Abstract
Toll-like receptors (TLRs) play key roles in sensing wide array of microbial signatures and induction of innate immunity. TLR2 in fish resembles higher eukaryotes by sensing peptidoglycan (PGN) and lipoteichoic acid (LTA) of bacterial cell wall and zymosan of yeasts. However, in fish TLR2, no study yet describes the ligand binding motifs in the leucine rich repeat regions (LRRs) of the extracellular domain (ECD) and important amino acids in TLR2-TIR (toll/interleukin-1 receptor) domain that could be engaged in transmitting downstream signaling. We predicted these in a commercially important freshwater fish species rohu (Labeo rohita) by constructing 3D models of TLR2-ECD, TLR2-TIR, and MyD88-TIR by comparative modeling followed by 40 ns (nanosecond) molecular dynamics simulation (MDS) for TLR2-ECD and 20 ns MDS for TLR2-TIR and MyD88-TIR. Protein (TLR2-ECD)–ligands (PGN, LTA, and zymosan) docking in rohu by AutoDock4.0, FlexX2.1, and GOLD4.1 anticipated LRR16–19, LRR12–14, and LRR20-CT as the most important ligand binding motifs. Protein (TLR2-TIR)—protein (MyD88-TIR) interaction by HADDOCK and ZDOCK predicted BB loop, αB-helix, αC-helix, and CD loop in TLR2-TIR and BB loop, αB-helix, and CD loop in MyD88-TIR as the critical binding domains. This study provides ligands recognition and downstream signaling.
1. Introduction
The innate immune response elicited by a variety of pattern recognition receptors (PRRs) is an immediate nonspecific and first line of defense of the host against invading various pathogens [1]. Toll-like receptors (TLRs) are a component of PRR and play a critical role by sensing organisms ranging from protozoa to bacteria and were involved in many infectious diseases [2]. They recognize a wide array of microbe associated molecular patterns (MAPMs) and activate downstream signaling to induce innate immunity [3]. The number of TLRs varied in different organisms and among these most TLRs are located on cell surface except for TLR3, TLR7, TLR8, and TLR9 [4, 5].
Toll-like receptor 2 (TLR2) was shown to be the principal mediator of macrophages activation. It functions as homodimer [6] or heterodimer with TLR1 or TLR6 [7] to recognize diverse bacterial products [8] and activation of MyD88-dependent signaling pathway. In this pathway, the TIR domain of MyD88 interacts with the TIR domain of TLR [9] and transmits downstream signals to induce innate immune genes expression.
PGN is a highly complex structural component and an important derivative of both Gram-positive and Gram-negative bacterial cell wall, and is the target of the innate immune system [10]. It is composed of alternating β-(1–4)-linked N-acetyl-glucosamine and N-acetyl-muramic acid residues cross-linked by peptide bridges [11]. It is recognized by various families of PRRs, including TLRs, nucleotide-binding oligomerization domain-containing proteins (NLRs), and peptidoglycan recognition proteins (PGRPs) [12]. In monocytes and macrophages, PGN binds to extracellular domain of TLR2 and activates signaling to induce inflammatory cytokines [13–15]. Structurally, PGN of most Gram-positive bacteria contains lysine at third position, and in Gram-negative and most rod-shaped Gram-positive bacteria lysine is replaced by DAP [16]. Nascent PGN of bacterial cell wall is poorly recognized by TLR2. However, after its autolysin the remodeled PGN binds TLR2 with high affinity [16]. LTA is an amphiphilic, negatively charged glycolipid [17] component of Gram-positive bacteria cell wall. TLR2 binds LTA and activates signaling cascade to induce TNF-α, IL-6, and IL-8 gene expression [18–20]. Zymosan is the cell wall derivative of Saccharomyces cerevisiae. It comprised mainly polysaccharides, of which β-glucan and mannan are the major constituents. It was widely used as a model to study fungus-mediated inflammation, phagocytosis, and the production of inflammatory cytokines and chemokines [21, 22]. TLR2 recognizes it directly or in coaction with CD14 and TLR6 [23, 24] to induce TNF-α gene expression [25]. TLR2 is the major pathway of proinflammatory signaling by zymosan interaction and is needed for the development of specific immune responses against pathogens [26].
Various studies on TLR2 have also been reported in zebrafish [27, 28], Japanese flounder [29], puffer fish [30], channel catfish [31], and in orange-spotted grouper [32, 33]. In European common carp, inductive over expression of TLR2 in macrophages was observed in response to PGN and LTA [34]. In the Indian major carps, modulation of TLR2 expression was reported in PGN, zymosan, and LTA treatment [35, 36].
India is the major supplier of fish in the world and ranks 3rd in freshwater fish production (FAO). Among various freshwater fishes, rohu (Labeo rohita) is the most commercially important and highly favored fish in the Indian subcontinent. TLR2 was characterized in rohu and the ligands that stimulate TLR2 signaling were also reported [35]. However, no studies have reported yet describing the structural characteristics of TLR2 in rohu and their key domains that binds to the specific ligands to stimulate cytokine expression. Furthermore, the key amino acids in the TLR2-TIR domains that interact with adapter molecule MyD88 to induce down-stream signaling were still unclear across the species.
To elucidate the structural scaffold in rohu TLR2, we report the 3D-model of extracellular domain of rohu TLR2 along with its key domains that are predicted to be involved in recognizing PGN, LTA and zymosan, and the critical region of interaction between TIR domains of TLR2 and MyD88. This is the first report across the fish species.
2. Materials and Methods
2.1. Domain Identification
Rohu TLR2 protein (GenBank ID: ADQ74644) with N-terminal extracellular domain (ECD), transmembrane domain, and C-terminal cytoplasmic TIR domain [35] was subjected to SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) and NetNGlyc1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) to predict the signal peptide and N-glycosylation sites respectively. The TIR domain in common carp MyD88 (GenBank ID: ADQ08685) was predicted by SMART (http://smart.embl-heidelberg.de/) and CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) domain finding programs and was verified with published report of MyD88-TIR domains in zebrafish (Q5XJ85) and puffer fish (A8QMS7) in UniProt database (http://www.uniprot.org/).
2.2. Sequence Alignment, Template Identification, and Comparative Modeling of ECD and TIR Domain in Rohu TLR2, and MyD88-TIR Domain in Common Carp
Amino acid sequence of rohu TLR2 was aligned by MegAlign [37] in DNASTAR-Lasergene program with the amino acid sequences of TLR2 in other species derived from UniProtKB database. The TIR domains of rohu TLR2 and common carp MyD88 were aligned by MegAlign with amino acid sequences of TIR domains in other species deduced from UniProtKB database. The secondary structures of TLR2- and MyD88-TIR domains were predicted by PSIPRED program (http://bioinf.cs.ucl.ac.uk/psipred/).
Template search for TLR2-ECD (561 aa), TLR2-TIR (146 aa), and MyD88-TIR (137 aa) domains in PDB database identified mouse TLR1-TLR2 heterodimer (PDB ID: 2Z81), TIR domain of human TLR2 (PDB ID: 1O77), and TIR domain of human MyD88 (PDB ID: 2Z5V) as the best homologous structures with top identity score. To ascertain the sensitivity and accuracy of the selected templates, FUGUE [38] program was used to perform sequence-structure comparison between the target and the template and was represented by JOY annotation program [39]. For each three domains (TLR2-ECD, TLR2-TIR, and MyD88-TIR) a set of twenty 3D models were generated by Modeller9v10 program [40]. Among these 20 models (for each domain), the model with lowest discrete optimized protein energy (DOPE) score was considered for further studies. The lowest DOPE models of TLR2-ECD, TLR2-TIR, and MyD88-TIR were subjected for loop modeling and refinement in Accelrys DS 2.5 (San Diego, Accelrys) under CHARMM force field. The long BB loops and DD loops in TLR2-TIR and MyD88-TIR models after loop refinement were notably analyzed, and changes were marked by superimposing them with their respective templates. The refined models were subjected to energy minimization by DS 2.5.
2.3. Molecular Dynamics Simulation
Molecular dynamics (MD) simulations were carried out for the modeled systems using the GROMACS 4.5.5 program [41]. Homology models were set for MDS under GROMOS54a7 force field. The 3D models were placed in a cubic box maintaining a distance of 10 Å for TLR2-ECD, 9 Å for TLR2-TIR, and 9 Å for MyD88-TIR between the box edges and the protein surface. The systems were solvated in simple point charge (SPC) models and were neutralized by adding counter ions. In order to remove spurious contacts energy minimization of the solvated systems was done using the steepest descent integrator. The bond lengths and geometry of water molecules were constrained. All of the three restrained models were subjected to position-restrained MD under NPT conditions for 1 ns (nanosecond). Finally, 40 ns production MD run was carried out for TLR2-ECD and 20 ns for TLR2-TIR and MyD88-TIR models using particle mesh Ewald (PME) electrostatics method under NPT conditions. Snapshots of the trajectory were taken in every 0.5 picoseconds. GROMACS and VMD 1.9.1 (http://www.ks.uiuc.edu/Research/vmd/) routines were utilized to check trajectories and the quality of the simulations. The graphs of trajectory analysis were created using Xmgr 4.1.2 (http://plasma-gate.weizmann.ac.il/Xmgr/).
