Abstract
5-Methylcytosine (5mC) in DNA can be oxidized stepwise to 5-hydroxymethylcytosine (5hmC), 5- formylcytosine (5fC), and 5-carboxylcytosine (5caC) by the TET family proteins. Thymine DNA glycosylase can further remove 5fC and 5caC, connecting 5mC oxidation with active DNA demethylation. Here we present a chemical modification-assisted bisulfite sequencing (CAB-Seq) that can detect 5caC with single-base resolution in DNA. We optimized 1-ethyl-3- [3-dimethylaminopropyl]carbodiimide hydrochloride (EDC)- catalyzed amide bond formation between the carboxyl group of 5caC and a primary amine group. We found that the modified 5caC can survive the bisulfite treatment without deamination. Therefore, this chemical labeling coupled with bisulfite treatment provides a base-resolution detection and sequencing method for 5caC.
5-Methylcytosine (5mC), the fifth base in the mammalian genome encoding critical epigenetic information, significantly impacts cellular biological functions and human diseases.1 While the mechanism of cytosine methylation has been well studied, active demethylation pathways have remained elusive. 2 Recent studies have reported the accumulation of 5- hydroxymethylcytosine (5hmC) in certain mammalian cells and tissues,3,4 and the ten-eleven translocation (TET) family of enzymes that can oxidize 5mC stepwise to 5- hydroxymethylcytosine (5hmC),4 5-formylcytosine (5fC), and finally 5-carboxylcytosine (5caC).4–7 The oxidation products 5fC and 5caC can be recognized and excised by mammalian DNA glycosylase (TDG) which generates an abasic site that can be further transformed to normal cytosine through base excision repair (BER), completing an active demethylation process.7–9
Quantification of these cytosine modifications in mouse embryonic stem (ES) cells showed decreased abundances from 5hmC to 5fC and further to 5caC, which are roughly 1300 ppm, 20 ppm, and 5 ppm per cytosine, respectively.5,6,10,11 The importance and the limited abundance of 5hmC and its further oxidized derivatives present challenges for effective methods to selectively label and sequence these modifications in genomic DNA.11–14 The developments of labeling and profiling technologies for 5hmC have significantly contributed to the study of 5hmC.11,13,15–17 Patterns of 5hmC distribution unique from those of 5mC were revealed.15,18 In order to further understand 5mC and the subsequent active demethylation, methods to profile and/or sequence 5fC and 5caC are