Abstract
Non-small cell lung cancer (NSCLC) cells harboring activating mutations of the epidermal growth factor receptor (EGFR) tend to display elevated activity of several survival signaling pathways. Surprisingly, these mutations also correlate with reduced phosphorylation of ERK and SHP2, a protein tyrosine phosphatase required for complete ERK activation downstream of most receptor tyrosine kinases. Since ERK activity influences cellular response to EGFR inhibition, altered SHP2 function could play a role in the striking response to gefitinib witnessed with EGFR mutation. Here, we demonstrate that impaired SHP2 phosphorylation correlates with diminished SHP2 function in NSCLC cells expressing mutant, versus wild-type, EGFR. In NSCLC cells expressing wild-type EGFR, SHP2 knockdown decreased ERK phosphorylation, basally and in response to gefitinib, and increased cellular sensitivity to gefitinib. In cells expressing EGFR mutants, these effects of SHP2 knockdown were less substantial, but expression of constitutively active SHP2 reduced cellular sensitivity to gefitinib. In cells expressing EGFR mutants, which do not undergo efficient ligand-mediated endocytosis, SHP2 was basally associated with GAB1 and EGFR, and SHP2′s presence in membrane fractions was dependent on EGFR activity. Whereas EGF promoted a more uniform intracellular distribution of initially centrally localized SHP2 in cells expressing wild-type EGFR, SHP2 was basally evenly distributed and did not redistribute in response to EGF in cells with EGFR mutation. Thus, EGFR mutation may promote association of a fraction of SHP2 at the plasma membrane with adapters which promote SHP2 activity. Consistent with this, SHP2 immunoprecipitated from cells with EGFR mutation was active, and EGF treatment did not change this activity. Overall, our data suggest that a fraction of SHP2 is sequestered at the plasma membrane in cells with EGFR mutation in a way that impedes SHP2′s ability to promote ERK activity and identify SHP2 as a potential target for co-inhibition with EGFR in NSCLC.
Keywords: oncogene addiction, extracellular signal-regulated kinase, gefitinib, endocytosis, GRB2-associated binder-1
Introduction
In non-small cell lung cancer (NSCLC), tumor response to the EGFR inhibitors gefitinib and erlotinib is generally limited to the 10-20% of NSCLCs carrying kinase-activating EGFR mutations.1-3 NSCLC cells harboring these mutations often display elevated phosphorylation of EGFR, AKT, signal transducer and activator of transcription 3/5 (STAT3/5), ERBB3, and MET.2,4-6 Recently, it was shown that these EGFR mutations also surprisingly result in impaired EGFR-mediated phosphorylation of both ERK, an important determinant of cell response to gefitinib, and the protein tyrosine phosphatase SHP2,7 which is required for complete ERK activation by most receptor tyrosine kinases.8 Thus, the striking responsiveness of tumors with EGFR mutation to EGFR inhibition may result from an imbalance in EGFR oncogenic signaling wherein activating mutations promote some signaling pathways while simultaneously impairing others.
Activation of receptor tyrosine kinases, including EGFR, results in SHP2 phosphorylation at Y542, which is required for normal SHP2-mediated ERK activation in response to many growth factors.9 Receptor activation and phosphorylation also results in SHP2 recruitment to receptors via direct binding or through adapters, which activates SHP2 through relief of auto-inhibitory intramolecular interactions.8 SHP2 is recruited to EGFR through binding to phosphorylated adapter proteins including GRB2-associated binder 1 (GAB1),10 whose association with EGFR is mediated by GRB2 upon EGFR phosphorylation at Y1068 and Y1086.11 Downstream of EGFR, SHP2 is primarily associated with promoting ERK activity by regulating RAS.12 SHP2-activating mutations have been identified in Noonan syndrome, juvenile myelomonocytic leukemia, and acute myelogenous leukemia.13,14 SHP2-activating mutations have also been found in lung cancer, although the consequences of these mutations are not fully understood.15
The aforementioned differences in SHP2 and ERK phosphorylation in NSCLC cells with EGFR mutation suggest SHP2 function may be perturbed in this setting. However, the role of SHP2 in NSCLC has not been thoroughly evaluated. In previous studies, HeLa cells expressing dominant-negative dynamin,16 a GTPase required for clathrin-mediated EGFR endocytosis, displayed diminished EGF-mediated phosphorylation of SHP2 and ERK.7 Since the EGFR-activating mutations observed in NSCLC result in impaired EGFR endocytosis,7,17 differential EGFR trafficking may explain the defects in SHP2 and ERK phosphorylation in NSCLC cells expressing EGFR mutants. SHP2 localization could also be altered in the context of EGFR mutation via association with internalization-impaired EGFR.
In this study, we find diminished SHP2 function in NSCLC cells expressing mutant versus wild-type EGFR. In cells expressing wild-type EGFR, SHP2 knockdown reduced ERK phosphorylation and increased cellular sensitivity to gefitinib. In cells expressing EGFR mutants, the effects of SHP2 knockdown were less pronounced, but expression of constitutively active SHP2 reduced cellular sensitivity to gefitinib. In cells expressing EGFR mutants, SHP2 was basally associated with GAB1 and EGFR, and the presence of SHP2 in membrane fractions was dependent on EGFR activity. In cells expressing wild-type EGFR, EGF promoted redistribution of initially centrally localized SHP2, but SHP2 was basally evenly distributed and did not redistribute in response to EGF in cells expressing EGFR mutants. SHP2 was catalytically active in cells expressing EGFR mutants, consistent with the finding that SHP2 association with adapters was not impaired, but rather basally elevated, in these cells. Overall, our data suggest that a fraction of SHP2 is sequestered at the plasma membrane in cells with EGFR mutation in a way that interferes with SHP2-mediated ERK activation and promotes cellular sensitivity to EGFR inhibition.
