Summary
An important feature of mechanism-based pharmacokinetic/pharmacodynamic (PK/PD) models is the identification of drug- and system-specific factors that determine the intensity and time-course of pharmacological effects. This provides an opportunity to integrate information obtained from in vitro bioassays and preclinical pharmacological studies in animals to anticipate the clinical and adverse responses to drugs in humans. The fact that contemporary PK/PD modeling continues to evolve and seeks to emulate systems level properties should provide enhanced capabilities to scale-up pharmacodynamic data. Critical steps in drug discovery and development, such as lead compound and first in human dose selection, may become more efficient with the implementation and further refinement of translational PK/PD modeling. In this review, we highlight fundamental principles in pharmacodynamics and the basic expectations for in vitro bioassays and traditional allometric scaling in PK/PD modeling. Discussion of PK/PD modeling efforts for recombinant human erythropoietin is also included as a case study showing the potential for advanced systems analysis to facilitate extrapolations and improve understanding of inter-species differences in drug responses.
Keywords: allometric scaling; cell life span models; mechanism-based modeling; pharmacodynamics, PD; pharmacokinetics, PK; receptor occupancy; recombinant human erythropoietin, rHuEpo; target-mediated drug disposition, TMDD
Introduction
The extrapolation of in silico, in vitro, and preclinical animal studies to predict the likely pharmacokinetic properties of drugs in humans now appears within reach, largely due to advancements in physiologically-based pharmacokinetic (PBPK) modeling.1,2) Whereas traditional allometry continues to prove useful under certain conditions for inter-species scaling of PK properties, significant progress has been achieved by transitioning from models of data (e.g., classic compartmental models) to those of biological systems. The PBPK modeling approach provides a framework for integrating drug-specific calculated parameters (e.g., octanol:water and blood:tissue partition coefficients) and in vitro measurements (e.g., plasma protein binding and hepatocyte intrinsic clearance) with physiological system-specific parameters (e.g., tissue volumes and blood flows). Given the relative success of anticipating human exposures to drugs and toxicants, 3,4) there is considerable interest in the development of techniques for the scaling of pharmacodynamic systems. Although drug responses are considerably more complex than processes controlling pharmacokinetics, the shift from empirical to mechanism-based PK/PD modeling5,6) should provide the best means for translating in vitro and animal data to human clinical pharmacology. 7)
In this review, we discuss the basic tenets of pharmacodynamics, namely 1) pharmacokinetics or drug exposure as the driving function, 2) capacity-limitation of drug-receptor interactions, and 3) physiological turnover processes and functional adaptation or homeostatic feedback mechanisms. As with PBPK models, these basic components identify drug and system specific properties that might be anticipated using in vitro assays, allometry, and/or preclinical animal experiments. A case study showing how human responses to recombinant human erythropoietin (rHuEpo) can be predicted from scaling a mathematical model developed in rats is provided as an example of utilizing mechanism-based PK/PD models to scale complex pharmacological systems.
Basic Principles of Pharmacodynamics
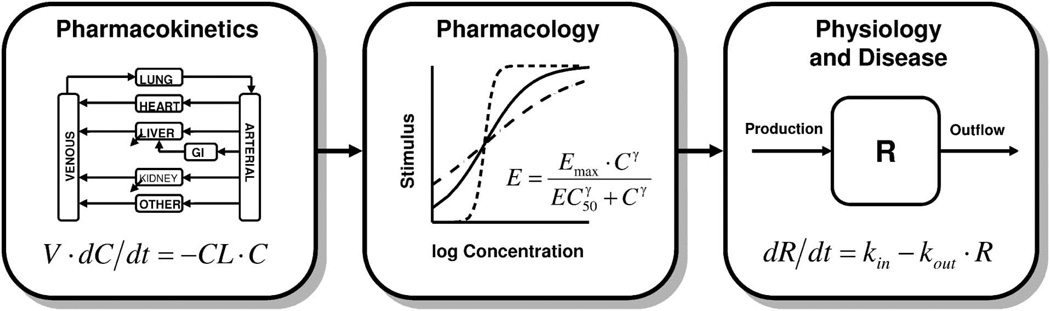
The basic tenets of pharmacokinetics (PK), pharmacology, and physiology continue to form the basis for contemporary pharmacodynamic systems analysis (Fig. 1). Pharmacokinetics, or the processes controlling the time-course of drug concentrations in relevant biological fluids, tissues, and sites of action (biophase), is the driving force for subsequent pharmacological and most toxicological effects. Although mammillary plasma clearance models (simple linear compartmental models) and area/moment analysis are the most commonly applied techniques for characterizing the absorption and disposition (distribution and elimination) properties of drugs, PBPK models provide a comprehensive platform for describing the major processes influencing the concentration time-course and net exposure of drugs in various fluids and tissues (Fig. 1, left panel). Each tissue of interest is anatomically arranged and described by a series of mass balance differential equations. Fick’s law of perfusion/diffusion and drug partitioning are featured along with a capacity-limited function for various drug binding, transport, and elimination processes. This approach provides insights into expected drug concentrations in important tissues, and potentially sites of action, and the intrinsic scalability of predictions across species and molecular drug properties is unparalleled. Whereas traditional PBPK model development has relied on destructive sampling in preclinical studies, advances in noninvasive imaging (such as positron emission tomography and magnetic resonance imaging) and microdialysis may eventually provide even finer details of in vivo drug disposition.8,9)
Fig. 1.
Major components of mechanism-based PK/PD models.