2.4. Model Validation
The final snapshot obtained at the end of each MDS was considered to represent the structures of the TLR2-ECD, TLR2-TIR, and MyD88-TIR models. These simulated models were set for validation by SAVES (http://nihserver.mbi.ucla.edu/SAVES/), WHAT IF [42], MolProbity [43], ProQ [44], ModFOLD [45], and MetaMQAP [46] servers. The simulated models were superimposed with their respective templates to examine the deflections by PyMOL (http://www.pymol.org/). Cross-check validation was carried out using model as template and the primary amino acids of the respective template as target.
2.5. Molecular Docking of PGN, LTA, and Zymosan with Rohu TLR2-ECD
Three different 2D structures of PGN [16], that is, (i) MurNac-L-Ala-i-D-Glu-L-Lys (PGN-I), (ii) MurNac-L-Ala-i-D-Glu-L-Lys-D-Ala-Gly (PGN-II), and (iii) MurNac-L-Ala-i-D-Glu-L-DAP-D-Ala (PGN-DAP), were generated by Chemsketch (http://www.acdlabs.com/resources/freeware/chemsketch/). The 2D structure of zymosan (CID: 11375554) was obtained from the NCBI PubChem database (http://pubchem.ncbi.nlm.nih.gov/) and LTA was from KEGG (KEGG: C06042) ligand database (http://www.genome.jp/ligand/). The 3D structures of all these compounds were generated using PRODRG2 server (http://davapc1.bioch.dundee.ac.uk/prodrg/) subjecting to chirality, full charges with energy minimization. The generated 3D structures were subjected to DS 2.5 for ligand minimization. The probable ligand binding pockets in TLR2-ECD were predicted by metaPocket finder [47] and Q-site finder [48]. The LTA binding site in mouse TLR2 (PDB ID: 3A7B) was also considered for docking. Molecular docking was carried out using AutoDock 4.0 [49], FlexX 2.1 [50], and GOLD 4.1 [51] following previously described methods [52] with receptor and ligand flexibility. In this, the important neighbouring residues at the predicted binding sites were set to flexible that covered all the active site residues and allowed for the flexible rotation of the ligand. Docking of previously reported PGN binding sites in other species [53] was also carried out. In AutoDock, the lowest-energy solution of the ligand all-atom RMSD cluster was taken to calculate the binding energy. The predicted interacting residues obtained by AutoDock were matched with the predicted binding pocket amino acids of metaPocket finder and Q-site finder, and these binding pockets were referred for docking in FlexX and GOLD. The docking poses with H-bond forming amino acids were graphically represented by PyMOL and DS 2.5.
2.6. Protein-Protein Interaction
Rohu and common carp (Cyprinus carpio) belong to the Cyprinidae family and are very closely related. Till date, rohu MyD88 gene has not been cloned. Therefore, to understand the TLR2 and MyD88 interaction, we considered common carp MyD88 (GenBank ID: ADQ08685). The interface residues for TLR2-TIR and MyD88-TIR domains were predicted with reference to their template proteins structural and functional properties. Protein-Protein Interaction Site Predictor (cons-PPISP) (http://pipe.scs.fsu.edu/ppisp.html), InterProSurf (http://curie.utmb.edu/prosurf.html), and PatchDock server (http://bioinfo3d.cs.tau.ac.il/PatchDock/) were used to find the interacting residues in TLR2-TIR and MyD88-TIR. Docking was performed using HADDOCK [54] and ZDOCK [55] web servers. Intermolecular contacts were analyzed with DIMPLOT, a part of LIGPLOT software package (http://www.ebi.ac.uk/thornton-srv/software/LigPlus/) using default parameters.
2.7. Structural Refinement and Stability Evaluation of Complexes
The best protein-ligand complexes obtained from docking studies of PGN, LTA, and zymosan with TLR2-ECD were subjected to MDS using the previously defined parameters in GROMACS. To gain insight into the structural stability of the protein-ligand and protein-protein complexes, MD simulations were performed for PGN-I-TLR2-ECD, PGN-II-TLR2-ECD, PGN-DAP-TLR2-ECD, LTA-TLR2-ECD, zymosan-TLR2-ECD, and TLR2-TIR-MyD88-TIR complex for different time periods of MDS. A production MD run for 10 ns was carried out for TLR2-ECD ligand complexes and protein-protein complex. The existence of H-bonds in the complex in different periods of MDS was analyzed.
2.8. In Silico Site-Directed Mutagenesis
To identify the key amino acids among interacting amino acid residues in TLR2-ECD, TLR2-TIR, and MyD88-TIR domains, site-directed mutagenesis was carried out in DS 2.5 under build mutant protocol. Redocking was performed to calculate the fitness score in GOLD after mutation, and docking scores were cross-checked with previous fitness scores. Protein-protein interaction hot spots were predicted after mutagenesis by HADDOCK.
3. Results and Discussion
3.1. Domain Analysis
The full length TLR2 protein is constituted of 792 amino acids including a signal peptide of 30 amino acids (1–30 aa). The mature TLR2 protein ECD, trans-membrane (TM) and TIR domain constituted of 34–590, 595–612, and 645–790 amino acids respectively. The alignment of TLR2 amino acids with other species revealed their good conservation across the species (See Figure S1 in supplementary material available online at doi: http://dx.doi.org/10.1155/2013/185282). Among them, rohu TLR2 showed highest identity with common carp (88.1%) and lowest identity with mouse (40.5%). N-glycosylation site prediction server predicted 10 glycosylation sites, out of which 9 were in the ECD and 1 in the TIR domain. Among these 10 N-glycosylation sites, 8 were potential with a value greater than threshold value (0.5) and the remaining two were below the threshold. Single N-glycosylation site was present each at LRR1, 3, 8, 14, 16, 18, 21, and TIR domain, and the remaining two were at LRR6. The multiple sequence alignment of TLR2-TIR with other species (Figure 1(a)) and secondary structure analysis (Figure 1(b)) showed well-conserved α-helices, β-sheets, and biologically most important BB and DD loops [56]. In MyD88-TIR domain the multiple sequence alignment and secondary structure prediction (Figures 2(a) and 2(b)) also revealed a good conservation among the phylogenetically divergent species.
Figure 1.
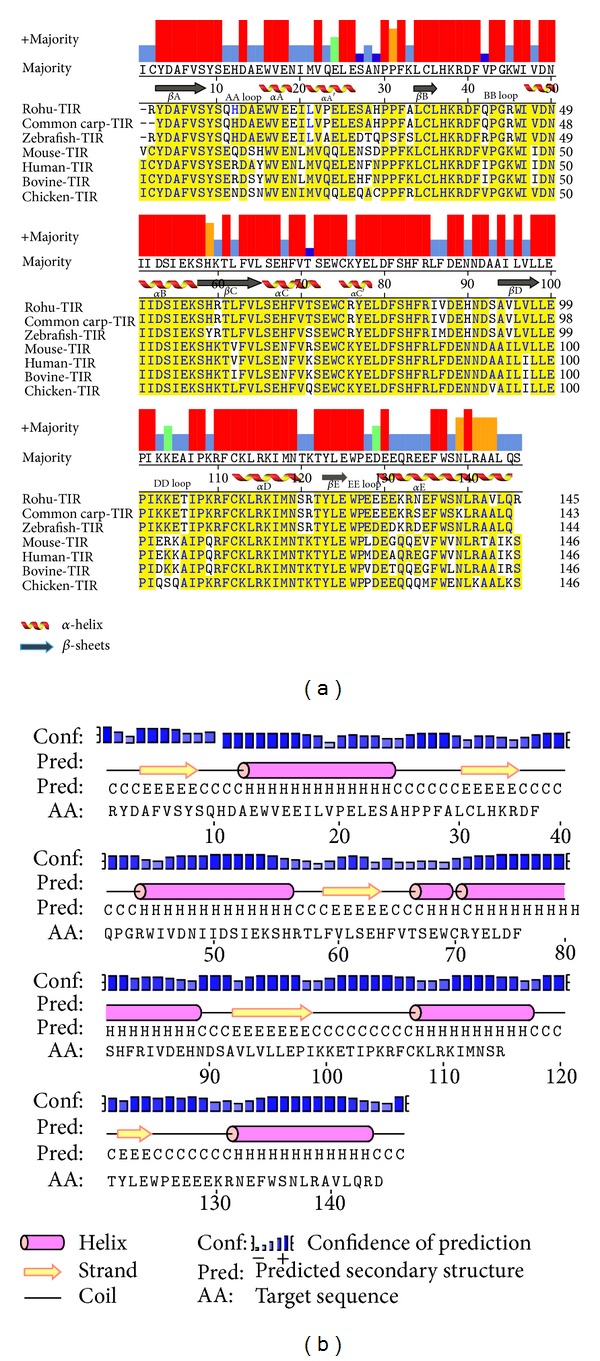
Multiple sequence alignment and secondary structure prediction of TLR2-TIR domain. (a) Multiple sequence alignment of TLR2-TIR domain of rohu with others by MegAlign program. Conserved residues were shown in yellow. Consensus residues are shown in the majority axis. (b) Secondary structure representation of TLR2-TIR domain by PSIPRED. Helices denoted as “H,” beta strands as “E,” and loops as “C.”
