Abstract
Insulin secretion plays a critical role in glucose homeostasis, and failure to secrete sufficient insulin is a hallmark of type 2 diabetes. Genome-wide association studies (GWAS) have identified loci contributing to insulin processing and secretion1,2; however, a substantial fraction of the genetic contribution remains undefined. To examine low-frequency (minor allele frequency (MAF) 0.5% to 5%) and rare (MAF<0.5%) nonsynonymous variants, we analyzed exome array data in 8,229 non-diabetic Finnish males. We identified low-frequency coding variants associated with fasting proinsulin levels at the SGSM2 and MADD GWAS loci and three novel genes with low-frequency variants associated with fasting proinsulin or insulinogenic index: TBC1D30, KANK1, and PAM. We also demonstrate that the interpretation of single-variant and gene-based tests needs to consider the effects of noncoding SNPs nearby and megabases (Mb) away. This study demonstrates that exome array genotyping is a valuable approach to identify low-frequency variants that contribute to complex traits.
Exome sequencing studies have discovered many low-frequency and rare coding variants3 that have yet to be examined systematically for association with complex traits. To determine the role of low-frequency coding variants in traits reflecting pancreatic beta-cell function, insulin sensitivity, and glycemia, we evaluated putative functional coding variants selected from exome sequences of >12,000 individuals (see Online Methods for a description of exome array design and content). We successfully genotyped 9,660 Finnish participants in the population-based Metabolic Syndrome in Men (METSIM) study4 for 247,870 variants on the Illumina HumanExome Beadchip. Clinical characteristics of 8,229 analyzed non-diabetic study participants are summarized in Supplementary Table 1. Among 242,071 variants passing quality control, 89,864 (38.1%) were variable in the studied individuals; of these, 71,077 were nonsynonymous, nonsense, or located in splice sites (Supplementary Table 2). We tested 59,029 variants with MAF>0.05% for association with insulin processing, secretion, and glycemic traits assuming additive allelic effects and using a linear mixed model to account for relatedness among study participants5.
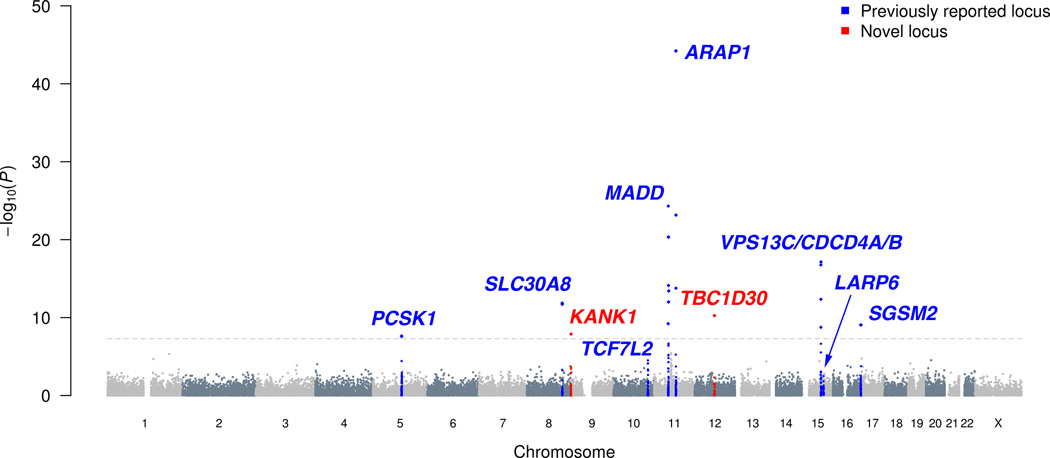
We first evaluated rare and low-frequency coding variants at the nine signals previously identified by GWAS for fasting proinsulin level adjusted for fasting insulin (hereafter referred to as fasting proinsulin)1. To recognize independent association signals, we carried out conditional analysis adjusting for the known GWAS variants, all of which were represented on the exome array and replicated in METSIM (P<.01; Figure 1, Supplementary Table 3). Coding low-frequency variants at the known SGSM2 and MADD loci showed strong evidence of association (P<5×10−8; Table 1, Supplementary Figures 1 and 2). Previous studies highlighted several possible candidate genes at these loci1,6,7.
Figure 1.
Manhattan plot for the fasting proinsulin analysis. Association results of the single-variant analysis (−log10 P values) are plotted against genomic position (NCBI Build 37). Previously identified loci are denoted in blue and loci identified by the current study in red. Fasting proinsulin levels were log-transformed and adjusted for fasting insulin, body mass index, age, and age2.
Table 1.