At the biophase, the law of mass action and the limited concentration of pharmacological targets often manifest as nonlinear, capacity-limited systems.10) The rate of change of a drug-receptor complex (RC) can be defined as:
| (1) |
where Rtot is the maximum receptor concentration, C is the drug concentration at the site of action, and kon and koff are the second-order association and first-order dissociation rate constants. Assuming equilibrium conditions, this equation can be rearranged to yield the general binding equation:
| (2) |
where KD is the equilibrium dissociation constant (koff/kon). Based on Clark’s theory of receptor occupancy, the stimulus or drug effect (E) can be directly proportional to the fraction of occupied receptors, such that E=α · RC, thus deriving a classic form of the Hill equation or sigmoid Emax model:
| (3) |
where Emax is the maximum effect, γ (or Hill coefficient) is a slope term that reflects the steepness of the effect-concentration curve, and the EC50 is a sensitivity parameter representing the drug concentration producing 50% of Emax. The typical stimulus/effect-log concentration relationship is thus curvilinear, and typical profiles for varying values of γ are shown in the center panel of Figure 1.
In contrast to the linear transduction of receptor occupancy (Eq. 3), Black and Leff introduced the operational model of agonism to provide a mechanistic interpretation of concentration-effect curves.11) The stimulus or effect is assumed to be nonlinearly related to the drug-receptor complex:
| (4) |
where KE is the RC value producing half-maximal effect. Combining Equations 2 and 4 yields:
| (5) |
where Emax is a system maximum and ( represents a transducer or efficacy function (Rtot/KE). This model can accommodate complex relationships, such as partial agonism, where observed capacity and sensitivity properties are actually hybrid terms composed of drug specific (KD and τ) and system specific (Emax) parameters. Regardless of whether linear or nonlinear transduction is operational, capacity-limitation is a hallmark property of pharmacology, and consequentially, a suitable range of dose-levels (or concentrations) are required to define the parameters of such systems. In addition, the implementation of Equation 5 requires pharmacodynamic data, or at least prior information, on the properties of a full agonist to identify the maximal system response.
Physiological turnover processes and homeostatic feedback mechanisms represent the third major component of pharmacodynamics (Fig. 1, right panel). An open system for a biological substance, R, with zero-order production (kin) and first-order removal (kout) can be defined by the following differential equation:
| (6) |
Assuming a time-invariant baseline or steady-state, the initial or baseline value (R0) can be defined as the ratio of the production and loss terms: R0=kin/kout. A family of basic indirect response models apply to many drugs where interaction with the pharmacological target (Eq. 3) serves to inhibit or stimulate either kin or kout.12) A series of transit compartments can also be factored into such models to emulate time-dependent transduction processes that often exhibit significant onset delays and exposure-response hysteresis.13) Knowledge of the turnover rates for physiological system components is important for the identification of the rate-limiting steps for specific pharmacological responses and might impact study design. Such information might also facilitate the characterization of feedback mechanisms that might result in tolerance and/or rebound phenomena.6) As both drugs and diseases often interfere with normal physiological processes, the turnover aspect of both indirect response models14) and transduction models15) renders them well suited for the simultaneous consideration of these factors in the time-course of disease progression.
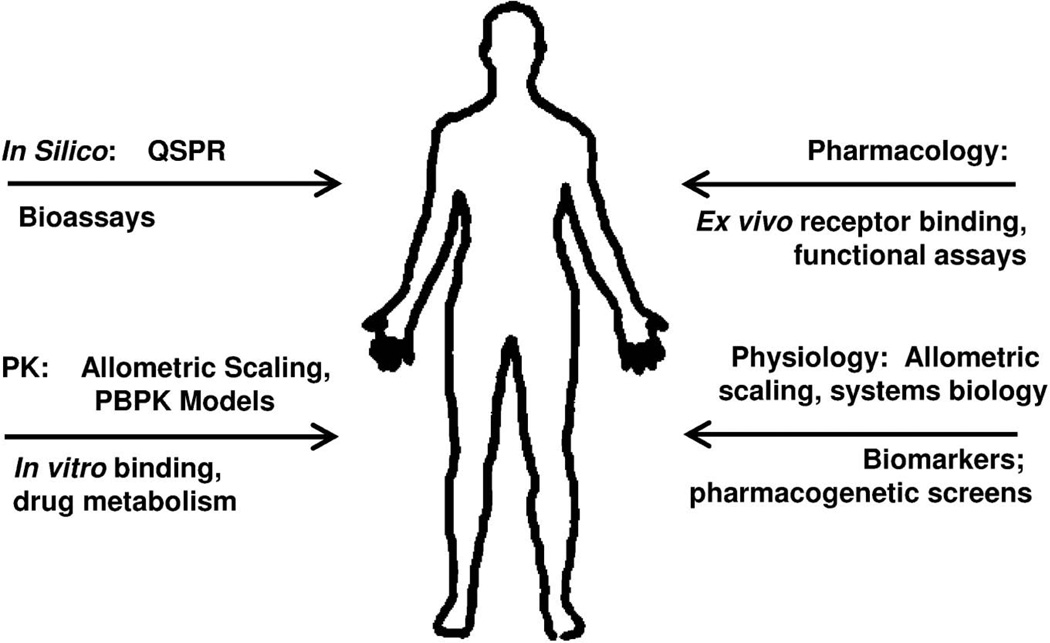
Mechanism-based models seek to integrate these basic components to identify critical pharmacological and (patho)-physiological system properties as well as the rate-limiting steps in responses to drugs.6,16) Useful models with a potential for translational medicine also provide a structural framework for incorporating in silico, in vitro, and preclinical PK/PD measurements to predict the effects of new drugs in humans and across levels of biological organization (Fig. 2). A discussion of all these methods is beyond the scope of this review, which will focus on in vitro assays and allometric principles in the context of mechanistic PK/PD models. The derivation of quantitative structure-PK/PD relationships (in silico modeling) to predict the exposure-response profiles of new chemical entities has been recently reviewed.2)
Fig. 2.
Sources of information that may be integrated into mechanism-based PK/PD models for scaling to human pharmacodynamics.
Predictive techniques (top of arrows) can be augmented by selective measurements (bottom of arrows).
