Abstract
Interferon-α (IFNα) and IFNβ, collectively known as type I IFNs, are the major effector cytokines of the host immune response against viral infections. However, the production of type I IFNs is also induced in response to bacterial ligands of innate immune receptors and/or bacterial infections, indicating a broader physiological role for these cytokines in host defence and homeostasis than was originally assumed. The main focus of this Review is the underappreciated immunomodulatory functions of type I IFNs in health and disease. We discuss their function in the regulation of innate and adaptive immune responses, the response to bacterial ligands, inflammasome activation, intestinal homeostasis and inflammatory and autoimmune diseases.
The inhibitory effect that one viral infection often exerts on the infectivity of a different virus was first observed in 1804 by Edward Jenner, who reported that herpetic infections could prevent the development of vaccinia lesions1. In retrospect, this observation might be the first documented description of what was later coined as the viral interference phenomenon. This phenomenon was first described in detail for plant viruses in the early 1930s2. After that, similar observations were made with bacteriophages3 and with animal viruses4. In 1954, Nagano and Kojima reported the inhibition of viral growth in areas of rabbit skin that had been previously inoculated with ultraviolet-inactivated vaccinia virus, and in 1957, during a study of the interference produced by heat-inactivated influenza virus, Isaacs and Lindenmann identified that a secreted factor was responsible for this phenomenon and they termed it interferon (IFN)5.
Type I IFNs belong to a family of cytokines that attracted much attention owing to their protective role against viral infection. IFNs are widely expressed cytokines that possess strong antiviral and immunomodulatory properties. The IFN family can be classified into three main types of cytokines — type I, type II and type III IFNs. In humans and mice, the type I IFN family is composed of 16 members, namely 12 IFNα subtypes, IFNβ, IFNε, IFNκ and IFNω6. By contrast, the type II IFN family includes only one cytokine: IFNγ, which also exhibits antiviral activities. The third type of IFNs is the IFNλ family, which includes IFNλ1 (also known as IL-29), IFNλ2 (also known as IL-28A) and IFNλ3 (also known as IL-28B). On the basis of protein sequence and structure, type III IFNs are markedly different from type I and type II IFNs and are more similar to members of the interleukin-10 (IL-10) family; however, they provoke antiviral responses and induce the activation of IFN-stimulated genes (ISGs)7.
Recent evidence has uncovered new roles for this family of cytokines beyond their well-known function in viral interference. This article highlights the function of type I IFNs in modulating immune responses. We map the molecular signalling pathways activated by type I IFNs, and describe the function of these cytokines in the response to bacterial ligands and their role in inflammasome activation. In addition, we discuss the role of type I IFNs in intestinal homeostasis and in inflammatory and autoimmune diseases such as coeliac disease, psoriasis, multiple sclerosis and cancer.
Type I IFN production and signalling
Induction of type I IFNs by bacterial ligands
Type I IFNs can be produced by almost every cell type, including leukocytes, fibroblasts and endothelial cells. The signalling pathways that lead to the induction of type I IFNs differ depending on the stimulus and the responding cell types, but they ultimately lead to the activation of some common signalling molecules, including TNF receptor-associated factor 3 (TRAF3)8 and the transcription factors IFN regulatory factor 3 (IRF3) and IRF7. Dimerized IRF3 and IRF7 translocate to the nucleus and, concomitantly with the transcription factor nuclear factor-κB (NF-κB), bind to both the IFNA and IFNB promoters9 to initiate the transcription of these IFN genes. Although the most important function of type I IFNs is typically considered to be in the induction of an antiviral immune response, these cytokines are also induced in response to many bacterial pathogens or their products, mainly through Toll-like receptor (TLR)-dependent pathways10,11.
TLRs are the key sensors of microbial invasion in mammals12, and they activate an innate defence programme that is crucial for host survival. Each TLR senses a particular subset of microbial signature molecules. Most TLRs that recognize bacterial products are linked to the induction of type I IFNs10; these TLRs include TLR3, TLR4, TLR7 and TLR9. Signalling through TLR3 and TLR4 induces type I IFN production in a broad range of cell types in a manner dependent on TIR-domain-containing adaptor protein inducing IFNβ (TRIF). By contrast, TLR7, TLR8 and TLR9 induce type I IFN production in dendritic cells (DCs) — mainly plasmacytoid DCs (pDCs) — via a pathway dependent on myeloid differentiation primary-response protein 88 (MYD88) (TABLE 1).
Table 1.
Inducers of type I IFNs and responding cells
| Inducer | Source | Receptor | Localization | Responding cell |
|---|---|---|---|---|
| ssRNA, dsRNA | Viruses | RIG-I and MDA5 | Cytoplasm | Multiple cell types |
| Cytosolic DNA | Viruses or bacteria | STING, DAI and RNA polymerase III |
Cytoplasm | Multiple cell types |
| dsRNA | Viruses | TLR3–TRIF | Endosomes | Macrophages, cDCs and epithelial cells |
| LPS | Gram-negative bacteria | TLR4–TRIF | Plasma membrane | Macrophages and cDCs |
| Viral glycolipids | Viruses | TLR4–TRIF | Plasma membrane | Macrophages and cDCs |
| ssRNA | Viruses or damaged host cells |
TLR7–MYD88 | Endosomes | pDCs, cDCs and macrophages |
| Imiquimod | Synthetic | TLR7–MYD88 | Endosomes | pDCs, cDCs and macrophages |
| ssRNA | Viruses | TLR8–MYD88 | Endosomes | cDCs |
| CpG DNA | Bacteria or viruses | TLR9–MYD88 | Endosomes | pDCs, cDCs and macrophages |
cDC, conventional DC; DAI, DNA-dependent activator of IRFs; DC, dendritic cell; dsRNA, double-stranded RNA; IFN, interferon; LPS, lipopolysaccharide; MDA5, melanoma differentiation-associated gene 5; MYD88, myeloid differentiation primary-response protein 88; pDC, plasmacytoid DC; RIG-I, retinoic-acid-inducible gene I; ssRNA, single-stranded RNA; STING, stimulator of IFN genes; TLR, Toll-like receptor; TRIF, TIR-domain-containing adaptor protein inducing IFNβ.
However, it became evident that TLR-deficient animals can still produce type I IFNs in response to RNA and DNA ligands13,14. These TLR-independent pathways include the cytoplasmic sensors retinoic-acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5). In addition, stimulator of IFN genes (STING) and DNA-dependent activator of IRFs (DAI; also known as DLM1 and ZBP1) have been reported to induce type I IFNs in response to cytosolic DNA15,16. STING is an endoplasmic reticulum-associated protein that has been shown to respond to DNA from various pathogens — including Listeria monocytogenes and the DNA virus herpes simplex virus 1 (HSV-1) — in macrophages, DCs and epithelial cells15. DAI, which was the first cytosolic DNA receptor to be described, recognizes viral, bacterial and mammalian double-stranded DNA and induces type I IFN production through the activation of TANK-binding kinase 1 (TBK1), which subsequently phosphorylates IRF3 (REF. 16). In addition, DAI stimulates the production of pro-inflammatory cytokines — such as IL-6 and tumour necrosis factor (TNF) — through the activation of the kinase receptor-interacting protein 1 (RIP1), which leads to the phosphorylation of NF-κB inhibitor-α (IκBα) and the subsequent activation of NF-κB17. However, DAI-deficient cells can still induce type I IFN production in response to foreign DNA18, suggesting the existence of additional mechanisms that contribute to type I IFN production in response to cytosolic DNA. Indeed, a recent study showed that DNA-dependent RNA polymerase III can use cytosolic DNA as a template to synthesize RNA containing a 5′-triphosphate group, and that this RNA activates the RIG-I–IPS1 (IFNB-promoter stimulator 1; also known as MAVS) signalling pathway19.
Signalling pathways activated by type I IFNs
Type I IFNs signal through a common heterodimeric receptor, known as the IFNα/β receptor (IFNAR), which is expressed by nearly all cell types. This receptor consists of two subunits — IFNAR1 and IFNAR2 (REF. 20) — that are constitutively associated with Janus kinase 1 (JAK1) and non-receptor tyrosine kinase 2 (TYK2)21. Activation of JAK1 and TYK2 results in the tyrosine phosphorylation and activation of several signal transducer and activator of transcription (STAT) family members; in most cells these include STAT1, STAT2, STAT3 and STAT5, but in lymphocytes type I IFNs also activate STAT4 and STAT6 (REFS 22,23).