Results
Effects of SHP2 knockdown on ERK phosphorylation
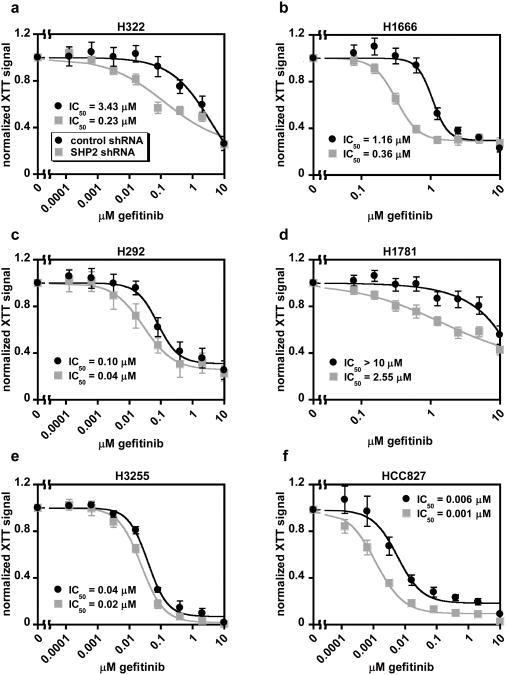
To assess the signaling role of SHP2 in NSCLC cells, we examined the effects of SHP2 knockdown on ERK phosphorylation in response to gefitinib in a panel of cell lines (Figure 1 and Supplementary Figure S1). In H322 and H292 cells (EGFRWT), SHP2 knockdown reduced ERK phosphorylation in untreated cells by more than 90%. In H1666 and H1781 cells (EGFRWT), SHP2 knockdown resulted in notable, but more modest, reductions in baseline ERK phosphorylation of ∼60% and 20%, respectively, as well as reductions in the gefitinib IC50 values for ERK phosphorylation. In H3255 cells (EGFRL858R), which display impaired EGF-mediated SHP2 phosphorylation relative to cells expressing wild-type EGFR (7; Supplementary Figure S2), SHP2 knockdown had no substantial effect on ERK phosphorylation at any concentration of gefitinib. In HCC827 cells (EGFRdelE746-A750), which also display impaired SHP2 phosphorylation (7; Supplementary Figure S2), there was a reduction in baseline ERK phosphorylation with SHP2 knockdown compared to controls, but the effect was less substantial than that observed in cells expressing wild-type EGFR, other than H1781. There was also no enhancement in gefitinib's ability to inhibit ERK phosphorylation in SHP2-depleted HCC827 cells relative to controls. Relative to ERK phosphorylation, SHP2 knockdown produced smaller changes in AKT and STAT3 phosphorylation in H1666 and H292 cells (Supplementary Figure S3).
Figure 1.
SHP2 knockdown reduces ERK phosphorylation more in NSCLC cells expressing wild-type EGFR than in those expressing EGFR mutants. H322 (a), H1666 (b), H292 (c), H1781 (d), H3255 (e), and HCC827 (f) cells expressing SHP2-targeting or non-targeting control shRNA were treated with 0-10 μM gefitinib for 48 hrs, and lysates were analyzed by Western blotting with antibodies against phosphorylated and total ERK. Densitometry data are represented as mean ± s.e.m. (n = 3); * denotes p < 0.05 relative to controls.
Effects of SHP2 knockdown on cellular response to gefitinib
In H322 and H1666 cells, SHP2 knockdown reduced gefitinib IC50 values for cell proliferation by 15- and 3-fold, respectively (Figure 2). In H1781 cells, a gefitinib-resistant cell line, the IC50 was reduced from > 10 μM to 2.55 μM. SHP2-depleted H292 cells were only modestly sensitized to gefitinib but were significantly growth inhibited in the absence of gefitinib (Supplementary Figure S4a). Thus, gefitinib may have been unable to enhance the already striking effects of SHP2 knockdown on H292 proliferation. In contrast, H3255 cells showed virtually no effect of SHP2 knockdown on sensitivity to gefitinib (Figure 2e). HCC827 cells displayed a small shift in sensitivity to gefitinib in response to SHP2 knockdown (Figure 2f), but we measured no proliferative effect in the absence of gefitinib (Supplementary Figure S4b).
Figure 2.
Knockdown of SHP2 enhances cellular sensitivity to gefitinib in subsets of NSCLC cells. H322 (a), H1666 (b), H292 (c), H1781 (d), H3255 (e), and HCC827 (f) cells expressing SHP2-targeting or non-targeting control shRNA were treated with 0-10 μM gefitinib for four days, and cell proliferation was measured by XTT assay. Normalized XTT signal values (y-axis) were computed at a given gefitinib concentration by dividing absorbances by those measured for cells treated with DMSO as a control. Data are represented as mean ± s.e.m. for three experiments with three replicate wells in each experiment (n = 3).
To ensure the measured effects were specific for SHP2, we knocked down SHP2 in a representative cell line expressing wild-type EGFR using an independent hairpin targeting the 3′ untranslated region of SHP2 and reconstituted cells with SHP2WT or SHP2Y542F (Figure 3). As before, SHP2-depleted cells displayed impaired ERK phosphorylation and enhanced sensitivity to gefitinib. These effects were partially rescued by reconstitution with SHP2WT or SHP2Y542F.
Figure 3.
Observed effects of SHP2 knockdown on ERK phosphorylation and gefitinib response are specific to SHP2. H1666 cells expressing SHP2-targeting shRNA or an empty pSicoR vector were transduced with SHP2WT, SHP2Y542F, or an empty pBabe vector. (a) Cells were treated with 0-10 μM gefitinib for 48 hrs, and lysates were analyzed by Western blotting with antibodies against indicated proteins. Images are representative of three sets of biological replicates. Densitometry data for ERK are represented as mean ± s.e.m. (n = 3); * denotes p < 0.05 relative to pSicoR and pBabe controls. (b) Cells were treated with 0-10 μM gefitinib for four days, and cell proliferation was measured by XTT assay. Normalized XTT signal values (y-axis) were computed at a given gefitinib concentration by dividing absorbances by those measured for cells treated with DMSO as a control. Data points are represented as mean ± s.e.m. for three replicate wells from at least three experiments.
