHR/OlaIpcv (STAR) vs. SHR/NCrl (German) | 9,137 | 9,174 | 0.40 |
| SHR/OlaIpcv (STAR) vs. SHR/NCrl-1 (STAR) | 9,217 | 9,242 | 0.27 |
| WKY/NCrl (STAR) vs. WKY/Nmna (STAR) | 9,309 | 9,319 | 0.11 |
| WKY/NCrl (US) vs. WKY/Nmna (STAR) | 9,249 | 9,272 | 0.25 |
| WKY/NCrl (STAR) vs. WKY/NCrl (US) | 9,248 | 9,273 | 0.27 |
| WKY/Ztm (STAR) vs. WKY/NHsd (UK) | 9,165 | 9,196 | 0.34 |
| WKY/NCrl (US) vs. WKY/NHsd (UK) | 9,049 | 9,294 | 2.64 |
| WKY/Nmna (STAR) vs. WKY/Ztm (STAR) | 8,999 | 9,235 | 2.56 |
| WKY/NCrl (STAR) vs. WKY/Ztm (STAR) | 8,991 | 9,228 | 2.57 |
Genetic comparison of four unique strains.
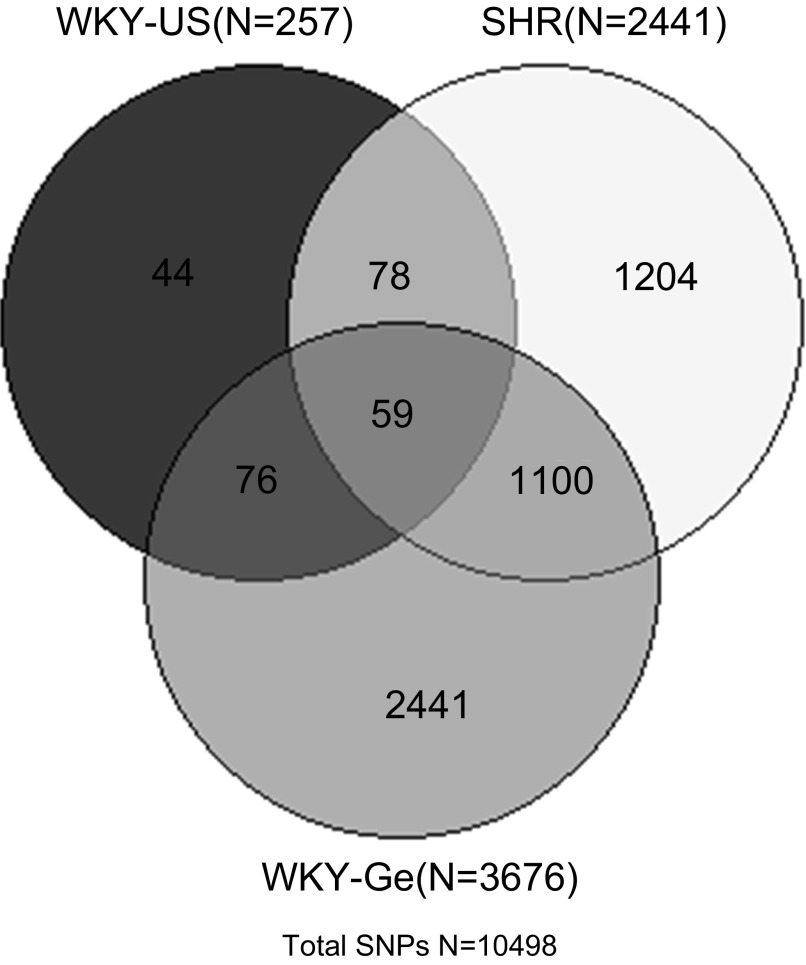
Based on the above analysis and our previous observation of ADHD behaviors in certain strains, we selected four individual lines for further comparison, validation, and functional annotation studies. These four strains were the German WKY/NCrl rats, a unique WKY strain and model for inattentive ADHD (26), and the US WKY/NCrl, SHR/NCrl, and WKY/NHsd rats, each representing one of the three core clusters that we identified from our analyses. Using the WKY/NHsd strain as the reference strain, because it is normotensive and showed normal behavior in testing for ADHD phenotypes (26), we further compared the other three strains in terms of their genetic differences from WKY/NHsd rats. Figure 4 shows the SNPs whose genotypes differed in the three strains compared with the WKY/NHsd rats. Each strain has some unique SNPs that were different from the WKY/NHsd strain. Three strains also shared 59 common SNPs that are divergent from the WKY/NHsd rats. Based on the number of discordant SNP genotypes, we also computed the genetic discordance among these four strains as the percentage of discordant SNPs: WKY/NHsd and WKY/NCrl-US, 2.45%; WKY/NCrl-US and WKY/NCrl-Ge, 34.9%; WKY/NHsd and WKY/NCrl-Ge, 35%; WKY/NHsd and SHR/NCrl, 23.3%; SHR/NCrl and WKY/NCrl-Ge, 36.2%. The genetic differences between the German WKY/NCrl, SHR/NCrl, and WKY/NHsd rats were similar to our previous report based on a less-dense genotyping array (26). The annotated functional consequences