Abstract
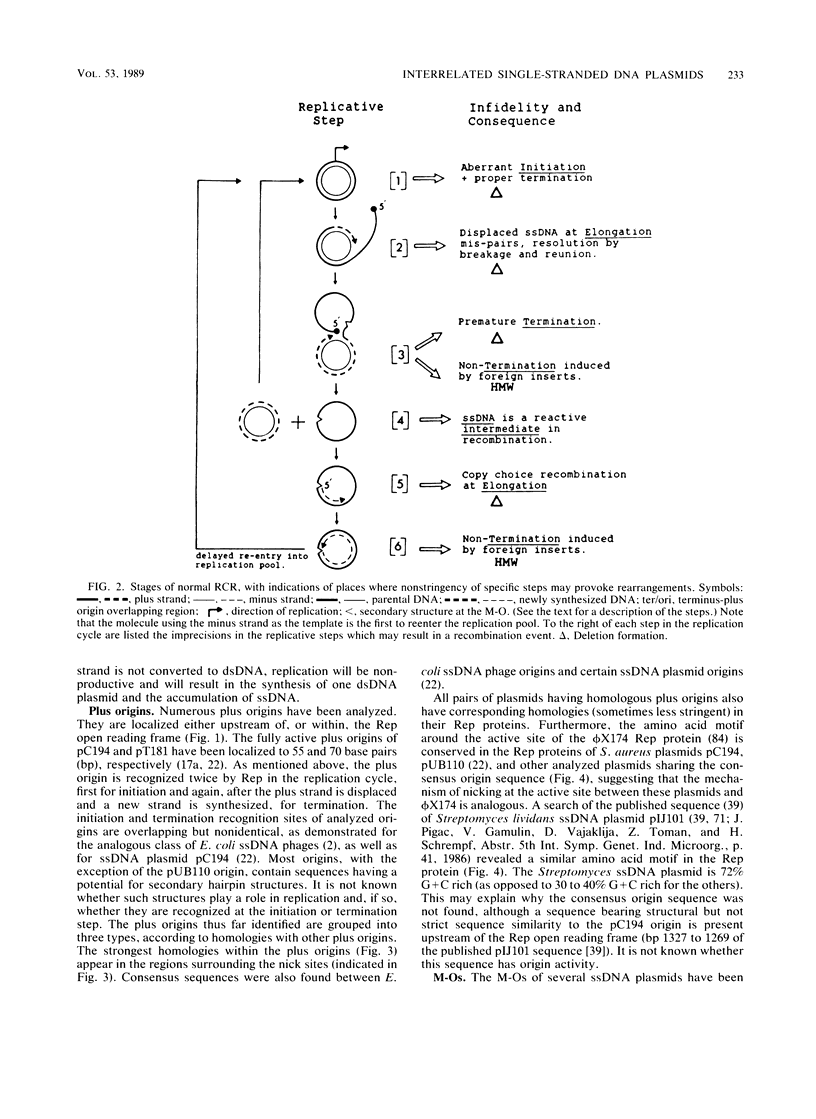
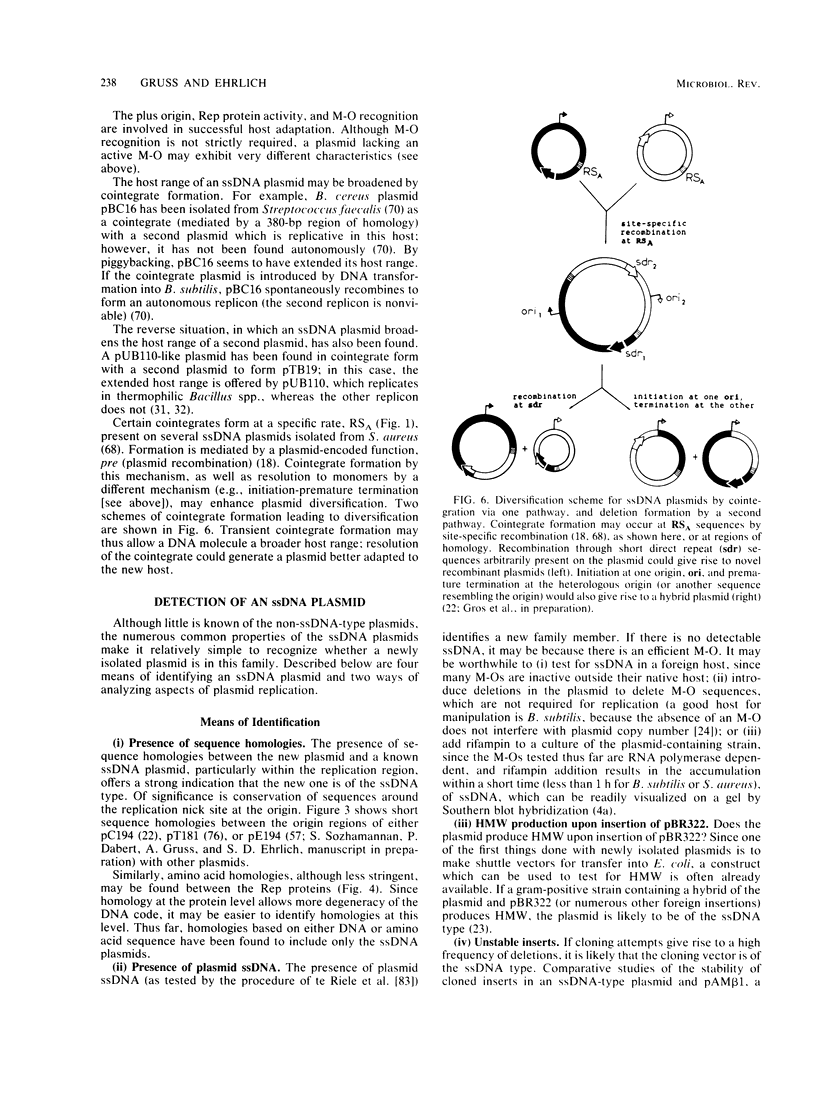
Many plasmids from gram-positive bacteria replicate via a single-stranded deoxyribonucleic acid (ssDNA) intermediate, most probably by a rolling-circle mechanism (these plasmids are referred to in this paper as ssDNA plasmids). Their plus and minus origins are physically separated, and replicative initiations are not simultaneous; it is this feature that allows visualization of ssDNA replication intermediates. The insertion of foreign DNA into an ssDNA plasmid may provoke a high frequency of deletions, changes of replicative products to high-molecular-weight forms, segregational loss, and decreased plasmid copy numbers. When an ssDNA plasmid is inserted into the chromosome, both deletions and amplifications may be induced. Both the mode of replication and the copy control mechanism affect the fate of inserted foreign material, usually selecting for its loss. Thus, after having tasted various morsels of DNA, the resulting plasmid stays trim. The features of the ssDNA plasmids seem to be beneficial for their viability and propagation, but not for their use as cloning vectors. However, plasmids replicating via ssDNA intermediates are being exploited to yield insights into the mechanisms of recombination and amplification.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso J. C., Lüder G., Trautner T. A. Requirements for the formation of plasmid-transducing particles of Bacillus subtilis bacteriophage SPP1. EMBO J. 1986 Dec 20;5(13):3723–3728. doi: 10.1002/j.1460-2075.1986.tb04706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988 Oct 7;55(1):113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- Bergemann A. D., Whitley J. C., Finch L. R. Homology of mycoplasma plasmid pADB201 and staphylococcal plasmid pE194. J Bacteriol. 1989 Jan;171(1):593–595. doi: 10.1128/jb.171.1.593-595.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Gros M. F., te Riele H., Ehrlich S. D., Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989 Jun;171(6):3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron S., Bosma P., van Belkum M., Luxen E. Stability function in the Bacillus subtilis plasmid pTA 1060. Plasmid. 1987 Jul;18(1):8–15. doi: 10.1016/0147-619x(87)90073-4. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E. Segregational instability of pUB110-derived recombinant plasmids in Bacillus subtilis. Plasmid. 1985 Nov;14(3):235–244. doi: 10.1016/0147-619x(85)90007-1. [DOI] [PubMed] [Google Scholar]
- Brunier D., Michel B., Ehrlich S. D. Copy choice illegitimate DNA recombination. Cell. 1988 Mar 25;52(6):883–892. doi: 10.1016/0092-8674(88)90430-8. [DOI] [PubMed] [Google Scholar]