Novel low frequency variants at fasting proinsulin loci previously identified by genome-wide association studies
| SNP | Gene | Variant | Chr | Position | Minor/major allele |
MAF | β̂ ± SE | Effect size in SD units ± SE |
Proportion of trait variance explained |
P value | Conditional P value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs61741902 | SGSM2 | V996I | 17 | 2,282,779 | A/G | .014 | .126 ± .021 | .41 ± .07 | .0047 | 8.7 × 10−10 | 4.8 × 10−10 |
| rs35233100 | MADD | R766X | 11 | 47,306,630 | T/C | .037 | −.100 ± .013 | −.32 ± .04 | .0075 | 7.6 × 10−15 | .0001 |
Chr, chromosome; MAF, minor allele frequency; SE, standard error; SD, standard deviation. Positions are from NCBI Build 37 with allele labels from the forward strand. Fasting proinsulin was log-transformed and adjusted for fasting insulin, BMI, age, and age2. Effects are reported for the minor allele.β̂ coefficient units are ln(pmol/l). Conditional P values are reported after adjusting for the lead SNPs from GWAS signals (rs4790333 at SGSM2; rs7944584 and rs1051006 at MADD). Full results of conditional analysis are provided in Supplementary Table 4. For rs35233100, effect size in SD units (± SE) and proportion of trait variance explained after adjusting for rs7944584 and rs1051006, are −0.17 (± .05) and 0.0007, respectively. Based on analysis of 8,224 non-diabetic males.
At SGSM2, rs61741902 (MAF=1.4%, P=8.9×10−10) encodes Val996Ile and is independent of GWAS variant rs4790333 (Pcond=4.8×10−10, r2=.001; Table 1, Figure 1, Supplementary Table 4). SGSM2 (small G protein signaling modulator 2) is a GTPase activating protein (GAP) that interacts with members of the Rab and Rap small G protein pathways and may act in a cascade of Rab-mediated steps in insulin secretory vesicle transport8–10. At rs61741902, the reference valine is well-conserved across vertebrates, and the isoleucine substitution is predicted to be damaging (Supplementary Table 5) Each additional copy of the minor allele was associated with an average increase of 0.41 standard deviations (SD) in fasting proinsulin (Table 1, Supplementary Figure 2). Still, the proportion of the trait variability explained is modest (0.47%; 95% CI = 0.22–0.82%) due to the low minor allele frequency. Identification of an independent and plausibly functional variant suggests that SGSM2 is the causal gene underlying the common fasting proinsulin GWAS signal.
At MADD, rs35233100 (MAF=3.7%, P=7.6×10−15) creates stop codon Arg766Ter and is in modest linkage disequilibrium (LD) with the lead GWAS variant rs7944584 (Pcond=.0001, r2=.17), and independent of the second GWAS variant rs1051006 (Pcond=5.0 × 10−16, r2=.02). The nonsense allele of rs35233100, associated with decreased proinsulin, is observed only on haplotypes containing the proinsulin-decreasing allele of rs7944584. Adjusting for one variant in a conditional analysis decreased, but did not eliminate, association for the other (P=4.9 × 10−25 and Pcond=5.7 × 10−15 for rs7944584; Table 1, Supplementary Table 4), suggesting biological contributions from the nonsense variant and an additional causal variant tagged by rs7944584. Of note, the trait-decreasing alleles of the two common GWAS-identified variants rs7944584 and rs1051006 tend to occur on different haplotypes, causing the evidence of association for either SNP to become dramatically more significant when adjusting for the other (rs1051006, P=0.033 and Pcond=2.7 × 10−8; rs7944584, P=4.9 × 10−25 and Pcond=8.3 × 10−31, Supplementary Table 4). Although the conditional association for the nonsense variant only achieves suggestive significance (P=.0001), it provides an especially plausible functional effect. The MADD nonsense variant is located in exon 13 of 36, suggesting that the mRNA would be targeted for nonsense-mediated decay11. MADD can act as a guanine nucleotide exchange factor for RAB3 proteins including RAB3A and RAB3B12, which are critical for insulin exocytosis13,14. Identification of a nonsense variant that contributes to the evidence of association suggests that MADD is a causal gene underlying the common GWAS signals.