required. Recently, we have developed selective chemical labeling and profiling methods for 5fC, indicating its function in TDG-mediated demethylation at gene regulatory elements.19 Similar conclusions were also obtained using an antibody-based approach.20 Under typical bisulfite conditions, both 5fC and 5caC are deaminated and read as T, making them indistinguishable from unmodified cytosine. We showed that the labeling of 5fC by hydroxylamine protects it from bisulfite-mediated deamination, thereby allowing detection and sequencing of 5fC with base resolution.19 We present here a chemical modification-assisted bisulfite sequencing method that allows base-resolution detection of 5caC in DNA.
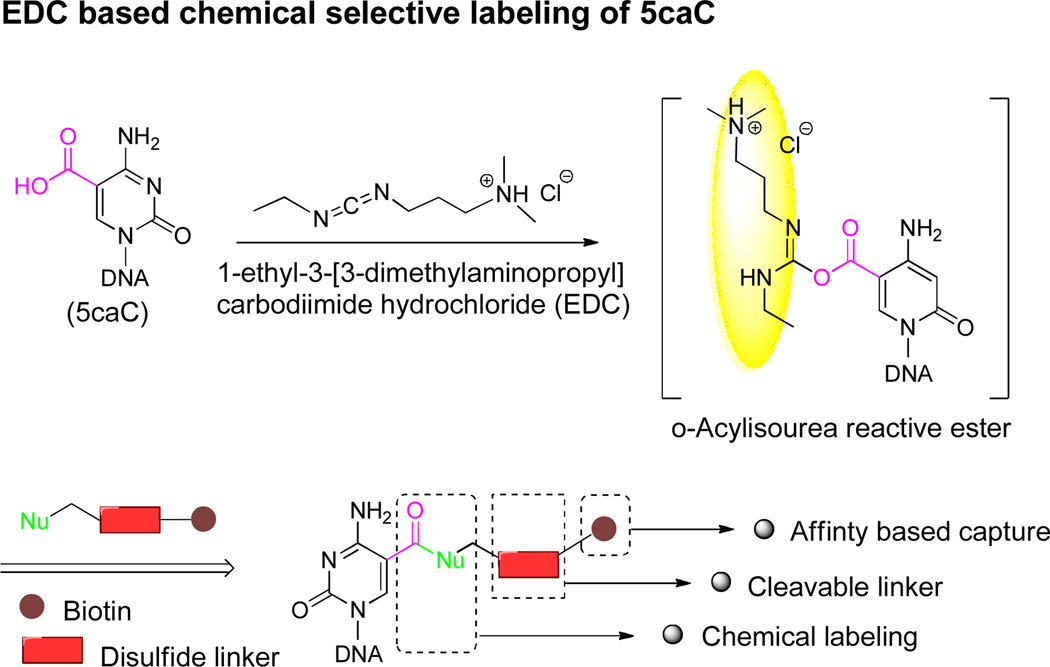
We chose the 1-ethyl-3-[3- dimethylaminopropyl]carbodiimide hydrochloride (EDC)- catalyzed amide bond formation between carboxyl group and primary amine group as our chemical selective labeling method. The EDC-based coupling reaction has been well developed and widely used.21–23 The reaction can be performed in aqueous solution under relatively mild conditions around neutral pH which prevents DNA degradation. The carboxylic acid group can react with EDC to form an o-acylisourea ester intermediate that can then be attacked by a primary amine (Scheme 1). Since the 3’ and 5’ terminal phosphate groups can be easily removed through treatment with phosphatase, the carboxyl group of 5caC will be the most reactive functional group with EDC. After the coupling reaction, a biotin-modified amine may be installed onto the 5caC-containing DNA (Scheme 1). Then, the streptavidin coated beads can be employed to capture the 5caC-containing fragments for subsequent enrichment. The designed disulfide bond between amine group and biotin can be smoothly cleaved by DTT treatment (see Figure S1). As a result, the 5caC-containing DNA may be pulled down, enriched, and subjected to high-throughput sequencing. The key to this strategy is to optimize the labeling efficiency and selectivity.
Scheme 1. EDC-catalyzed chemical labeling of 5caC.a.
aDesigned labeling strategy for 5caC. The carboxyl group of 5caC in duplex DNA can be labeled by EDC-catalyzed coupling reaction to form biotin-S-S-5caC using biotin-S-S-amine.
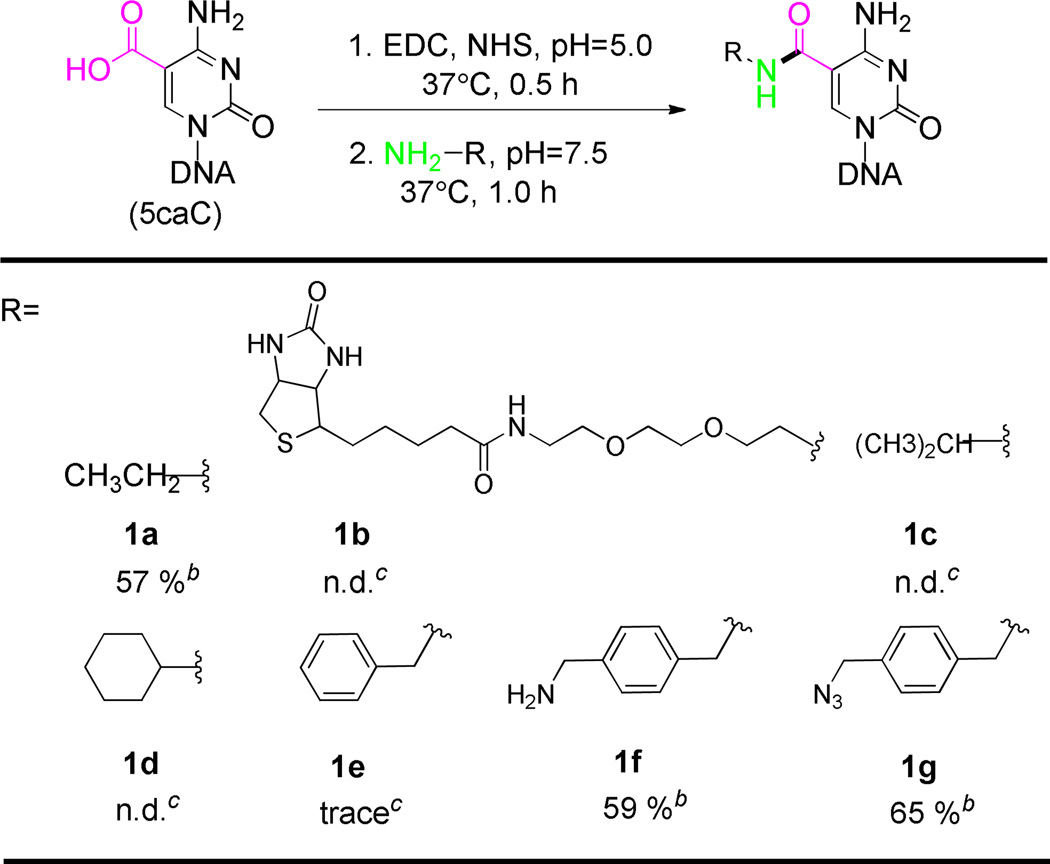