Figure 2.
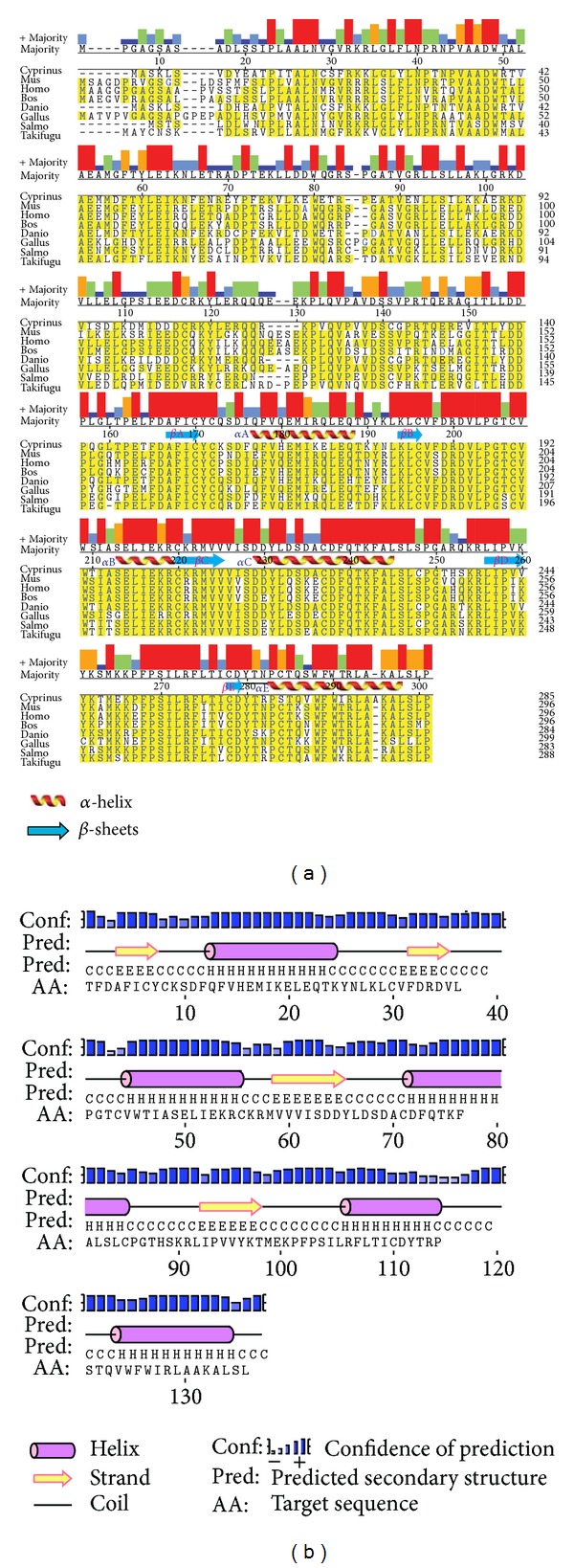
Multiple sequence alignment and secondary structure prediction of MyD88-TIR domain. (a) Multiple sequence alignment of MyD88-TIR domain of common carp with others by MegAlign program. Conserved residues were shown in yellow. Consensus residues are shown in the majority axis. (b) Secondary structure representation of MyD88-TIR domain by PSIPRED. Helices denoted as “H,” beta strands as “E,” and loops as “C.”
3.2. Structural Analysis of TLR2-ECD, TLR2-TIR, and MyD88-TIR Domains
The BLAST search analysis showed that the ligand recognizing LRR regions in TLR2-ECD shared the close structural relationship with mouse TLR1-TLR2 heterodimer (PDB ID: 2Z81) having 35% and 52% sequence identities and similarities, respectively. The TLR2-TIR and MyD88-TIR domains shared 71% and 78% sequence identities with their respective templates (PDB ID: 1O77 and 2Z5V). The sequence-structure alignment by FUGUE revealed a good conservation of secondary structures (α-helices and β-sheets) between the target and template. The identity scores between the LRR regions of TLR2-ECD and 2Z81 were presented in Table 1. The sequence identities between the important biological regions as reported in human and mouse TIR domains with their respective templates were given in Tables 2 and 3. The structure-structure alignment between the lowest DOPE score models and their respective templates showed good structural conservation across the domains. The TLR2-ECD took a horseshoe shape with 23 LRR domains including LRRNT and LRRCT. Most of the LRR domains consisted of β-strands connected by long loop and some with α-helices. The β-strands faced towards the concave surface in TLR2-ECD model, and the α-helices were present at the convex surface. There were five α-helices and five β-sheets in TLR2-TIR domain and four α-helices and five β-sheets in MyD88-TIR domain.
Table 1.
Sequence identities between rohu TLR2-ECD (target) and mouse TLR2-ECD (template).
| LRR | Identity (%) | LRR | Identity (%) |
|---|---|---|---|
| 1 | 47.82 | 12 | 27.77 |
| 2 | 29.16 | 13 | 20 |
| 3 | 63.63 | 14 | 33.33 |
| 4 | 52 | 15 | 37.93 |
| 5 | 34.46 | 16 | 34.78 |
| 6 | 35.57 | 17 | 50 |
| 7 | 9.52 | 18 | 52.38 |
| 8 | 15.38 | 19 | 50 |
| 9 | 28.57 | 20 | 40.9 |
| 10 | 26.31 | 21 | 29.16 |
| 11 | 28 |
Table 2.
Sequence identities between rohu TLR2-TIR (target) and human TLR2-TIR (template).
| Position | Identity (%) | Position | Identity (%) |
|---|---|---|---|
| βA | 100 | CD loop | 46.66 |
| AA loop | 66.67 | βD | 60 |
| αA | 58.33 | DD loop | 69.23 |
| βB | 42.85 | αD | 88.88 |
| BB Loop | 100 | DE loop | 50 |
| αB | 90.9 | βE | 100 |
| βC | 75 | EE loop | 50 |
| αC | 69.23 | αE | 50 |
Table 3.
Sequence identities between common carp MyD88-TIR (target) and human MyD88-TIR (template).
| Position | Identity (%) | Position | Identity (%) |
|---|---|---|---|
| βA | 100 | CC loop | 66.66 |
| AA loop | 80 | βD | 60 |
| αA | 69.23 | DD loop | 70.58 |
| βB | 100 | βE | 100 |
| BB Loop | 86.66 | EE loop | 66.66 |
| αB | 100 | αE | 68.75 |
| βC | 85.71 |
3.3. Molecular Dynamics of Homology Models
The stability and MD properties were observed up to 40 ns for TLR2-ECD and up to 20 ns for TLR2-TIR and MyD88-TIR domains, and the RMSD values over time were shown in Figure 3. The MD analysis in TLR2-ECD showed that the RMSD trajectory rose from the beginning to 12 ns with an average RMSD of 4.23 Å. It attained an approximately stable plateau with an average RMSD of 4.673 Å till the end of simulation. The RMSD trajectories of TLR2-TIR and MyD88-TIR were observed to be stable after 5 ns with an average RMSD of 2.74 Å and 3.68 Å, respectively. The RMSF of Cα atoms of each amino acid in TLR2-ECD identified LRR7-11 as the most flipped region (Figure 4(a)). These regions are constituted of six β-sheets, one α-helix, and long loops. In higher vertebrates, this region was reported as lipopeptide binding region [57]. The flexible long loops (BB and DD loops) in TLR2-TIR and MyD88-TIR domains showed major fluctuations in the RMSF graph and were expected to be engaged in protein-protein interaction (Figures 4(b) and 4(c)). Secondary structure analysis from the trajectory in TLR2-ECD showed little variation of α-helices and β-sheets. However, the coil regions much varied with respect to simulation period. In TLR2-TIR, no major secondary structural changes were observed during MDS.
Figure 3.