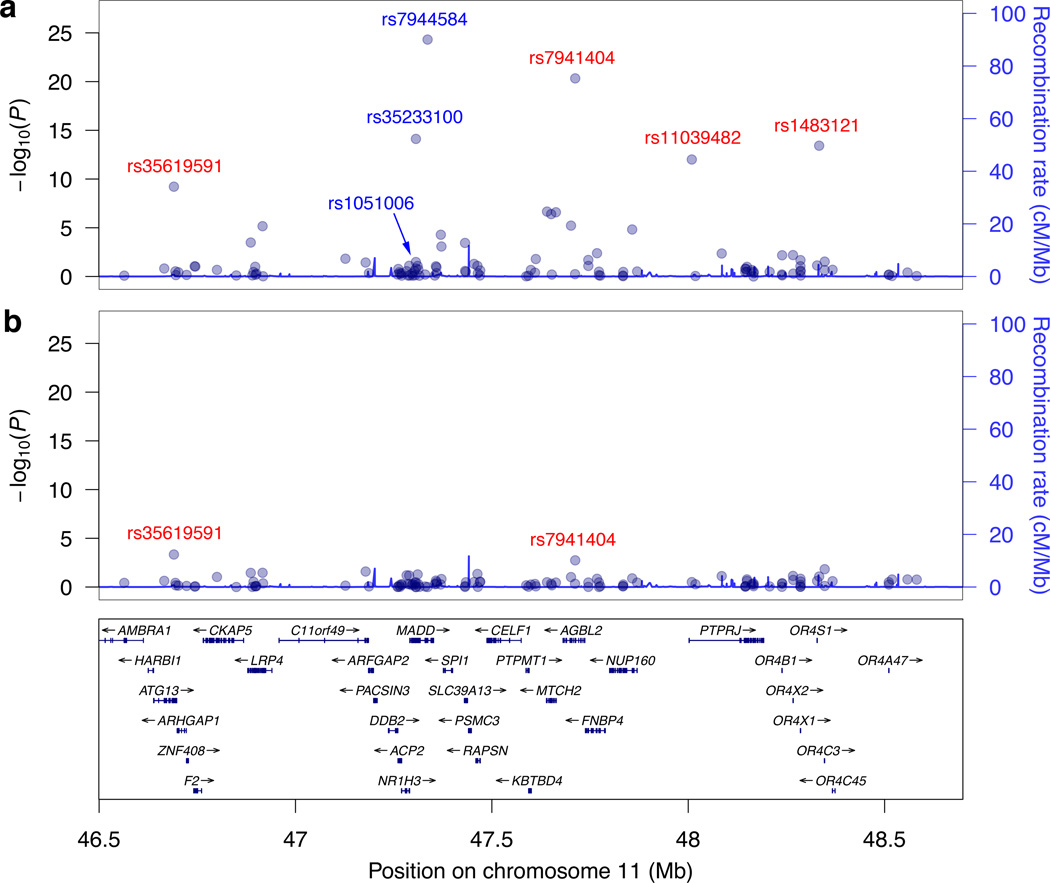
LD at chromosome 11 from 46–57 megabases (Mb) and encompassing MADD has been reported to extend long distances15. Consistent with this, we noted significant or suggestive (P<1×10−5) fasting proinsulin association with nonsynonymous variants up to ~9 Mb away from the lead (noncoding) GWAS variant. Proinsulin-associated variants included rs628524, located ~9 Mb away and encoding Ser171Asn in the olfactory receptor OR5M11 (P=3.7×10−6 for fasting proinsulin; P as low as 5.0×10−10 for related traits) and rs7941404, located 376 kb away and encoding Arg349His in AGBL2 (MAF=11.8%; P=4.7×10−21). After adjusting for the three MADD variants (rs7944584, rs1051006, rs35233100), association significance for the distant variants was reduced by orders of magnitude (Supplementary Table 6, Figure 2). The fact that these associations were not eliminated suggests that additional variant(s) in this region await identification or that we may be adjusting for imperfect proxies of causal variants. These results also demonstrate that LD should be considered when interpreting GWAS results in this region. For example, the recently reported16 novel fasting glucose locus at OR4S1, represented by rs1483121, is in LD (r2=.19) with the lead and nonsense MADD SNPs ~1 Mb away (Supplementary Table 6).
Figure 2.
The MADD gene is located in a region of unusually high linkage disequilibrium on chromosome 11 from 46 to 57 Mb. Regional association results of the single-variant analysis (−log10 P values) are plotted against genomic position (NCBI Build 37) for fasting proinsulin before (a) and after (b) adjustment of the lead SNPs for the common GWAS signals (rs7944584 and rs1051006) and the nonsense variant rs35233100 (MAF 3.7%) at MADD. Fasting proinsulin levels were log-transformed and adjusted for fasting insulin, body mass index, age, and age2. The conditioning SNPs are indicated in blue. For clarity, only a portion of the 11 Mb region and a subset of the genes are shown.
Next, we tested coding variants across the genome for association with 19 traits measuring pancreatic beta-cell function, insulin sensitivity, and glucose levels. We identified two genes harboring low-frequency nonsynonymous variants with novel associations for fasting proinsulin levels: rs150781447 encoding TBC1D30 Arg279Cys (MAF=2.0%, P=5.5×10−11) and rs3824420 encoding KANK1 Arg667His (MAF=3.0%, P=1.3×10−8). The TBC1D30 variant was most strongly associated with late-phase proinsulin-to-insulin conversion (proinsulin AUC30–120; P=1.3×10−16) and the KANK1 variant with early-phase proinsulin-to-insulin conversion (proinsulin AUC0–30; P=1.6×10−9) (Table 2, Supplementary Figure 1). The TBC1D30 variant effect is large, with each additional copy of the minor allele resulting in an average increase of 0.50 SD in proinsulin AUC30–120 (Table 2, Supplementary Figure 2). This variant explained 0.94% of the trait variability (95% CI = 0.55–1.44%). We also observed a novel locus for insulin secretion as measured by the insulinogenic index, represented by nonsynonymous SNPs in PAM (smallest P=1.9×10−8) and PPIP5K2, located 200 kb apart, both with MAF=5.3%, and in near-perfect LD (r2=0.997) (Table 2, Supplementary Figures 1,2, and 3).
Table 2.