SHP2 association with GAB1 and EGFR and subcellular compartmentalization
To investigate the mechanism underlying apparent differential SHP2 function in cells with or without EGFR mutation, we examined SHP2 association with GAB1 and EGFR. In H3255 and HCC827 cells, SHP2 was basally associated with GAB1 and phosphorylated EGFR to a significantly greater degree than in either H322 or H1666 cells (Figure 4a). In H3255 cells, these associations were diminished by gefitinib (Figure 4b). EGF enhanced SHP2 association with GAB1 and EGFR in all cell lines, but the fold increases in association were generally smaller in H3255 and HCC827 cells (Figure 4a). Since EGFR mutants fail to undergo efficient EGF-mediated endocytosis,7,17 we interpreted these findings as indicating that a fraction of SHP2 was sequestered at the plasma membrane in cells with EGFR mutation. To further substantiate this, we analyzed SHP2′s distribution in a subset of these cells by subcellular fractionation. In H1666 and H3255 cells, the majority of SHP2 was cytosolic. Only in H3255 cells, however, did gefitinib reduce SHP2 levels in crude membrane fractions, suggesting that SHP2 was membrane-localized in an EGFR-dependent manner (Figure 4c). The EGFR activity-independent presence of SHP2 in H1666 membrane fractions could be explained by SHP2 localization to membrane compartments which settle with plasma membrane in the crude membrane fraction generated by our protocol. This possibility is suggested by previous studies.18
Figure 4.
GAB1 and EGFR are basally associated with SHP2 in NSCLC cells expressing EGFR mutants and are induced to associate with SHP2 by EGF in NSCLC cells expressing wild-type EGFR. Serum-starved cells treated with or without 10 ng/mL EGF for 5 min (a), or serum-starved H3255 cells treated with or without 5 μM gefitinib for 15 min (b), were lysed. Lysates were immunoprecipitated with either an SHP2 or control antibody, and immunoprecipitates were analyzed by Western blotting using antibodies against the indicated proteins. Images are representative of three sets of biological replicates. Densitometry data in (a) are represented as mean ± s.e.m. (n = 3); * denotes p < 0.05 relative to wild-type EGFR-expressing cells not stimulated with EGF. (c) Subcellular fractions were prepared from H1666 and H3255 cells treated with or without gefitinib, as described in Materials and Methods, and equivalent relative amounts of the fractions for both cell lines were analyzed by Western blotting using antibodies against indicated proteins. To improve signals, membrane fractions were 10× more concentrated than cytosolic fractions in terms of the relative amount of total lysate loaded. Images are representative of three sets of biological replicates. Blots were quantified to determine the relative difference in membrane-localized SHP2 in H1666 versus H3255 cells. For each condition, SHP2 signal from the membrane fraction was divided by the SHP2 signal from the cytosol fraction to determine a membrane/cytosol SHP2 signal. Data are represented as mean ± s.e.m. (n = 4); * denotes p < 0.05 relative to untreated cells.
Intracellular distribution of SHP2
We further examined EGF's ability to alter the intracellular distribution of SHP2 in H1666 and H3255 cells by immunofluorescence. In H1666 cells, the distribution shifted from one where SHP2 was concentrated around the cell center to one where some SHP2 moved toward the cell periphery and SHP2 was distributed more uniformly (Figure 5). Similar changes were noted by confocal microscopy, including movement of SHP2 to membrane ruffles (Supplementary Figure S5). EGF also caused the formation of EGFR- and RAB5-positive endocytic vesicles in H1666 cells (Supplementary Figure S6). In H3255 cells, SHP2 was basally uniformly distributed. EGF did not alter this distribution or generate endocytic vesicles (Figure 5; Supplementary Figures S5 and S6), consistent with previous reports of impaired EGFR internalization in these cells.
Figure 5.
Intracellular redistribution of SHP2 in response to EGF is observed in H1666 cells, but not in H3255 cells. (a) Serum-starved H1666 and H3225 cells were treated with 10 ng/mL EGF for up to 15 min, fixed, and stained with Hoechst (blue) and antibodies against EGFR (red) and SHP2 (green). Images are representative of three biological replicates. (b) As described in Materials and Methods, intracellular SHP2 pixel intensities were quantified as a function of normalized radial distance from cell centers (x = 0) to the cell periphery (x = 1). Three images for each condition were used for this analysis, in which all cells entirely contained with the image were analyzed. Data are represented as mean (solid line) ± s.e.m. (shaded area; n ≥ 7 cells).
Role of GAB1 in SHP2 localization and EGF-mediated effects
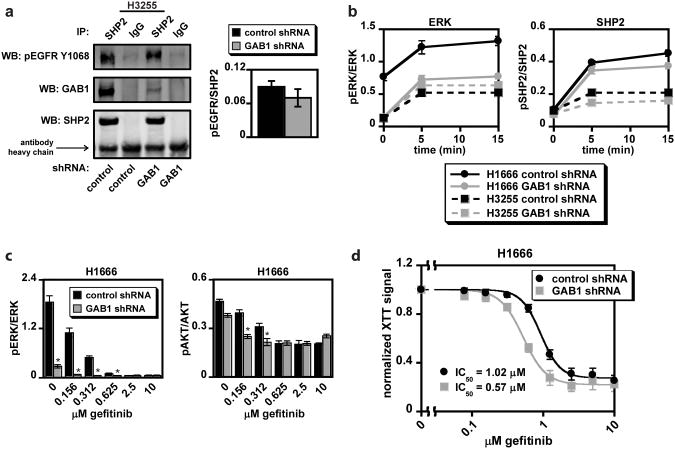
GAB1 knockdown (Supplementary Figure S7a) did not alter basal association of SHP2 with Y1068-phosphorylated EGFR in H3255 cells (Figure 6a), nor did it alter SHP2′s intracellular distribution in either H1666 or H3255 cells (Supplementary Figure S7b), suggesting that recruitment of SHP2 to EGFR and the cell periphery can be accomplished independent of GAB1 binding, potentially through GAB2. GAB1 knockdown did, however, diminish EGF-mediated ERK and SHP2 phosphorylation, reduce ERK and AKT phosphorylation in response to gefitinib, and increase cellular sensitivity to gefitinib in H1666 cells (Figures 6b-d and Supplementary Figure S7c-d). Although GAB1-depleted H3255 cells displayed a modest reduction in SHP2 phosphorylation, there was no effect on ERK phosphorylation (Figure 6b), suggesting that the mechanism by which SHP2 function is impaired in these cells may simultaneously perturb GAB1 function.