We started by screening reactions on the short double-stranded 5caC-containing oligonucleotides and monitoring the reaction by mass spectrometry, quantifying the yield by HPLC (Table 1). To our delight, ethylamine can react with 5caC DNA with moderate labeling efficiency (Table 1a). This approach also showed high selectivity to 5caC among other cytosine modifications such as 5hmC and 5fC (see Figure S2). The side reaction between the DNA phosphate backbone and ethylamine is undetectable on mass spectrometry. A two-step EDC coupling strategy, which generates the N-hydroxysuccinimide (NHS) ester intermediate at pH = 5.0 and then formation of the amide bond at pH = 7.5, showed higher reactivity than the direct coupling at pH = 6.0. Unfortunately, when the biotin-PEG2-amine was employed under the optimized reaction conditions, the coupling reaction failed as a result of the increase in steric hindrance (1b in Table 1). Subsequently, a series of primary amines were tested in order to optimize labeling efficiency. As expected, the bulky amines greatly inhibited the labeling reaction (1c-1e in Table 1).
Table 1.
EDC-mediated 5caC coupling with different amines.a
 |
General reaction conditions: double strand 5caC DNA (10 µmol), NHS (20 mM), EDC (2 mM), Mes (pH=5.0, 75 mM) were incubated in aqueous solution at 37°C for 0.5 h. Buffer exchange to sodium phosphate (pH=7.5, 100 mM), NaCl (150 mM), amine (10 mM), incubated at 37 °C for 1.0 h.
Reaction yield based on HPLC analysis (see Figure S5).
Undetectable product by mass spectrometry. (see Figure S6).
Further screening revealed that the xylene group significantly facilitates this transformation on DNA. The xylenediamine showed high reaction activity comparable to ethylamine (1f in Table 1). Notably, the benzylamine only gave trace amounts of coupling product (1e in Table 1). Subsequently, we synthesized (4-aminomethyl)benzylazide which possesses the xylene structure but also contains an azide group for the well-developed bio-orthogonal coupling of the biotin tag.24,25 As expected, the (4-aminomethyl)benzylazide showed as good reactivity as xylenediamine (1g in Table 1). We show that the azide group can be coupled with a strained cyclooctyne bearing a disulfide linker to a biotin.26,27 After a simple DTT treatment, the disulfide bond can be selectively cleaved with high efficiency. The high reaction activity and selectivity during each step was monitored by mass spectrometry (see Figure S3). The biotin-labeled 5caC nucleoside was further confirmed with high resolution mass spectrometry after DNA digestion (see Figure S4). This chemical labeling step has no noticeable sequence bias.