Novel loci for insulin processing and secretion
| SNP | Gene | Variant | Chr | Position | Minor/major allele |
MAF | Lead trait | β̂ ±SE | Effect size in SD units ± SE |
Proportion of trait variance explained |
P value |
| Identified by low-frequency variants | |||||||||||
| rs150781447 | TBC1D30 | R279C | 12 | 65,224,220 | T/C | .020 | Proinsulin AUC30–120 | .204 ± .025 | .50 ± .06 | .0094 | 1.3 × 10−16 |
| rs3824420 | KANK1 | R667H | 9 | 712,766 | A/G | .029 | Proinsulin AUC0–30 | .107 ± .018 | .28 ± .05 | .0045 | 1.6 × 10−9 |
| rs35658696 | PAM | D563G | 5 | 102,338,811 | G/A | .053 | Insulinogenic index | −.152 ± .027 | −.21 ± .04 | .0044 | 1.9 × 10−8 |
| rs36046591 | PPIP5K2 | S1228G | 5 | 102,537,285 | G/A | .053 | Insulinogenic index | −.152 ± .027 | −.21 ± .04 | .0043 | 2.3 × 10−8 |
| Identified by common variants | |||||||||||
| rs2650000 | HNF1A | intergenic | 12 | 121,388,962 | A/C | .455 | Insulinogenic index | −.076 ± .012 | −.10 ± .02 | .0054 | 5.0 × 10−10 |
| rs505922 | ABO | intronic | 9 | 136,149,229 | C/T | .471 | Disposition index | −.038 ± .006 | −.09 ± 02 | .0043 | 3.8 × 10−9 |
| rs60980157 | GPSM1 | S391L | 9 | 139,235,415 | T/C | .300 | Insulinogenic index | .072 ± .013 | .10 ± .02 | .0041 | 1.4 × 10−8 |
Chr, chromosome; MAF, minor allele frequency; SE, standard error; SD, standard deviation; CI, confidence interval. Positions are from NCBI Build 37 with allele labels from the forward strand. The lead trait is the trait with smallest P value. Traits were log-transformed and adjusted for BMI, age, and age2. Effects are reported for the minor allele. The SNPs at PAM and PPIP5K2 are tightly linked (D′=0.999, r2=0.997). Based on analysis of 8,103-8,191 non-diabetic males.
Common SNPs at GPSM1, HNF1A, and ABO, previously associated with other traits, are here associated with insulin secretion or beta-cell function in non-diabetic individuals (Table 2, Supplementary Figure 3). GPSM1 Ser391Leu is in LD with noncoding rs3829109 (r2=.69), previously associated with fasting glucose2. At ABO, the T allele of rs505922 is a proxy for the O blood group and has been associated with diverse phenotypes, including decreased pancreatic cancer risk17 and increased risk for duodenal ulcer18. Near HNF1A, rs2650000 was previously associated with LDL-cholesterol19 and C-reactive protein20; other HNF1A variants are associated with MODY3 (MIM #600496) and type 2 diabetes risk21.
TBC1D30 and KANK1 both function in G protein signaling and are strong biological candidates. TBC1D30 (TBC 1 domain family, member 30) encodes a GAP protein that likely regulates activity of specific Rab GTPases including RAB3A22 and RAB8A23. Rab3A knockout mice show a severe decrease in glucose-induced first phase insulin release and a 75% decrease in plasma insulin levels, without insulin resistance24. The reference arginine at rs150781447 is well-conserved across vertebrate species and the cysteine substitution is predicted to be damaging25 (Supplementary Table 5). The variant is located within a Rab-GAP domain and within the Kozak sequence of one TBC1D30 isoform and may alter translation initiation.
KANK1 (KN motif and ankyrin repeat domain-containing protein 1) plays a role in cytoskeleton formation by regulating actin polymerization26 and negatively regulates Rac1 and RhoA G protein signaling, pathways that have been implicated in insulin secretion27,28. At rs3824420, the reference arginine is not well conserved across species and the protein structure is predicted to tolerate the histidine substitution without an effect on function; this variant may still affect KANK1 or may tag another nearby variant. While rs3824420 has low frequency in Europeans (MAF=2.9% in Finns), it is common in East Asians (MAF=16%; Supplementary Table 7).
PAM encodes peptidylglycine alpha-amidating monooxygenase, an essential secretory granule membrane enzyme that catalyzes α-amidation of peptide hormones such as proinsulin29. Older mice heterozygous for Pam deficiency exhibit glucose intolerance30. At rs35658696, the reference aspartic acid is well conserved across vertebrates and located in one of the catalytic domains, and the glycine substitution is predicted to be damaging (Supplementary Table 5). The nearby gene PPIP5K2 is involved in cell signaling but has no known connection to insulin pathways. At rs36046591 in PPIP5K2, in near-perfect LD (r2=0.997) with rs35658696, the glycine substitution is predicted to be tolerated and the reference serine is not well conserved across species. This difference suggests that the PAM variant is causal at that locus, rather than the PPIP5K2 variant, but it is impossible to dissect them genetically.
Next, we carried out gene-based tests to investigate further the role of rare and low-frequency variants in insulin secretion and processing. Gene-based tests offer an alternative to single-variant tests, which are often underpowered to detect association with rare variants. Tests were performed on trait residuals adjusted for relatedness and covariates (see Online Methods). To address the impact of less common and rare variants, we considered only SNPs with MAF<3% or MAF<1%. In total, we tested 10,515 genes having at least two such variants using the SKAT-O test31.