of the 2.45% divergent SNPs between the WKY/NHsd and WKY/NCrl-US rats are listed in the Supplementary File, which shows that these SNPs are distributed throughout all rat chromosomes assayed by the array. In addition, 55.5% of the SNPs are intergenic, and 14.5% are upstream or downstream to known genes. Among the 30.1% SNPs within gene regions, only 3.5% are coding variants and 1.17% change the amino acid (Supplementary File).
Fig. 4.
Genome-wide SNP genotyping results were compared across 3 different rat strains: US WKY/NCrl, German WKY/NCrl, and SHR/NCrl compared with WKY/NHsd. Ge, German vendor; US, US vendor. The numbers of discordant SNPs are shown for each comparison in the Venn diagram.
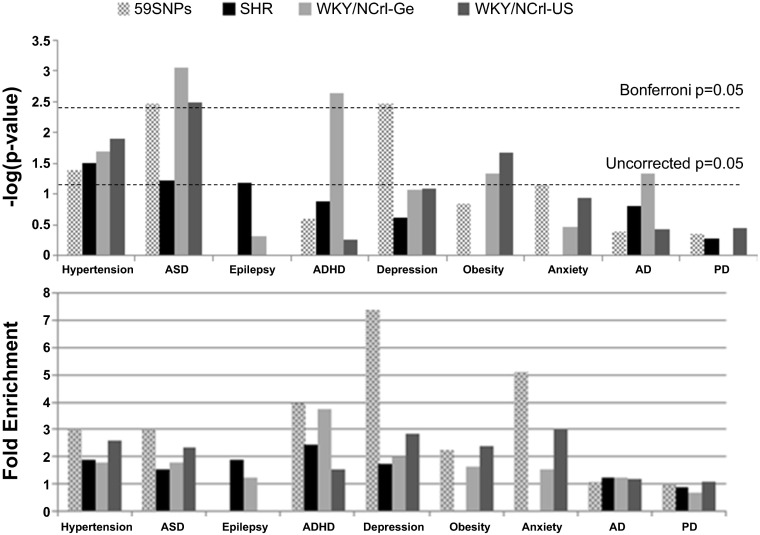
To further explore potential functional consequences of this genetic divergence, we retrieved the known rat genes within a 665 Kb region surrounding each of these divergent SNPs. These genes may contain genetic variations that distinguish them from the WKY/NHsd. We tested for the enrichment of the divergent genes in nine different diseases or phenotypes for all three strains (the two WKY/NCrl and the SHR rats) compared with the WKY/NHsd rats. Figure 5, top, plots the negative log P values for each test. The threshold for achieving significance following Bonferroni correction is illustrated (to correct for the tests of 9 different diseases). We found significant enrichment for autism genes for both WKY/NCrl rats and all three strain shared genes (genes tagged by the 59 common discordant SNPs). The German WKY/NCrl rats were enriched for ADHD genes, and the three strain shared genes for all three strains were enriched for depression genes. Several other gene lists that showed nominal significance (uncorrected P < 0.05) and a more than twofold increased enrichment included obesity and hypertension genes for the US WKY/NCrl rats and the hypertension genes for the three strain shared genes. There is a weak trend for enrichment of epilepsy genes for SHR rats. We found no evidence of enrichment of the Alzheimer's and Parkinson's disease gene lists.
Fig. 5.
