BRCA1 deficiency results in impaired Nrf2-mediated antioxidant responses followed by cell death, with estradiol rescuing the effect by inducing Nrf2 stabilization.
Abstract
Oxidative stress plays an important role in cancer development and treatment. Recent data implicate the tumor suppressor BRCA1 in regulating oxidative stress, but the molecular mechanism and the impact in BRCA1-associated tumorigenesis remain unclear. Here, we show that BRCA1 regulates Nrf2-dependent antioxidant signaling by physically interacting with Nrf2 and promoting its stability and activation. BRCA1-deficient mouse primary mammary epithelial cells show low expression of Nrf2-regulated antioxidant enzymes and accumulate reactive oxygen species (ROS) that impair survival in vivo. Increased Nrf2 activation rescues survival and ROS levels in BRCA1-null cells. Interestingly, 53BP1 inactivation, which has been shown to alleviate several defects associated with BRCA1 loss, rescues survival of BRCA1-null cells without restoring ROS levels. We demonstrate that estrogen treatment partially restores Nrf2 levels in the absence of BRCA1. Our data suggest that Nrf2-regulated antioxidant response plays a crucial role in controlling survival downstream of BRCA1 loss. The ability of estrogen to induce Nrf2 posits an involvement of an estrogen-Nrf2 connection in BRCA1 tumor suppression. Lastly, BRCA1-mutated tumors retain a defective antioxidant response that increases the sensitivity to oxidative stress. In conclusion, the role of BRCA1 in regulating Nrf2 activity suggests important implications for both the etiology and treatment of BRCA1-related cancers.
Reactive oxygen species (ROS) have a complex role in cancer development and progression (Cairns et al., 2011). Redox homeostasis is fundamental to maintaining normal cellular functions and ensuring cell survival of cancer cells with aberrant metabolism. Although elevated ROS levels can be protumorigenic and induce tumor formation through their mutagenic properties (Shibutani et al., 1991), high ROS levels can also limit tumor formation. As such, reduced intracellular ROS levels through the action of antioxidant signaling have been demonstrated to promote cell transformation and tumorigenic phenotypes. In vitro antioxidant exposure increases cell survival and anchorage-independent growth in premalignant mammary epithelial cells (MECs; Schafer et al., 2009). In cancer cell lines, the antioxidant genes GCLC and HMOX1, identified through a genome-wide shRNA screen, are essential for cell survival (Marcotte et al., 2012). In addition, the antioxidant enzymes SOD2, GLS2, and SEPHS1 are required for in vivo mammary tumorigenesis (Possemato et al., 2011).
ROS detoxifying enzymes are predominantly driven by the transcription factor Nrf2 (NF-E2–related factor 2), which is tightly regulated by Keap1 (KELCH-like ECH-associated protein 1)-mediated ubiquitination (Kobayashi et al., 2004). Oncogenic events that regulate Nrf2 transcription or alter its protein stability would presumably provide a survival advantage. Indeed, oncogenic K-Ras or c-Myc transcriptionally regulates Nrf2 to activate an antioxidant program and lower intracellular ROS that is required for cell proliferation and tumorigenesis (DeNicola et al., 2011). Nrf2 has also been implicated in controlling cell proliferation downstream of the hyperactivated oncogenic PI3K/Akt pathway (Mitsuishi et al., 2012). In breast cancer, deregulation of NRF2 stability by KEAP1 silencing through promoter hypermethylation has been recently reported in a subset of tumors that are more frequently estrogen receptor (ER) positive and human epidermal growth factor receptor 2 negative (Barbano et al., 2013). Although KEAP1 mutations are rare, genome-wide sequencing of human breast cancers identified a KEAP1 mutation (C23Y) that disrupts its interaction with NRF2, leading to increased NRF2 protein stability and antioxidant signaling in a subset of human breast cancers (Sjöblom et al., 2006; Nioi and Nguyen, 2007).
Loss-of-function mutations in the tumor suppressor gene BRCA1 account for 5–10% of breast cancer cases in the Western world and confer increased risk for development of ovarian cancer (Narod and Foulkes, 2004). Because these tumors are characterized by high genomic instability, lack of DNA repair as the result of BRCA1 inactivation is considered the main cause of tumor formation. However, new functions of BRCA1 such as the regulation of the oncogenic microRNA 155 (Chang et al., 2011), the maintenance of heterochromatin structure (Zhu et al., 2011), and the modulation of oxidative stress (Vurusaner et al., 2012) have been recently discovered. In the context of oxidative stress, BRCA1 overexpression in human breast cancer cells up-regulates several antioxidant genes and reduces H2O2-induced DNA damage and apoptosis (Bae et al., 2004; Saha et al., 2009). Although Brca1 loss-of-function in mouse embryonic fibroblasts from Brca1Δ11/Δ11 mutant mice shows higher ROS levels than cells from Brca1 WT mice and is more sensitive to apoptosis induced by oxidative stress (Cao et al., 2007), the mechanism by which BRCA1 regulates oxidative stress and its impact in BRCA1-associated tumorigenesis has not been fully uncovered.