Activation of STAT1 and STAT2 leads to the recruitment of IRF9 and the formation of a STAT1–STAT2–IRF9 complex, which is known as the IFN-stimulated gene factor 3 (ISGF3) complex. This complex then migrates to the nucleus and binds to IFN-stimulated response elements (ISREs) in the promoters of ISGs to initiate gene transcription. Other STAT complexes that do not recruit IRF9, including STAT1 homodimers, bind to IFNγ-activated site (GAS) enhancer elements in the promoters of ISGs24,25. Both type I and type II IFNs can induce the activation of GAS elements through, for example, the formation of STAT1 homodimers. However, in contrast to type I IFNs, IFNγ cannot induce the formation of ISGF3 complexes and therefore is not able to promote the engagement of ISRE sites to activate those genes that have only ISREs in their promoters21 (FIG. 1).
Figure 1. Signalling pathways activated by type I and type II IFNs.
Different signal transducer and activator of transcription (STAT) family members can be activated by interferons (IFNs). STAT1 homodimers can be formed in response to both type I IFNs and type II IFN (IFNγ). These homodimers bind to IFNγ-activated site (GAS) enhancer elements in the promoters of IFN-stimulated genes, and this results in the induction of genes encoding pro-inflammatory cytokines and apoptotic factors. Type I and type II IFNs can also activate STAT3 homodimers, and this can result in the production of both pro-inflammatory cytokines and anti-inflammatory cytokines (such as interleukin-10 (IL-10)), although the underlying mechanisms are not known. STAT1–STAT2 heterodimers, which are activated by type I IFNs, bind to IFN regulatory factor 9 (IRF9) in the cytosol to form the IFN-stimulated gene factor 3 (ISGF3) complex, which in turn migrates to the nucleus to bind to IFN-stimulated response elements (ISREs) and activate antiviral and antibacterial genes. In addition, type I IFNs stimulate IL-10 production either through the phosphoinositide 3-kinase (PI3K)–AKT pathway or through STAT3 homodimers. Finally, in a STAT-independent manner, type I IFNs activate both p38, which is an upstream activator of several genes regulated by ISREs and GAS elements, and mammalian target of rapamycin (mTOR), which regulates mRNA translation. CREB, cAMP-responsive-element-binding protein; IFNAR, IFNα/β receptor; IFNGR, IFNγ receptor; JAK, Janus kinase; SBE, STAT3-binding element; TBX21, T box 21; TYK2, non-receptor tyrosine kinase 2.
In addition, both type I and type II IFNs can induce the recruitment and phosphorylation of STAT3 (REFS 21,26). Following its phosphorylation, STAT3 forms homodimers that translocate to the nucleus, where they bind to STAT3-binding elements (SBEs). STAT1 and STAT3 have a high level of sequence similarity and can heterodimerize and activate similar genes in vitro27. However, in vivo, the sets of genes that are activated by these two factors are very different28 and depend on the cell type and the activating cytokine, and in many physiological contexts STAT1 and STAT3 exert opposing effects. For example, in most cell types, STAT1 activates several proapoptotic and anti-proliferative genes29,30, whereas STAT3 inhibits apoptosis and promotes proliferation through the induction of anti-apoptotic genes of the B cell lymphoma (BCL) family and of oncogenes such as MYC31,32.
STAT1 and STAT3 also have opposite roles in inflammation. IFNγ-dependent STAT1 activation usually mediates a pro-inflammatory response that favours the recruitment of immune cells to the site of inflammation33, and this increases the production of pro-inflammatory mediators34 and enhances antigen processing and presentation by MHC class I and II molecules35,36. By contrast, STAT3 is a key mediator of IL-10 signalling, which negatively regulates pro-inflammatory responses by activated macrophages and DCs and can directly inhibit STAT1 activation37. Interestingly, other receptors, such as the IL-6 receptor, can activate STAT3, but this does not result in an anti-inflammatory response, possibly owing to the participation of additional regulatory factors, such as members of the suppressor of cytokine signalling (SOCS) family38,39. This dual ability of STAT3 to differentially regulate inflammation in different scenarios might explain some of the controversial roles that type I IFNs have in different human conditions (discussed below). Several studies also suggest that cross-regulation between STAT1 and STAT3 and their relative abundance in a cell are the defining factors that determine the biological effects of their upstream activators, such as IL-6 and IFNs40,41.
Type I IFNs were also shown to promote the production of IL-10 by lipopolysaccharide (LPS)-stimulated macrophages42 and by LPS-stimulated human peripheral blood mononuclear cells (PBMCs)43. This effect was initially attributed to the activation of STAT3 and IRF1 in IFNα-stimulated human monocytes44. However, another study indicated that IFNβ stimulates IL-10 production independently of STAT3, by activating a signalling pathway mediated by JAK1 and phosphoinositide 3-kinase (PI3K). This pathway increases the phosphorylation and nuclear translocation of cAMP-responsive-element-binding protein (CREB), which promotes IL-10 production in human DCs45 (FIG. 1). It is not clear why IL-10 production is mediated by different signalling pathways in response to IFNα (the STAT3 pathway) and IFNβ (the PI3K pathway). However, the differential activation of STAT3 versus STAT1, the ability to induce IL-10 production and/or the participation of additional regulatory elements may explain the diverse effects of type I IFNs in several pathological conditions.
In addition to the JAK–STAT signalling pathways, there is evidence that type I IFNs activate other (non-STAT) signalling pathways that have crucial roles in their different biological properties. For example, it has been shown that type I IFNs activate signalling pathways mediated by mitogen-activated protein kinases (MAPKs), specifically p38 and extracellular signal-regulated kinase 1 (ERK1) or ERK2. p38 activity is required for type I IFN-dependent transcription of several genes that are regulated by ISREs and GAS elements in an STAT-independent manner46–48. The functional relevance of this signalling pathway in the biological effects of type I IFNs was evidenced by several studies indicating that p38 is required for the growth-inhibitory and antiviral effects of type I IFNs49,50. In addition to the induction of the p38 signalling cascade, the MAPK/ERK kinase (MEK)–ERK pathway is activated by type I IFNs51 and participates in the response to viral infection52. Finally, as mentioned above, type I IFNs can also induce the activation of the PI3K signalling pathway in a STAT-independent manner53,54. The PI3K pathway has an important role in mediating gene transcription in response to both type I and type II IFNs and is essential for mediating their antiviral effects against the encephalomyocarditis virus in vitro55. Activation of the PI3K signalling cascade controls the activation of mammalian target of rapamycin (mTOR), which regulates mRNA translation. Notably, it was shown that type I IFNs activate pathways for the initiation of mRNA translation that are downstream of mTOR, such as the pathway that involves the activation of p70 S6 kinase and the subsequent phosphorylation of the S6 ribosomal protein56,57. The activation of mTOR by type I IFNs was unrelated to the activation of STAT family members and had no effect on gene transcription56,57, indicating that mTOR selectively regulates IFN-induced mRNA translation. In terms of biological relevance, the activation of mTOR signalling has been shown to mediate the antiviral effects of IFNα against the hepatitis C virus58.
Modulating immune responses by type I IFNs
Type I IFNs and suppression of intestinal inflammation
The intestinal microbiota influences the development of local and systemic immune responses and the proliferation and barrier functions of the intestinal epithelium, in part via the activation of host TLRs59. Over the last few years, data from our laboratory and others have shown that the systemic administration of TLR9 ligands reduces the severity of colonic injury and inflammation in models of experimental colitis60–62, in part through the TLR9-induced production of type I IFNs by DCs60. In addition, IFNα enhances the barrier function of intestinal epithelial cells in vitro by activating STAT3, which maintains the expression of several key tight junction molecules, such as members of the claudin family (J.L., unpublished observations). Similarly, activation of TLR3 by the synthetic viral RNA polyinosinic–polycytidylic acid (polyI:C), which induces type I IFN production in many cell types, was shown to protect mice from experimental colitis62. Furthermore, Ifnar−/− mice were extremely susceptible to dextran-sulphate sodium (DSS)-induced colitis60,63, and the administration of recombinant IFNβ to DSS-treated wild-type mice mimicked the anti-inflammatory effects of TLR9 ligands60.