For each of the 3 strains (SHR, WKY/NCrl-Ge, and WKY/NCrl-US), genes in the divergent regions from the WKY/NHsd rats were retrieved and tested for enrichment in candidate gene lists for 9 different diseases/phenotypes. Top: the negative log of the Fisher's exact test P values. Bottom: fold enrichments.
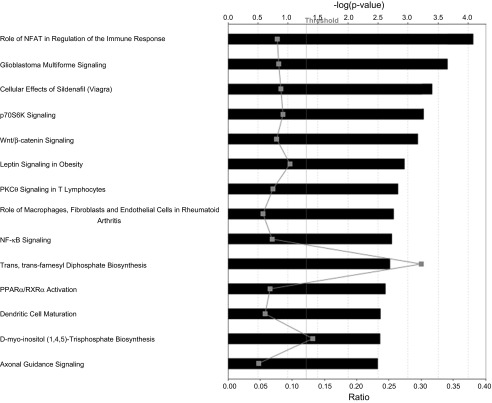
We also examined the “canonical pathways” that are implicated between the two closest WKY clusters, represented by the US WKY/NCrl and the WKY/NHsd substrains. A total of 257 SNPs differed between the two strains, among them 59 SNPs were shared with the German WKY/NCrl and the SHR/NCrl rats (see Fig. 4). A total of 747 known rat genes were mapped within the 665 Kb region of the 257 divergent SNPs, 698 of which could be mapped to the IPA database for analysis. Top ranked canonical pathways included those involved in immune response, nervous system development and signaling, and obesity (Fig. 6).
Fig. 6.
The top 14 most significant canonical pathways implicated in WKY/NHsd and US WKY/NCrl rat comparisons were plotted. The top y-axis displays the −log of P value, which is calculated by Fisher's exact test right-tailed. The gray points represent the ratio (bottom y-axis), which is calculated by dividing the number of genes in a given pathway that meet cutoff criteria by total number of genes that make up that pathway. The cutoff criterion is an enrichment P value < 0.05, which is denoted as the line marked “Threshold.”
Sequence prediction and validation.
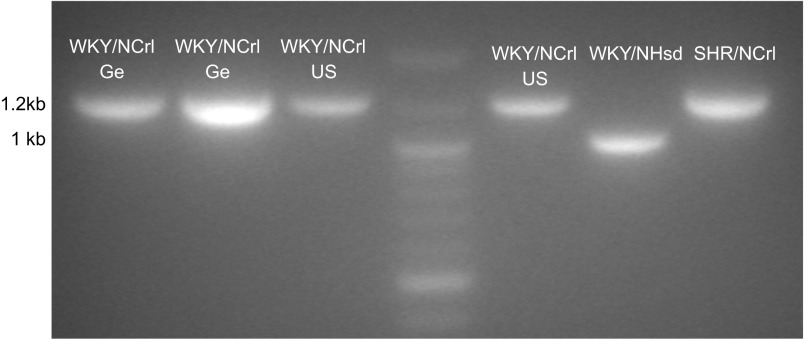
There are known genetic variations in the dopamine transporter gene (Slc6a3) for the SHR and German WKY/NCrl rats and known genetic variations in Slc9a9 for the WKY/NCrl-Ge rats compared with WKY/NHsd rats (see Table 5; Refs. 15, 37). To test if we could predict the sequences of these known variations according to their closest SNP genotypes, we amplified and sequenced these known regions. We confirmed these predicted genetic variations. For Slc9a9, we found that the US WKY/NCrl rats have the V512G and K534R double mutations in exon 16, which we previously found in the German WKY/NCrl rats (37)(Table 5). For Slc6a3, we found that the US WKY/NCrl rats also have the synonymous single base change (T→C) within the coding sequence of exon 4 (not shown) and a 160 bp insertion immediately upstream of exon 4, which were both previously reported in the SHR and the German WKY/NCrl rats (15) (Fig. 7). Notably, we also amplified the cDNA sequence of Slc6a3 in these rats using primers in the far upstream and downstream exons. Both gel analysis and sequencing showed the same splicing status of exon 4 for all four strains (data not shown), indicating that these two variants do not affect the alternative splicing of Slc6a3 in this region.
Table 5.