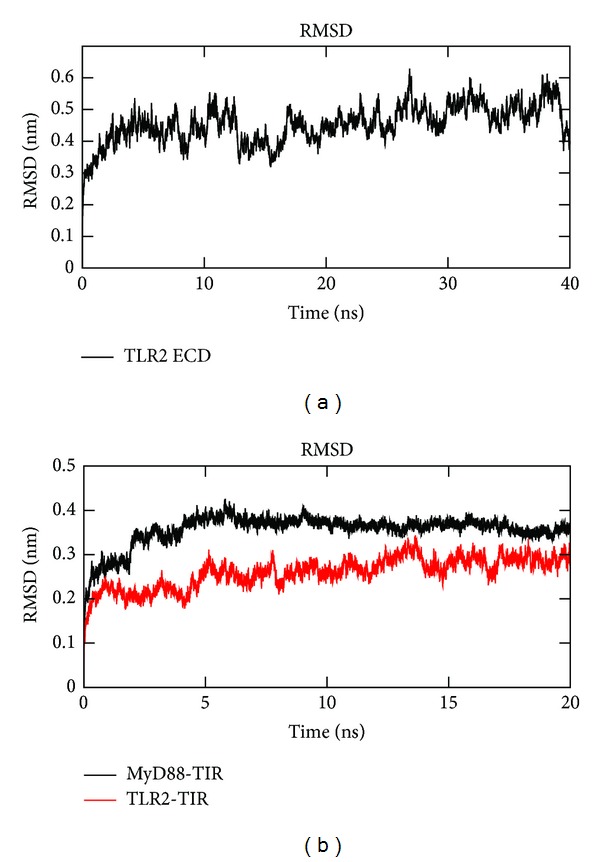
Root mean square deviation (RMSD) analysis. RMSD of (a) TLR2-ECD up to 40 ns and (b) MyD88-TIR and TLR2-TIR domains up to 20 ns MD simulation.
Figure 4.
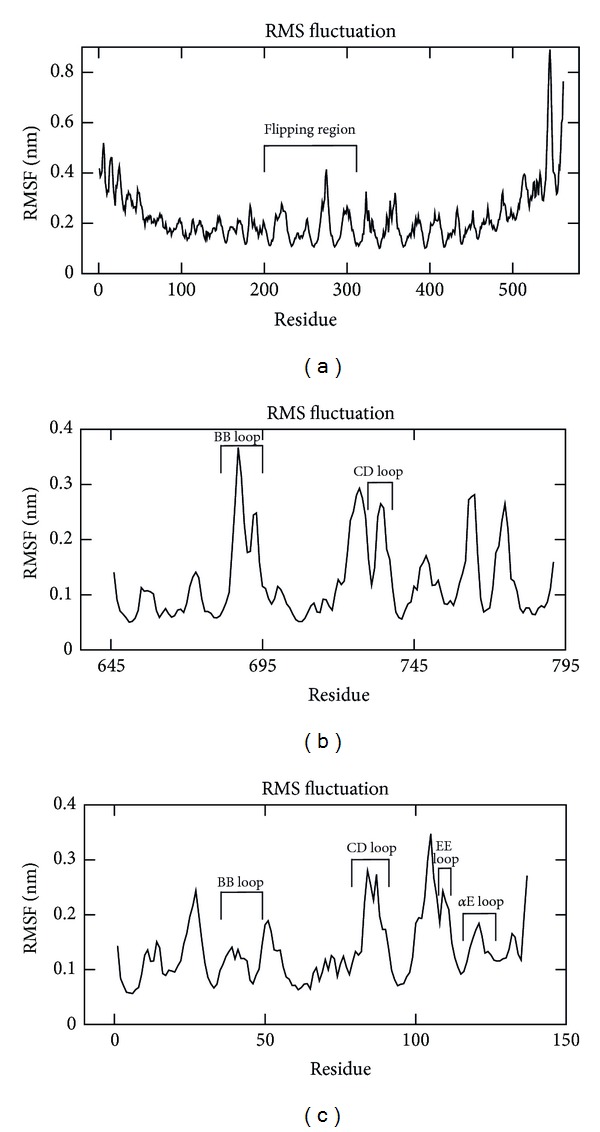
Root mean square fluctuation (RMSF) analysis for homology models. RMSF per residue over the dynamics was shown in graph. (a) TLR2-ECD; (b) TLR2-TIR; (c) MyD88-TIR.
3.4. Validation of Homology Models
The PROCHECK analysis at SAVES of three models (TLR2-ECD, TLR2-TIR, and MyD88-TIR) showed that the phi-psi angles of most of the residues were in the allowed regions of Ramachandran plot (Figure S2). The SAVES results (Table 4(a)) of all models were within the cut-off range suggesting the reliability of our proposed models. The protein stereochemical quality analysis by ProQ, ModFOLD, and MetaMQAP servers showed an acceptable score of all models (Table 4(b)). The average coarse packing quality, planarity, and the collision with symmetry axis, bond lengths, and bond angles obtained by the WHAT IF server of all models revealed the satisfactory acceptance of the models. The results of MolProbity server of all models were also within the range. The RMSD value for all atoms and Cα atoms by superimposing target models with their respective templates showed very low deviation along the significant biological domains. The low deviation between the target-template structures suggested the acceptance of the proposed models. The cross-check validation report indicated acceptability between the experimental structures (PDB ID: 2Z81, 1O77, and 2Z5V) and their models. The RMSD values calculated by PyMOL superimposition program for Cα atoms between the PDB coordinates and the lowest DOPE score models of 2Z81, 1O77, and 2Z5V generated by Modeller were 1.13 Å, 0.518 Å, and 0.705 Å, respectively. The comparison of Ramachandran plot analysis of the homology models of 2Z81, 1O77, and 2Z5V showed similar results for both experimental and hypothetical models (Table S1). The cross-check validation fortified the acceptance of TLR2-ECD, TLR2-TIR, and MyD88-TIR models.
Table 4.
Validation report for TLR2-ECD, TLR2-TIR, and MyD88-TIR homology models.
(a) Validation by SAVES server
| Ramachandran plot (PROCHECK) | TLR2-ECD | TLR2-TIR | MyD88-TIR |
|---|---|---|---|
| Residue (%) | Residue (%) | Residue (%) | |
| Most favored regions | 66.7 | 78.7 | 73.2 |
| Additionally allowed regions | 30.0 | 20.6 | 24.4 |
| Generously allowed regions | 1.8 | 0.0 | 1.6 |
| Disallowed regions | 1.6 | 0.7 | 0.8 |
| Verify3D score | 95.37 | 97.28 | 87.6 |
| ERRAT | 61.059 | 86.364 | 86.325 |
| PROVE (mean Z-score) | 1.609 | 1.48 | 1.63 |
(b) Stereochemical quality of homology models by ProQ, ModFOLD, and MetaMQAP server
| TLR2-ECD | TLR2-TIR | MyD88-TIR | |
|---|---|---|---|
| ProQ (LG/MX) | 7.062/0.432 | 6.401/0.772 | 7.067/0.847 |
| ModFOLD (Q/P) | 0.6326/0.0065 | 0.7473/0.00038 | 0.5787/0.0022 |
| MetaMQAP (GDT/RMSD) | 68.93/2.137 | 78.767/1.523 | 72.628/2.319 |
*ProQ-LG: >1.5 fairly good; >2.5 very good; >4 extremely good. ProQ-MX: >0.1 fairly good; >0.5 very good; >0.8 extremely good. ModFOLD-Q: >0.5 medium confidence; >0.75 high confidence. ModFOLD-P: <0.05 medium confidence; <0.01 high confidence. MetaMQAP-GDT/RMSD: an ideal model has a GDT score over 59 and an RMSD around 2.0 Å.
3.5. Binding Site Analysis of PGN, LTA, and Zymosan with TLR2-ECD
For docking analysis, the predicted top seven (B1 to B7) probable ligand binding pockets in TLR2-ECD (close to the N-glycosylation sites) were considered (Table S2) including previously reported LTA binding site in mouse TLR2 [58]. Interaction of PGN with TLR2-ECD in AutoDock revealed PGN binding sites at B5, and it was in agreement with the previous observation [53]. Both PGN-I and PGN-DAP showed good interactions at B5 regions (Figures 5(a) and 5(b)). The lists of interacting amino acids were presented in Table 5(a). Docking at B6 site also presented good binding score for PGN ligands (Figures 5(c) and 5(d)). AutoDock identified Bmouse and B7 with highest binding affinities for LTA and zymosan, respectively (Figures 5(e) and 5(f)).
Figure 5.