Different types of haematopoietic cells contribute to DSS-induced colonic inflammation. The depletion of CD11chi DCs after DSS administration showed that type I IFNs induced by TLR9-activated CD11chi DCs attenuate the severity of colitis in this model64. This study also showed that, depending on their mode of activation, DCs can enhance or inhibit acute DSS-induced colitis by secreting various cytokines (such as TNF, IL-6, IL-10 and type I IFNs), by regulating the production of chemokines that affect the composition of the cellular infiltrate, and by affecting the rate of resolution of inflammation in the colon64. In addition, a recent study using a genetically modified strain of Lactobacillus acidophilus that constitutively expresses IFNβ reported that the local expression of IFNAR1 by CD103+ DCs in the Peyer’s patches is necessary for protection against DSS-induced intestinal inflammation63. In this study, saturating expression of IFNβ before the induction of colitis resulted in the transient suppression of IFNAR1 expression on intestinal CD103+ DCs; this prevented IFNβ from signalling during the inflammatory period, mimicking the phenotype of Ifnar−/− mice with DSS-induced colitis63.
Regulation of adaptive immune responses by type I IFNs
In addition to the T cell-independent protective effects of TLR9-mediated type I IFN induction, the role of TLR9 agonist-induced protection in T cell-dependent intestinal inflammation has also been evaluated. TLR9 signalling in DCs in the lamina propria of the small intestine was shown to regulate the functions of regulatory T (TReg) cells and effector T cells65. In this study, TReg cell induction was inhibited by DNA derived from commensal bacteria through TLR9, and this was shown to favour the development of protective mucosal immune responses; however the role of type I IFNs in these effects was not clarified65. In a T cell-transfer model of colitis, treatment with a TLR9 ligand in the absence of type I IFN signalling failed to induce functionally suppressive CD4+CD62L+ T cells, whereas pretreatment of the donor mice with recombinant IFNβ and subsequent T cell transfer resulted in reduced intestinal inflammation and decreased secretion of pro-inflammatory cytokines66. Indeed, a role for type I IFNs in TLR9-induced protection in T cell-dependent colitis was previously shown in germ-free mice; in this study, type I IFNs were shown to induce the expression of regulatory markers on CD4+CD62L+ T cells67. These data correlate with previous reports that type I IFNs induce regulatory capacities in T cells68,69. Therefore, type I IFNs seem to exert a protective effect in the intestinal mucosa in both acute and chronic models of colitis.
Type I IFNs have been shown to have an important role in the differentiation of both CD4+ and CD8+ T cells. Initial studies found that IL-12 signalling in CD4+ T cells and the subsequent activation of STAT4, which in turn promotes T-bet expression, are crucial events in T helper 1 (TH1) cell differentiation70. In addition to IL-12, type I IFNs were shown to induce the tyrosine phosphorylation and DNA binding of STAT4 (REF. 71) and to act directly on human T cells, but not mouse T cells, to drive TH1 cell development72. However, it was later reported that type I IFNs activate STAT4 directly and that this activation is required for IFNγ production during viral infections in mice73,74. Thus, phosphorylation of STAT4 has been detected in response to type I IFN treatment of both human and mouse T cells74–76 but, in contrast with IL-12, type I IFNs were unable to sustain STAT4 phosphorylation and therefore were not sufficient to drive TH1 cell commitment in vitro75,77. However, type I IFNs regulate TH1 cell differentiation and effector functions in vivo by synergizing with other cytokines, such as IL-18 and IL-21 (REFS 78–80). Moreover, type I IFNs have been associated with the suppression of TH2 and TH17 cell-mediated responses. In human cells, type I IFNs reversed TH2 cell commitment by suppressing the expression of the TH2 cell-associated transcription factor GATA-binding protein 3 (GATA3)78. Similarly, type I IFNs also suppress the differentiation of TH17 cells in both mice81 and humans82.
Forkhead box P3 (FOXP3)+ TReg cells have a pivotal role in maintaining immunological tolerance and homeostasis. IFNβ treatment markedly improved the frequency and suppressive function of TReg cells in patients with relapsing–remitting multiple sclerosis9,83 and in patients with chronic hepatitis C virus infection84, and also increased Foxp3 mRNA expression in PBMCs from patients with relapsing–remitting multiple sclerosis85. FOXP3+ TReg cells show functional and phenotypical plasticity in response to environmental cues and are capable of secreting pro-inflammatory cytokines, for example during severe inflammation86,87. Recently, a higher frequency of IFNγ-secreting TReg cells has been reported in untreated patients with relapsing–remitting multiple sclerosis compared with the frequency in control individuals88. By contrast, in patients treated with IFNβ, the frequency of IFNγ+FOXP3+ T cells was similar to that in healthy controls88. In addition, ongoing studies in our laboratory indicate that type I IFN signalling is essential for the maintenance of FOXP3 expression in vivo and for the suppressive activity of TReg cells in a model of T cell-mediated colitis in mice (S. E. Lee, J.M.G.-N. and E.R., unpublished observations). These data suggest a novel role for type I IFNs in TH cell differentiation, as well as in the suppressive function of TReg cells.
Type I IFNs and inflammasome activation
Four different initiator components that activate different types of inflammasome in response to different stimuli have been identified: NOD-, LRR- and pyrin domain-containing 1 (NLRP1), NLRP3, NOD-, LRR- and CARD-containing 4 (NLRC4; also known as IPAF) and absent in melanoma 2 (AIM2)89.
Recently, an inhibitory role for type I IFNs in the activation of the inflammasome has been reported90. In this study, type I IFNs inhibited IL-1β production through two different mechanisms. First, IFNβ signalling directly inhibited the NLRP1 and NLRP3 inflammasomes (but not the NLRC4 and AIM2 inflammasomes) in a STAT1-dependent manner. Second, type I IFNs induced the production of IL-10, which in turn activated the transcription factor STAT3 in an autocrine manner to reduce the levels of pro-IL-1α and pro-IL-1β90 (FIG. 2a). Furthermore, type I IFNs increased the susceptibility of mice to Candida albicans infection, an effect that was attributed to the reduction in IL-1β production90. Therefore, these data suggested an inhibitory role of type I IFNs in inflammasome activation.
Figure 2. Type I IFNs regulate inflammasome activation.
a | Type I interferons (IFNs) inhibit the production of IL-1β by the inflammasome through two different mechanisms. First, they activate signal transducer and activator of transcription 1 (STAT1), which directly inhibits the NOD-, LRR- and pyrin domain-containing 1 (NLRP1) and NLRP3 inflammasomes but not the absent in melanoma 2 (AIM2) or NOD-, LRR- and CARD-containing 4 (NLRC4) inflammasomes. Second, type I IFNs induce the production of interleukin-10 (IL-10), which binds to the IL-10 receptor (IL-10R) in an autocrine manner and activates STAT3, which in turn reduces the levels of the precursors pro-IL-1α and pro-IL-1β. b | Type I IFN signalling is required for efficient activation of the AIM2 inflammasome in response to Francisella tularensis. After the bacterium enters the phagosome, the phagosome is rapidly acidified. Acidification causes the release of bacterial DNA into the cytosol, and this DNA activates an unidentified DNA sensor, which in turn activates IFN regulatory factor 3 (IRF3) to initiate the production of type I IFNs. IFNβ then binds to the IFNα/β receptor (IFNAR) in an autocrine manner to enhance the activation of the AIM2 inflammasome, possibly by increasing phagosomal acidification and/or bactericidal activity, thereby favouring the release of more bacterial DNA. CREB, cAMP-responsive-element-binding protein; TLR, Toll-like receptor.
By contrast, previous work reported a positive effect of type I IFNs in inflammasome activation during Francisella tularensis infection91. The cytosolic pathogens F. tularensis and L. monocytogenes were both shown to induce a type I IFN response that was essential for caspase 1 activation and IL-1β production. Consistent with this, F. tularensis DNA released into the cytosol activated the production of type I IFNs via IRF3, and this was necessary for the activation of the DNA-sensing AIM2 inflammasome92 (FIG. 2b). The discrepancies regarding the role of type I IFN signalling in the production of IL-1β might be related to the type of inflammasome; that is, type I IFNs mediate the inhibition of the NLRP3 inflammasome but the activation of the AIM2 inflammasome. Of particular interest, however, is the case of L. monocytogenes, which has been shown to activate the NLRP3 inflammasome89 and to increase IL-1β production in an IFNβ-dependent manner.