We found significant associations between fasting proinsulin and TBC1D30, SGSM2, and ATG13 when using a MAF upper bound of 3% (Table 3, Supplementary Figure 4); by conditioning on the low-frequency variants detected by single-variant analysis, we demonstrated that these signals are driven by low-frequency variants. After adjusting for the common and nonsense variant signals at MADD, significance for ATG13, ~609 kb away, decreased by orders of magnitude (Table 3), showing that this signal is partially driven by the MADD variants and suggesting that other variants in this region await identification or that we may be adjusting for imperfect proxies of the causal variant. No additional associations were detected with other traits, including type 2 diabetes (data not shown).
Table 3.
Genes associated with fasting proinsulin identified by gene-based tests of aggregated low-frequency nonsynonymous variants with MAF < 3%
| Gene | Number of Variants |
Variants (minor allele counts) | P value | Conditional P value |
|---|---|---|---|---|
| TBC1D30 | 2 | R279C(324), P746L(427) | 3.3 × 10−9 | .75a |
| SGSM2 | 3 | Y416C(78), T789P(3), V996I(236) | 2.0 × 10−9 | .68b |
| ATG13 | 7 | L5V(20), I131V(1), Q249P(3), R392W(1), | 1.8 × 10−8 | .0055d |
| L427Q(3), G434R(488), X406G(200)c | 37e |
Fasting proinsulin was log-transformed and adjusted for fasting insulin, BMI, age, and age2. Residuals were adjusted for relatedness and gene-based testing was carried out using the SKAT-O test (see Online Methods). This analysis was based on 8,224 participants. Reported associations were significant after Bonferroni correction for testing 19 traits for 10,515 genes (significance threshold: 2.5 × 10−7).
After adjusting for the low-frequency nonsynonymous variant R279C (rs150781447) at TBC1D30 (MAF 2.0%).
After adjusting for the low-frequency nonsynonymous variant V996I (rs61741902) at SGSM2 (MAF 1.4%).
Annotation relative to a non-canonical (longer) isoform.
After adjusting for lead SNPs of common GWAS signals (rs7944584 and rs1051006) and nonsense variant rs35233100 at MADD (MAF 3.7%).
After adjusting for the low-frequency nonsynonymous variant G434R (rs35619591) at ATG13 (MAF 3.0%).
In summary, we identified two low-frequency coding variants in genes at known loci and three novel genes with low-frequency variants associated with insulin processing or secretion. At least four of these genes play roles in G-protein signaling (Supplementary Figure 5). We show that the interpretation of both single-variant and gene-based tests needs to consider the effects of distant common SNPs, an especially important consideration when exome sequence data are analyzed without data on the surrounding noncoding regions. Although regions of long-range LD are unusual, at least 24 have been reported15 to extend >1 Mb in Europeans, a distance frequently used to claim independence of association signals in GWAS meta-analyses. Several of the identified exome array variants are plausibly functional, although ~25% and ~28% of low-frequency nonsynonymous variants on the exome array were annotated as conserved and plausibly damaging, respectively (Supplementary Table 2), and the exome array does not provide complete coverage of all functional variants at each locus. By the content of the exome array, this study was also limited in its ability to look at very rare variants. While sequencing will still be required to completely assess variants associated with insulin processing, secretion, and glycemic traits, this study provides proof-of-principle that exome array genotyping is a powerful approach to identify low-frequency functional variants and fine-map GWAS-identified loci in complex traits.
ONLINE METHODS
Study participants
We attempted exome array genotyping of 9,717 participants in the Metabolic Syndrome in Men (METSIM) study4. Male study participants were randomly selected from the population register of Kuopio, Eastern Finland (population 95,000). Participants undertook a 1-day outpatient visit to the Clinical Research Unit at the University of Kuopio. Participants with diagnosed type 1 diabetes or type 2 diabetes (previously diagnosed, on diabetes medication, with fasting glucose ≥ 7 mmol/l, or with 2-hour glucose ≥ 11.1 mmol/l) were excluded from quantitative trait analysis. Clinical characteristics of non-diabetic study participants are provided in Supplementary Table 1. The study was approved by the ethics committee of the University of Kuopio and Kuopio University Hospital; informed consent was obtained from all study participants.
OGTT and laboratory measurements
Clinical testing was performed following a 12-h overnight fast. A 2-h oral 75 g glucose tolerance test (OGTT) was performed with blood samples drawn at 0, 30, and 120 min, for measurement of plasma proinsulin, insulin, and glucose levels. Plasma specific proinsulin (Human Proinsulin RIA kit; Linco Research, St. Charles, MO; no cross-reaction with insulin or C-peptide) and insulin (ADVIA Centaur Insulin IRI, No. 02230141; Siemens Medical Solutions Diagnostics, Tarrytown, NY; minimal cross-reaction with proinsulin or C-peptide) were measured by immunoassay, and plasma glucose by enzymatic hexokinase photometric assay (Konelab System Reagents, Thermo Fisher Scientific, Vantaa, Finland).