- Burdett V. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1980 Nov;18(5):753–760. doi: 10.1128/aac.18.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton S., Projan S. J., Highlander S. K., Moghazeh S. M., Novick R. P. Control of pT181 replication II. Mutational analysis. EMBO J. 1984 Oct;3(10):2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Chang S. Y., Gray O. Structural and genetic analyses of a par locus that regulates plasmid partition in Bacillus subtilis. J Bacteriol. 1987 Sep;169(9):3952–3962. doi: 10.1128/jb.169.9.3952-3962.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T., Hopwood D. A. "Strong incompatibility" between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet. 1988 Oct;214(2):286–294. doi: 10.1007/BF00337723. [DOI] [PubMed] [Google Scholar]
- Devine K. M., Hogan S. T., Higgins D. G., McConnell D. J. Replication and segregational stability of Bacillus plasmid pBAA1. J Bacteriol. 1989 Feb;171(2):1166–1172. doi: 10.1128/jb.171.2.1166-1172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Iordanescu S., Novick R. P., Murray R. W., Steck T. R., Khan S. A. Functional organization of the plasmid pT181 replication origin. J Mol Biol. 1989 Jan 20;205(2):355–362. doi: 10.1016/0022-2836(89)90346-x. [DOI] [PubMed] [Google Scholar]
- Gennaro M. L., Kornblum J., Novick R. P. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol. 1987 Jun;169(6):2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Novick R. P. An enhancer of DNA replication. J Bacteriol. 1988 Dec;170(12):5709–5717. doi: 10.1128/jb.170.12.5709-5717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Novick R. P. cmp, a cis-acting plasmid locus that increases interaction between replication origin and initiator protein. J Bacteriol. 1986 Oct;168(1):160–166. doi: 10.1128/jb.168.1.160-166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goze A., Ehrlich S. D. Replication of plasmids from Staphylococcus aureus in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7333–7337. doi: 10.1073/pnas.77.12.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. Insertion of foreign DNA into plasmids from gram-positive bacteria induces formation of high-molecular-weight plasmid multimers. J Bacteriol. 1988 Mar;170(3):1183–1190. doi: 10.1128/jb.170.3.1183-1190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Dubnau D. Analysis of plasmid deletional instability in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1014–1023. doi: 10.1128/jb.162.3.1014-1023.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka M., Akiyama M., Horiuchi T. A consensus sequence of three DNA replication terminus sites on the E. coli chromosome is highly homologous to the terR sites of the R6K plasmid. Cell. 1988 Nov 4;55(3):467–475. doi: 10.1016/0092-8674(88)90033-5. [DOI] [PubMed] [Google Scholar]