In this study, we investigated the link between BRCA1 and oxidative stress both in normal MECs and in breast tumors. We used a Brca1 conditional knockout mouse (Liu et al., 2007) to specifically delete the Brca1 gene in the mammary gland. Although Brca1 deletion under the control of K14- or K6a-driven cre recombinase does not lead to any observable changes in the mammary gland (Liu et al., 2007; Smart et al., 2011), our study of primary and immortalized mouse and human BRCA1-deficient MECs shows that BRCA1 deficiency results in ROS accumulation in these cells. This effect is the result of impaired Nrf2-driven antioxidant signaling. We demonstrated that BRCA1 is a novel Nrf2-binding protein that affects Keap1-mediated Nrf2 ubiquitination activity, thereby controlling Nrf2 stability and activation. In BRCA1-deficient cells, up-regulation of the Nrf2-mediated antioxidant pathway through inactivation of Keap1 rescues cell survival defects and ROS levels induced by BRCA1 loss-of-function. Interestingly, human BRCA1-mutated breast tumor cells also retain a defective Nrf2 antioxidant response that impinges on their sensitivity to oxidative stress and cisplatin treatment. Our work establishes a physiological function of BRCA1 in ROS regulation in the mammary gland and sheds light on the role of oxidative stress in BRCA1-mediated tumor suppression with important therapeutic implications.
RESULTS
BRCA1 loss-of-function in MECs causes ROS accumulation
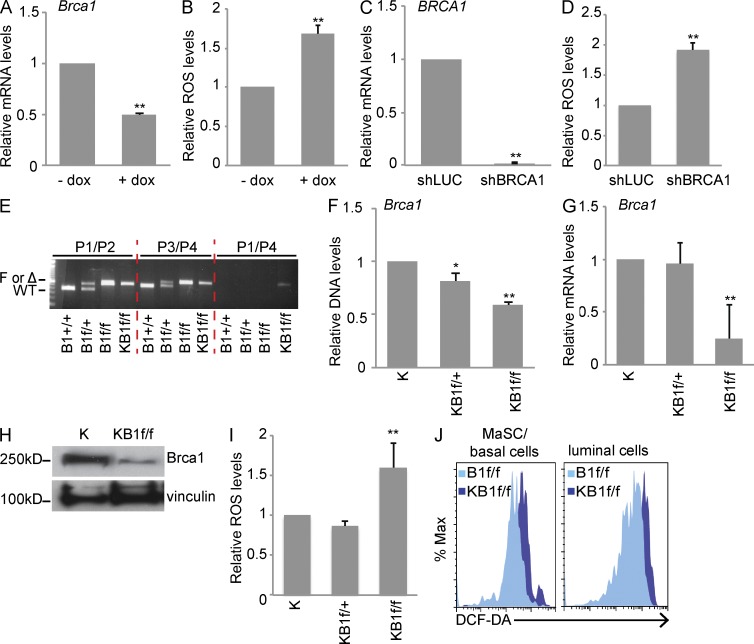
To investigate the link between BRCA1 expression and ROS levels in MECs, we down-regulated Brca1 in immortalized mouse mammary COMMA-1D cells using a doxycycline (dox)-inducible lentiviral shRNA (Fig. 1 A). As shown in Fig. 1 B, cells with low Brca1 expression (+dox) had high ROS levels compared with dox-untreated cells (−dox). Consistent with this result, ROS also accumulated in human MECs (HMECs) infected with a lentiviral shRNA against human BRCA1 (shBRCA1) compared with cells expressing an shRNA directed toward Luciferase (Luc; shLUC) used as control (Fig. 1, C and D).
Figure 1.