The cytokines IL-1α and IL-1β both bind to IL-1 receptor type 1 (IL-1R1), leading to the activation of NF-κB, MAPKs and certain IRFs93,94. In a recent study, we showed that IL-1R1 is necessary for the TLR9-dependent activation of a type I IFN and IL-10 response95. The mechanism by which IL-1R1 signalling modulates type I IFN production involves changes in the ubiquitylation profile of TRAF3, an E3 ubiquitin ligase that interacts with both MYD88 and TRIF, and the type of ubiquitylation determines whether type I IFNs or pro-inflammatory cytokines are produced8,96. Lysine 48 (K48)-linked polyubiquitylation of TRAF3 leads to its proteosomal degradation and the activation of MAPKs and pro-inflammatory cytokines, whereas K63-linked polyubiquitylation of TRAF3 results in the activation of IRFs and subsequent type I IFN production96,97. K63-linked, but not K48-linked, polyubiquitylation of TRAF3 is greatly reduced in the absence of IL-1 signalling. This effect is mediated by deubiquitylating enzyme A (DUBA; also known as OTUD5)95, which specifically cleaves the K63-linked polyubiquitin chain on TRAF3 (REF. 97). Together, these data suggested a model in which IL-1R1 positively regulates TLR-induced type I IFN production (FIG. 3). This mechanism may explain previous observations of IL-1R1-mediated protection against intestinal damage from Citrobacter rodentium infection98 and DSS-induced colitis98,99.
Figure 3. IL-1R1 signalling regulates type I IFN production.

Interleukin-1 receptor 1 (IL-1R1) signalling positively modulates the production of type I interferons (IFNs) through the differential ubiquitylation of TNF receptor-associated factor 3 (TRAF3). Lysine 48 (K48)-linked polyubiquitylation of TRAF3 leads to its proteosomal degradation and the production of pro-inflammatory cytokines. By contrast, K63-linked polyubiquitylation of TRAF3 triggers the activation of IFN regulatory factor 3 (IRF3) and the subsequent production of type I IFNs. The absence of IL-1 signalling results in increased levels of deubiquitylating enzyme A (DUBA), which cleaves the K63-linked but not the K48-linked ubiquitin chains on TRAF3. LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor-κB; TLR, Toll-like receptor.
Type I IFNs and bacterial infections
Although type I IFNs are induced by bacterial pathogens, the role of type I IFNs in the context of bacterial infections is not completely understood. Bacterial induction of type I IFNs can be mediated through the TLR-dependent recognition of bacterial products, such as LPS, or through the TLR-independent recognition of bacterial ligands that are delivered to the host cytosol. Many investigators have reported a variety of beneficial but also detrimental immune functions for type I IFNs during bacterial infection. For example, type I IFNs have an important role in mediating the pathology of LPS-induced septic shock84. By contrast, type I IFNs impair the clearance of L. monocytogenes100,101, Mycobacterium tuberculosis102 and Chlamydia muridarum83,103, and they are detrimental to host survival after infection with F. tularensis104.
Two mechanisms have been proposed to explain these phenomena. The first mechanism suggests that microorganisms induce type I IFN production as a strategy to induce apoptosis in lymphocytes, resulting in the suppression of adaptive immune responses105. The second mechanism proposes that type I IFNs suppress the production of IL-17A and IL-17F, which are necessary for neutrophil-mediated bacterial clearance104. By contrast, type I IFNs are necessary for host resistance against other bacterial pathogens. Mice deficient in IFNAR showed decreased survival and increased bacterial burdens after infection with Streptococcus pneumoniae or Escherichia coli106, and this was attributed to the reduced expression of certain cytokines, such as TNF or IFNγ. Moreover, type I IFNs were shown to play an important part in restricting the growth of Legionella pneumophila in macrophages107. Taken together, these data show that type I IFNs have a wide range of immunomodulatory effects in response to bacterial infections, and this clearly expands the old notion that type I IFNs serve only as antiviral cytokines.
Type I IFNs in autoimmune disease
The connection between type I IFNs and several autoimmune and inflammatory disorders is well known, although there is considerable variation in the precise mechanisms and in the role of these cytokines in each condition. Some autoimmune diseases (such as psoriasis and systemic lupus erythematosus (SLE)) are improved by the inhibition of type I IFNs or their upstream regulators. By contrast, other conditions that are characterized by strong TH1 and/or TH17 cell responses — such as arthritis, inflammatory bowel disease (IBD) and multiple sclerosis — benefit from the administration of type I IFNs (FIG. 4). All these inflammatory conditions have an important impact on public health and are the focus of numerous analyses that are readily available in the literature. Therefore, they are only briefly discussed here.
Figure 4. Type I IFNs in human diseases.
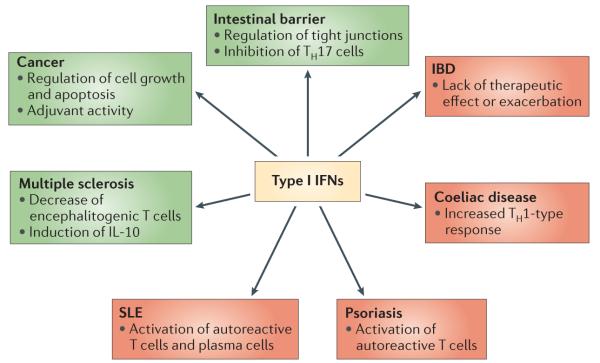
Type I interferons (IFNs) are implicated in different human diseases, although their role in each condition varies. Type I IFNs usually have a beneficial impact in inflammatory syndromes, such as inflammatory bowel disease (IBD) and multiple sclerosis. By contrast, some autoimmune diseases, such as psoriasis and systemic lupus erythematosus (SLE), are improved by type I IFN inhibition. This figure summarizes the positive (green) and negative (red) roles of type I IFNs in different human conditions. IL-10, interleukin-10; TH, T helper.
Inflammatory bowel disease
IBD encompasses two major clinical entities: Crohn’s disease and ulcerative colitis. Conventional therapies for IBD include 5-aminosalicylic acid (5-ASA), antibiotics (such as ciprofloxacin and metronidazole), corticosteroids, immunosuppressants and TNF-specific antibodies (such as infliximab and adalimumab)108. However, poor efficacy in some cases (with 5-ASA or antibiotic treatment) or the development of serious side effects (with corticosteroid treatment) led to the search for other biological therapies. Given their mechanisms of action and their beneficial effects on intestinal homeostasis in animal models, type I IFNs have been tested as a treatment for IBD. Although a few clinical studies of IFNα or IFNβ therapy in patients with ulcerative colitis initially showed promising results109–111, most of these studies failed to demonstrate a beneficial therapeutic effect112–115. Furthermore, documented cases of the exacerbation of ulcerative colitis during IFNα therapy for chronic hepatitis C infection116,117 and of the development of ulcerative colitis in patients with multiple sclerosis following treatment with IFNβ1a118 call into question whether type I IFNs have a therapeutic role in IBD.
Coeliac disease
Coeliac disease has a strong genetic component, as it is highly associated with HLA-DQ2 and HLA-DQ8 alleles119. The pathogenesis of coeliac disease involves the polarization of T cells into TH1 cells and the production of high levels of IFNγ in response to gluten120. TH1 cell polarization is driven by many cytokines, including IL-21, IL-18 and IFNα121, the levels of which are usually elevated in the mucosa of untreated patients with coeliac disease122. Interestingly, the addition of IFNα-specific, but not IL-18-specific, blocking antibodies to biopsy specimens from patients with coeliac disease inhibits IFNγ production in ex vivo organ cultures123. Activation of gluten-specific CD4+ T cells requires that gluten antigens be presented by antigen-presenting cells (APCs) that express HLA-DQ2 or HLA-DQ8 (REFS 124,125). The expression of these alleles is relatively low in normal mucosa, but can be upregulated on APCs by type I and type II IFNs, and this facilitates the activation of other inflammatory cells, thereby favouring an inflammatory response to gluten peptides122,126,127. Notably, enteric viruses can induce the production of type I IFNs, leading to the upregulation of HLA-DQ2 or HLA-DQ8 on APCs, and this would result in the perfect milieu for the activation of gluten-specific TH1 cells. The development of coeliac disease in patients undergoing treatment with recombinant type I IFNs, such as patients with hepatitis C virus infection122,128, also supports this hypothesis.
Psoriasis
Psoriasis is a T cell-mediated chronic inflammatory skin disease that is characterized by the production of large amounts of type I IFNs by pDCs that are recruited to and accumulate in the dermis. High levels of type I IFNs are responsible for the local activation and expansion of pathogenic T cell populations, which in turn trigger the abnormal proliferation and differentiation of keratinocytes and the development of diffuse epidermal hyperplasia, known as acanthosis. The activation of TH17 cells has been associated with this phenomenon129, and the blockade of type I IFN signalling by IFNAR-specific antibodies effectively inhibited the activation of autoreactive T cells and the development of skin lesions in a xenograft model of psoriasis130. In addition, antibodies specific for blood DC antigen 2 (BDCA2; also known as CLEC4C) suppressed the production of type I IFNs by pDCs through the activation of SRC family protein tyrosine kinases, and this also inhibited the activation of autoreactive T cells and the development of skin lesions131.