Extrapolation of In Vitro Bioassays
Pharmacodynamic modeling of several systems has revealed that properties of drug interactions with pharmacological targets measured in vitro may be correlated with specific model parameters often reflective of drug potency. Shimada and colleagues developed an ion-channel binding model based on in vitro binding data of calcium channel antagonists, which demonstrate relatively slow rates of association and dissociation.17) The pharmacologic effect was assumed to be proportional to the concentration of the drug-receptor complex and, as an extension of Equation 1, the rate of change of the effect was defined as:
| (7) |
The inclusion of the binding parameters was sufficient to explain the hysteresis observed between the PK and antihypertensive effect of eight calcium channel antagonists in Japanese patients. The calculated KD values based on estimates of kon and koff were shown to be significantly correlated with those obtained from in vitro experiments. These results suggest that PK and in vitro binding data alone could be used to predict the pharmacodynamic profile of future drugs in this class. Kalvass and colleagues18) performed extremely insightful PK/PD studies with seven opioids in mice showing the importance of time-course of brain distribution and binding in determining their antinociceptive effects. The EC50 of unbound drugs in brain showed excellent correlation with in vitro receptor binding affinities (KD). From a drug development perspective, these examples demonstrate how in vitro assays may be coupled with useful PK/PD models to anticipate the outcomes of similar compounds and may guide lead compound selection.
Relative receptor affinity has been shown to be correlated with in vivo estimates of drug potency for several drugs, and in vitro measurements could be used in scaling of EC50 values across species. In a 5-way randomized placebo-controlled crossover study aimed at evaluating the dosing equivalency of four systemically administered corticosteroids, mechanism-based PK/PD models were used to estimate EC50 values for several immunomodulatory effects, including cortisol suppression, lymphocyte and neutrophil trafficking, and ex vivo inhibition of lymphocyte proliferation.19,20) The estimated potencies for all of these responses were highly correlated with relative receptor affinity (in vitro KD values normalized to dexamethasone). Differences in protein homology and other genetic sources of variability may result in altered drug binding affinity among species. Chien and colleagues corrected an EC50 value for a competitor drug measured in humans, using several factors including receptor binding, to predict the in vivo human EC50 for a new chemical entity (NCE):21)
| (8) |
where δfu and δKD are correction factors for differences in the free fraction in plasma (fu) and binding affinity (KD). For example,
| (9) |
The scaled EC50 from animal and in vitro data (Eq. 8) was coupled with other projected parameters to simulate a dose-response curve (Eq. 3 with an added baseline) for a new antihypertensive agent, relative to a competitor, in the preclinical phase of development. Monte Carlo simulations included a relatively large confidence interval about expected outcomes; however, data from clinical studies would eventually be used to confirm and update the model.
Traditional Allometric Scaling in PK/PD
Although the structural nature of physiologically-based models makes them uniquely suited for scaling and predicting human drug exposures, the extrapolation of PK-PD models from animals to humans is primarily based on classic allometric principles.22) There is a general expectation that many physiological processes and organ sizes (θ) tend to obey a power law:23)
| (10) |
with W representing body weight and a and b as drug/process coefficients. The exponent, b, tends to be around 0.75 for clearance processes, 1.0 for organ sizes or physiological volumes, and 0.25 for physiological times or the duration of physiological events (e.g., heart-beat and breath duration, cell lifespans, and turnover times of endogenous substances or processes).24) A theoretical basis for allometric scaling has been proposed by West and colleagues based on the fractal nature of biological systems and energy conservation principles.25) Empirical models have also been coupled with allometric relationships and in vitro metabolism experiments using nonlinear mixed effects modeling to improve the scalability of such models.26,27)
The basic expectations in pharmacodynamics are that physiological turnover rate constants of most general structures and functions should be predictable among species based on allometric principles, whereas capacity (Emax) and sensitivity (EC50) parameters tend to be similar across species. Brodie and colleagues were the first to examine some PK-PD properties across species, revealing inter-species differences in duration of action and biological half-life, but similarity in plasma concentrations on awakening (i.e., concentration producing a standard response analogous to an EC50), following hexobarbital administration.28) There has long been a general belief that the plasma drug concentration required to elicit a certain (intensity of) action is often similar in experimental animals and humans.29) While interspecies differences in relative receptor affinity and plasma protein binding occur (Eqs. 8 and 9),21) there are several examples that show reasonable agreement of such properties between rats and humans for chemically-related series of drugs. Ito and colleagues demonstrated a linear correlation between the logarithm of KD values of benzodiazepines in rat and human cerebral cortex tissue over several orders of magnitude.30) Cox and coworkers also showed good agreement for the EC50 values of four synthetic opioids between these same species.31) Mechanistic modeling was applied to PK/PD data for S(+)-ketoprofen obtained from several species, and estimated parameters further support these basic expectations. Pharmacokinetic parameters were shown to scale proportionally to body weight (albeit with unusual power coefficients), and anti-inflammatory PD parameters exhibited limited ranges that were essentially independent of body weight.32)
Mechanism-Based PK/PD Modeling of rHuEpo
To demonstrate the use of scaling principles within mechanism-based PK/PD models, we present here scaled pharmacodynamic responses using a rat model of rHuEpo PK/PD to anticipate the time-course of several biomarkers in humans. This drug is clinically indicated for the treatment of specific types of anemia, and binding of this endogenous protein to its biological receptor (EPOR) expressed by progenitor cells in bone marrow elicits proliferation and differentiation of erythroid cells, thereby increasing reticulocytes, red blood cells, and hemoglobin concentrations in blood. Erythropoietin exhibits a high degree of homology among mammals, which explains the conserved biological activity of rHuEpo in various species.