Figure 6.
GAB1 knockdown alters EGF-mediated ERK phosphorylation and response to gefitinib in H1666 cells. In H1666 and H3255 cells expressing GAB1-targeting or non-targeting control shRNA: (a) Serum-starved H3255 cells were lysed, and lysates were immunoprecipitated with either an SHP2 or control antibody. Immunoprecipitates were analyzed by Western blotting using antibodies against indicated proteins. Images are representative of three sets of biological replicates, and densitometry data are represented as mean ± s.e.m. (n = 3). (b) Serum-starved cells were treated with or without 10 ng/mL EGF for up to 15 min, and lysates were analyzed by Western blotting using antibodies against phosphorylated and total ERK and SHP2. Densitometry data are represented as mean ± s.e.m. (n = 3). (c) H1666 cells were treated with 0-10 μM gefitinib for 48 hrs, and lysates were analyzed by Western blotting with antibodies against phosphorylated and total ERK and AKT. Densitometry data are represented as mean ± s.e.m. (n = 3); * denotes p < 0.05 relative to controls. (d) H1666 cells were treated with 0-10 μM gefitinib for four days, and cell proliferation was measured by XTT assay. Normalized XTT signal (y-axis) represents the normalization of values obtained from cells at a given gefitinib concentration by dividing these values by those obtained from cells treated with DMSO as a control. Data are represented as mean ± s.e.m. for three experiments with three replicate wells in each experiment (n = 3).
SHP2 activity
SHP2 knockdown in H1666 and H3255 cells resulted in fractional reductions of measured phosphatase activities comparable to the reductions in immunoprecipitated SHP2 levels (Figure 7a), indicating that SHP2 was active in both cell lines. In response to EGF, SHP2 activity increased in H1666 cells, with p = 0.08 for this comparison (Figure 7b). EGF elicited no change in SHP2 activity in H3255 cells (p = 0.50). Note that comparison of activity between cell lines is not straightforward because more SHP2 was immunoprecipitated from H3255 lysates, lysates were not controlled for cell numbers due to proliferation differences, and only a fraction of SHP2 was adapter-bound in each cell line. Thus, the lower apparent normalized SHP2 activity in H3255 versus H1666 cells may not necessarily reflect a lower total SHP2 activity level on a per cell basis.
Figure 7.
SHP2 is active in H1666 and H3255 cells, and EGF increases SHP2 activity in H1666 cells. H1666 and H3255 cells expressing SHP2-targeting or non-targeting control shRNA (a), and serum-starved H1666 and H3255 cells treated with or without 10 ng/mL EGF for 5 min (b) were lysed, and SHP2 was immunoprecipitated from whole cell lysates. Half of each immunoprecipitate was used to determine phosphatase activity, as described in Materials and Methods, while the remainder was used to determine SHP2 levels by immunoblot. Data are represented as mean ± s.e.m. (n = 3); AU, arbitrary units. Blot images in (a) are representative of three sets of biological replicates. Values reported in (b) were determined by dividing AU values from the phosphatase activity assay by the quantified SHP2 levels obtained from immunoblots.
Effects of SHP2 mutation
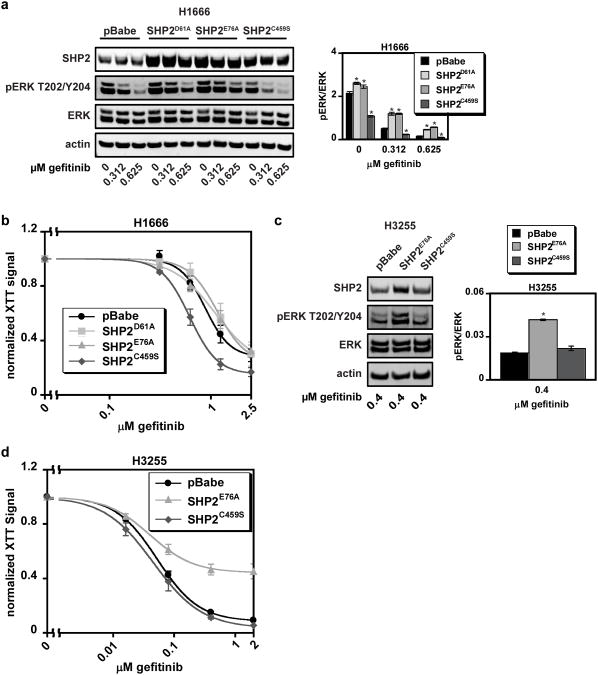
In H1666 cells, expression of constitutively active SHP2D61A or SHP2E76A mitigated gefitinib-mediated reductions in ERK phosphorylation, and expression of catalytically-inactive SHP2C459S reduced ERK phosphorylation basally and in response to gefitinib (Figure 8a). Despite increased ERK phosphorylation in H1666 cells expressing SHP2D61A or SHP2E76A, there was no change in gefitinib sensitivity in these cells (Figure 8b), suggesting that the parental cell line's capacity to activate ERK was at a threshold level for maintaining cell survival. However, H1666 cells expressing SHP2C459S were more responsive to gefitinib, mirroring the effects of SHP2 knockdown. In H3255 cells, expression of SHP2E76A augmented ERK phosphorylation in the presence of gefitinib and substantially decreased cellular sensitivity to gefitinib (Figure 8c and d). As expected, SHP2C459S expression had little effect on ERK phosphorylation or gefitinib response in H3255s.
Figure 8.