Summarized SNP genotypes and known sequence variations for Slc6a3 and Slc9a9. WKY/NCrl-US rats are predicted and confirmed to have Slc6a3 insertion and Slc9a9 double mutations
| SNP Genotypes |
Known Genetic Variations |
Prediction & Sequence Verification |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr: Position | Allele | SHR/NCrl (STAR) | WKY/NMna (STAR) Or/NCrl (US) | WKY/NCrl (Ge) | WKY/NHsd Or/Ztm (STAR) | SHR/NCrl | WKY/NHsd | WKY/NCrl (Ge) | Ref. No. | WKY/NCrl (Ge) | WKY/NCrl (US) |
| 1: 30153588 | G/A | AA | AA | GG | Slc6a3 Insertion | no insertion | (15) | Slc6a3 Insertion | Slc6a3 Insertion | ||
| 8: 99572345 | A/G | AA | GG | GG | AA | Slc9a9 512V and 534K | Slc9a9 512V and 534K | Slc9a9 512G and 534R | (37) | Slc9a9 512G and 534R | |
| 8: 100139524 | G/C | GG | CC | GG | |||||||
Fig. 7.
Agarose (2%) gel analysis of PCR amplifying region flanking Slc6a3 exon 4. SHR/NCrl and both US and Germany WKY/NCrl contain a 160 bp insertion upstream of exon 4, compared with WKY/NHsd rats.
DISCUSSION
SHR rats have been a valuable model for studies of hypertension and ADHD. However, genetic heterogeneity among inbred WKY control strains has confounded the results of many studies. Our study is the first to examine the genome-wide genetic relationships among all commonly used SHR and WKY strains. We found surprisingly large genetic variation among them. The most striking finding was that the German WKY/NCrl strain was distinctly different from all other WKY and SHR strains, including the WKY/NCrl strain from US and UK breeders. While the German WKY/NCrl strain was shown to be a model of inattentive ADHD (26), the US and UK WKY/NCrl have not been validated for ADHD phenotypes, although the US WKY/NCrl rats have been used for their depressive and stress-responsive phenotypes (4, 21, 29, 35). According to our genetic results, the US and UK WKY/NCrl strains are very similar and maintained the characteristics of WKY strains, whereas the German WKY/NCrl has evolved to be a distinct strain. Therefore, the sources of the WKY/NCrl rats should always be clearly noted.
The genetic variations among most of the substrains can be explained by their breeding histories. The SHR rats were bred from outbred WKY rats in Kyoto School of Medicine from 1963 and sent to the National Institutes of Health (NIH) in 1966 at F13, prior to being fully inbred (13). Most of the commercial breeders outside of Japan received SHR rats from NIH after F20 was available, including Charles River Laboratories (US), which received NIH SHR rats at F32 in 1973 (27). The SHR rats bred at Kyoto University achieved F20 in 1969. Therefore the Japanese substrains such as SHR/KYO and SHR/Izm appeared to be more divergent from the NIH-derived SHR substrains including SHR/NCrl, SHR/Olapcv, and SHR/molbbb. Harlan Laboratories received the SHR from NIH in 1982 (communication with the Harlan Laboratories), much later than the Charles River Laboratories, supporting the larger differences between the SHR/NHsd and SHR/NCrl. The inbreeding for a normotensive control line from outbred Wistar rats was commenced in 1971 in Japan, long after the SHR been fully inbred, leading to large overall genetic difference between the SHR and WKY strains. Similarly large genetic divergences existed between the Japan- and NIH-derived lines. The 2.5% genetic difference between the two smallest WKY clusters are likely because NIH distributed WKY rats before the strain was fully inbred. For example, Harlan Laboratories received WKY rats from NIH in 1982, much later than the Charles River Laboratories, which received NIH-WKY in 1971 (12). However, the reason for the genetic and behavioral divergence of the German WKY/NCrl strains is not known and cannot be explained by the reported breeding history or genetic drift. It is possible that this strain may have been accidentally contaminated by other strains and subsequently allowed to be bred as a distinct inbred line within the German facility. Indeed, in the current study, we found that German WKY/NCrl rats have a very low level of heterozygous genotypes (<0.2%), similar to all other WKY and SHR inbred substrains including the WKY/NCrl rats from the US supplier and lower than that of the SHR/NHsd rats. In contrast, outbred SD rat have 8.61% heterozygous SNPs. This observation suggests that the German WKY/NCrl rat is a new strain with a high degree of inbreeding. Over the years from 2004 to 2010, we received multiple shipments of tissue of the German WKY/NCrl rats from Dr. Sagvolden, including those used in the previously published study using 5K arrays (26) and those in the current study using 10K arrays. We have found a consistent genetic profile for the German WKY/NCrl rats over this time period, confirming a stable inbred line. It is not known, however, whether this line will be continued, because Charles River Laboratories do implement “clone back” strategies to preserve inbred lines, although we did not observe any change over the period our studies covered. Researchers who are interested in using this line as an inattentive ADHD rat model should be aware of the genetic background and genotype their rats to verify the genetic identity. The genome-wide SNP genotyping result of the German WKY/NCrl rat from our study can be used to design genotyping assays.