Systemic lupus erythematosus
The activation of pDCs by nucleic acid-containing immune complexes leads to the production of large amounts of type I IFNs, and elevated levels of IFNα are detected in the blood of patients with SLE132,133. Type I IFNs in the serum of patients with SLE were responsible for the differentiation of monocytes into DCs and the expression of MHC class II molecules and the co-stimulatory molecules CD80 and CD86 by these DCs. In addition, normal monocytes cultured with serum from patients with SLE, but not those cultured with autologous serum, could present self antigens to autoreactive T cells134. In another study, type I IFNs produced by pDCs, together with IL-6, were also shown to induce plasma cell differentiation135. Patients with SLE are treated with various medications, including non-steroidal anti-inflammatory drugs, glucocorticoids and immunosuppressants. Many of these therapies are associated with severe adverse effects. Therefore, there is a medical need for more-specific and safer therapies that target selective pathways involved in the pathogenesis of SLE. Recent data from a Phase I clinical trial showed that the administration of neutralizing antibodies specific for IFNα effectively inhibited the overexpression of IFNα-inducible genes in skin lesions of patients with SLE136.
Multiple sclerosis
It is generally considered that both TH1 and TH17 cells are involved in the pathogenesis of multiple sclerosis and experimental autoimmune encephalomyelitis (EAE; an animal model of multiple sclerosis). Human recombinant IFNβ therapy is widely prescribed for certain stages of multiple sclerosis, particularly relapsing–remitting multiple sclerosis137. Two human recombinant IFNβ forms are currently used: IFNβ1a, which is produced in Chinese hamster ovary cells; and IFNβ1b, which is produced in E. coli138. The exact mechanism behind the beneficial effect of type I IFNs in multiple sclerosis and EAE is not known, but recombinant IFNβ reduces the attack frequency and severity of multiple sclerosis138,139. However, not all patients with relapsing–remitting multiple sclerosis respond to treatment, and the development of IFNβ-specific autoantibodies may cause relapses and side effects.
IFNβ-deficient mice are more susceptible to EAE development140, and type I IFNs can attenuate EAE in mice141, suggesting that IFNβ modulates both multiple sclerosis and EAE. In addition, the conditional deletion of Ifnar1 showed that specific myeloid cells, such as monocytes and macrophages142 or DCs143, modulate autoimmune inflammation of the central nervous system in an IFNAR-dependent manner. The expression of IFNAR by microglial cells (brain-endogenous macrophages) also modulates the severity of EAE142. IFNβ reduces the ability of microglial cells to present antigens, and this in turn reduces the recruitment and effector functions of encephalitogenic T cells144. Another mechanism that was proposed to explain the immunomodulatory effects of IFNβ in patients with multiple sclerosis involves the induction of IL-10. Indeed, upregulation of IL-10 production is a hallmark of IFNβ treatment in patients with multiple sclerosis145.
Cancer
Type I IFNs have been extensively used for the treatment of several types of cancer, including haematological malignancies (for example, hairy cell leukaemia and some B or T cell lymphomas) and solid tumours (for example, melanoma, renal cell carcinoma and Kaposi’s sarcoma)146,147.
IFNα1-producing tumour cells were shown to be less tumorigenic and less able to metastasize when transferred into mice, and this was attributed to the activation of several host antitumour mechanisms at the tumour site148. These studies suggested that type I IFNs could act as an adjuvant in cancer vaccines133. Indeed, IFNα showed an effective adjuvant activity in patients with melanoma149. In addition to their putative role as adjuvants in antitumour vaccines, type I IFNs inhibit angiogenesis and act on DCs to enhance the ability of these cells to cross-present apoptotic antigens to CD8+ T cells150 to trigger a cytotoxic T lymphocyte response151,152.
The main pitfall for the use of type I IFNs as an anti-tumour therapy is the frequent severe side effects, which decrease the enthusiasm for the application of type I IFNs in this clinical setting.
Concluding remarks
Our understanding of the functions of type I IFNs and the mechanisms that control these functions is continuously evolving. Since their discovery, type I IFNs have been associated with host defence responses to viral infections, and it is only recently that their functions in bacterial infections and in immune-mediated and inflammatory disorders have been appreciated. The molecular mechanisms by which type I IFNs exert their immunomodulatory functions and the reasons why they restrain the development of some immunopathologies while increasing the severity of others are still largely unknown. One possible explanation for these opposing effects could be found in the differential regulation of STAT family members in different tissues or organs under different physiological and/or inflammatory conditions. A better understanding of the underlying pathophysiology is mandatory for the design of effective treatments with type I IFNs or with type I IFN-specific neutralizing antibodies.
Acknowledgements
We thank S. Herdman for his editorial assistance. This work was supported by US National Institutes of Health grants AI068685, AI077989, AI095623 and DK35108; grants from the Crohn’s and Colitis Foundation of America; and grant CP10/00417 from Instituto de Salud Carlos III, Madrid, Spain.
Glossary
- Viral interference
The antagonistic or inhibitory effect induced by one virus or its components on the propagation of another virus.
- IFN-stimulated genes (ISGs)
These genes contain promoters that are responsive to interferon (IFN) signalling, and they are responsible for the antiviral and immunomodulatory properties of IFNs. Over 400 such genes have been identified by microarray analyses. Some, such as RNA-activated protein kinase, ribonuclease L, MX1 (myxovirus resistance 1) and ISG15 (IFN-stimulated gene of 15 kDa), have well-documented antiviral activities, but the precise biological function of most of these genes is unknown.
- Inflammasome
A cytosolic multiprotein complex that activates caspase 1 and regulates the release of IL-1β and IL-18 in response to exogenous pathogens and endogenous danger signals. This complex minimally consists of a danger-sensing initiator component and the effector component, which is mature caspase 1.
- Coeliac disease
An immune-mediated enteropathy triggered by intolerance to dietary ingestion of glutamine- and proline-rich proteins, collectively known as gluten, which is present in wheat, barley, rye and other grains. This disease results in gastrointestinal symptoms such as diarrhoea, nutrient malabsorption and weight loss.
- Multiple sclerosis
A chronic inflammatory disease of the central nervous system that causes the progressive destruction of the myelin sheaths around axons in any area of the brain, optic nerve and spinal cord. This results in slower nerve impulses.
- Peyer’s patches
Collections of lymphoid tissue that are located in the mucosa of the small intestine, with an outer epithelial layer that consists of specialized epithelial cells called M cells.
- Systemic lupus erythematosus (SLE)
An autoimmune disease characterized by the presence of circulating immune complexes that contain antinuclear antibodies bound to self nucleic acids and other nuclear antigens.
- Inflammatory bowel disease (IBD)
A chronic inflammatory condition that affects the intestinal tract. The proposed pathogenesis of IBD involves a complex model that includes abnormalities of innate immune function and their relationship with the commensal microbiota, inappropriate release of pro-inflammatory cytokines and other mediators, alterations of the intestinal epithelial barrier, and a cytokine imbalance that promotes the pro-inflammatory activity of adaptive immune cells.
Footnotes
Competing interests statement The authors declare no competing financial interests.
References
- 1.Jenner E. Letter to the Editor. Med. Phys. J. 1804;12:97–102. [PMC free article] [PubMed] [Google Scholar]
- 2.Salaman RN. Protective inoculation against a plant virus. Nature. 1933;131:468. [Google Scholar]
- 3.White PB. Lysogenic strains of V. cholerae and the influence of lysozyme on cholera phage activity. J. Pathol. Bacteriol. 1937;44:276–278. [Google Scholar]
- 4.Hoskins M. A protective action of neurotropic against viscerotropic yellow fever virus in Macacus rhesus. Am. J. Trop. Med. 1935;15:675–680. [Google Scholar]
- 5.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 6.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 7.Gad HH, et al. Interferon-λ is functionally an interferon but structurally related to the interleukin-10 family. J. Biol. Chem. 2009;284:20869–20875. doi: 10.1074/jbc.M109.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. This study showed that TRAF3 is essential for both type I IFN and IL-10 production, but dispensable for the production of pro-inflammatory cytokines.