Phenotypes
Association results are reported for five traits: fasting proinsulin (adjusted for fasting insulin), early-phase (ProinsAUC0–30) and late-phase (ProinsAUC30–120) glucose-stimulated proinsulin-to-insulin conversion measured as proinsulin area under the curve (AUC) during the first 30 min and remaining 90 min of an OGTT, insulin secretion assessed by the insulinogenic index32, and a disposition index measure of β-cell compensation for insulin resistance defined as InsAUC0–30/GluAUC0–30 × Matsuda index of insulin sensitivity (Matsuda ISI)4,33. The reported associations were discovered by analyzing a total of 19 traits. Other measures of β-cell function included: oral glucose-stimulated proinsulin-to-insulin conversion during the first 30 min (ProinsAUC0–30/InsAUC0–30), and 30–120 min (ProinsAUC30–120/InsAUC30–120) of the OGTT, unadjusted fasting proinsulin, fasting proinsulin/insulin ratio, HOMA-β34, fasting insulin, insulin at 120 min, insulin AUC during the first 30 min (InsAUC0–30) and during 30–120 min (InsAUC30–120), and early-phase glucose-stimulated insulin release (InsAUC0–30/GluAUC0–30) adjusted for Matsuda ISI35. Indices of insulin sensitivity included HOMA-IR34 and the Matsuda ISI36. Associations with fasting and 120 min glucose were also tested. Supplementary Figure 6 shows correlations among traits. We calculated AUC measures using the trapezoid rule.
Exome array
The Illumina HumanExome-12v1_A Beadchip includes 247,870 markers focused on protein-altering variants selected from >12,000 exome and genome sequences representing multiple ethnicities and complex traits. Nonsynonymous variants had to be observed three or more times in at least two studies, splicing and stop-altering variants two or more times in at least two studies. Additional array content includes variants associated with complex traits in previous GWAS, HLA tags, ancestry informative markers, markers for identity-by-descent estimation, and random synonymous SNPs. Details about SNP content and selection strategies can be found at the exome array design webpage (see URLs).
Genotyping and quality control
9,717 study samples, 104 blind duplicate samples, and 116 HapMap samples of different ethnicities were genotyped at the Genetic Resources Core Facility (GRCF) at the Johns Hopkins Institute of Genetic Medicine. Genotype calling was carried out using Illumina's GenTrain version 1.0 clustering algorithm in GenomeStudio version 2011.1. Cluster boundaries were determined using study samples. After clustering, 5,574 non-autosomal and 3,379 autosomal variants identified through filtering strategies developed at GRCF were manually reviewed and clusters edited as necessary. After technical failure and marker-level quality control, 242,458 of 247,870 (97.8%) attempted markers were successfully genotyped and had call rate >95% (average call rate 99.95%).
We evaluated genotyping quality using concordance rates for HapMap samples genotyped in our study and (a) sequenced by Complete Genomics or the 1000 Genomes Project (on-target regions of integrated phase 1 release; see URLs) or (b) genotyped on the Illumina HumanOmni2.5 Beadchip by the 1000 Genomes Project. These comparisons were based on 60,574, 117,063, and 39,056 overlapping variants and 17, 49, and 86 individuals, respectively. Overall concordance rates were 99.933%, 99.972%, and 99.956% for Complete Genomics and 1000 Genomes sequence data, and HumanOmni2.5 Beadchip data, respectively. Considering the external data as truth, concordance rates for homozygous genotypes were 99.982%, 99.987%, and 99.974%, and for heterozygous genotypes were 99.678%, 99.529% and 99.886%, respectively.
In total, 9,660 of 9,717 (99.4%) individuals were successfully genotyped (call rate > 98%). For the 242,458 SNPs that passed quality control, genotype concordance among the 104 blind duplicate sample pairs was 99.998%. Three sex-mismatched individuals were identified and excluded from subsequent analyses. One individual per pair of 6 known twin pairs and 6 unexplained apparent duplicates were excluded.
We carried out principal components analysis (PCA) twice, once excluding HapMap samples to identify population outliers, and then including HapMap samples to help interpret outliers. To avoid artifactual results due to family relatedness37, we computed principal components using SNP loadings estimated from a subset of 7,304 not-close-relatives. We defined close relatives as ones for whom estimated genome-wide identical-by-descent (IBD) proportion of alleles shared was >0.10. We estimated IBD sharing using PLINK's "--genome" option38 and carried out PCA using SMARTPCA37 on a linkage-disequilibrium-pruned set of 22,464 autosomal SNPs obtained by removing large scale high-LD regions15,39, SNPs with a MAF < 0.01, or SNPs with HWE P value < 10−6, and carrying out LD pruning using the PLINK option: "--indep-pairwise 50 5 0.2". Inspecting the first 10 PCs, we identified 12 population outliers, 9 of whom had self-reported non-Finnish ancestry; we excluded these 12 individuals from subsequent analysis. After further removal of 25 individuals with diagnosed type 1 diabetes, 1,376 with type 2 diabetes, and 3 with missing phenotypes, 8,229 individuals remained for quantitative trait analysis.