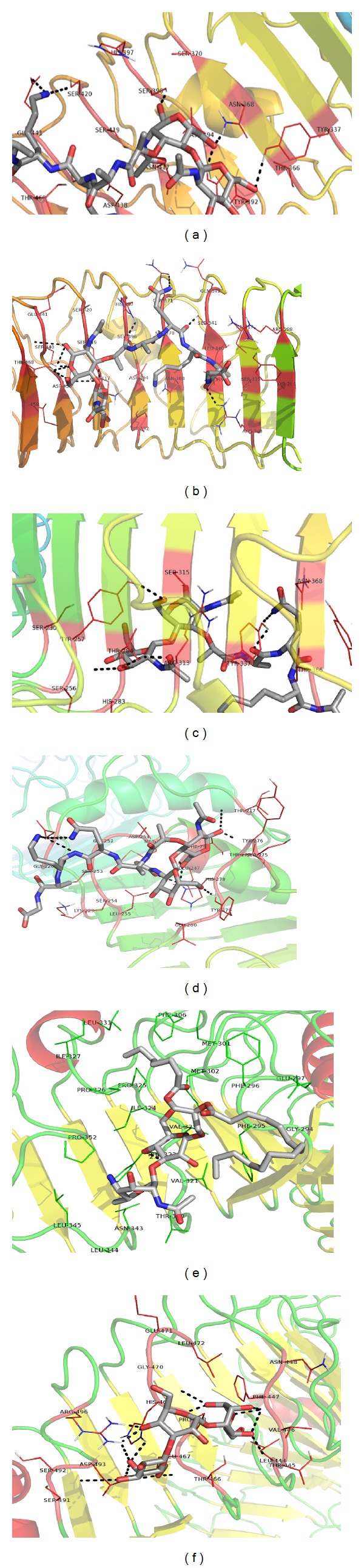
Illustration of the interaction of PGN, LTA, and zymosan with the modeled 3D structure of rohu TLR2-ECD by AutoDock 4.0 program. At B5 region, interaction of PGN-I with TLR2-ECD (a) and PGN-DAP with TLR2-ECD (b); at B6 region, interaction of PGN-I with TLR2-ECD (c) and PGN-DAP with TLR2-ECD (d); interaction of LTA with TLR2-ECD (e) and zymosan with TLR2-ECD (f). The TLR2-ECD was shown in ribbon and ligands (PGN, LTA, and zymosan) were shown in solid form. Amino acid number depicted in the figure was shown as per the matured protein (after removal of the signal peptide).
Table 5.
Molecular interaction of rohu TLR2-ECD with ligands.
(a) Docking analysis of TLR2-ECD with PGN, LTA, and zymosan by AutoDock 4.0
| Grid centre and ligand | Interacting residues | Binding energy kcal/mol | No. of H-bondsa |
|---|---|---|---|
| PGN-I (B5) | Tyr366, Thr395, Asn397, Ser399, Tyr421, Asn423, Ser425, His426, Asn446, Ser448, Ser449, Asp467, Glu470, Thr497 | −4.29 | 6 |
| PGN-I (B6) | Ser259, Ser285, Tyr286, His312, Thr313, Arg342, Ser344, Tyr366 | −4.33 | 6 |
| PGN-DAP (B5) | Asp368, Leu369, Ser370, Gln371, Asn397, Ser399, Gln400, Tyr421, Asp423, Ser425, His426, Asn446, Ser448, Ser449, Asp467, Ser469, Glu470, Thr489 | −3.19 | 11 |
| PGN-DAP (B6) | Thr246, Glu254, Gly255, Lys258, Leu276, Thr277, Met279, Asp280, Gly281, Ser282, Ser283, Leu284, Ser304, Tyr305, Thr306, His307, Tyr308, Glu309 | −3.38 | 7 |
| LTA sites | Glu323, Phe324, Phe325, Glu326, Met330, Met331, Phe335, Thr349, Val350, Phe351, Val352, Ile353, Pro354, Pro355, Ile356, Leu360, Asn372, Leu373, Leu374, Pro381 | −1.92 | 1 |
| Zymosan | Leu473, Thr474, Val475, Phe476, Asn477, Thr495, Leu496, Pro497, His498, Gly499, Glu500, Leu501, Ser520, Ser521, Asp522, Arg525 | −7.55 | 13 |
aHydrogen bonds.
(b) Docking analysis of TLR2-ECD with PGN, LTA, and zymosan by FlexX 2.1
| Ligands | Interacting residues | Binding energy kcal/mol | No. of H-Bondsa |
|---|---|---|---|
| PGN-I | Asn397, Ser399, Gln400, Asn401, His426, Ser428, Lys451, Glu470 | −18.85 | 18 |
| PGN-II | Ser428, Phe429, Val430, Ser448, Lys451, Arg453, Lys454, Asp472, Ser469, Gln470 | −12.80 | 14 |
| PGN-DAP | Gln400, Asn401, His426, Ser428, Ser449, Lys451 | −15.47 | 15 |
| LTA | Asn318, Leu319, Asp320, Ile321, Phe324, Asn347, Gly348, Thr349, Val350, Gln371 | −6.92 | 13 |
| Zymosan | Arg492, Leu493, Met494, Leu496, Arg516, Met517, Ser520, Asp522 | −13.81 | 10 |
aHydrogen bonds.
(c) Docking analysis of TLR2-ECD with PGN, LTA, and zymosan by GOLD 4.1
| Ligands | Interacting residues | GOLD fitness score | No. of H-Bondsa |
|---|---|---|---|
| PGN-I (B5) | Ile394, Asn397, Ser399, Gln400, Tyr421, Asp423, Ser425, His426, Asn446, Ser448, Ser449, Asp467, Ser469, Glu470, Thr489, Gly490, Glu511, Arg512 | 42.38 | 17 |
| PGN-II (B5) | Tyr366, Asp368, Ser370, Gln371, Asn397, Ser399, Gln400, Tyr421, Ser425, Asn446, Ser448, Val465, Asp467, Ser469, Glu470, Thr489, Gly490, Arg512 | 44.01 | 20 |
| PGN-DAP (B5) | Asn446, Ser448, Ser449, Asp467, Ser469, Glu470, Ile487, Thr489, Gly490, Gln511, Arg512 | 40.55 | 13 |
| PGN-II (B6) | Thr246, Glu247, Pro248, Phe249, Lys250, Thr252, Thr277, Asp280, Ser304, Tyr305, Thr306, His307, Tyr308, | 23.00 | 8 |
| LTA | Leu319, Asp320, Ile321, Phe324, Phe327, Met330, Met331, Phe335, Gly348, Thr349, Val350, Phe351, Glu380, Pro381 | 44.65 | 4 |
| Zymosan | Arg492, Leu493, Met494, Thr495, Leu496, Ala514, Leu515, Arg516, Met517, Phe518, Asn519, Ser520, Ser521, Asp522, Arg525 | 39.71 | 8 |
aHydrogen bonds.
In FlexX docking, the binding sites B3 and B4 revealed high positive FlexX score for all ligands and, hence, excluded from further studies. The B5 site (LRR16–19) resulted in good FlexX score −18.85, −12.80, and −15.47 for PGN-I, PGN-II, and PGN-DAP, respectively. Docking at B6 site (LRR8-10) also resulted a satisfactory FlexX score for PGN-II (−13.23) and comparatively a low FlexX score was predicted for PGN-I and PGN-DAP. The rest of the binding pockets were found to be irrelevant for PGN interaction with very little positive and negative FlexX score. The interaction of LTA with TLR2-ECD with highest FlexX score (−6.92) was obtained at Bmouse region (LRR12–14). Interaction of LTA at other binding sites yielded irrelevant FlexX score. Zymosan interacted with TLR2-ECD at B7 (LRR20-CT) region effectively with a FlexX score of (−13.81), and other binding sites were found to be very less interactive. The FlexX scores at different binding regions were presented in Table 5(b).
The GOLD scores for PGN-I, PGN-II, and PGN-DAP at B5 and B6 sites, for LTA at Bmouse and for zymosan at B7 site were given in Table 5(c). The GOLD fitness score was found to be highest in the proposed binding sites of FlexX program in comparison to other binding sites (Figures 6(a)–6(f)). The GOLD docking score of PGN-I, PGN-II, and PGN-DAP was predicted to be less at B6 site as compared to B5 site (Figures 6(a)–6(c)). Interaction of PGN-II at B6 site also showed a good GOLD fitness score (Figure 6(d)). Docking of zymosan (Figure 6(e)) and LTA (Figure 6(f)) in GOLD also ensured B7 and Bmouse as the potential binding sites.
Figure 6.