- 9.Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 10.Monroe KM, McWhirter SM, Vance RE. Induction of type I interferons by bacteria. Cell. Microbiol. 2010;12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trinchieri G. Type I interferon: friend or foe? J. Exp. Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akira S. Toll-like receptors and innate immunity. Adv. Immunol. 2001;78:1–56. doi: 10.1016/s0065-2776(01)78001-7. [DOI] [PubMed] [Google Scholar]
- 13.Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. A description of STING as an important mediator in the host defence response against DNA-containing pathogens and in the adjuvant activity of DNA-based vaccines.
- 16.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. The first identification of DAI as a new cytosolic DNA sensor that enhances the induction of type I IFN genes and other genes involved in innate immunity.
- 17.Kaiser WJ, Upton JW, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-κB activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 19.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 21.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. An excellent review of the signalling mechanisms of type I IFNs, including the classical JAK–STAT pathways, as well as the MAPK and mTOR pathways.
- 22.Fasler-Kan E, Pansky A, Wiederkehr M, Battegay M, Heim MH. Interferon-α activates signal transducers and activators of transcription 5 and 6 in Daudi cells. Eur. J. Biochem. 1998;254:514–519. doi: 10.1046/j.1432-1327.1998.2540514.x. [DOI] [PubMed] [Google Scholar]
- 23.Matikainen S, et al. Interferon-α activates multiple STAT proteins and upregulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–1991. [PubMed] [Google Scholar]
- 24.Aaronson DS, Horvath CM. A road map for those who don’t know JAK–STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 25.Platanias LC, Fish EN. Signaling pathways activated by interferons. Exp. Hematol. 1999;27:1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 26.Sato T, Selleri C, Young NS, Maciejewski JP. Inhibition of interferon regulatory factor-1 expression results in predominance of cell growth stimulatory effects of interferon-γ due to phosphorylation of Stat1 and Stat3. Blood. 1997;90:4749–4758. [PubMed] [Google Scholar]
- 27.Heinrich PC, Behrmann I, Muller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker SJ, Rane SG, Reddy EP. Hematopoietic cytokine receptor signaling. Oncogene. 2007;26:6724–6737. doi: 10.1038/sj.onc.1210757. [DOI] [PubMed] [Google Scholar]
- 29.Regis G, Pensa S, Boselli D, Novelli F, Poli V. Ups and downs: the STAT1:STAT3 seesaw of interferon and gp130 receptor signalling. Semin. Cell Dev. Biol. 2008;19:351–359. doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Stephanou A, Brar BK, Knight RA, Latchman DS. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ. 2000;7:329–330. doi: 10.1038/sj.cdd.4400656. [DOI] [PubMed] [Google Scholar]
- 31.Azare J, et al. Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin β6. Mol. Cell. Biol. 2007;27:4444–4453. doi: 10.1128/MCB.02404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dechow TN, et al. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc. Natl Acad. Sci. USA. 2004;101:10602–10607. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dustin ML, Singer KH, Tuck DT, Springer TA. Adhesion of T lymphoblasts to epidermal keratinocytes is regulated by interferon γ and is mediated by intercellular adhesion molecule 1 (ICAM-1) J. Exp. Med. 1988;167:1323–1340. doi: 10.1084/jem.167.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brucet M, Marques L, Sebastian C, Lloberas J, Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon γ is mediated by STAT1 and IRF-1. Genes Immun. 2004;5:26–35. doi: 10.1038/sj.gene.6364035. [DOI] [PubMed] [Google Scholar]
- 36.Marques L, Brucet M, Lloberas J, Celada A. STAT1 regulates lipopolysaccharide- and TNF-α-dependent expression of transporter associated with antigen processing 1 and low molecular mass polypeptide 2 genes in macrophages by distinct mechanisms. J. Immunol. 2004;173:1103–1110. doi: 10.4049/jimmunol.173.2.1103. [DOI] [PubMed] [Google Scholar]
- 37.Ito S, et al. Interleukin-10 inhibits expression of both interferon α- and interferon γ-induced genes by suppressing tyrosine phosphorylation of STAT1. Blood. 1999;93:1456–1463. [PubMed] [Google Scholar]
- 38.Yasukawa H, et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature Immunol. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 39.El Kasmi KC, et al. General nature of the STAT3-activated anti-inflammatory response. J. Immunol. 2006;177:7880–7888. doi: 10.4049/jimmunol.177.11.7880. [DOI] [PubMed] [Google Scholar]
- 40.Tanabe Y, et al. Cutting edge: role of STAT1, STAT3, and STAT5 in IFN-αβ responses in T lymphocytes. J. Immunol. 2005;174:609–613. doi: 10.4049/jimmunol.174.2.609. [DOI] [PubMed] [Google Scholar]
- 41.Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-γ. J. Biol. Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- 42.Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J. Immunol. 2007;178:6705–6709. doi: 10.4049/jimmunol.178.11.6705. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Chen M, Wandinger KP, Williams G, Dhib-Jalbut S. IFN-β-1b inhibits IL-12 production in peripheral blood mononuclear cells in an IL-10-dependent mechanism: relevance to IFN-β-1b therapeutic effects in multiple sclerosis. J. Immunol. 2000;165:548–557. doi: 10.4049/jimmunol.165.1.548. [DOI] [PubMed] [Google Scholar]
- 44.Ziegler-Heitbrock L, et al. IFN-α induces the human IL-10 gene by recruiting both IFN regulatory factor 1 and Stat3. J. Immunol. 2003;171:285–290. doi: 10.4049/jimmunol.171.1.285. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, et al. The role of glycogen synthase kinase 3 in regulating IFN-β-mediated IL-10 production. J. Immunol. 2011;186:675–684. doi: 10.4049/jimmunol.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uddin S, et al. Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem. 1999;274:30127–30131. doi: 10.1074/jbc.274.42.30127. [DOI] [PubMed] [Google Scholar]
- 47.Uddin S, et al. The Rac1/p38 mitogen-activated protein kinase pathway is required for interferon α-dependent transcriptional activation but not serine phosphorylation of Stat proteins. J. Biol. Chem. 2000;275:27634–27640. doi: 10.1074/jbc.M003170200. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, et al. Role of p38α Map kinase in type I interferon signaling. J. Biol. Chem. 2004;279:970–979. doi: 10.1074/jbc.M309927200. [DOI] [PubMed] [Google Scholar]
- 49.Mayer IA, et al. The p38 MAPK pathway mediates the growth inhibitory effects of interferon-α in BCR-ABL-expressing cells. J. Biol. Chem. 2001;276:28570–28577. doi: 10.1074/jbc.M011685200. [DOI] [PubMed] [Google Scholar]
- 50.Ishida H, et al. Involvement of p38 signaling pathway in interferon-α-mediated antiviral activity toward hepatitis C virus. Biochem. Biophys. Res. Commun. 2004;321:722–727. doi: 10.1016/j.bbrc.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 51.David M, et al. Requirement for MAP kinase (ERK2) activity in interferon α- and interferon β-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 52.Wang F, et al. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nature Immunol. 2004;5:1266–1274. doi: 10.1038/ni1132. [DOI] [PubMed] [Google Scholar]
- 53.Uddin S, et al. Interferon-α engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J. Biol. Chem. 1995;270:15938–15941. doi: 10.1074/jbc.270.27.15938. [DOI] [PubMed] [Google Scholar]
- 54.Kaur S, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc. Natl Acad. Sci. USA. 2008;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaur S, et al. Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J. Immunol. 2008;181:7316–7323. doi: 10.4049/jimmunol.181.10.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lekmine F, et al. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J. Biol. Chem. 2003;278:27772–27780. doi: 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- 57.Lekmine F, et al. Interferon-γ engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp. Cell Res. 2004;295:173–182. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 58.Matsumoto A, et al. Interferon-α-induced mTOR activation is an anti-hepatitis C virus signal via the phosphatidylinositol 3-kinase–Akt-independent pathway. J. Gastroenterol. 2009;44:856–863. doi: 10.1007/s00535-009-0075-1. [DOI] [PubMed] [Google Scholar]
- 59.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katakura K, et al. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J. Clin. Invest. 2005;115:695–702. doi: 10.1172/JCI22996. This study demonstrated that the protective effect of TLR9 signalling in colonic injury is mediated by type I IFNs.