The disposition of rHuEpo in several species is polyexponential and nonlinear, and typical PK profiles have been described using a two-compartment model with a concentration-dependent Michaelis-Menten elimination function operating in parallel with a linear nonsaturable clearance pathway.33–35) Target-mediated drug disposition (TMDD) represents a likely explanation for the capacity-limited elimination of erythropoietin; a condition where a significant proportion of the drug (relative to dose) is bound to its pharmacological target, such that this interaction influences the PK properties of the drug.36,37) Receptor-mediated endocytosis is a major clearance mechanism for many protein drugs,38) and this saturable process can result in nonlinear drug disposition.39) The binding of erythropoietin to EPOR is specific and results in saturable internalization of the drug-receptor complex. 40) Chapel and colleagues demonstrated that bone marrow ablation in sheep produced a significant decrease in erythropoietin clearance, providing experimental evidence that target binding and transport plays a major role in the in vivo disposition of erythropoietin.41) Interestingly, the simultaneous modeling of PK profiles of rHuEpo from a wide-range of intravenous dose levels in rats, monkeys, and humans revealed that full and reduced TMDD models42,43) well characterized rHuEpo disposition and provided a basis for linking an established pharmacodynamic model.44)
Woo and Jusko have provided a comparison of interspecies PK/PD properties of rHuEpo.45) Although the prospective use of allometric scaling can be limited,46) it is generally considered that peptide and protein drugs are more likely to exhibit allometric PK relationships than small molecules owing to the relative species conservation of mechanisms that control the biodistribution and elimination of such compounds.47–49) Despite the non-linear disposition of rHuEpo, total systemic clearance and the steady-state volume of distribution show good correlation with body weight. The exponent for clearance (0.708) was close to the expected value of 0.75; however, the exponent for the volume of distribution (0.853) was slightly lower than the expected value (1.0). Pharmacokinetic model specific parameters, such as Michaelis-Menten capacity or Vmax, the central volume of distribution, and a first-order rate constant of absorption, also scaled to body weight with exponents of 0.504, 0.983, and −0.349, respectively (based on rat, monkey, and human data). As anticipated, the pharmacological capacity and sensitivity parameters were essentially species-independent.
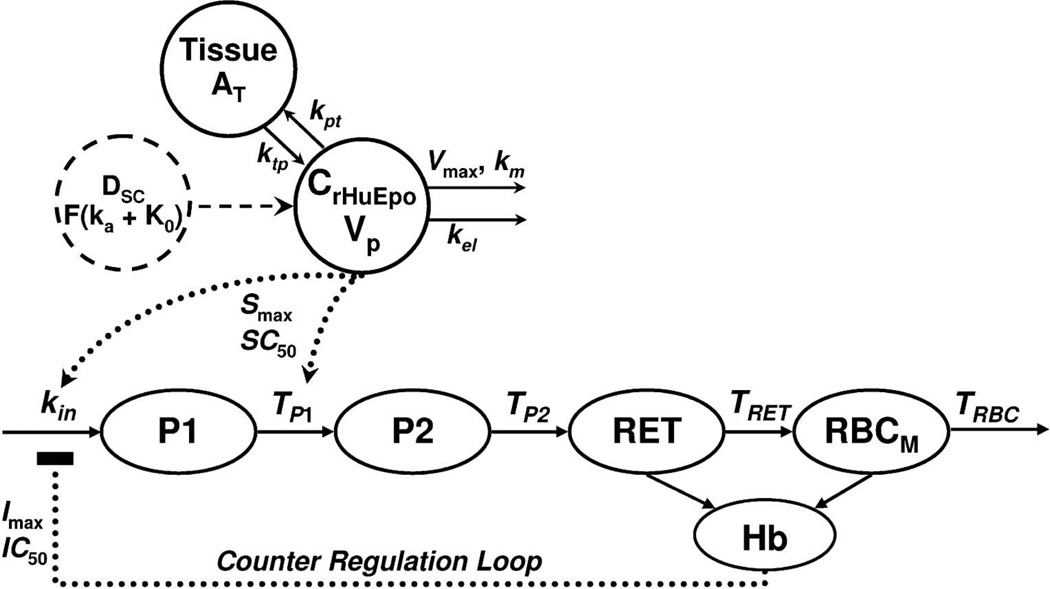
We sought to predict the time-course of reticulocytes, red blood cells, and hemoglobin concentrations in humans after rHuEpo administration from an established PK/PD model developed in rodents. Pharmacodynamic data were extracted from a clinical study in which healthy male volunteers were given 150 IU/kg subcutaneously (SC) three times weekly for 4 weeks.34) The general structure of the PK/PD model for rHuEpo developed from rat preclinical data is shown in Figure 3.35) The PK component of the model can be described by:
| (11) |
| (12) |
where Ap and At represent the amounts of rHuEpo in the central and tissue compartments, Vmax and Km are Michaelis-Menten parameters, Vp is the volume of the central rHuEpo compartment, kel is first-order elimination rate constant, and kpt and ktp are first-order distribution rate constants. The initial conditions of Equations 11 and 12 are zero, and the input function after SC drug administration is defined as:
| (13) |
where F is bioavailability, fr is the fraction of the dose undergoing first-order absorption (ka), and τ is the time period of zero-order input. This input function is based on the complex absorption profile due in part to the significant role of the lymphatics in the uptake of proteins administered subcutaneously.50,51)
Fig. 3.
PK/PD model diagram for the absorption and disposition of rHuEpo and drug effects on reticulocytes (RET), red blood cells (RBC), and hemoglobin concentrations (Hb)
Model is described by Equations 11–17 and symbols are defined in the text.
The catenary PD model (Fig. 3) contains two precursor compartments (P1 and P2) linked to reticulocyte (RET), red blood cell (RBC), and hemoglobin (Hb) compartments. This model mimics the process of erythropoiesis from bone marrow to blood, and is based on cell life span concepts integrated into indirect response models for drugs that alter the generation of natural cells.52) Cells are assumed to be produced at a constant rate, circulate for a specific duration of time (Ti), and are then eliminated from the system not by a first-order process, but at the same rate as the input, delayed by the cell life span (senescence or conversion to another cell type). The precursor compartments represent early progenitor cells and erythroblasts, and TP1 and TP2 are the average times taken for cells to differentiate. The rates of change of the reticulocyte (RET) and mature RBC (RBCM) counts are described by:
| (14) |
| (15) |
with TRET and TRBC as the average life span of these cells, and kin is the zero-order production rate constant. The initial conditions for Equations 14 and 15 are RET0 and RBC0–RET0, where RET0 and RBC0 are baseline measurements. The stimulation function S(t) was defined as:
| (16) |
where Smax is the maximal stimulation factor and SC50 is the rHuEpo concentration resulting in 50% of Smax. Hemoglobin concentrations were calculated as the product of the mean corpuscular hemoglobin (MCH; measured) and sum of RET and RBCM (Eqs. 14, 15). A counter regulation feedback loop is also included, I(t), driven by the difference in Hb from baseline values, and was defined as:
| (17) |
where the maximal inhibition factor (Imax) was fixed to 1, and IC50 is the Hb difference from baseline producing 50% feedback inhibition.