- Highlander S. K., Novick R. P. Plasmid repopulation kinetics in Staphylococcus aureus. Plasmid. 1987 May;17(3):210–221. doi: 10.1016/0147-619x(87)90029-1. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Ano T., Fujii M., Aiba S. Two replication determinants of an antibiotic-resistance plasmid, pTB19, from a thermophilic bacillus. J Gen Microbiol. 1984 Jun;130(6):1399–1408. doi: 10.1099/00221287-130-6-1399. [DOI] [PubMed] [Google Scholar]
- Iordanescu S. Effect of the deletion of a fragment dispensable for the autonomous maintenance of plasmid pT181 on the competition between incompatible plasmids. Plasmid. 1986 May;15(3):191–198. doi: 10.1016/0147-619x(86)90037-5. [DOI] [PubMed] [Google Scholar]
- Iordanescu S. Staphylococcus aureus chromosomal mutation specifically affecting the copy number of Inc3 plasmids. Plasmid. 1983 Sep;10(2):130–137. doi: 10.1016/0147-619x(83)90065-3. [DOI] [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M., Della Latta P., Novick R. Incompatibility and molecular relationships between small Staphylococcal plasmids carrying the same resistance marker. Plasmid. 1978 Sep;1(4):468–479. doi: 10.1016/0147-619x(78)90005-7. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Jannière L., Ehrlich S. D. Recombination between short repeated sequences is more frequent in plasmids than in the chromosome of Bacillus subtilis. Mol Gen Genet. 1987 Nov;210(1):116–121. doi: 10.1007/BF00337766. [DOI] [PubMed] [Google Scholar]
- Kendall K. J., Cohen S. N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988 Oct;170(10):4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Murray R. W., Koepsel R. R. Mechanism of plasmid pT181 DNA replication. Biochim Biophys Acta. 1988 Dec 20;951(2-3):375–381. doi: 10.1016/0167-4781(88)90109-1. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J Bacteriol. 1982 Feb;149(2):642–649. doi: 10.1128/jb.149.2.642-649.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler T. M., Thorne C. B. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J Bacteriol. 1987 Nov;169(11):5271–5278. doi: 10.1128/jb.169.11.5271-5278.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Cleavage of single-stranded DNA by plasmid pT181-encoded RepC protein. Nucleic Acids Res. 1987 May 26;15(10):4085–4097. doi: 10.1093/nar/15.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Murray R. W., Rosenblum W. D., Khan S. A. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W., Chopra I. Genetic studies of a multi-resistant strain of Staphylococcus aureus. J Med Microbiol. 1974 May;7(2):285–297. doi: 10.1099/00222615-7-2-285. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Lopez P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986 Dec 20;192(4):753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Parisi J. T. Nucleotide sequence of the constitutive macrolide-lincosamide-streptogramin B resistance plasmid pNE131 from Staphylococcus epidermidis and homologies with Staphylococcus aureus plasmids pE194 and pSN2. J Bacteriol. 