The difference between the SHR and the WKY groups are large (20–30%), which probably involves genes beyond those involved in hypertension and ADHD. Considering that WKY inbreeding was initiated 10 yr later than that of the SHR strains, and many early releases of both SHR and WKY rats have resulted in numerous distinct substrains as observed in our analyses, many researchers have questioned the validity of using the WKY as the control for the SHR. Indeed, results from different studies can vary if animals from different sources are used. However, when compared with outbred and other inbred strains, WKY rats are still the closest genetic controls for the SHR rats, although care should be taken in selecting the most appropriate control strains. For example, the behaviorally normal WKY/NHsd substrain should be used when study the ADHD behavior (27). It may be helpful to include a phenotypically normal outbred strain, such as SD rats (26), in addition to the WKY/NHsd rats when studying the ADHD behavior of the SHR rats or the inattentive German WKY/NCrl rats.
The genetic variations within the main SHR and the WKY strains are consistent with previous observations and their breeding histories (7, 10–13, 16). However, we found one subgroup of SHR rats, and two different subgroups of WKY rats had negligible genetic differences within groups (<0.5%), which is equivalent to the residual minimum heterozygosity in each strain. Strains within these three clusters may also exhibit minimum phenotypic differences and thus be suitable for use interchangeably. Interestingly, between the two WKY subgroups, there is a relatively small, yet nonnegligible genetic difference (∼2.5%). Examining the genes that are represented by this 2.5% genetic divergence, we confirmed that these two WKY groups may represent two phenotypically different inbred strains and provide unique opportunities for modeling human neuropsychiatric disorders. This is especially advantageous given their small genetic differences, which means that any phenotypic or behavioral differences could be traced to a very small number of genes. For example, Pare (21) observed that the US WKY/NCrl and the WKY/NHsd were highly similar in their stress responsive and depressive behaviors compared with Taconic outbred WKY rats and Wistar rats. However, the WKY/NCrl rats were significantly more immobile in the open field and more prone to develop stress-induced ulcers than the WKY/NHsd rats. Despite this report, studies of depression using WKY rats as a model system have used both the US WKY/NCrl rats and the WKY/NHsd (UK and US sources) equally, producing results that are not readily comparable. According to our analyses, WKY/NCrl rats may differ from WKY/NHsd rats in a number of depression and anxiety related genes, particularly in the genes tagged by the shared 59 SNPs (Fig. 5). With whole genome and exon sequencing techniques, it is increasingly easy to obtain the complete genomic sequence, which could allow one to identify stress-depression causal genetic variants that distinguish the US WKY/NCrl rats and the WKY/NHsd rats.
Without genome sequence data, our study relied on only 9,407 SNPs. However, the genetic comparisons of strains were largely consistent with our previous report using approximately half that number of SNPs from a less dense array (26). The comparison results were also consistent with that of the STAR project, which had twice the number of SNPs. However, the pathway and disease gene analyses were based on predicted genes within 1.3 Mb (±650 Kb) regions centered on each target SNP, which could lead to false positives but also miss genes that are further away from the tagging SNPs. In addition, we may have missed genes from regions not well covered by the 7,409 SNPs. With this limitation in mind, our pathway and disease gene analyses still confirmed several known phenotypic features of the strains, such as the significant enrichment of ADHD, depression, and obesity genes in some strains, and revealed no enrichment for Alzheimer and Parkinson disease genes.