- 61.Rachmilewitz D, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520–528. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 62.Vijay-Kumar M, et al. Activation of Toll-like receptor 3 protects against DSS-induced acute colitis. Inflamm. Bowel Dis. 2007;13:856–864. doi: 10.1002/ibd.20142. [DOI] [PubMed] [Google Scholar]
- 63.McFarland AP, et al. Localized delivery of interferon-β by Lactobacillus exacerbates experimental colitis. PloS ONE. 2011;6:e16967. doi: 10.1371/journal.pone.0016967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abe K, et al. Conventional dendritic cells regulate the outcome of colonic inflammation independently of T cells. Proc. Natl Acad. Sci. USA. 2007;104:17022–17027. doi: 10.1073/pnas.0708469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofmann C, et al. T cell-dependent protective effects of CpG motifs of bacterial DNA in experimental colitis are mediated by CD11c+ dendritic cells. Gut. 2010;59:1347–1354. doi: 10.1136/gut.2009.193177. [DOI] [PubMed] [Google Scholar]
- 67.Bleich A, et al. CpG motifs of bacterial DNA exert protective effects in mouse models of IBD by antigen-independent tolerance induction. Gastroenterology. 2009;136:278–287. doi: 10.1053/j.gastro.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 68.Bilsborough J, George TC, Norment A, Viney JL. Mucosal CD8α+ DC, with a plasmacytoid phenotype, induce differentiation and support function of T cells with regulatory properties. Immunology. 2003;108:481–492. doi: 10.1046/j.1365-2567.2003.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levings MK, et al. IFN-α and IL-10 induce the differentiation of human type 1 T regulatory cells. J. Immunol. 2001;166:5530–5539. doi: 10.4049/jimmunol.166.9.5530. [DOI] [PubMed] [Google Scholar]
- 70.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-γ production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 71.Cho SS, et al. Activation of STAT4 by IL-12 and IFN-α: evidence for the involvement of ligand-induced tyrosine and serine phosphorylation. J. Immunol. 1996;157:4781–4789. [PubMed] [Google Scholar]
- 72.Rogge L, et al. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J. Immunol. 1998;161:6567–6574. [PubMed] [Google Scholar]
- 73.Cousens LP, et al. Two roads diverged: interferon α/β- and interleukin 12-mediated pathways in promoting T cell interferon γ responses during viral infection. J. Exp. Med. 1999;189:1315–1328. doi: 10.1084/jem.189.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen KB, et al. Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 75.Berenson LS, Farrar JD, Murphy TL, Murphy KM. Frontline: absence of functional STAT4 activation despite detectable tyrosine phosphorylation induced by murine IFN-α. Eur. J. Immunol. 2004;34:2365–2374. doi: 10.1002/eji.200324829. [DOI] [PubMed] [Google Scholar]
- 76.Farrar JD, Smith JD, Murphy TL, Murphy KM. Recruitment of Stat4 to the human interferon-α/β receptor requires activated Stat2. J. Biol. Chem. 2000;275:2693–2697. doi: 10.1074/jbc.275.4.2693. [DOI] [PubMed] [Google Scholar]
- 77.Berenson LS, Gavrieli M, Farrar JD, Murphy TL, Murphy KM. Distinct characteristics of murine STAT4 activation in response to IL-12 and IFN-α. J. Immunol. 2006;177:5195–5203. doi: 10.4049/jimmunol.177.8.5195. [DOI] [PubMed] [Google Scholar]
- 78.Huber JP, Farrar JD. Regulation of effector and memory T-cell functions by type I interferon. Immunology. 2011;132:466–474. doi: 10.1111/j.1365-2567.2011.03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matikainen S, et al. IFN-α and IL-18 synergistically enhance IFN-γ production in human NK cells: differential regulation of Stat4 activation and IFN-γ gene expression by IFN-α and IL-12. Eur. J. Immunol. 2001;31:2236–2245. [PubMed] [Google Scholar]
- 80.Strengell M, Julkunen I, Matikainen S. IFN-α regulates IL-21 and IL-21R expression in human NK and T cells. J. Leukoc. Biol. 2004;76:416–422. doi: 10.1189/jlb.1003488. [DOI] [PubMed] [Google Scholar]
- 81.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nature Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 82.Moschen AR, Geiger S, Krehan I, Kaser A, Tilg H. Interferon-α controls IL-17 expression in vitro and in vivo. Immunobiology. 2008;213:779–787. doi: 10.1016/j.imbio.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 83.Qiu H, et al. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J. Immunol. 2008;181:2092–2102. doi: 10.4049/jimmunol.181.3.2092. [DOI] [PubMed] [Google Scholar]
- 84.Karaghiosoff M, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nature Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 85.Vandenbark AA, et al. Interferon-β-1a treatment increases CD56bright natural killer cells and CD4+CD25+ Foxp3 expression in subjects with multiple sclerosis. J. Neuroimmunol. 2009;215:125–128. doi: 10.1016/j.jneuroim.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 86.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nature Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oldenhove G, et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nature Med. 2011;17:673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 90.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. An important study showing that type I IFN signalling inhibits NLRP1 and NLRP3 inflammasomes in a two-step process through STAT1 and IL-10.
- 91.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fujita T, et al. Induction of the transcription factor IRF-1 and interferon-β mRNAs by cytokines and activators of second-messenger pathways. Proc. Natl Acad. Sci. USA. 1989;86:9936–9940. doi: 10.1073/pnas.86.24.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rivieccio MA, et al. The cytokine IL-1β activates IFN response factor 3 in human fetal astrocytes in culture. J. Immunol. 2005;174:3719–3726. doi: 10.4049/jimmunol.174.6.3719. [DOI] [PubMed] [Google Scholar]
- 95.Gonzalez-Navajas JM, et al. Interleukin 1 receptor signaling regulates DUBA expression and facilitates Toll-like receptor 9-driven antiinflammatory cytokine production. J. Exp. Med. 2010;207:2799–2807. doi: 10.1084/jem.20101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tseng PH, et al. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nature Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. An elegant study showing the different effects of K63- and K48-linked polyubiquitylation on the biological functions of TRAF3.
- 97.Kayagaki N, et al. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–1632. doi: 10.1126/science.1145918. The first report that DUBA binds to TRAF3 and selectively cleaves the K63-linked polyubiquitin chains, thereby dampening type I IFN production.
- 98.Lebeis SL, Powell KR, Merlin D, Sherman MA, Kalman D. Interleukin-1 receptor signaling protects mice from lethal intestinal damage caused by the attaching and effacing pathogen Citrobacter rodentium. Infect. Immun. 2009;77:604–614. doi: 10.1128/IAI.00907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kojouharoff G, et al. Neutralization of tumour necrosis factor (TNF) but not of IL-1 reduces inflammation in chronic dextran sulphate sodium-induced colitis in mice. Clin. Exp. Immunol. 1997;107:353–358. doi: 10.1111/j.1365-2249.1997.291-ce1184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 103.Nagarajan UM, et al. Type I interferon signaling exacerbates Chlamydia muridarum genital infection in a murine model. Infect. Immun. 2008;76:4642–4648. doi: 10.1128/IAI.00629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Henry T, et al. Type I IFN signaling constrains IL-17A/F secretion by γδ T cells during bacterial infections. J. Immunol. 2010;184:3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carrero JA, Unanue ER. Lymphocyte apoptosis as an immune subversion strategy of microbial pathogens. Trends Immunol. 2006;27:497–503. doi: 10.1016/j.it.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 106.Mancuso G, et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- 107.Coers J, Vance RE, Fontana MF, Dietrich WF. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell. Microbiol. 2007;9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 108.Burger D, Travis S. Conventional medical management of inflammatory bowel disease. Gastroenterology. 2011;140:1827–1837. doi: 10.1053/j.gastro.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 109.Musch E, Andus T, Malek M, Chrissafidou A, Schulz M. Successful treatment of steroid refractory active ulcerative colitis with natural interferon-β — an open long-term trial. Z. Gastroenterol. 2007;45:1235–1240. doi: 10.1055/s-2007-963378. [DOI] [PubMed] [Google Scholar]