1986 Sep;167(3):888–892. doi: 10.1128/jb.167.3.888-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. L., Blaschek H. P. Transformation of Heat-Treated Clostridium acetobutylicum Protoplasts with pUB110 Plasmid DNA. Appl Environ Microbiol. 1984 Oct;48(4):737–742. doi: 10.1128/aem.48.4.737-742.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell R. W., Sweeney H. M., Cohen S. Conjugational transfer of gentamicin resistance plasmids intra- and interspecifically in Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1983 Jan;23(1):151–160. doi: 10.1128/aac.23.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden J. S., Marians K. J. Replication of pBR322 DNA in vitro with purified proteins. Requirement for topoisomerase I in the maintenance of template specificity. J Biol Chem. 1985 Aug 5;260(16):9316–9325. [PubMed] [Google Scholar]
- Minton N. P., Oultram J. D., Brehm J. K., Atkinson T. The replication proteins of plasmids pE194 and pLS1 have N-terminal homology. Nucleic Acids Res. 1988 Apr 11;16(7):3101–3101. doi: 10.1093/nar/16.7.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R. E., Ano T., Imanaka T., Aiba S. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1 and its copy mutants. Mol Gen Genet. 1986 Jan;202(1):169–171. doi: 10.1007/BF00330534. [DOI] [PubMed] [Google Scholar]
- Naidoo J. Interspecific co-transfer of antibiotic resistance plasmids in staphylococci in vivo. J Hyg (Lond) 1984 Aug;93(1):59–66. doi: 10.1017/s0022172400060939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Jannière L., Ehrlich S. D. Recombination between repeated DNA sequences occurs more often in plasmids than in the chromosome of Bacillus subtilis. Mol Gen Genet. 1984;197(1):46–54. doi: 10.1007/BF00327921. [DOI] [PubMed] [Google Scholar]
- Noirot P., Petit M. A., Ehrlich S. D. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):39–48. doi: 10.1016/0022-2836(87)90509-2. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Projan S. J., Carleton S., Highlander S. K., Gruss A., Khan S. A., Iordanescu S. Control of pT181 replication I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984 Oct;3(10):2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Edelman I., Lofdahl S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J Mol Biol. 1986 Nov 20;192(2):209–220. doi: 10.1016/0022-2836(86)90360-8. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kumar C. C., Carleton S., Gruss A., Highlander S. K., Kornblum J. Replication control for pT181, an indirectly regulated plasmid. Basic Life Sci. 1985;30:299–320. doi: 10.1007/978-1-4613-2447-8_24. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Rosenblum W., Edelman I. Staphylococcal plasmid cointegrates are formed by host- and phage-mediated general rec systems that act on short regions of homology. Mol Gen Genet. 1984;195(1-2):374–377. doi: 10.1007/BF00332777. [DOI] [PubMed] [Google Scholar]