The parameter values, their sources, and scaled-up values in humans used for the PK/PD model simulations are listed in Table 1. Inter-individual variability (IIV) for each parameter used in the Monte Carlo simulations is also reported. Volume, clearance, and first-order rate constants were scaled with allometric exponents of 1, 0.75, and −0.25. The baseline values for RET, RBC, and MCH (and their respective IIV) were considered drug and species independent and were set to literature values for humans.53) Life span parameters were scaled using an allometric exponent of 0.124 which was previously estimated using RBC data obtained from over 20 species.45) Only nominal variability was assigned to PK terms (10% CV%), whereas CV% values were set to 20% for Smax and 30% for sensitivity parameters (SC50 and IC50).
Table 1.
Pharmacokinetic and pharmacodynamic parameters of rHuEpo scaled from rats to humans
| Parameter | Value | Origin | b (exponent) |
Predicted Human Parameter |
IIV (%CV) |
|---|---|---|---|---|---|
|
Pharmacokinetics | |||||
| Vmax (IU/hr) | 0.69 | Rat | 0.75 | 37 | 10 |
| Km (mIU/mL) | 67.3 | Rat | n.s. | 67.3 | 10 |
| Vp (L) | 0.0212 | Rat | 1 | 4.28 | 10 |
| kel (1/hr) | 0.224 | Rat | −0.25 | 0.0594 | 10 |
| ka (1/hr) | 0.146 | Rat | −0.25 | 0.0387 | 10 |
| F | 0.586 | Rat | n.s. | 0.586 | 10 |
| fr | 0.322 | Rat | n.s. | 0.322 | 0 |
| kpt (1/hr) | 0.171 | Rat | −0.25 | 0.0453 | 10 |
| ktp (1/hr) | 0.148 | Rat | −0.25 | 0.0392 | 10 |
| τ (hr) | 13.5 | Rat | n.s. | 13.50 | 0 |
| Pharmacodynamics | |||||
| Smax | 1.87 | Rat | n.s. | 1.87 | 20 |
| SC50 (mIU/mL) | 65.4 | Rat | n.s. | 65.4 | 30 |
| IC50 (g/dL) | 1.79 | Rat | n.s. | 1.79 | 30 |
| RET0 (×109/L) | 48 | Human | n/a | 48 | 20 |
| TP1 (hr) | 43 | Rat | 0.124 | 83 | 0 |
| TP2 (hr) | 3.02 | Rat | 0.124 | 5.83 | 10 |
| TRET (hr) | 72.3 | Rat | 0.124 | 140 | 10 |
| TRBC (hr) | 1440 | Rat | 0.124 | 2780 | 10 |
| RBC0 (×109/L) | 4500 | Human | n/a | 4500 | 4 |
| MCH (pg/cell) | 30 | Human | n/a | 30 | 1.4 |
n/a, not applicable; n.s., not scaled; RET0 and RBC0 are baseline measurements
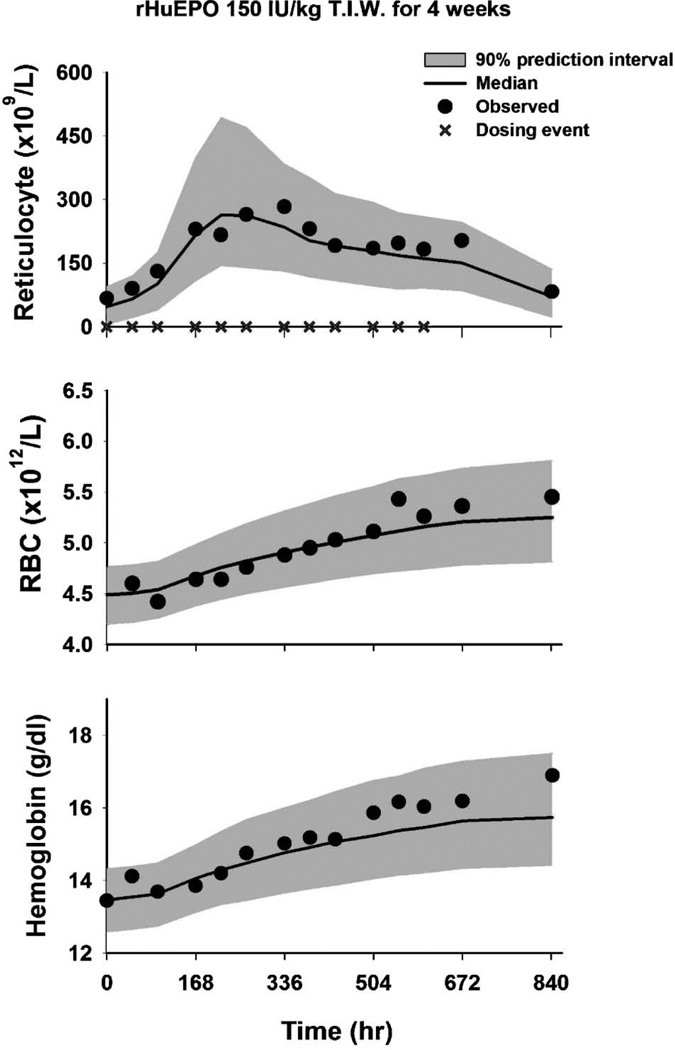
Monte Carlo simulations were conducted using ADAPT II (Biomedical Simulation Resource, USC, Los Angeles), and mean observed data and model predicted profiles are shown in Figure 4. The predicted values of the three biomarkers are in good agreement with observed data, which fall well within the 90% prediction interval (gray areas). The successful scaling of the rat PK/PD model of rHuEpo to human responses demonstrates how basic allometric principles and preclinical data may be integrated using mechanism-based models to make useful predictions. It is important to recognize that the biomarkers of drug activity and preclinical PK/PD models must be meaningful across species. The likelihood of these appears to be greater for macromolecules as compared to small molecules;54) however, a similar interspecies scaling approach was shown to apply to two 5-HT1A receptor agonists.55)
Fig. 4.