Ectopic expression of SHP2 mutants alters cellular response to gefitinib in H1666 and H3255 cells. The following experiments were carried out with H1666 and H3255 cells transduced with SHP2D61A (H1666 only), SHP2E76A, SHP2C459S, or an empty pBabe vector: (a and c) H1666 and H3255 cells were treated with the indicated concentrations of gefitinib for 48 hrs, and lysates were analyzed by Western blotting with antibodies against indicated proteins. Images for (a) and (c) are representative of three sets of biological replicates. Densitometry data for ERK are represented as mean ± s.e.m. (n = 3); * denotes p < 0.05 relative to controls. (b and d) H1666 and H3255 cells were treated with up to 2.5 μM gefitinib for four days, and cellular proliferation was measured by XTT assay. Normalized XTT signal values (y-axis) were computed at a given gefitinib concentration by dividing absorbances by those measured for cells treated with DMSO as a control. Data are represented as mean ± s.e.m. for three experiments with three replicate wells in each experiment (n = 3).
Importance of SHP2-mediated effects downstream of MET
Since SHP2-GAB1 association is required for sustained ERK activation downstream of MET,19 we hypothesized that SHP2 could play a role in hepatocyte growth factor (HGF)-mediated resistance to EGFR inhibition in NSCLC cells by maintaining GAB1-mediated signaling in the presence of gefitinib.20,21 To explore this idea, we treated SHP2-depleted H1666 and HCC827 cells with gefitinib in the presence or absence of HGF. Although HGF sustained phosphorylated GAB1 Y627, a SHP2 binding site, in the presence of gefitinib, SHP2 knockdown did not affect HGF-mediated rescue of ERK phosphorylation in either cell line (Supplementary Figure S8).
Discussion
Magnitudes of SHP2-mediated effects
Other than in H292 cells, where SHP2 knockdown substantially inhibited cell growth, SHP2 depletion in cells expressing wild-type EGFR increased sensitivity to gefitinib by 3- to 15-fold, as measured by XTT assay. Gefitinib IC50 values for cells expressing wild-type or mutant EGFR typically differ by a factor of ten or more.22 Thus, SHP2 depletion in cells expressing wild-type EGFR generally produced an effect consistent with differences on the lower end of what is observed among NSCLC cells with or without EGFR mutation. Of course, other factors contribute to differences in NSCLC cellular sensitivity to gefitinib, including differential regulation of phosphatidylinositol 3-kinase (PI3K)/AKT and STAT3/5.2,4,5 Our study appears to be the first, however, to identify a mechanism wherein a survival signaling pathway is impaired by EGFR-activating mutations in a way which impacts cellular response to EGFR inhibition.
We also found that SHP2 depletion most strongly impaired ERK phosphorylation in cells expressing wild-type versus mutant EGFR. However, H1781 (EGFRWT) cells were an outlier in terms of the relatively modest effect of SHP2 knockdown on ERK phosphorylation. Despite this, there was a substantial effect of SHP2 knockdown on cellular response to gefitinib. H1781 cells express a constitutively active HER2 mutant (VC insertion at G776) and are dependent on HER2 for ERK and AKT phosphorylation.23 Since HER2 can sequester EGFR at the plasma membrane,24,25 the possibility exists for SHP2 to be sequestered with EGFR in these cells as well. Indeed, this appeared to occur (Supplementary Figure S9). However, as SHP2 promotes RAS activity downstream of HER2,26 HER2-mediated SHP2 function may contribute to the modest effect of SHP2 knockdown on ERK phosphorylation in these cells. Further studies are needed to parse the effects of SHP2 downstream of EGFR and HER2 in these cells.
Differential roles of SHP2 and GAB1
A previous study demonstrated impaired SHP2 phosphorylation in NSCLC cells expressing EGFR mutants,7 but the phenotypic implications of SHP2 phosphorylation status were not directly evaluated. Our finding that reconstitution of SHP2Y542F in SHP2-depleted H1666 cells rescued ERK phosphorylation as efficiently as SHP2WT suggests that SHP2 Y542 phosphorylation is dispensable for SHP2-mediated activation of ERK, consistent with previous findings regarding EGF-mediated ERK activation in 3T3 fibroblasts.9 Thus, impaired SHP2 phosphorylation with EGFR mutation may not be the cause, but rather a result, of a mechanism whereby SHP2 function (but not activity) is diminished by SHP2 sequestration. We also note that despite the lack of an effect of SHP2 knockdown in H3255 cells (EGFRL858R), there were small effects of SHP2 knockdown in HCC827 cells (EGFRE746_A750del). This difference between H3255 and HCC827 cells could reflect a functional difference between the two EGFR mutants or a differential role of receptors such as MET, which is basally phosphorylated to a higher degree in HCC827 cells.6
Based on our studies of GAB1 knockdown, GAB1 function may also be perturbed by EGFR mutation. GAB1 also appears to be an important determinant of cellular response to gefitinib in an NSCLC cell line expressing wild-type EGFR. This could be due to the function of GAB1 upstream of SHP2 in regulating ERK phosphorylation, the function of GAB1 in promoting AKT phosphorylation by recruiting PI3K, or both. Additional work is needed to clarify the role and regulation of GAB1.