Statistical analysis
Single-variant analysis
We tested for trait-SNP association assuming an additive genetic model using a linear mixed model to correct for relatedness using EMMAX5. We excluded SNPs with MAF < 0.05% or Hardy-Weinberg equilibrium (HWE) P value < 10−6. To reduce the impact of outliers, we log-transformed traits with skewed distributions and then Winsorized all traits at 5 standard deviations from the mean. All traits were adjusted for BMI, age, and age2 prior to association testing. We analyzed both untransformed residuals and rank-based inverse-normal transformed residuals to assess robustness of association results to distributional assumptions. Since no appreciable differences were observed between the two analyses, we report results for untransformed residuals. Finally, we visually inspected genotype cluster plots and checked HWE P values for all described variants. The lowest HWE P value for a reported novel associated variant was 0.09.
Population stratification
To correct for population stratification, we modeled population structure as part of the random effects, indistinguishable from the relatedness effect5. To investigate residual population stratification, we calculated genomic control inflation factors40 and inspected quantile-quantile (QQ) plots for test statistics both before and after removal of established and newly discovered loci (2 Mb segments centered on lead SNPs) (Supplementary Figure 7).
Conditional analysis
To identify additional association signals after accounting for the effects of known and newly discovered trait loci, we carried out conditional analyses where we included the allele count at the lead SNP(s) at the conditioning loci as covariate(s). To allow discovery of more than two association signals per locus, we used a stepwise procedure where additional SNPs were added to the model according to their conditional P value, as programmed in EMMAX5. We estimated LD metrics r2 and D' using 9,633 METSIM individuals who passed genotyping quality control. LD with SNPs not included on the exome array was determined based on whole-genome sequence data for 1,479 Northern European individuals.
Gene-based analysis
For gene-based testing we used the SKAT-O31 test which encompasses burden tests and SKAT41 as special cases. SKAT-O has been shown to perform well under a range of scenarios, including scenarios in which protective, deleterious, and null variants are present, and in which a large number of variants are causal and associated in the same direction31. To account for relatedness, we adopted an approach similar to GRAMMAR42 by first obtaining trait residuals adjusted for relatedness using GenABEL43 and then carrying out gene-based testing. We performed analyses using default weights31 and MAF upper bounds of 1% and 3% for the combination of nonsynonymous, stop-altering, and splice-site variants. In total, 10,515 genes with at least two variants were tested. The results of the naive SKAT-O analysis and the analysis adjusted for relatedness were highly correlated (Supplementary Figure 8). To evaluate whether common or low-frequency SNPs associated with the trait in the single-variant analysis can account for a gene-based test signal, we also carried out conditional analyses by including the allele count at such SNP(s) as covariate(s).
Statistical significance
We declared a single variant-trait association significant if the nominal P value was < 4.46 × 10−8, corresponding to a Bonferroni correction for 1,121,551 tests (19 phenotypes × 59,029 variants). We declared a gene-based test association significant if the nominal P value was < 2.50 × 10−7, corresponding to a Bonferroni correction for 199,785 tests (19 phenotypes × 10,515 genes).
Annotation
We annotated variants relative to GENCODE version 7 coding transcripts44 using in-house developed software (unpublished). Amino acid substitution positions are relative to the canonical UniProt protein sequence45.
URLs
Exome array design, http://genome.sph.umich.edu/wiki/Exome_Chip_Design;
Complete Genomics 69 Genomes Data, http://www.completegenomics.com/public-data/69-Genomes;
The 1000 Genomes Project, www.1000genomes.org;
PLINK, http://pngu.mgh.harvard.edu/~purcell/plink;
SMARTPCA, http://genetics.med.harvard.edu/reich/Reich_Lab/Software.html;
EMMAX, http://genetics.cs.ucla.edu/emmax;
GenABEL, http://www.genabel.org
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Academy of Finland (contract no. 124243), The Finnish Heart Foundation, The Finnish Diabetes Foundation, TEKES (contract no. 1510/31/06), the Commission of the European Community (HEALTH-F2-2007-201681), and National Institutes of Health (NIH) grants DK093757, DK072193, DK062370, and 1Z01 HG000024. Genotyping was conducted at the Genetic Resources Core Facility (GRCF) at the Johns Hopkins Institute of Genetic Medicine.