Simulated profiles for reticulocyte, red blood cell (RBC), and hemoglobin concentrations (Hb) in response to rHuEpo given as 150 IU/kg SC three times weekly to healthy male volunteers
The symbols represent original data from Ramakrishnan et al. 34) Solid lines are median predicted profiles using a PK/PD model developed in rats (Fig. 3) and allometrically scaled parameters (Table 1). Shaded regions represent the 90% prediction intervals.
In summary, the scaling of pharmacodynamic data relies heavily on the ability to predict and integrate the fundamental processes controlling drug exposure (pharmacokinetics), drug action (pharmacology), and interactions with physiological systems. Preclinical data and in vitro bioassays can provide important insights into these properties, especially as pharmacodynamic parameters tend to be species independent; however, it is important to verify whether measurements of drug effects are meaningful across species. Notwithstanding the limitations of prospective allometry, such power law relation-ships have proven useful in scaling-up physiological turnover processes and PK properties for many drugs. New techniques are needed to identify conditions under which allometric scaling may or may not be appropriate in PK/PD models. Physiologically-based PK models will likely become commonplace given their intrinsic potential for projecting human PK properties from in vitro and in silico measurements and data obtained in other species. Animal studies can provide preliminary data for the development of mechanism-based PK/PD models, which will continue to evolve toward efficient descriptions of pharmacological systems. Such models offer the best approach toward effectively combining and interpreting the major determinants of drug action across species.
Acknowledgments
This research was supported in part by Grant GM 57980 from the National Institutes of Health (to W.J.J.), and a New Investigator Grant from the American Association of Pharmaceutical Scientists (to D.E.M.).
Footnotes
Full text of this paper is available at http://www.jstage.jst.go.jp/browse/dmpk
References
- 1.Nestorov I. Whole-body physiologically based pharmacokinetic models. Expert Opin. Drug. Metab. Toxicol. 2007;3:235–249. doi: 10.1517/17425255.3.2.235. [DOI] [PubMed] [Google Scholar]
- 2.Mager DE. Quantitative structure-pharmacokinetic/pharmacodynamic relationships. Adv. Drug Deliv. Rev. 2006;58:1326–1356. doi: 10.1016/j.addr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Beliveau M, Lipscomb J, Tardif R, Krishnan K. Quantitative structure-property relationships for interspecies extrapolation of the inhalation pharmacokinetics of organic chemicals. Chem. Res. Toxicol. 2005;18:475–485. doi: 10.1021/tx049722k. [DOI] [PubMed] [Google Scholar]
- 4.Parrott N, Paquereau N, Coassolo P, Lave T. An evaluation of the utility of physiologically based models of pharmacokinetics in early drug discovery. J. Pharm. Sci. 2005;94:2327–2343. doi: 10.1002/jps.20419. [DOI] [PubMed] [Google Scholar]
- 5.Danhof M, de Jongh J, De Lange ECM, Della Pasqua O, Ploeger BA, Voskuyl RA. Mechanism-based pharmacokinetic-pharmacodynamic modeling: Biophase distribution, receptor theory, and dynamical systems analysis. Annu. Rev. Pharmacol. Toxicol. 2007;47:21.21–21.44. doi: 10.1146/annurev.pharmtox.47.120505.105154. [DOI] [PubMed] [Google Scholar]
- 6.Mager DE, Wyska E, Jusko WJ. Diversity of mechanism-based pharmacodynamic models. Drug Metab. Dispos. 2003;31:510–518. doi: 10.1124/dmd.31.5.510. [DOI] [PubMed] [Google Scholar]
- 7.Mager DE, Jusko WJ. Development of translational pharmacokinetic-pharmacodynamic models. Clin. Pharmacol. Ther. 2008;83:909–912. doi: 10.1038/clpt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunner M, Langer O. Microdialysis versus other techniques for the clinical assessment of in vivo tissue drug distribution. AAPS J. 2006;8:E263–E271. doi: 10.1007/BF02854896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liefaard LC, Ploeger BA, Molthoff CF, Boellaard R, Lammertsma AA, Danhof M, Voskuyl RA. Population pharmacokinetic analysis for simultaneous determination of B (max) and K (D) in vivo by positron emission tomography. Mol. Imaging. Biol. 2005;7:411–421. doi: 10.1007/s11307-005-0022-3. [DOI] [PubMed] [Google Scholar]
- 10.Ariens EJ. Affinity and intrinsic activity in the theory of competitive inhibition. Arch. Int. Pharmacodyn. Ther. 1954;99:32–49. [PubMed] [Google Scholar]
- 11.Black JW, Leff P. Operational models of pharmacological agonist. Proc. R. Soc. Lond. B. Biol. Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 12.Dayneka NL, Garg V, Jusko WJ. Comparison of four basic models of indirect pharmacodynamic responses. J. Pharmacokinet. Biopharm. 1993;21:457–478. doi: 10.1007/BF01061691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mager DE, Jusko WJ. Pharmacodynamic modeling of time-dependent transduction systems. Clin. Pharmacol. Ther. 2001;70:210–216. doi: 10.1067/mcp.2001.118244. [DOI] [PubMed] [Google Scholar]