Mechanistic insights
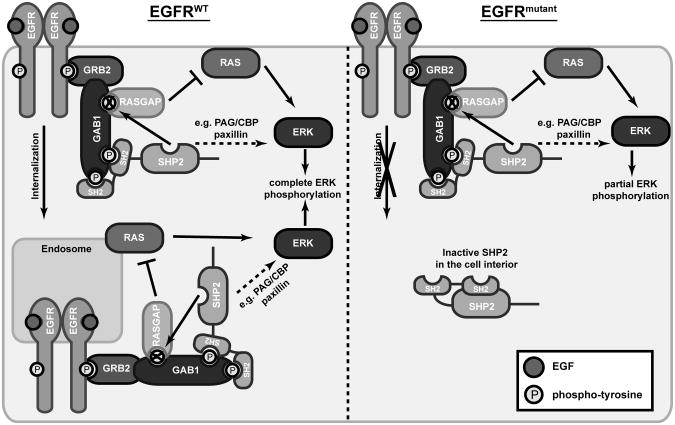
Our data suggest that EGFR mutation promotes constitutive binding of a fraction of SHP2 to EGFR through GAB1 and other adapters. Since adapter engagement of SHP2′s SH2 domains promotes SHP2 activity, it is perhaps not surprising that SHP2 was biochemically active in cells with EGFR mutation. Given these findings, our immunofluorescence microscopy and subcellular fractionation results, and previous findings that EGFR mutants are endocytosis-impaired, EGFR mutation appears to result in sequestration of at least some SHP2 at the plasma membrane in a state where it should be biochemically active. The finding that not all SHP2 was sequestered at the plasma membrane in cells with EGFR mutation (as observed by immunofluorescence and fractionation) could be a stoichiometric effect. Indeed, in A431 cells, with over 3 × 106 EGFR/cell, a more complete redistribution of SHP2 to the cell periphery was observed in response to EGF than we observed.27 Moreover, in EGFR mutant cells, the SHP2 which is not sequestered, and less likely to be adapter-bound, should be at a lower activity and therefore less functionally relevant. This model is consistent with our findings that in EGFR mutant cells, where only a fraction of SHP2 was sequestered, SHP2 depletion had relatively small effects, but expression of constitutively active SHP2 rescued ERK phosphorylation and sensitivity to gefitinib. The reason why SHP2 sequestration may impede SHP2′s ability to promote ERK phosphorylation could be related to previous findings that normal EGFR endocytosis is required for complete ERK activation in some cellular contexts.16 This coupling between endocytosis and ERK could involve a role for SHP2 localization wherein normal trafficking of SHP2-containing complexes promotes ERK activity by allowing complex access to substrates in the cell interior (Figure 9).
Figure 9.
Sequestration of SHP2 at the plasma membrane may enhance gefitinib response in cells expressing EGFR mutants by reducing ERK activity. In the proposed mechanism, EGFR activation and phosphorylation leads to recruitment of GRB2, which is constitutively bound with GAB1 via an SH3 domain-mediated interaction. Phosphorylated GAB1 (and possibly other adapters) recruits SHP2, whose activity leads to ERK activation through dephosphorylation of a RASGAP binding site on GAB1 (as shown) or through other mechanisms (as depicted by dotted arrow), such as dephosphorylation of CSK binding sites on PAG/CBP and paxillin. The function of SHP2 in this complex at both the plasma membrane and the cell interior leads to complete ERK phosphorylation. When SHP2 is sequestered at the plasma membrane in complex with internalization-impaired mutant EGFR (e.g. EGFRL858R), ERK phosphorylation is impaired. Due in part to impaired ERK activity resulting from sequestration of SHP2, cells with EGFR mutants display increased sensitivity to EGFR inhibition.
Therapeutic implications
SHP2 knockdown in an NSCLC cell line was previously shown to slow xenograft growth in mice.28 In addition, SHP2-activating mutations have been found in solid tumors, including NSCLC.15 As far as we are aware, however, the effects of SHP2 expression and mutation on cellular response to EGFR inhibition have not previously been evaluated. Our finding that SHP2 knockdown in NSCLC cells expressing wild-type EGFR enhanced cellular response to gefitinib suggests that combined inhibition of EGFR and SHP2 may improve response in tumors that are unresponsive to EGFR inhibition alone. The largest effects of SHP2 knockdown on enhancing response to EGFR inhibition in cells with wild-type EGFR tended to occur at or below 1 μM gefitinib, the maximum achievable plasma concentration at a clinically relevant dose.29 Thus, it is conceivable that such a co-inhibition strategy could have clinical impact. Our finding that expression of constitutively active SHP2 mutants mitigated the effects of gefitinib on ERK phosphorylation in H1666 and H3255 cells suggests that SHP2 activity can maintain the activity of ERK in the presence of EGFR inhibitors. Although we noted no major effect of ectopic expression of these mutants on sensitivity to gefitinib in H1666 cells, H3255 cells expressing SHP2E76A displayed decreased sensitivity to gefitinib. It would therefore be interesting to explore the implications for drug resistance in cells with SHP2 mutation.
Summary
Our findings point to SHP2 as a potential target to be co-inhibited with EGFR in the treatment of NSCLC cells expressing wild-type EGFR. Expanding these studies to an in vivo model would be helpful in determining if a clinical benefit for combined SHP2/EGFR inhibition exists, although such studies would be hampered by the present lack of effective and specific SHP2 inhibitors. Our findings also highlight the non-intuitive possibility for activating mutations of receptors such as EGFR to impair the function of specific signaling pathways in ways which promote cellular response to receptor-targeting therapeutics.
Materials and Methods
Cell culture
H1666 and H3255 cells were maintained in ACL4.7 All others were maintained in RPMI 1640 supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 1 mM L-glutamine, and 10% fetal bovine serum (FBS). H1666 cells were obtained from the American Type Culture Collection. H3255, H322, and H1781 cells were provided by Dr. Pasi Jänne (Dana-Farber Cancer Institute). H292 and HCC827 cells were provided by Dr. Eric Haura (Moffitt Cancer Center). For serum starvation, cells were switched to media containing 0.1% FBS for 16-18 hrs.
Cell proliferation assay
Proliferative response to gefitinib was measured by XTT assay according to manufacturer's specifications (Roche, Indianapolis, IN, USA). Cells seeded in 96-well plates were treated with up to 10 μM gefitinib for 4 days. Subsequently, fresh media and XTT reagent were added to wells, and plates were incubated for 2-4 hrs at 37°C to maximize signal-to-background. Wells containing only media were used for background correction. Each experiment was performed at least three times with each condition plated in three replicate wells on each day.
shRNA and expression constructs
Sequences encoding short hairpins targeting human SHP2 and GAB1 were cloned in the pSicoR vector (Tyler Jacks, MIT; 30). The SHP2 shRNA targeted nucleotides 1780-1798 of SHP2 mRNA (GGACGTTCATTGTGATTGA) or, for reconstitution experiments, 5890-5908 (GTATTGTACCAGAGTATTA). The GAB1 shRNA targeted nucleotides 987-1005 of GAB1 mRNA (gaaacagactgcaatgata). Lentivirus was produced by calcium phosphate-mediated transfection of 293FT cells (Invitrogen, Carlsbad, CA, USA) with vector and the packaging plasmids pCMV-VSVG, pMDL-gp-RRE, and pRSV-Rev (Marilyn Farquhar, UCSD). Virus was harvested 48 and 72 hrs post-transfection and used to infect target cells, which were selected in puromycin.