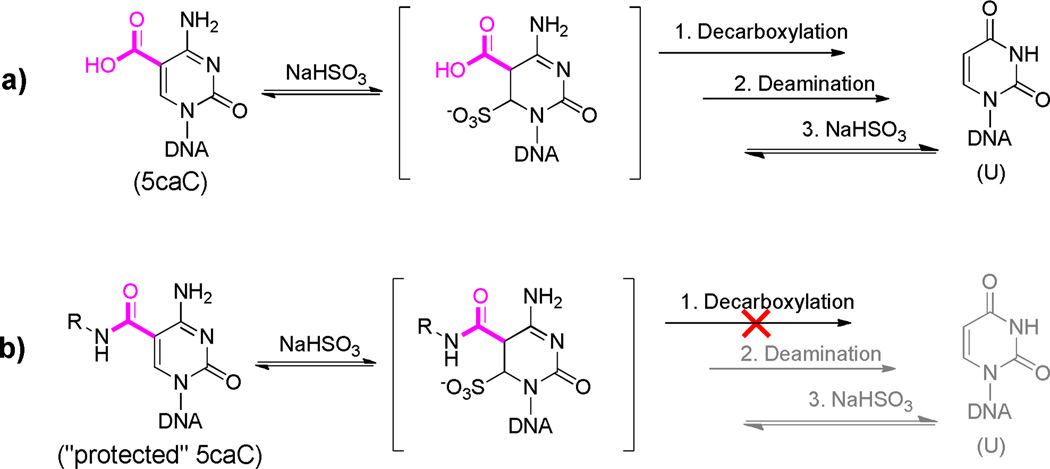
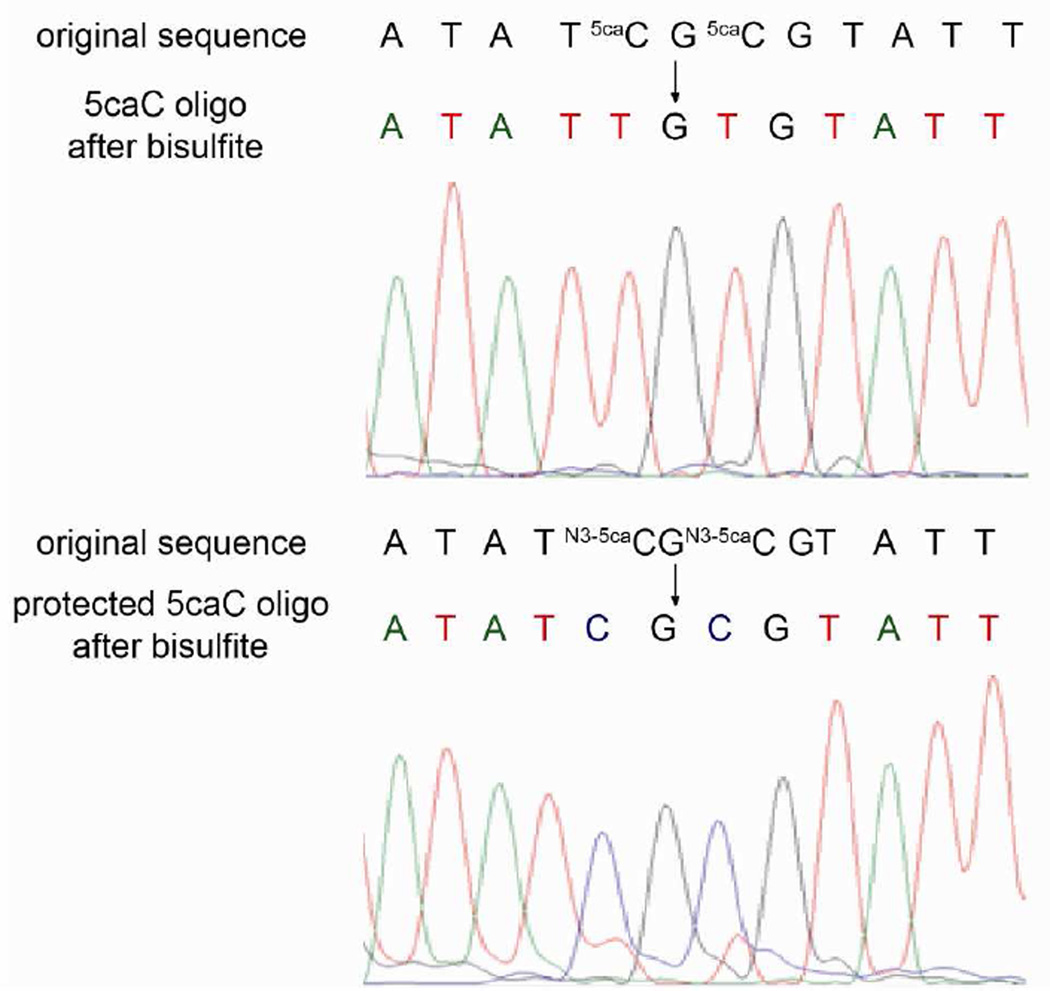
With this efficient labeling method in hand we tested if the protection of the carboxyl group could prevent deamination of 5caC under typical bisulfite treatment. It has been shown that under typical bisulfite conditions 5caC will be deaminated and read as T after subsequent PCR amplification and sequencing.7 This mechanism likely goes through a process of decarboxylation followed by deamination (a in Scheme 2).28 Protection of the carboxyl group as an amide bond could block the decarboxylation step and significantly slow down the bisulfite-mediated deamination (b in Scheme 2). In order to probe this mechanism, we performed preliminary studies on 5caC single nucleoside monitored by HPLC. We found that after bisulfite treatment, 5caC forms the same bisulfite adduct intermediate as normal cytosine, which will convert to the uridine bisulfite adduct, supporting decarboxylation as a potentially critical step (see Figure S7). To further confirm this observation, we labeled a synthetic 76mer 5caC-containing DNA with (4- aminomethyl)benzylazide and applied the labeled DNA to bisulfite treatment. The labeled DNA did not block DNA polymerase and can be successfully amplified with PCR under common conditions. No PCR amplification bias was detected after the EDC-mediated labeling of 5caC (Figure S8). After Sanger sequencing, the 5caC sites were mainly read as cytosine (C) instead of thymine (T) while the unlabeled 5caCs were still read as T (Figure 1). Thus, the 5caC can be “protected” from bisulfite-based deamination after the EDC-mediated labeling reaction. The different behaviors of 5caC before and after EDC labeling in bisulfite sequencing provide the single-base resolution detection of 5caC in DNA. In real biological applications, combined with high-throughput sequencing technology or colony picking, the modified cytosine can be detected at very low abundance through modified bisulfite sequencing as we have demonstrated recently.15,19 Together with enrichment-based approach,20 this chemical method has potential to gain base-resolution information of 5caC genome-wide.