- 14.Post TM, Freijer JI, DeJongh J, Danhof M. Disease system analysis: basic disease progression models in degenerative disease. Pharm. Res. 2005;22:1038–1049. doi: 10.1007/s11095-005-5641-5. [DOI] [PubMed] [Google Scholar]
- 15.Earp JC, Dubois DC, Molano DS, Pyszczynski NA, Keller CE, Almon RR, Jusko WJ. Modeling corticosteroid effects in a rat model of rheumatoid arthritis I: mechanistic disease progression model for the time course of collagen-induced arthritis in Lewis rats. J. Pharmacol. Exp. Ther. 2008;326:532–545. doi: 10.1124/jpet.108.137372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy G. Mechanism-based pharmacodynamic modeling. Clin. Pharmacol. Ther. 1994;56:356–358. doi: 10.1038/clpt.1994.149. [DOI] [PubMed] [Google Scholar]
- 17.Shimada S, Nakajima Y, Yamamoto K, Sawada Y, Iga T. Comparative pharmacodynamics of eight calcium channel blocking agents in Japanese essential hypertensive patients. Biol. Pharm. Bull. 1996;19:430–437. doi: 10.1248/bpb.19.430. [DOI] [PubMed] [Google Scholar]
- 18.Kalvass JC, Olson ER, Cassidy MP, Selley DE, Pollack GM. Pharmacokinetics and pharmacodynamics of seven opioids in P-glycoprotein-competent mice: assessment of unbound brain EC50,u and correlation of in vitro, preclinical, and clinical data. J. Pharmacol. Exp. Ther. 2007;323:346–355. doi: 10.1124/jpet.107.119560. [DOI] [PubMed] [Google Scholar]
- 19.Hong Y, Mager DE, Blum RA, Jusko WJ. Population pharmacokinetic/pharmacodynamic modeling of systemic corticosteroid inhibition of whole blood lymphocytes: modeling interoccasion pharmacodynamic variability. Pharm. Res. 2007;24:1088–1097. doi: 10.1007/s11095-006-9232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mager DE, Lin SX, Blum RA, Lates CD, Jusko WJ. Dose equivalency evaluation of major corticosteroids: pharmacokinetics and cell trafficking and cortisol dynamics. J. Clin. Pharmacol. 2003;43:1216–1227. doi: 10.1177/0091270003258651. [DOI] [PubMed] [Google Scholar]
- 21.Chien JY, Friedrich S, Heathman MA, de Alwis DP, Sinha V. Pharmacokinetics/pharmacodynamics and the stages of drug development: Role of modeling and simulation. AAPSJ. 2005;7:E544–E559. doi: 10.1208/aapsj070355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedrick RL. Animal scale-up. J. Pharmacokinet. Biopharm. 1973;1:435–461. doi: 10.1007/BF01059667. [DOI] [PubMed] [Google Scholar]
- 23.Adolph EF. Quantitative relations in the physiological constitutions of mammals. Science. 1949;109:579–585. doi: 10.1126/science.109.2841.579. [DOI] [PubMed] [Google Scholar]
- 24.Boxenbaum H. Interspecies scaling, allometry, physiological time, and the ground plan of pharmacokinetics. J. Pharmacokinet. Biopharm. 1982;10:201–227. doi: 10.1007/BF01062336. [DOI] [PubMed] [Google Scholar]
- 25.West GB, Brown JH, Enquist BJ. A general model for the origin of allometric scaling laws in biology. Science. 1997;276:122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 26.Cosson VF, Fuseau E, Efthymiopoulos C, Bye A. Mixed effect modeling of sumatriptan pharmacokinetics during drug development. I. Interspecies allometric scaling. J. Pharmacokinet. Biopharm. 1997;25:149–167. doi: 10.1023/a:1025728028890. [DOI] [PubMed] [Google Scholar]
- 27.Proost JH, Beljaars L, Olinga P, Swart PJ, Kuipers ME, Reker-Smit C, Groothuis GM, Meijer DK. Predictions of the pharmacokinetics of succinylated human serum albumin in man from in vivo disposition data in animals and in vitro liver slice incubations. Eur. J. Pharm. Sci. 2006;27:123–132. doi: 10.1016/j.ejps.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Quinn GP, Axelrod J, Brodie BB. Species, strain and sex difference in metabolism of hexobarbitone, amidopyrine, antipyrine and aniline. Biochem. Pharmacol. 1958;1:152–159. [Google Scholar]
- 29.Levy G. The case for preclinical pharmacodynamics. In: Yacobi A, Skelly JP, Shah VP, Benet LZ, editors. Integration of Pharmacokinetics, Pharmacodynamics, and Toxicokinetics in Rational Drug Development. New York: Plenum; 1993. pp. 7–13. [Google Scholar]
- 30.Ito KK, Asakura A, Yamada Y, Nakamura K, Sawada Y, Iga T. Prediction of the therapeutic dose for benzodiazepine anxiolytics based on receptor occupancy theory. Biopharm. Drug Dispos. 1997;18:293–303. doi: 10.1002/(sici)1099-081x(199705)18:4<293::aid-bdd24>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Cox EH, Langemeijer MWE, Gubbens-Stibbe JM, Muir KT, Danhof M. The comparative pharmacodynamics of remifentanil and its metabolite, GR90291, in a rat EEG model. Anesthesiol. 1999;90:535–544. doi: 10.1097/00000542-199902000-00030. [DOI] [PubMed] [Google Scholar]
- 32.Lepist EI, Jusko WJ. Modeling and allometric scaling of s(+)-ketoprofen pharmacokinetics and pharmacodynamics: A retrospective analysis. J. Vet. Pharmacol. Ther. 2004;27:211–218. doi: 10.1111/j.1365-2885.2004.00579.x. [DOI] [PubMed] [Google Scholar]
- 33.Ramakrishnan R, Cheung WK, Farrell F, Joffee L, Jusko WJ. Pharmacokinetic and pharmacodynamic modeling of recombinant human erythropoietin after intravenous and subcutaneous dose administration in cynomolgus monkeys. J. Pharmacol. Exp. Ther. 2003;306:324–331. doi: 10.1124/jpet.102.047191. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan R, Cheung WK, Wacholtz MC, Minton N, Jusko WJ. Pharmacokinetic and pharmacodynamic modeling of recombinant human erythropoietin after single and multiple doses in healthy volunteers. J. Clin. Pharmacol. 2004;44:991–1002. doi: 10.1177/0091270004268411. [DOI] [PubMed] [Google Scholar]