- 110.Nikolaus S, et al. Interferon β-1a in ulcerative colitis: a placebo controlled, randomised, dose escalating study. Gut. 2003;52:1286–1290. doi: 10.1136/gut.52.9.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Madsen SM, et al. An open-labeled, randomized study comparing systemic interferon-α-2A and prednisolone enemas in the treatment of left-sided ulcerative colitis. Am. J. Gastroenterol. 2001;96:1807–1815. doi: 10.1111/j.1572-0241.2001.03875.x. [DOI] [PubMed] [Google Scholar]
- 112.Pena Rossi C, et al. Interferon β-1a for the maintenance of remission in patients with Crohn’s disease: results of a phase II dose-finding study. BMC Gastroenterol. 2009;9:22. doi: 10.1186/1471-230X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pena-Rossi C, et al. Clinical trial: a multicentre, randomized, double-blind, placebo-controlled, dose-finding, phase II study of subcutaneous interferon-β-1a in moderately active ulcerative colitis. Aliment. Pharmacol. Ther. 2008;28:758–767. doi: 10.1111/j.1365-2036.2008.03778.x. [DOI] [PubMed] [Google Scholar]
- 114.Gasche C, et al. Prospective evaluation of interferon-α in treatment of chronic active Crohn’s disease. Dig. Dis. Sci. 1995;40:800–804. doi: 10.1007/BF02064982. [DOI] [PubMed] [Google Scholar]
- 115.Musch E, et al. Interferon-β-1a for the treatment of steroid-refractory ulcerative colitis: a randomized, double-blind, placebo-controlled trial. Clin. Gastroenterol. Hepatol. 2005;3:581–586. doi: 10.1016/s1542-3565(05)00208-9. [DOI] [PubMed] [Google Scholar]
- 116.Mitoro A, et al. Exacerbation of ulcerative colitis during α-interferon therapy for chronic hepatitis C. Intern. Med. 1993;32:327–331. doi: 10.2169/internalmedicine.32.327. [DOI] [PubMed] [Google Scholar]
- 117.Watanabe T, et al. A case of exacerbation of ulcerative colitis induced by combination therapy with PEG-interferon α-2b and ribavirin. Gut. 2006;55:1682–1683. doi: 10.1136/gut.2006.105197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schott E, et al. Development of ulcerative colitis in a patient with multiple sclerosis following treatment with interferon β1a. World J. Gastroenterol. 2007;13:3638–3640. doi: 10.3748/wjg.v13.i26.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Freeman HJ, Chopra A, Clandinin MT, Thomson AB. Recent advances in celiac disease. World J. Gastroenterol. 2011;17:2259–2272. doi: 10.3748/wjg.v17.i18.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.MacDonald TT, Spencer J. Evidence that activated mucosal T cells play a role in the pathogenesis of enteropathy in human small intestine. J. Exp. Med. 1988;167:1341–1349. doi: 10.1084/jem.167.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nature Med. 2002;8:567–573. doi: 10.1038/nm0602-567. [DOI] [PubMed] [Google Scholar]
- 122.Monteleone G, Pender SL, Wathen NC, MacDonald TT. Interferon-α drives T cell-mediated immunopathology in the intestine. Eur. J. Immunol. 2001;31:2247–2255. doi: 10.1002/1521-4141(200108)31:8<2247::aid-immu2247>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 123.Di Sabatino A, et al. Evidence for the role of interferon-alfa production by dendritic cells in the Th1 response in celiac disease. Gastroenterology. 2007;133:1175–1187. doi: 10.1053/j.gastro.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 124.Molberg O, et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nature Med. 1998;4:713–717. doi: 10.1038/nm0698-713. [DOI] [PubMed] [Google Scholar]
- 125.Molberg O, et al. Gliadin specific, HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scand. J. Immunol. 1997;46:103–109. doi: 10.1046/j.1365-3083.1997.d01-93.x. [DOI] [PubMed] [Google Scholar]
- 126.Kagnoff MF. Celiac disease: pathogenesis of a model immunogenetic disease. J. Clin. Invest. 2007;117:41–49. doi: 10.1172/JCI30253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 128.Cammarota G, Cuoco L, Cianci R, Pandolfi F, Gasbarrini G. Onset of coeliac disease during treatment with interferon for chronic hepatitis C. Lancet. 2000;356:1494–1495. doi: 10.1016/S0140-6736(00)02880-4. [DOI] [PubMed] [Google Scholar]
- 129.Zheng Y, et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 130.Nestle FO, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-α production. J. Exp. Med. 2005;202:135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dzionek A, et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon α/β induction. J. Exp. Med. 2001;194:1823–1834. doi: 10.1084/jem.194.12.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54:1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 133.Kirou KA, et al. Activation of the interferon-α pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 134.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-α in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 135.Jego G, et al. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 136.Yao Y, et al. Neutralization of interferon-α/β-inducible genes and downstream effect in a phase I trial of an anti-interferon-α monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785–1796. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 137.Ann Marrie R, Rudick RA. Drug insight: interferon treatment in multiple sclerosis. Nature Clin. Pract. Neurol. 2006;2:34–44. doi: 10.1038/ncpneuro0088. [DOI] [PubMed] [Google Scholar]
- 138.Bendtzen K. Critical review: assessment of interferon-β immunogenicity in multiple sclerosis. J. Interferon Cytokine Res. 2010;30:759–766. doi: 10.1089/jir.2010.0091. [DOI] [PubMed] [Google Scholar]
- 139.The IFNB Multiple Sclerosis Study Group Interferon β-1b is effective in relapsing-remitting multiple sclerosis. I. Clinical results of a multicenter, randomized, double-blind, placebo-controlled trial. Neurology. 1993;43:655–661. doi: 10.1212/wnl.43.4.655. [DOI] [PubMed] [Google Scholar]
- 140.Teige I, et al. IFN-β gene deletion leads to augmented and chronic demyelinating experimental autoimmune encephalomyelitis. J. Immunol. 2003;170:4776–4784. doi: 10.4049/jimmunol.170.9.4776. [DOI] [PubMed] [Google Scholar]
- 141.Brod SA, Burns DK. Suppression of relapsing experimental autoimmune encephalomyelitis in the SJL/J mouse by oral administration of type I interferons. Neurology. 1994;44:1144–1148. doi: 10.1212/wnl.44.6.1144. [DOI] [PubMed] [Google Scholar]
- 142.Prinz M, et al. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 2008;28:675–686. doi: 10.1016/j.immuni.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 143.Guo B, Chang EY, Cheng G. The type I IFN induction pathway constrains Th17-mediated autoimmune inflammation in mice. J. Clin. Invest. 2008;118:1680–1690. doi: 10.1172/JCI33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Teige I, Liu Y, Issazadeh-Navikas S. IFN-β inhibits T cell activation capacity of central nervous system APCs. J. Immunol. 2006;177:3542–3553. doi: 10.4049/jimmunol.177.6.3542. [DOI] [PubMed] [Google Scholar]
- 145.Rudick RA, et al. Interferon β induces interleukin-10 expression: relevance to multiple sclerosis. Ann. Neurol. 1996;40:618–627. doi: 10.1002/ana.410400412. [DOI] [PubMed] [Google Scholar]
- 146.Ferrantini M, Capone I, Belardelli F. Interferon-α and cancer: mechanisms of action and new perspectives of clinical use. Biochimie. 2007;89:884–893. doi: 10.1016/j.biochi.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 147.Rizza P, Moretti F, Belardelli F. Recent advances on the immunomodulatory effects of IFN-α: implications for cancer immunotherapy and autoimmunity. Autoimmunity. 2010;43:204–209. doi: 10.3109/08916930903510880. [DOI] [PubMed] [Google Scholar]
- 148.Ferrantini M, Belardelli F. Gene therapy of cancer with interferon: lessons from tumor models and perspectives for clinical applications. Semin. Cancer Biol. 2000;10:145–157. doi: 10.1006/scbi.2000.0333. [DOI] [PubMed] [Google Scholar]
- 149.Di Pucchio T, et al. Immunization of stage IV melanoma patients with Melan-A/MART-1 and gp100 peptides plus IFN-α results in the activation of specific CD8+ T cells and monocyte/dendritic cell precursors. Cancer Res. 2006;66:4943–4951. doi: 10.1158/0008-5472.CAN-05-3396. [DOI] [PubMed] [Google Scholar]
- 150.Lapenta C, et al. IFN-α-conditioned dendritic cells are highly efficient in inducing cross-priming CD8+ T cells against exogenous viral antigens. Eur. J. Immunol. 2006;36:2046–2060. doi: 10.1002/eji.200535579. [DOI] [PubMed] [Google Scholar]
- 151.Lapenta C, et al. Potent immune response against HIV-1 and protection from virus challenge in hu-PBL-SCID mice immunized with inactivated virus-pulsed dendritic cells generated in the presence of IFN-α. J. Exp. Med. 2003;198:361–367. doi: 10.1084/jem.20021924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Santodonato L, et al. Monocyte-derived dendritic cells generated after a short-term culture with IFN-α and granulocyte-macrophage colony-stimulating factor stimulate a potent Epstein-Barr virus-specific CD8+ T cell response. J. Immunol. 2003;170:5195–5202. doi: 10.4049/jimmunol.170.10.5195. [DOI] [PubMed] [Google Scholar]