SHP2 cDNAs encoding wild-type, D61A, E76A, Y542F, and C459S SHP2 (Ben Neel, Ontario Cancer Institute) were inserted at the EcoRI site of the pBabe vector. Retrovirus was produced by calcium phosphate-mediated transfection of amphotropic Phoenix cells (Gary Nolan, Stanford University) with vector. Virus was harvested 24, 30, and 48 hrs post-transfection and used to infect target cells, which were selected in puromycin or hygromycin.
Constructs were validated by sequencing. SHP2 and GAB1 knockdowns were validated by Western blot and qPCR, respectively.
Immunoblotting
Cell lysates were prepared using cell extraction buffer (Invitrogen; #FNN0011) supplemented with 1 mM PMSF, additional protease inhibitors (Sigma, St. Louis, MO, USA), and phosphatase inhibitors (Sigma). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes, which were blocked in Odyssey Blocking Buffer (OBB; LI-COR, Lincoln, NE, USA) and stripped with 0.2 M NaOH as needed. Images were obtained using a LI-COR Odyssey Infrared Imaging System.
Immunoprecipitation
Cell lysates were prepared with immunoprecipitation lysis buffer (Cell Signaling Technology, Danvers, MA, USA; #9803) supplemented with 1 mM PMSF, additional protease inhibitors, and phosphatase inhibitors. 500 μg of total protein was precleared with Dynabeads (Invitrogen) for 4 hrs and subsequently incubated with Dynabeads conjugated to SHP2 or control antibody at 4°C overnight. Beads were washed with lysis buffer, re-suspended in LDS sample buffer (Invitrogen), and boiled before SDS-PAGE.
SHP2 activity assay
500 μg of total protein from cell lysates was incubated overnight with agarose beads conjugated to an SHP2 antibody. Beads were washed with lysis buffer and split into two equal fractions. One fraction was reserved for immunoblotting. Beads from the other fraction were washed with assay buffer (Millipore, Billerica, MA, USA; #20-180) and resuspended in assay buffer containing 100 μM 6,8-difluoro-4-methylumbelliferyl phosphate (Invitrogen). The reaction was performed at 37°C for 30 min with occasional mixing, and reaction product fluorescence was measured at excitation and emission wavelengths of 360 nm and 460 nm, respectively. Linearity of signal with respect to time and protein concentration was validated for both cell lines.
Immunofluorescence
Serum starved cells on coverslips were treated with EGF, fixed in 4% paraformaldehyde in PBS for 20 min, and permeabilized with 0.25% Triton X-100 for 5 min. Coverslips were incubated with primary antibodies diluted in OBB (EGFR, SHP2) or 1% BSA/0.3% Triton X-100 in PBS (RAB5) for 3 hrs at 37°C. Coverslips were washed with 0.1% Tween-20 in PBS and incubated with Alexa Fluor 488- and 594-conjugated secondary antibodies and Hoechst (Invitrogen) in the same diluents used for primary antibodies for 1 hr at 37°C. Coverslips were washed again, mounted on glass slides, and treated with Prolong Gold antifade (Invitrogen). Specificity of the SHP2 antibody was confirmed by comparison with SHP2 knockdown cells. Epifluorescence images were obtained with a Zeiss Axiovert 40 CFL microscope (100× objective). Confocal images were obtained with a Nikon Eclipse TE-300 microscope (60× objective).
To analyze individual cells from these images, MATLAB was used to determine pixel intensities as a function of distance from the cell center. This was done by outlining individual cells, locating cell centers, and generating lines from the center to the cell periphery in all angular directions, along which pixel intensities were quantified. Data were averaged and normalized to obtain a vector of intensities versus normalized distances from the cell center.
Subcellular fractionation
Serum starved cells were treated with 0 or 5 μM gefitinib, washed, and collected in hypotonic buffer (10 mM Tris-HCl, pH 7.4, 1 mM MgCl2, 1 mM EDTA) supplemented with 1 mM PMSF, additional protease inhibitors, and phosphatase inhibitors. Crude lysates were generated with a Dounce homogenizer and centrifuged at 3000 and 9300 rpm, for 5 min at each speed, to remove nuclei and mitochondria, respectively. Cleared lysates were centrifuged at 100 000 g for 60 min to separate membrane and cytosol fractions. Membrane pellets were washed with PBS, resuspended in hypotonic buffer, and centrifuged again at 100 000 g. After additional washes, membrane pellets were resuspended in immunoprecipitation lysis buffer to solubilize proteins before SDS-PAGE.
Quantitative polymerase chain reaction (qPCR)
Cellular RNA was isolated using an RNEasy kit (Qiagen, Valencia, CA, USA), and cDNA was transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosciences, Foster City, CA, USA). Using the cDNA as a template, qPCR was performed with previously developed primers for GAB1 mRNA using SYBR Green Master Mix (Applied Biosciences).31 Reactions were monitored on a Model 7300 Real Time PCR System (Applied Biosciences). RNA polymerase II mRNA was quantified as a normalization control using 5′-GCACCACGTCCAATGACAT-3′ as the forward primer and 5′-GTGCGGCTGCTTCCATAA-3′ as the reverse primer.
Antibodies and other reagents
EGFR (immunoblotting; #2232), pAKT S473 (#9271), AKT (#9272), pERK T202/Y204 (#4377), ERK (#4695), RAB5 (#3547), pSTAT3 Y705 (#9138), and pGAB1 Y627 (#3233) antibodies were from Cell Signaling Technology. SHP2 (sc-280) and EGFR (immunofluorescence; sc-81449) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Actin (MAB 1501) and GAB1 (#06-579) antibodies were from Millipore. pEGFR Y1068 (#1727) and pSHP2 Y542 (#2184) antibodies were from Epitomics (Burlingame, CA, USA), unless otherwise noted. Infrared dye- and Alexa Fluor-conjugated secondary antibodies were from Rockland Immunochemicals (Gilbertsville, PA, USA) and Invitrogen, respectively.