BRCA1 loss-of-function induces high ROS levels in MECs. (A) Brca1 mRNA expression in COMMA-1D cells infected with dox-inducible Brca1 shRNA and treated (+dox) or not (−dox) with dox. (B) Quantitation of ROS in COMMA-1D cells as processed in A. (C) BRCA1 mRNA levels in HMECs infected with Luc shRNA (control) or BRCA1 shRNA. (D) Quantitation of ROS in HMEC as treated in C. (A–D) Data represent the mean ± SEM of three biological replicates. (E) Representative PCR with genomic DNA isolated from B1+/+, B1f/+, B1f/f, and KB1f/f pMECs using specific primers for detection of Brca1 WT allele, loxP site in intron 3 (F), or cre-mediated deleted allele (Δ). Primers are described in Liu et al. (2007) and Table S1. (F) qPCR with genomic DNA from K, KB1f/+, and KB1f/f pMECs using specific primers directed against Brca1 WT allele as reported in Table S1. (G) BRCA1 mRNA levels in K, KB1f/+, and KB1f/f pMECs. (H) Representative analysis of BRCA1 protein levels in K and KB1f/f pMECs. Vinculin was used as a loading control. (I) ROS levels in K, KB1f/+, and KB1f/f pMECs. (F, G, and I) Data represent the mean ± SEM of n = 5 mice of each genotype. (J) Representative FACS profile of ROS levels in MaSC/basal and luminal cell subpopulations in B1f/f and KB1f/f pMECs stained with DCF-DA. *, P < 0.05; **, P < 0.01.
We next determined ROS levels in Brca1-deficient primary MECs (pMECs) using a previously generated Brca1 conditional knockout mouse (B1f/f) carrying a cre-inducible deletion of Brca1 exons 5–13 (Liu et al., 2007). Targeted Brca1 deletion to the mammary gland epithelium was achieved by crossing B1f/f mice to K14cre mice (Jonkers et al., 2001) to obtain K14cre;Brca1f/f mice (KB1f/f) along with K14cre (K) and K14cre;Bf/+ (KB1f/+) control mice. K14 is mainly expressed in mammary stem cells (MaSCs) and basal cells (Shackleton et al., 2006). However, analysis of pMECs isolated from females obtained by crossing K14cre mice with a transgenic strain expressing the red fluorescent protein Tomato (Muzumdar et al., 2007) showed that K14cre was active in both MaSCs/basal and luminal cells (not depicted). PCR with genomic DNA from pMECs isolated from #4 mammary glands of B1+/+, B1f/+, B1f/f, and KB1f/f mice showed cre-mediated deletion of Brca1 exons 5–13 only in KB1f/f samples (Fig. 1 E). Quantitative PCR (qPCR) with genomic DNA confirmed a significant reduction of Brca1 WT allele in KB1f/f pMECs (Fig. 1 F), which correlated with low Brca1 mRNA expression in KB1f/f pMECs compared with K and KB1f/+controls (Fig. 1 G). Consequently, KB1f/f pMECs also had low Brca1 protein levels (Fig. 1 H). Intracellular ROS staining showed that KB1f/f pMECs had higher ROS than K and KB1f/+ pMECs (Fig. 1 I).
Mammary glands contain distinct cell subpopulations that vary in their repopulating capacity and differentiation markers (Shackleton et al., 2006; Stingl et al., 2006). Immunostaining of pMECs with antibodies (Abs) specific for the epithelial surface markers CD24 and CD49f (Joshi et al., 2010) revealed that both KB1f/f MaSC-enriched basal cells (CD24+CD49fhigh) and luminal cells (CD24+CD49flow) showed elevated ROS compared with B1f/f controls (Fig. 1 J). Together, these results demonstrate that in MECs, both in vitro and in vivo down-regulation of BRCA1 expression induce accumulation of intracellular ROS levels.
BRCA1-deficient cells have a reduced Nrf2-mediated antioxidant response
Intracellular ROS levels can be regulated by the activity of antioxidant enzymes whose expression depends on the transcription factor Nrf2 (Kensler and Wakabayashi, 2010). Although Nrf2 activation is mainly controlled by protein stability (Li and Kong, 2009), recent data suggest that Nrf2 mRNA expression is also important in regulating Nrf2 activity (Kwak et al., 2002; DeNicola et al., 2011; Yang et al., 2011).