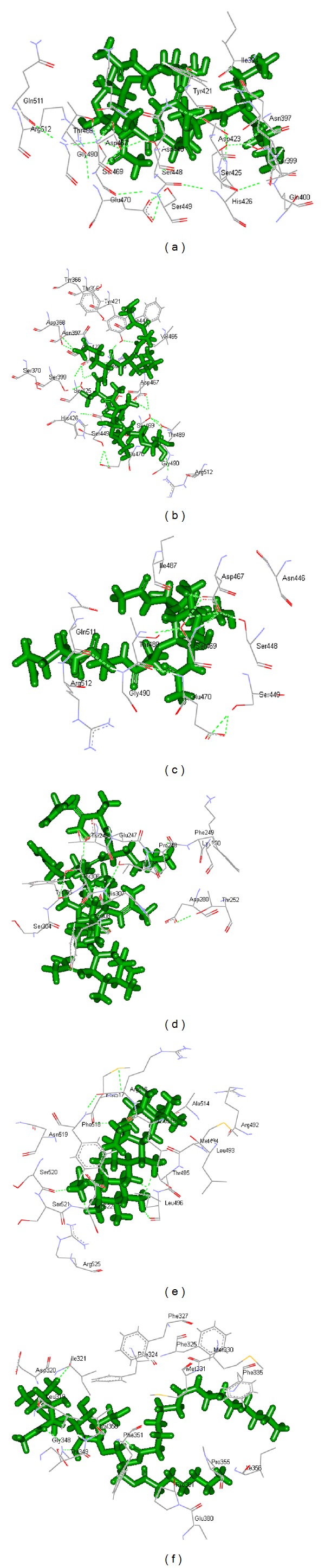
Illustration of the interaction of PGN, LTA, and zymosan with the modeled 3D structure of rohu TLR2-ECD by GOLD 4.1 program. Interaction of (a) PGN-I and TLR2-ECD at LRR16-19; (b) PGN-II and TLR2-ECD at LRR16-19; (c) PGN-DAP and TLR2-ECD at LRR16-19; (d) PGN-II and TLR2-ECD at LRR8-10; (e) zymosan and TLR2-ECD at LRR20-CT; (f) LTA and TLR2-ECD at LRR12-14. Rohu TLR2-ECD model was shown as line and ligands (PGN, LTA, and zymosan) are shown as stick.
3.6. TLR2-TIR and MyD88-TIR Interaction
The MyD88 functions as an adaptor molecule that transmits signal to downstream molecules from ligand activated TLRs by interacting with the TIR domains. The predicted interface residues in cons-PPISP, InterProSurf, and PatchDock were observed to be present in the most flexible regions of TLR2-TIR and MyD88-TIR domains (Table S3). Majority of the interacting amino acids were distributed in BB loop, αB, αC, and CD loop in TLR2-TIR and BB loop, αB helix, and CD loop in MyD88-TIR (Table S3). The best cluster obtained in HADDOCK showed a very low RMSD (1.2 ± 0.7 Å) and intermolecular energy (−722.9 kcal mol-1) with a buried surface area of 2029.2 Å2. The binding orientation and amino acid interactions generated by DS 2.5 were presented in Figure 7(a). The phylogenies of interacted amino acids identified the strongly bonded residues and were highlighted in Figure 7(b). DIMPLOT analysis of the complex showed that BB loop, αC, and αC′ helix residues of TLR2-TIR domain were mostly interacted with AA, BB loops and αB, αC helices of MyD88-TIR domain (Figure S3(a)). The protein-protein complex (Rank-1) in ZDOCK yielded approximately same interacting amino acids residues (as HADDOCK) for TLR2-TIR and MyD88-TIR domains (Figure S3(b)). Amino acid residues in TLR2-TIR and MyD88-TIR domain involved in hydrogen bonding and hydrophobic interactions were presented in Table 6.
Figure 7.
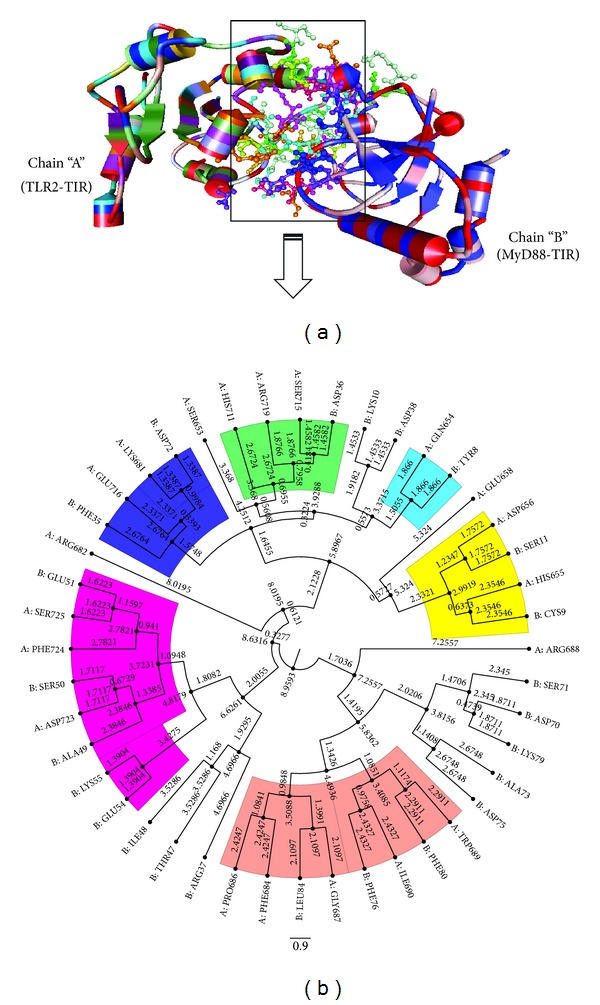
Interaction of TLR2-TIR and MyD88-TIR in Discovery Studio 2.5. (a) TLR2-TIR model is labeled as chain “A” and MyD88-TIR model is labeled as chain “B.” Interface residues are shown inside a rectangle box in ball and stick representation. (b) Clustering of interface residues between TLR2-TIR and MyD88-TIR domain in tree format. Residues of TLR2-TIR are marked as chain “A” and MyD88-TIR as chain “B.” Strongly interacting residues are highlighted with different colors.
Table 6.
List of hydrogen bond forming and hydrophobic interacting residues between TLR2-TIR and MyD88-TIR domains.
| Hydrogen bonds | Hydrophobic interactions | |||
|---|---|---|---|---|
| Donor | Acceptor | Length (Å) | Donor | Acceptor |
| Trp689-NE1 | Leu84-O | 1.97 | Ala73 | Gln654 |
| Ser715-O | Ser50-N | 2.34 | Gln77 | His655 |
| Ser715-O | Ile48-O | 2.84 | Tyr8 | Asp656 |
| Ser715-OG | Asp36-OD2 | 2.78 | Ser71 | His680 |
| Asp723-OD2 | Lys55-NZ | 2.70 | Asp72 | Lys681 |
| Thr714-N | Asp36-OD2 | 2.62 | Cys9 | Arg682 |
| Thr714-O | Asp36-O | 2.72 | Phe76 | Phe684 |
| Ser83-OG | Gly687-N | 2.94 | Lys79 | Pro686 |
| Gln654-NE2 | Asp38-OD2 | 2.72 | Leu82 | Gly687 |
| Ser11-SG | Asp656-OD1 | 3.15 | Ser83 | Trp689 |
| Cys9-NZ | Asp656-OD1 | 2.70 | Phe80 | Ile690 |
| Asp72-N | Asp656-OD2 | 2.56 | Leu84 | Glu710 |
| Cys9-NZ | His655-O | 2.61 | Cys85 | His711 |
| Cys9-NZ | His655-ND1 | 2.78 | Pro86 | Val713 |
| Pro686-O | Ser50-OG | 2.14 | Asp36 | Thr714 |
| Arg688-NH2 | Glu51-OE2 | 2.75 | Ala73 | Gln654 |
| Asp692-OD1 | Lys10-NZ | 2.20 | Gln77 | His655 |
| Lys681-NZ | Asp72-OD2 | 2.73 | Tyr8 | Asp656 |
| His655-NE2 | Asp75-OD2 | 1.91 | Ser71 | His680 |
3.7. Stability Analysis of Protein Complexes by MDS
Previously it was reported that binding site B5 (LRR16–19) (corresponding to human) had the highest affinity for PGN recognition, and binding site B6 (LRR8–10) had a low potential for PGN recognition [53]. In this study, docking scores in B6 for PGN were comparatively lower than B5 region. However, the number of N-glycosylation sites closer to B6 was higher. This data may suggest the possibility of PGN interaction as it is comprised of N-acetylglucosamine. The H-bond analysis showed more number of H-bonds in B5 region (avg. 8 numbers) than B6 region (avg. 5 numbers) with highest binding affinity (Figures 8(a)–8(c)). The existence of H-bonds fluctuated at B6 region during different time periods of MD simulation (Figure 8(d)). Thus, both the docking analysis and MDS suggested B5 region (LRR16–19) as the possible PGN recognition site in rohu TLR2. H-bond analysis for LTA and zymosan at Bmouse (LRR12–14) and B7 (LRR20-CT) region depicted a stable orientation with conserved number of H-bonds throughout the simulation (Figures 8(e) and 8(f)). The DIMPLOT analysis of TLR2-TIR and MyD88-TIR complex generated after 10 ns of MDS showed that the amino acid residues that were involved in protein-protein interaction were retained after the MDS (Figure S4). This suggested that the predicted interacting loops and helices in TLR2-TIR and MyD88-TIR domains were significant for protein-protein interaction.
Figure 8.