Gefitinib (LC Laboratories, Woburn, MA, USA) was diluted in DMSO. Recombinant human EGF was from Peprotech (Rocky Hill, NJ, USA). Recombinant human HGF (R&D Systems, Minneapolis, MN, USA) was generously provided by Dr. Anil Rustgi (University of Pennsylvania).
Statistics
Statistical analyses were performed using a paired two-tailed student's t-test.
IC50 calculations
Gefitinib IC50 values were determined by fitting a four parameter logistic function to normalized data.
Supplementary Material
Acknowledgments
C.M.F. was supported in part by the University of Pennsylvania Cell and Molecular Biology Training Grant (T32 GM-07229), Training Program in Cancer Pharmacology (R25 CA101871-07), and a fellowship from the Ashton Foundation. A.M.R. received support from the University of Pennsylvania Institute for Regenerative Medicine. This project was funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. This work was also supported in part by laboratory startup funds from the University of Pennsylvania. We thank Dr. Ben Neel, Dr. Eric Haura, Dr. Pasi Jänne, Dr. Tyler Jacks, Dr. Marilyn Farquhar, Dr. Gary Nolan, Dr. Anil Rustgi, and Dr. Susan Margulies for generously providing reagents and instrumentation. We also thank Ms. Gladys Gray Lawrence, Dr. Ranganath Parthasarathy, Mr. Calixte Monast, and Ms. Alice Macdonald Walsh for technical assistance.
CMF was supported by training grant T32 GM-07229, training program R25 CA101871-07, and an Ashton Fellowship. AMR received support from the University of Pennsylvania Institute for Regenerative Medicine. This work was also supported by laboratory startup funds from the University of Pennsylvania.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
Supplementary Information: Supplementary information is available at Oncogene's website.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–7. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–46. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Akca H, Tani M, Hishida T, Matsumoto S, Yokota J. Activation of the AKT and STAT3 pathways and prolonged survival by a mutant EGFR in human lung cancer cells. Lung Cancer. 2006;54:25–33. doi: 10.1016/j.lungcan.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–93. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–7. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazzara MJ, Lane K, Chan R, Jasper PJ, Yaffe MB, Sorger PK, et al. Impaired SHP2-mediated extracellular signal-regulated kinase activation contributes to gefitinib sensitivity of lung cancer cells with epidermal growth factor receptor-activating mutations. Cancer Res. 2010;70:3843–50. doi: 10.1158/0008-5472.CAN-09-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–93. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 9.Araki T, Nawa H, Neel BG. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. J Biol Chem. 2003;278:41677–84. doi: 10.1074/jbc.M306461200. [DOI] [PubMed] [Google Scholar]
- 10.Gu H, Neel BG. The “Gab” in signal transduction. Trends Cell Biol. 2003;13:122–30. doi: 10.1016/s0962-8924(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 11.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 12.Shi ZQ, Yu DH, Park M, Marshall M, Feng GS. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol. 2000;20:1526–36. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan G, Kalaitzidis D, Neel BG. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 2008;27:179–92. doi: 10.1007/s10555-008-9126-y. [DOI] [PubMed] [Google Scholar]
- 14.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–92. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 15.Bentires-Alj M, Paez JG, David FS, Keilhack H, Halmos B, Naoki K, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–20. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 16.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–9. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 17.Hendriks BS, Griffiths GJ, Benson R, Kenyon D, Lazzara M, Swinton J, et al. Decreased internalisation of erbB1 mutants in lung cancer is linked with a mechanism conferring sensitivity to gefitinib. Syst Biol (Stevenage) 2006;153:457–66. doi: 10.1049/ip-syb:20050108. [DOI] [PubMed] [Google Scholar]
- 18.Salvi M, Stringaro A, Brunati AM, Agostinelli E, Arancia G, Clari G, et al. Tyrosine phosphatase activity in mitochondria: presence of Shp-2 phosphatase in mitochondria. Cell Mol Life Sci. 2004;61:2393–404. doi: 10.1007/s00018-004-4211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maroun CR, Naujokas MA, Holgado-Madruga M, Wong AJ, Park M. The tyrosine phosphatase SHP-2 is required for sustained activation of extracellular signal-regulated kinase and epithelial morphogenesis downstream from the met receptor tyrosine kinase. Mol Cell Biol. 2000;20:8513–25. doi: 10.1128/mcb.20.22.8513-8525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T, et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res. 2008;68:9479–87. doi: 10.1158/0008-5472.CAN-08-1643. [DOI] [PubMed] [Google Scholar]
- 22.Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, et al. Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4:521–8. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- 23.Wang SE, Narasanna A, Perez-Torres M, Xiang B, Wu FY, Yang S, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Offterdinger M, Bastiaens PI. Prolonged EGFR signaling by ERBB2-mediated sequestration at the plasma membrane. Traffic. 2008;9:147–55. doi: 10.1111/j.1600-0854.2007.00665.x. [DOI] [PubMed] [Google Scholar]
- 25.Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D. Coregulation of epidermal growth factor receptor/human epidermal growth factor receptor 2 (HER2) levels and locations: quantitative analysis of HER2 overexpression effects. Cancer Res. 2003;63:1130–7. [PubMed] [Google Scholar]
- 26.Zhou X, Agazie YM. Molecular mechanism for SHP2 in promoting HER2-induced signaling and transformation. J Biol Chem. 2009;284:12226–34. doi: 10.1074/jbc.M900020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Y, Chen Z, Chen L, Fang B, Win-Piazza H, Haura E, et al. Critical role of Shp2 in tumor growth involving regulation of c-Myc. Genes Cancer. 2010;1:994–1007. doi: 10.1177/1947601910395582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 30.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci U S A. 2004;101:10380–5. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickrema A, Uddin S, Sharma A, Chen F, Alsayed Y, Ahmad S, et al. Engagement of Gab1 and Gab2 in erythropoietin signaling. J Biol Chem. 1999;274:24469–74. doi: 10.1074/jbc.274.35.24469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.