Although SHR rats showed a more than twofold enrichment of ADHD genes, this enrichment was not statistically significant. In addition to the above-mentioned limitations on the predicted genes-based analysis, there might be several other reasons for the lack of enrichment of ADHD genes. First, although the genes on our ADHD list were selected based on prior literature, that literature is relatively weak, with no single gene showing evidence for genome-wide significance. Unlike the SFARI autism gene list, which was manually curated by the experts in the field, the ADHD gene lists and rankings from various databases were heavily skewed by publication biases due to the many candidate gene studies in the literature. Unbiased genome-wide association studies do not yet have a large enough sample size to detect risk genes with genome-wide significance, probably because many of the risk variants have only small effects. Secondly, the informative SNPs are often not causal variants themselves. Instead, they represent other underlying genetic differences within the LD regions, which are still unknown until we acquire sequencing data. Inclusion of all known genes within the LD regions for the enrichment test will include genes that do not have genetic variations. Finally, the ADHD phenotype of the SHR rats may be due to a small number of genetic variations, such as the ones we identified in Slc9a9 and Slc6a3. Yet the overall genetic differences between SHR rats and the WKY/NHsd rats are much larger, including many genes that do not play a role for the ADHD phenotype. In this regard, the gene list enrichment test is only a weak test of the validity of the SHR as a model for ADHD.
Similar reasons may also be responsible for the lack of significant enrichment of hypertension genes for the SHR rats. However, we found that all three strains, the German and US WKY/NCrl rats and the SHR/NCrl rats compared with the WKY/NHsd rats showed a nominally significant twofold enrichment of hypertension genes. It has been known that some WKY rats develop spontaneous hypertension at older ages with a lower increase in blood pressure than SHR rats (8). It is possible that some WKY rats still maintain some hypertension risk genes from their common ancestor, the Wistar rat, but a different set of risk genes are likely to be responsible for the early onset high blood pressure of the SHR.
Interestingly, the most significant finding among all nine different diseases tested was for ASD. Both lines of WKY/NCrl rats showed significant enrichment of ASD genes. IPA also showed that the top ranked canonical pathways in the US WKY/NCrl and WKY/NHsd comparisons were those involved in immune response, nervous system development and signaling, and obesity, all of which have been implicated in autism pathophysiology (3, 6, 17, 18, 31–33). Autistic behaviors have never been examined in any WKY strains. It would be intriguing to see if any strains show these traits. If true, the two closest WKY clusters may present a unique advantage for modeling autism because of their small genetic differences.
With the recent improvements in genetic manipulation, rats are rapidly regaining importance as model systems to study complex neuropsychiatric disorders because of their advantages over mice in their complex cognitive and social behavior and their ability to learn and perform complex tasks. Our study provides a clear answer to a decades-long question about the genetic heterogeneity of WKY and SHR strains and a comprehensive genetic relationship among most of the commonly used strains. This will facilitate the design of new research utilizing these models and will help to interpret previously published results. Furthermore, we identified two closely related WKY substrain clusters, which may provide a unique opportunity for modeling complex neuropsychiatric conditions such as autism and depression.
GRANTS