Scheme 2. Proposed deamination mechanism of 5caC under bisulfite treatment.
Figure 1.
Sanger sequencing results of unlabeled 76 mer 5caC (upper panel) and EDC-labeled 5caC (bottom panel) after bisulfite treatment. The original sequence and the sequence after bisulfite were shown above the corresponding peak. In the bottom panel, the labeled DNA was not further purified before bisulfite treatment. The small background peak of T comes from unlabeled cytosine, which reflects the labeling efficiency.
In summary, we show here an efficient chemical selective labeling of 5caC in DNA catalyzed by EDC using xylene-based primary amine. The xylene structure is critical for highly efficient labeling. Importantly, we showed that the chemically labeled 5caC could survive from bisulfite-mediated deamination and be differentiated at single-base resolution using this new version of CAB-Seq specific for 5caC. This effective method, communicated here, can help the broad community interested in 5caC biology to detect this modified base in various biological samples.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institutes of Health HG006827 (C.H.), NS079625, and HD073162 (P.J.). We thank S. F. Reichard, MA for editing the manuscript.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Complete experimental procedures, mass spectrum data, qPCR data and HPLC data for reactions, spectral data for new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Klose RJ, Bird AP. Trends Biochem. Sci. 2006;31:89. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Bhutani N, Burns DM, Blau HM. Cell. 2011;146:866. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriaucionis S, Heintz N. Science. 2009;324:929. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324:930. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. Angew. Chem. Int. Ed. 2011;50:7008. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 6.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Science. 2011;333:1300. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Science. 2011;333:1303. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiti A, Drohat AC. J. Biol. Chem. 2011;286:35334. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Nat. Chem. Biol. 2012;8:328. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Nat. Biotechnol. 2011;29:68. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song CX, Sun Y, Dai Q, Lu XY, Yu M, Yang CG, He C. Chembiochem. 2011;12:1682. doi: 10.1002/cbic.201100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Nature. 2011;473:394. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raiber EA, Beraldi D, Ficz G, Burgess H, Branco MR, Murat P, Oxley D, Booth MJ, Reik W, Balasubramanian S. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Cell. 2012;149:1368. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, Klungland A. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Science. 2012;336:934. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 18.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. Nature. 2011;480:490. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 19.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, Gao J, Liu P, Li L, Xu GL, Jin P, He C. Cell. 2013;153:678. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Cell. 2013;153:692. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panchaud A, Hansson J, Affolter M, Bel Rhlid R, Piu S, Moreillon P, Kussmann M. Mol. Cell. Proteomics. 2008;7:800. doi: 10.1074/mcp.M700216-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, Kushiro K, Graham NA, Asthagiri AR. Proc. Natl. Acad. Sci. 2009;106:11149. doi: 10.1073/pnas.0812651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naue N, Fedorov R, Pich A, Manstein DJ, Curth U. Nucleic Acids Res. 2011;39:1398. doi: 10.1093/nar/gkq988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Tornoe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 26.Ning X, Guo J, Wolfert MA, Boons GJ. Angew. Chem. Int. Ed. 2008;47:2253. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiesser S, Hackner B, Pfaffeneder T, Muller M, Hagemeier C, Truss M, Carell T. Angew. Chem. Int. Ed. 2012;51:6516. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.