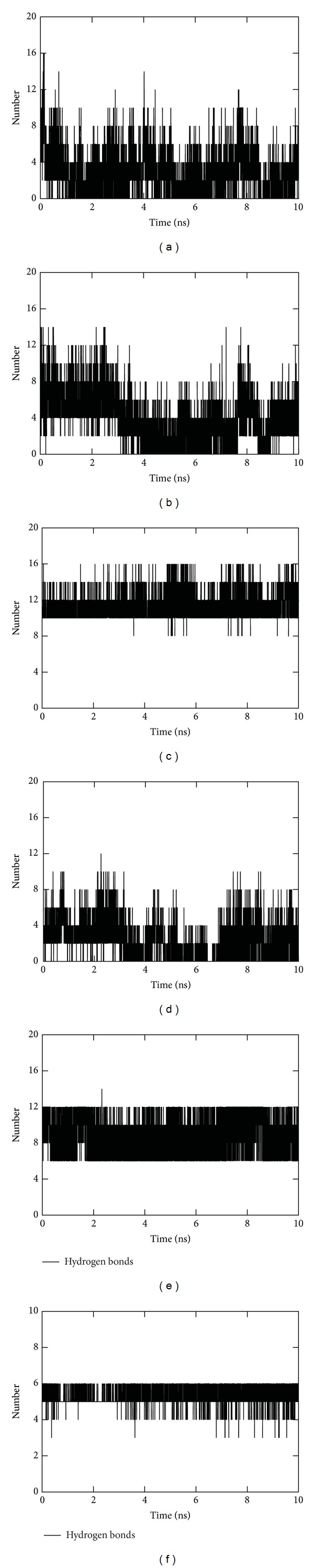
Hydrogen bond analysis of protein-ligand complexes. H-bond analysis of (a) PGN-I and TLR2-ECD complex, (b) PGN-II and TLR2-ECD complex, (c) PGN-DAP and TLR2-ECD complex, (d) PGN-II and TLR2-ECD complex at LRR8-10 region (B6), (e) LTA and TLR2-ECD complex at LRR20-CT, (f) and zymosan and TLR2-ECD complex at LRR12-14. X-axis represents time period of simulation and Y-axis represents number of hydrogen bonds.
3.8. Validation of Binding Domains by Site-Directed Mutagenesis
Alanine scanning of B5 regions of PGN binding domains showed loss of interaction due to absence of donors or acceptors in the active sites. But no single mutation of alanine for the interacting residues at this region showed a complete loss of docking. Proline and aspartic acid scanning of B5 region also ensued very low fitness score (9.6 and 12.8) in GOLD. Analysis of B5 region by mutating residues in pair, triplet, and quadruple combinations at a time indicated the viable importance of Asp394, Ser396, Asn397, and Ser399 as the fitness score attained a very minimum value in comparison to other GOLD runs.
Mutagenesis study at B7 residues in TLR2-ECD that formed H-bond with zymosan revealed a good fitness score for alanine scanning. However, proline scanning of all residues revealed loss of zymosan interaction with TLR2-ECD. Single proline mutation and acidic to basic mutation of different interacting residues followed by individual GOLD runs explored the importance of Ser520 and Asp522. A single mutation of Ser520-Pro520 and Asp522-Gln522 resulted in complete loss of zymosan interaction. Mutation of other residues altered the GOLD fitness score; however, in none of the cases docking loss was ensued. Alanine and proline scanning of LTA binding site resulted in low docking score emphasizing the role of key residues Asp320 and Phe324 in LTA recognition.
4. Conclusion
The proposed 3D model of rohu TLR2 describes the protein features and its important domains. It also accentuates the importance of predicting key amino acids and LRR regions that are responsible for the specific ligand interaction and TLR2 signaling in fish and depicts a residue-detailed structural theoretical model. In the absence of crystal structure of TLR2 in any fish, this study provides structural insight into the TLR2 domains architecture. In rohu fish, the peptides at LRR16–19 (at B5), LRR12-14 (at Bmouse) and LRR20-CT (at B7) are predicted to be the most important interacting regions for PGN, LTA, and zymosan interactions, respectively. The structural organization of TIR domains in fish TLR2 and adapter molecule MyD88 have also been described. The interaction between TLR2-TIR and MyD88-TIR domain highlighted the contribution of BB loop, αB, αC, and CD loop in TLR2-TIR and BB loop, αB helix, and CD loop in MyD88-TIR domain. The data generated in this study are likely to be helpful to conduct further in vivo study to develop the strategy of innate immune activation and disease prevention in fish.
Supplementary Material
Figure S1. Multiple sequence alignment of rohu TLR2 with other species by MegAlign program. N-termini, C-termini, LRR, trans-membrane (TM) and TIR domain regions are labeled. Conserved residues are shown inside yellow box. Majority axis represents the highest occurrence of a residue in a column.
Figure S2. Ramachandran plot analysis of (a) TLR2-ECD, (b) TLR2-TIR and (c) MyD88-TIR model. The plot was calculated in PROCHECK program.
Figure S3. DIMPLOT analysis of interaction between TLR2-TIR and MyD88-TIR domains (a) HADDOCK analysis and (b) ZDOCK analysis.
Figure S4. The DIMPLOT analysis of TLR2-TIR and MyD88-TIR complex after 10 ns of MDS.
Table S1. Cross-check validation report of homology models.
Table S2. List of predicted binding sites in rohu TLR2-ECD.
Table S3. The predicted interface residues in TLR2-TIR and MyD88-TIR domains.
Acknowledgments
This work was financially supported by the grant of National Agricultural Innovation Project (NAIP), Indian Council of Agricultural Research (ICAR) (Project Code C4-C30018). The authors express their gratitude to Dr. A. Bandyopadhyay and Dr. S. Kochar, National Coordinator, NAIP-Comp-4, for their valuable suggestions and help. The authors would like to express their deepest gratitude to Dr. G. C. Sahoo, HOD, Bioinformatics Division, RMRIMS, Patna, India, for his valuable suggestion. Part of the work related to Discovery Studio, FlexX, and GOLD was carried out at RMRIMS, Patna.The authors would like to thank Mr. Roman Laskowski for providing the academic license of Ligplot. They thank Mr. Mahesh Patra (Research Associate) and Mr. Sukanta Kumar Pradhan, HOD, Department of Bioinformatics, Orissa University of Agriculture and Technology, Bhubaneswar, Odisha, India, for their support and constructive suggestions in data analysis. They gratefully acknowledge the Bioinformatics Resources and Applications Facility (BRAF) at the Center for Development of Advanced Computing (C-DAC), Pune, India.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.de la Barrera S, Alemán M, Sasiain MDC. Toll-like receptors in human infectious diseases. Current Pharmaceutical Design. 2006;12(32):4173–4184. doi: 10.2174/138161206778743493. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nature Immunology. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R. Toll-like receptors and innate immunity. Nature Reviews Immunology. 2001;1(2):135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 5.Beutler BA. TLRs and innate immunity. Blood. 2009;113(7):1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buwitt-Beckmann U, Heine H, Wiesmüller K-H, et al. TLR1- and TLR6-independent recognition of bacterial lipopeptides. Journal of Biological Chemistry. 2006;281(14):9049–9057. doi: 10.1074/jbc.M512525200. [DOI] [PubMed] [Google Scholar]
- 7.Takeuchi O, Sato S, Horiuchi T, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. Journal of Immunology. 2002;169(1):10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 8.Lien E, Sellati TJ, Yoshimura A, et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. Journal of Biological Chemistry. 1999;274(47):33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R, Preston-Hurlburt P, Kopp E, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Molecular Cell. 1998;2(2):253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 10.Dziarski R. Recognition of bacterial peptidoglycan by the innate immune system. Cellular and Molecular Life Sciences. 2003;60(9):1793–1804. doi: 10.1007/s00018-003-3019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological Reviews. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan R, Mariuzza RA. Peptidoglycan recognition proteins of the innate immune system. Trends in Microbiology. 2007;15(3):127–134. doi: 10.1016/j.tim.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Iwaki D, Mitsuzawa H, Murakami S, et al. The extracellular toll-like receptor 2 domain directly binds peptidoglycan derived from Staphylococcus aureus . Journal of Biological Chemistry. 2002;277(27):24315–24320. doi: 10.1074/jbc.M107057200. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuzawa H, Wada I, Sano H, et al. Extracellular toll-like receptor 2 region containing Ser40-Ile64 but not Cys30-Ser39 is critical for the recognition of Staphylococcus aureus peptidoglycan. Journal of Biological Chemistry. 2001;276(44):41350–41356. doi: 10.1074/jbc.M104177200. [DOI] [PubMed] [Google Scholar]
- 15.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. Journal of Biological Chemistry. 1999;274(25):17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 16.Asong J, Wolfert MA, Maiti KK, Miller D, Boons G-J. Binding and cellular activation studies reveal that toll-like receptor 2 can differentially recognize peptidoglycan from gram-positive and gram-negative bacteria. Journal of Biological Chemistry. 2009;284(13):8643–8653. doi: 10.1074/jbc.M806633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutcliffe IC, Shaw N. Atypical lipoteichoic acids of gram-positive bacteria. Journal of Bacteriology. 1991;173(22):7065–7069. doi: 10.1128/jb.173.22.7065-7069.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deininger S, Traub S, Aichele D, et al. Presentation of lipoteichoic acid potentiates its inflammatory activity. Immunobiology. 2008;213(6):519–529. doi: 10.1016/j.imbio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Von Aulock S, Deininger S, Draing C, Gueinzius K, Dehus O, Hermann C. Gender difference in cytokine secretion on immune stimulation with LPS and LTA. Journal of Interferon and Cytokine Research. 2006;26(12):887–892. doi: 10.1089/jir.2006.26.887. [DOI] [PubMed] [Google Scholar]