This study was supported by National Institute of Mental Health Grant R01MH-066877 to S. V. Faraone and an NARSAD Young Investigator Award to Y. Zhang-James.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: Y.Z.-J. conception and design of research; Y.Z.-J. performed experiments; Y.Z.-J., F.A.M., and S.V.F. analyzed data; Y.Z.-J., F.A.M., and S.V.F. interpreted results of experiments; Y.Z.-J. and S.V.F. prepared figures; Y.Z.-J. drafted manuscript; Y.Z.-J., F.A.M., and S.V.F. edited and revised manuscript; Y.Z.-J., F.A.M., and S.V.F. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the work of Dr. Terje Sagvolden (1945–2011), who discovered the behavioral abnormality of the German WKY/NCrl rats and initiated the genetic investigation of the different WKY strains. Dr. Sagvolden also provided rat tissue for SNP genotyping from the German WKY/NCrl rats and UK WKY/NHsd rats. We thank Dr. Tania DasBanerjee and Karen Gentile for technical assistance, and Dr. Stephen Glatt for statistical advice.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Braw Y, Malkesman O, Dagan M, Bercovich A, Lavi-Avnon Y, Schroeder M, Overstreet DH, Weller A. Anxiety-like behaviors in pre-pubertal rats of the Flinders Sensitive Line (FSL) and Wistar-Kyoto (WKY) animal models of depression. Behav Brain Res 167: 261–269, 2006 [DOI] [PubMed] [Google Scholar]
- 3. Careaga M, Ashwood P. Autism spectrum disorders: from immunity to behavior. Methods Mol Biol 934: 219–240, 2012 [DOI] [PubMed] [Google Scholar]
- 4. Conti LH. Interactions between corticotropin-releasing factor and the serotonin 1A receptor system on acoustic startle amplitude and prepulse inhibition of the startle response in two rat strains. Neuropharmacology 62: 256–263, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. DasBanerjee T, Middleton FA, Berger DF, Lombardo JP, Sagvolden T, Faraone SV. A comparison of molecular alterations in environmental and genetic rat models of ADHD: a pilot study. Am J Med Genet B Neuropsychiatr Genet 147B: 1554–1563, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garay PA, McAllister AK. Novel roles for immune molecules in neural development: implications for neurodevelopmental disorders. Front Synapt Neurosci 2: 136, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hinojos CA, Boerwinkle E, Fornage M, Doris PA. Combined genealogical, mapping, and expression approaches to identify spontaneously hypertensive rat hypertension candidate genes. Hypertension 45: 698–704, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Howes LG, Summers RJ, Rowe PR, Louis WJ. The measurement of central noradrenergic activity in spontaneously hypertensive rats: a comparison of free 3,4-dihydroxyphenylethyleneglycol levels with FLA-63 induced noradrenaline depletion. J Hypertens 3: 237–242, 1985 [DOI] [PubMed] [Google Scholar]
- 8a. Institute of Laboratory Animal Resources Spontaneously Hypertensive (SHR) Rats: Guidelines for Breeding, Care, and Use. Washington, DC: National Academy of Sciences, 1976 [Google Scholar]
- 9. Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucl Acids Res 32: D493–D496, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katsuya T, Higaki J, Miki T, Nakura J, Ikegami H, Morishita R, Nagano M, Higashimori K, Nakamura F, Mikami H, Ogihara T. Substrain comparison of genetically hypertensive rats using DNA fingerprinting, and genetic analysis of blood pressure in the inbred rats. Tohoku J Exp Med 165: 253–260, 1991 [DOI] [PubMed] [Google Scholar]
- 11. Kurtz TW, Montano M, Chan L, Kabra P. Molecular evidence of genetic heterogeneity in Wistar-Kyoto rats: implications for research with the spontaneously hypertensive rat. Hypertension 13: 188–192, 1989 [DOI] [PubMed] [Google Scholar]
- 12. Kurtz TW, Morris RC., Jr Biological variability in Wistar-Kyoto rats. Implications for research with the spontaneously hypertensive rat. Hypertension 10: 127–131, 1987 [DOI] [PubMed] [Google Scholar]
- 13. Louis WJ, Howes LG. Genealogy of the spontaneously hypertensive rat and Wistar-Kyoto rat strains: implications for studies of inherited hypertension. J Cardiovasc Pharmacol 16, Suppl 7: S1–S5, 1990 [PubMed] [Google Scholar]
- 14. McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mill J, Sagvolden T, Asherson P. Sequence analysis of Drd2, Drd4, and Dat1 in SHR and WKY rat strains. Behav Brain Funct 1: 24, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nabika T, Nara Y, Ikeda K, Endo J, Yamori Y. Genetic heterogeneity of the spontaneously hypertensive rat. Hypertension 18: 12–16, 1991 [DOI] [PubMed] [Google Scholar]
- 17. Okerlund ND, Cheyette BN. Synaptic Wnt signaling-a contributor to major psychiatric disorders? J Neurodev Dis 3: 162–174, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Onore C, Careaga M, Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun 26: 383–392, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Overstreet DH. Modeling depression in animal models. Meth Mol Biol 829: 125–144, 2012 [DOI] [PubMed] [Google Scholar]