Footnotes
AUTHOR CONTRIBUTIONS
J.R.H. led statistical analysis, and J.R.H., A.U.J., H.M.S., X.S., L.Y., and C.F. performed statistical analysis. J.R.H., M.P.F., M.L.B., and P.S.C. performed bioinformatics analysis. A.S., H.C., J.K., and M.L. obtained and analyzed phenotype data. J.M.R., H.L., I.M., R.I., E.W.P., and K.F.D. generated genotype data. B.M.N., M.L.D., and G.R.A. designed the genotyping array. H.M.K. and G.R.A. developed statistical analysis tools. J.K. and M.L. designed and supervised the METSIM study. J.R.H., M.P.F., M.L.B., M.B., and K.L.M. drafted the manuscript and all authors reviewed the manuscript. J.R.H., A.U.J., M.P.F., M.L.B., A.S., H.M.S., X.S., L.Y., C.F., T.M.T., L.J.S., F.S.C., G.R.A., R.M.W., M.B., M.L., and K.L.M. contributed to discussion and interpreted the data. M.B., M.L., and K.L.M. designed and supervised this study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Strawbridge RJ, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott RA, et al. Large-scale association study using the Metabochip array reveals new loci influencing glycemic traits and provides insight into the underlying biological pathways. Nat Genet. 2012 doi: 10.1038/ng.2385. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiezun A, et al. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stančáková A, et al. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingelsson E, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Sasaki T, Minoshima S, Shimizu N. Identification of three novel proteins (SGSM1,2,3) which modulate small G protein (RAP and RAB)-mediated signaling pathway. Genomics. 2007;90:249–260. doi: 10.1016/j.ygeno.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Nottingham RM, Ganley IG, Barr FA, Lambright DG, Pfeffer SR. RUTBC1 protein, a Rab9A effector that activates GTP hydrolysis by Rab32 and Rab33B proteins. J Biol Chem. 2011;286:33213–33222. doi: 10.1074/jbc.M111.261115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutter GA, Hill EV. Insulin vesicle release: walk, kiss, pause ... then run. Physiology (Bethesda) 2006;21:189–196. doi: 10.1152/physiol.00002.2006. [DOI] [PubMed] [Google Scholar]
- 11.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppola T, et al. The death domain of Rab3 guanine nucleotide exchange protein in GDP/GTP exchange activity in living cells. Biochem J. 2002;362:273–279. doi: 10.1042/0264-6021:3620273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regazzi R, et al. Expression, localization and functional role of small GTPases of the Rab3 family in insulin-secreting cells. J Cell Sci. 1996;109((Pt 9)):2265–2273. doi: 10.1242/jcs.109.9.2265. [DOI] [PubMed] [Google Scholar]
- 14.Piper Hanley K, et al. In vitro expression of NGN3 identifies RAB3B as the predominant Ras-associated GTP-binding protein 3 family member in human islets. J Endocrinol. 2010;207:151–161. doi: 10.1677/JOE-10-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning AK, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amundadottir L, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41:986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanikawa C, et al. A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat Genet. 2012;44:430–434. S1–S2. doi: 10.1038/ng.1109. [DOI] [PubMed] [Google Scholar]
- 19.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishibashi K, Kanno E, Itoh T, Fukuda M. Identification and characterization of a novel Tre-2/Bub2/Cdc16 (TBC) protein that possesses Rab3A-GAP activity. Genes Cells. 2009;14:41–52. doi: 10.1111/j.1365-2443.2008.01251.x. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaekura K, et al. Insulin secretory deficiency and glucose intolerance in Rab3A null mice. J Biol Chem. 2003;278:9715–9721. doi: 10.1074/jbc.M211352200. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakinuma N, Zhu Y, Wang Y, Roy BC, Kiyama R. Kank proteins: structure, functions and diseases. Cell Mol Life Sci. 2009;66:2651–2659. doi: 10.1007/s00018-009-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowluru A. Friendly, and not so friendly, roles of Rac1 in islet beta-cell function: lessons learnt from pharmacological and molecular biological approaches. Biochem Pharmacol. 2011;81:965–975. doi: 10.1016/j.bcp.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammar E, Tomas A, Bosco D, Halban PA. Role of the Rho-ROCK (Rho-associated kinase) signaling pathway in the regulation of pancreatic beta-cell function. Endocrinology. 2009;150:2072–2079. doi: 10.1210/en.2008-1135. [DOI] [PubMed] [Google Scholar]
- 29.Rajagopal C, Mains RE, Eipper BA. Signaling from the secretory granule to the nucleus. Crit Rev Biochem Mol Biol. 2012;47:391–406. doi: 10.3109/10409238.2012.694845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czyzyk TA, et al. Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev Biol. 2005;287:301–313. doi: 10.1016/j.ydbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012 doi: 10.1093/biostatistics/kxs014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stumvoll M, Van Haeften T, Fritsche A, Gerich J. Oral glucose tolerance test indexes for insulin sensitivity and secretion based on various availabilities of sampling times. Diabetes Care. 2001;24:796–797. doi: 10.2337/diacare.24.4.796. [DOI] [PubMed] [Google Scholar]
- 33.Retnakaran R, et al. Hyperbolic relationship between insulin secretion and sensitivity on oral glucose tolerance test. Obesity (Silver Spring) 2008;16:1901–1907. doi: 10.1038/oby.2008.307. [DOI] [PubMed] [Google Scholar]
- 34.Matthews DR, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 35.Stančáková A, et al. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5,327 nondiabetic Finnish men. Diabetes. 2009;58:2129–2136. doi: 10.2337/db09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 37.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weale ME. Quality control for genome-wide association studies. Methods Mol Biol. 2010;628:341–372. doi: 10.1007/978-1-60327-367-1_19. [DOI] [PubMed] [Google Scholar]
- 40.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 41.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 44.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.