- 35.Woo S, Krzyzanski W, Jusko WJ. Pharmacokinetic and pharmacodynamic modeling of recombinant human erythropoietin after intravenous and subcutaneous administration in rats. J. Pharmacol. Exp. Ther. 2006;319:1297–1306. doi: 10.1124/jpet.106.111377. [DOI] [PubMed] [Google Scholar]
- 36.Levy G. Pharmacologic target-mediated drug disposition. Clin. Pharmacol. Ther. 1994;56:248–252. doi: 10.1038/clpt.1994.134. [DOI] [PubMed] [Google Scholar]
- 37.Mager DE. Target-mediated drug disposition and dynamics. Biochem. Pharmacol. 2006;72:1–10. doi: 10.1016/j.bcp.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 38.Lobo ED, Hansen RJ, Balthasar JP. Antibody pharmacokinetics and pharmacodynamics. J. Pharm. Sci. 2004;93:2645–2668. doi: 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama Y, Hanano M. Receptor-mediated transport of peptide hormones and its importance in the overall hormone disposition in the body. Pharm. Res. 1989;6:192–202. doi: 10.1023/a:1015905331391. [DOI] [PubMed] [Google Scholar]
- 40.Mufson RA, Gesner TG. Binding and internalization of recombinant human erythropoietin in murine erythroid precursor cells. Blood. 1987;69:1485–1490. [PubMed] [Google Scholar]
- 41.Chapel S, Veng-Pedersen P, Hohl RJ, Schmidt RL, McGuire EM, Widness JA. Changes in erythropoietin pharmacokinetics following busulfan-induced bone marrow ablation in sheep: evidence for bone marrow as a major erythropoietin elimination pathway. J. Pharmacol. Exp. Ther. 2001;298:820–824. [PubMed] [Google Scholar]
- 42.Mager DE, Jusko WJ. General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J. Pharmacokinet. Pharmacodyn. 2001;28:507–532. doi: 10.1023/a:1014414520282. [DOI] [PubMed] [Google Scholar]
- 43.Mager DE, Krzyzanski W. Quasi-equilibrium pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Pharm. Res. 2005;22:1589–1596. doi: 10.1007/s11095-005-6650-0. [DOI] [PubMed] [Google Scholar]
- 44.Woo S, Krzyzanski W, Jusko WJ. Target-mediated pharmacokinetic and pharmacodynamic model of recombinant human erythropoietin (rHuEPO) J. Pharmacokinet. Pharmacodyn. 2007;34:849–868. doi: 10.1007/s10928-007-9074-0. [DOI] [PubMed] [Google Scholar]
- 45.Woo S, Jusko WJ. Interspecies comparisons of pharmacokinetics and pharmacodynamics of recombinant human erythropoietin. Drug Metab. Dispos. 2007;35:1672–1678. doi: 10.1124/dmd.107.015248. [DOI] [PubMed] [Google Scholar]
- 46.Bonate PL, Howard D. Prospective allometric scaling: Does the emperor have clothes? J. Clin. Pharmacol. 2000;40:335–340. doi: 10.1177/00912700022009017. [DOI] [PubMed] [Google Scholar]
- 47.Mahmood I. Interspecies scaling of protein drugs: prediction of clearance from animals to humans. J. Pharm. Sci. 2004;93:177–185. doi: 10.1002/jps.10531. [DOI] [PubMed] [Google Scholar]
- 48.Mordenti J, Chen SA, Moore JA, Ferraiolo BL, Green JD. Interspecies scaling of clearance and volume of distribution data for five therapeutic proteins. Pharm. Res. 1991;8:1351–1359. doi: 10.1023/a:1015836720294. [DOI] [PubMed] [Google Scholar]
- 49.Tang H, Mayersohn M. A global examination of allometric scaling for predicting human drug clearance and the prediction of large vertical allometry. J. Pharm. Sci. 2006;95:1783–1799. doi: 10.1002/jps.20481. [DOI] [PubMed] [Google Scholar]
- 50.Porter CJ, Charman SA. Lymphatic transport of proteins after subcutaneous administration. J. Pharm. Sci. 2000;89:297–310. doi: 10.1002/(SICI)1520-6017(200003)89:3<297::AID-JPS2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 51.Radwanski E, Chakraborty A, Van Wart S, Huhn RD, Cutler DL, Affrime MB, Jusko WJ. Pharmacokinetics and leukocyte responses of recombinant human interleukin-10. Pharm. Res. 1998;15:1895–1901. doi: 10.1023/a:1011918425629. [DOI] [PubMed] [Google Scholar]
- 52.Krzyzanski W, Ramakrishnan R, Jusko WJ. Basic pharmacodynamic models for agents that alter production of natural cells. J. Pharmacokinet. Biopharm. 1999;27:467–489. doi: 10.1023/a:1023249813106. [DOI] [PubMed] [Google Scholar]
- 53.Woo S, Krzyzanski W, Duliege AM, Stead RB, Jusko WJ. Population pharmacokinetics and pharmacodynamics of peptidic erythropoiesis receptor agonist (ERA) in healthy volunteers. J. Clin. Pharmacol. 2008;48:43–52. doi: 10.1177/0091270007309702. [DOI] [PubMed] [Google Scholar]
- 54.Agoram BM, Martin SW, van der Graaf PH. The role of mechanism-based pharmacokinetic-pharmacodynamic (PK-PD) modelling in translational research of biologics. Drug Discov. Today. 2007;12:1018–1024. doi: 10.1016/j.drudis.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 55.Zuideveld KP, van der Graaf PH, Peletier LA, Danhof M. Allometric scaling of pharmacodynamic responses: Application to 5-Ht1A receptor mediated responses from rat to man. Pharm. Res. 2007;24:2031–2039. doi: 10.1007/s11095-007-9336-y. [DOI] [PubMed] [Google Scholar]