- 20. Pare WP. Open field, learned helplessness, conditioned defensive burying, and forced-swim tests in WKY rats. Physiol Behav 55: 433–439, 1994 [DOI] [PubMed] [Google Scholar]
- 21. Pare WP, Kluczynski J. Differences in the stress response of Wistar-Kyoto (WKY) rats from different vendors. Physiol Behav 62: 643–648, 1997 [DOI] [PubMed] [Google Scholar]
- 22. Perry GM, Sagvolden T, Faraone SV. Intra-individual variability in genetic and environmental models of attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 153B: 1094–1101, 2010 [DOI] [PubMed] [Google Scholar]
- 23. Perry GM, Sagvolden T, Faraone SV. Intraindividual variability (IIV) in an animal model of ADHD - the Spontaneously Hypertensive Rat. Behav Brain Funct 6: 56, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, Foglio M, Fujiyama A, Gut IG, Gauguier D, Guigo R, Guryev V, Heinig M, Hummel O, Jahn N, Klages S, Kren V, Kube M, Kuhl H, Kuramoto T, Kuroki Y, Lechner D, Lee YA, Lopez-Bigas N, Lathrop GM, Mashimo T, Medina I, Mott R, Patone G, Perrier-Cornet JA, Platzer M, Pravenec M, Reinhardt R, Sakaki Y, Schilhabel M, Schulz H, Serikawa T, Shikhagaie M, Tatsumoto S, Taudien S, Toyoda A, Voigt B, Zelenika D, Zimdahl H, Hubner N. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet 40: 560–566, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sagvolden T. Behavioral validation of the spontaneously hypertensive rat (SHR) as an animal model of attention-deficit/hyperactivity disorder (AD/HD). Neurosci Biobehav Rev 24: 31–39, 2000 [DOI] [PubMed] [Google Scholar]
- 26. Sagvolden T, Dasbanerjee T, Zhang-James Y, Middleton F, Faraone S. Behavioral and genetic evidence for a novel animal model of attention-deficit/hyperactivity disorder predominantly inattentive subtype. Behav Brain Funct 4: 56, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sagvolden T, Johansen EB, Woien G, Walaas SI, Storm-Mathisen J, Bergersen LH, Hvalby O, Jensen V, Aase H, Russell VA, Killeen PR, Dasbanerjee T, Middleton FA, Faraone SV. The spontaneously hypertensive rat model of ADHD–the importance of selecting the appropriate reference strain. Neuropharmacology 57: 619–626, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sagvolden T, Russell VA, Aase H, Johansen EB, Farshbaf M. Rodent models of attention-deficit/hyperactivity disorder. Biol Psychiatry 57: 1239–1247, 2005 [DOI] [PubMed] [Google Scholar]
- 29. Scholl JL, Renner KJ, Forster GL, Tejani-Butt S. Central monoamine levels differ between rat strains used in studies of depressive behavior. Brain Res 1355: 41–51, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. St Lezin E, Simonet L, Pravenec M, Kurtz TW. Hypertensive strains and normotensive ‘control’ strains. How closely are they related?. Hypertension 19: 419–424, 1992 [DOI] [PubMed] [Google Scholar]
- 31. Suda S, Iwata K, Shimmura C, Kameno Y, Anitha A, Thanseem I, Nakamura K, Matsuzaki H, Tsuchiya KJ, Sugihara G, Iwata Y, Suzuki K, Koizumi K, Higashida H, Takei N, Mori N. Decreased expression of axon-guidance receptors in the anterior cingulate cortex in autism. Mol Autism 2: 14, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sullivan EL, Nousen L, Chamlou K. Maternal high fat diet consumption during the perinatal period programs offspring behavior. Physiol Behav 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tanne JH. Maternal obesity and diabetes are linked to children's autism and similar disorders. BMJ 344: e2768, 2012 [DOI] [PubMed] [Google Scholar]
- 34. Wall DP, Pivovarov R, Tong M, Jung JY, Fusaro VA, DeLuca TF, Tonellato PJ. Genotator: a disease-agnostic tool for genetic annotation of disease. BMC Med Genom 3: 50, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang S, Tian Y, Song L, Lim G, Tan Y, You Z, Chen L, Mao J. Exacerbated mechanical hyperalgesia in rats with genetically predisposed depressive behavior: role of melatonin and NMDA receptors. Pain 153: 2448–2457, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamori Y, Igawa T, Tagami M, Kanbe T, Nara Y, Kihara M, Horie R. Humoral trophic influence on cardiovascular structural changes in hypertension. Hypertension 6: III27–III32, 1984 [DOI] [PubMed] [Google Scholar]
- 37. Zhang-James Y, Dasbanerjee T, Sagvolden T, Middleton FA, Faraone SV. Slc9a9 mutations, gene expression, and protein-protein interactions in rat models of attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 156B: 835–843, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.