- Peeters B. P., de Boer J. H., Bron S., Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988 Jun;212(3):450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. Streptococcus plasmid pAM alpha 1 is a composite of two separable replicons, one of which is closely related to Bacillus plasmid pBC16. J Bacteriol. 1983 Aug;155(2):607–615. doi: 10.1128/jb.155.2.607-615.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigac J., Vujaklija D., Toman Z., Gamulin V., Schrempf H. Structural instability of a bifunctional plasmid pZG1 and single-stranded DNA formation in Streptomyces. Plasmid. 1988 May;19(3):222–230. doi: 10.1016/0147-619x(88)90040-6. [DOI] [PubMed] [Google Scholar]
- Polak J., Novick R. P. Closely related plasmids from Staphylococcus aureus and soil bacilli. Plasmid. 1982 Mar;7(2):152–162. doi: 10.1016/0147-619x(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Kornblum J., Moghazeh S. L., Edelman I., Gennaro M. L., Novick R. P. Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol Gen Genet. 1985;199(3):452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Moghazeh S., Novick R. P. Nucleotide sequence of pS194, a streptomycin-resistance plasmid from Staphylococcus aureus. Nucleic Acids Res. 1988 Mar 25;16(5):2179–2187. doi: 10.1093/nar/16.5.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Monod M., Narayanan C. S., Dubnau D. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J Bacteriol. 1987 Nov;169(11):5131–5139. doi: 10.1128/jb.169.11.5131-5139.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan S. J., Novick R. Comparative analysis of five related Staphylococcal plasmids. Plasmid. 1988 May;19(3):203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- Ray D. S., Hines J. C., Kim M. H., Imber R., Nomura N. M13 vectors for selective cloning of sequences specifying initiation of DNA synthesis on single-stranded templates. Gene. 1982 Jun;18(3):231–238. doi: 10.1016/0378-1119(82)90160-3. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Zinder N. D. Hemimethylation prevents DNA replication in E. coli. Cell. 1987 Sep 25;50(7):1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Shafferman A., Kolter R., Stalker D., Helinski D. R. Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J Mol Biol. 1982 Oct 15;161(1):57–76. doi: 10.1016/0022-2836(82)90278-9. [DOI] [PubMed] [Google Scholar]
- Sioud M., Baldacci G., Forterre P., de Recondo A. M. Novobiocin induces accumulation of a single strand of plasmid pGRB-1 in the archaebacterium Halobacterium GRB. Nucleic Acids Res. 1988 Aug 25;16(16):7833–7842. doi: 10.1093/nar/16.16.7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Baas P. D., Jansz H. S. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986 May 27;14(10):4229–4238. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]