- 20.Lee I-T, Wang S-W, Lee C-W, et al. Lipoteichoic acid induces HO-1 expression via the TLR2/MyD88/c-Src/NADPH oxidase pathway and Nrf2 in human tracheal smooth muscle cells. Journal of Immunology. 2008;181(7):5098–5110. doi: 10.4049/jimmunol.181.7.5098. [DOI] [PubMed] [Google Scholar]
- 21.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and toll-like receptor 2. Journal of Experimental Medicine. 2003;197(9):1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown GD, Herre J, Williams DL, Willment JA, Marshall ASJ, Gordon S. Dectin-1 mediates the biological effects of β-glucans. Journal of Experimental Medicine. 2003;197(9):1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Underhill DM, Ozinsky A, Hajjar AM, et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401(6755):811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 24.Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato M, Sano H, Iwaki D, et al. Direct binding of toll-like receptor 2 to Zymosan, and Zymosan-induced NF-κB activation and TNF-α secretion are down-regulated by lung collectin surfactant protein A. Journal of Immunology. 2003;171(1):417–425. doi: 10.4049/jimmunol.171.1.417. [DOI] [PubMed] [Google Scholar]
- 26.Frasnelli ME, Tarussio D, Chobaz-Péclat V, Busso N, So A. TLR2 modulates inflammation in zymosan-induced arthritis in mice. Arthritis Research & Therapy. 2005;7(2):R370–379. doi: 10.1186/ar1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio . Molecular Immunology. 2004;40(11):759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Meijer AH, Gabby Krens SF, Medina Rodriguez IA, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Molecular Immunology. 2004;40(11):773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Hirono I, Takami M, Miyata M, et al. Characterization of gene structure and expression of two toll-like receptors from Japanese flounder Paralichthys olivaceus . Immunogenetics. 2004;56(1):38–46. doi: 10.1007/s00251-004-0657-2. [DOI] [PubMed] [Google Scholar]
- 30.Oshiumi H, Tsujita T, Shida K, Matsumoto M, Ikeo K, Seya T. Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics. 2003;54(11):791–800. doi: 10.1007/s00251-002-0519-8. [DOI] [PubMed] [Google Scholar]
- 31.Baoprasertkul P, Peatman E, Abernathy J, Liu Z. Structural characterisation and expression analysis of Toll-like receptor 2 gene from catfish. Fish and Shellfish Immunology. 2007;22(4):418–426. doi: 10.1016/j.fsi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Li Y-W, Luo X-C, Dan X-M, et al. Orange-spotted grouper (Epinephelus coioides) TLR2, MyD88 and IL-1β involved in anti-Cryptocaryon irritans response. Fish and Shellfish Immunology. 2011;30(6):1230–1240. doi: 10.1016/j.fsi.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Wei YC, Pan TS, Chang MX, et al. Cloning and expression of Toll-like receptors 1 and 2 from a teleost fish, the orange-spotted grouper Epinephelus coioides . Veterinary Immunology and Immunopathology. 2011;141(3-4):173–182. doi: 10.1016/j.vetimm.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro CMS, Hermsen T, Taverne-Thiele AJ, Savelkoul HFJ, Wiegertjes GF. Evolution of recognition of ligands from gram-positive bacteria: similarities and differences in the TLR2-mediated response between mammalian vertebrates and teleost fish. Journal of Immunology. 2010;184(5):2355–2368. doi: 10.4049/jimmunol.0900990. [DOI] [PubMed] [Google Scholar]
- 35.Samanta M, Swain B, Basu M, et al. Molecular characterization of toll-like receptor 2 (TLR2), analysis of its inductive expression and associated down-stream signaling molecules following ligands exposure and bacterial infection in the Indian major carp, rohu (Labeo rohita) Fish and Shellfish Immunology. 2012;32(3):411–425. doi: 10.1016/j.fsi.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 36.Basu M, Swain B, Sahoo BR, Maiti NK, Samanta M. Induction of toll-like receptor (TLR) 2, and MyD88-dependent TLR- signaling in response to ligand stimulation and bacterial infections in the Indian major carp, mrigal (Cirrhinus mrigala) Molecular Biology Reports. 2012;39(5):6015–6028. doi: 10.1007/s11033-011-1415-9. [DOI] [PubMed] [Google Scholar]
- 37.Clewley JP, Arnold C. MEGALIGN. The multiple alignment module of LASERGENE. Methods in Molecular Biology. 1997;70:119–129. [PubMed] [Google Scholar]
- 38.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. Journal of Molecular Biology. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 39.Mizuguchi K, Deane CM, Blundell TL, Johnson MS, Overington JP. JOY: protein sequence-structure representation and analysis. Bioinformatics. 1998;14(7):617–623. doi: 10.1093/bioinformatics/14.7.617. [DOI] [PubMed] [Google Scholar]
- 40.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. Journal of Computational Chemistry. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.Vriend G. WHAT IF: a molecular modeling and drug design program. Journal of Molecular Graphics. 1990;8(1):52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 43.Chen VB, Arendall WB, III, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D. 2010;66(1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallner B, Fang H, Elofsson A. Automatic consensus-based fold recognition using Pcons, ProQ, and Pmodeller. Proteins. 2003;53(6):534–541. doi: 10.1002/prot.10536. [DOI] [PubMed] [Google Scholar]
- 45.Mcguffin LJ. The ModFOLD server for the quality assessment of protein structural models. Bioinformatics. 2008;24(4):586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 46.Pawlowski M, Gajda MJ, Matlak R, Bujnicki JM. MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics. 2008;9, article 403 doi: 10.1186/1471-2105-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang B. Metapocket: a meta approach to improve protein ligand binding site prediction. OMICS A Journal of Integrative Biology. 2009;13(4):325–330. doi: 10.1089/omi.2009.0045. [DOI] [PubMed] [Google Scholar]
- 48.Laurie ATR, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 49.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. Journal of Computational Chemistry. 2007;28(6):1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 50.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. Journal of Molecular Biology. 1996;261(3):470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 51.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. Journal of Molecular Biology. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 52.Sahoo BR, Basu M, Swain B, et al. Structural insights of rohu TLR3, its binding site analysis with fish reovirus dsRNA, poly I:C and zebrafish TRIF. International Journal of Biological Macromolecules. 2012;51(4):531–543. doi: 10.1016/j.ijbiomac.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 53.Li Y, Efferson CL, Ramesh R, Peoples GE, Hwu P, Ioannides CG. A peptidoglycan monomer with the glutamine to serine change and basic peptides bind in silico to TLR-2 (403–455) Cancer Immunology, Immunotherapy. 2011;60(4):515–524. doi: 10.1007/s00262-010-0959-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Vries SJ, van Dijk M, Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nature Protocols. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 55.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 56.Basith S, Manavalan B, Govindaraj RG, Choi S. In silico approach to inhibition of signaling pathways of toll-like receptors 2 and 4 by ST2L. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0023989.e23989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Kang JY, Nan X, Jin MS, et al. Recognition of lipopeptide patterns by toll-like receptor 2-toll-like receptor 6 heterodimer. Immunity. 2009;31(6):873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multiple sequence alignment of rohu TLR2 with other species by MegAlign program. N-termini, C-termini, LRR, trans-membrane (TM) and TIR domain regions are labeled. Conserved residues are shown inside yellow box. Majority axis represents the highest occurrence of a residue in a column.
Figure S2. Ramachandran plot analysis of (a) TLR2-ECD, (b) TLR2-TIR and (c) MyD88-TIR model. The plot was calculated in PROCHECK program.
Figure S3. DIMPLOT analysis of interaction between TLR2-TIR and MyD88-TIR domains (a) HADDOCK analysis and (b) ZDOCK analysis.
Figure S4. The DIMPLOT analysis of TLR2-TIR and MyD88-TIR complex after 10 ns of MDS.
Table S1. Cross-check validation report of homology models.
Table S2. List of predicted binding sites in rohu TLR2-ECD.
Table S3. The predicted interface residues in TLR2-TIR and MyD88-TIR domains.