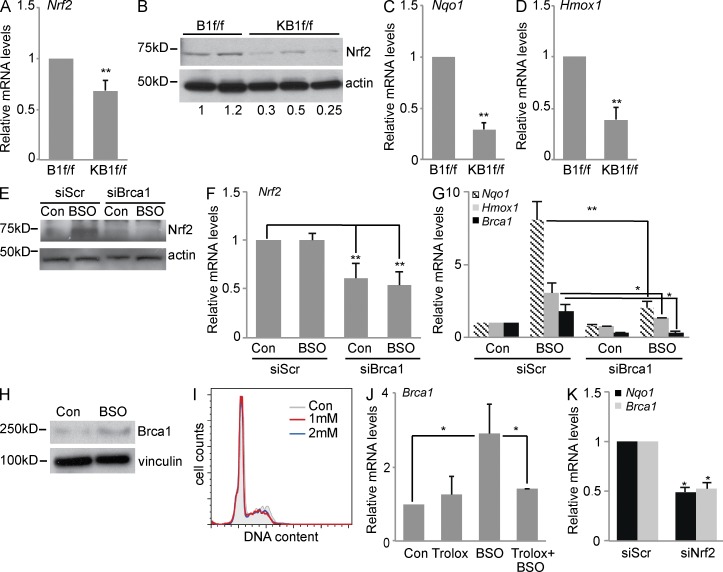
When we evaluated Nrf2 expression in KB1f/f pMECs, we found that both mRNA (Fig. 2 A) and protein levels (Fig. 2 B) were significantly lower in Brca1-deficient cells compared with control cells (B1f/f). These data are supported by previous findings that BRCA1 binds the Nrf2 promoter and regulates its transcription (Kang et al., 2011). Consistent with low Nrf2 expression, KB1f/f pMECs showed reduced mRNA levels of the Nrf2 transcriptional targets Nqo1 and Hmox1 compared with controls (Fig. 2, C and D).
Figure 2.
Brca1 expression is regulated by oxidative stress and controls Nrf2 abundance and transactivation activity. (A) Nrf2 mRNA levels in B1f/f and KB1f/f pMECs. (B) Representative immunoblot of Nrf2 protein levels in pMECs from B1f/f (n = 2) and KB1f/f (n = 3) mice. Western blot quantitation is shown below. (C and D) RT-PCR analysis of Nrf2 targets, Nqo1 (C) and Hmox1 (D), in B1f/f and KB1f/f pMECs. (A, C, and D) Data are the mean ± SEM of n = 5 mice of each genotype. (E) Representative immunoblot of Nrf2 protein levels in control (Con) or BSO-treated COMMA-1D cells after transfection with scrambled siRNA (siScr) or Brca1-specific siRNA (siBrca1). (F) Nrf2 mRNA levels in cells treated as in E. (G) Expression levels of Nqo1, Hmox1, and Brca1 mRNAs in cells treated as in E. (H) Representative immunoblot of Brca1 protein levels in control and BSO-treated COMMA-1D cells. Vinculin was used as a loading control. (I) Representative cell cycle profile by PI staining of control or BSO (1 or 2 mM)-treated COMMA-1D cells. (J) Brca1 mRNA levels in COMMA-1D cells that have been left untreated (Con) or after treatment with Trolox, BSO, and Trolox plus BSO for 48 h. (K) Nqo1 and Brca1 mRNA expression in untreated COMMA-1D cells transfected with scrambled siRNA or Nrf2-specific siRNA (siNrf2). (F, G, J, and K) Data represent the mean ± SEM of three biological replicates. *, P < 0.05; **, P < 0.01.
We next determined whether BRCA1 expression also regulated Nrf2 levels and activity in response to exogenous oxidative stress in vitro. COMMA-1D cells were treated with l-buthionine-sulfoximine (BSO), a specific inhibitor of glutathione synthesis (Griffith, 1999; Marí et al., 2009). BSO treatment resulted in ROS accumulation (not depicted) and Nrf2 protein accumulation (Fig. 2 E) with no change of Nrf2 mRNA levels (Fig. 2 F). Interestingly, Brca1 silencing by specific siRNAs (siBRCA1) negatively affected both BSO-induced Nrf2 protein increase (Fig. 2 E) and Nrf2 mRNA expression in both untreated (control) and BSO-treated cells (Fig. 2 F). Brca1 inactivation also resulted in down-regulation of the Nrf2 target genes Nqo1 and Hmox1 after BSO treatment (Fig. 2 G).
We found that oxidative stress induced by BSO also increased Brca1 mRNA (Fig. 2 G) and protein (Fig. 2 H) levels. BRCA1 expression has been shown to be regulated in a cell cycle–dependent manner (Chen et al., 1996). However, Brca1 up-regulation by BSO was not caused by cell cycle changes (Fig. 2 I), but rather by high ROS levels because cotreatment of BSO with a vitamin E–derived antioxidant, Trolox (Davies et al., 1988), abrogated Brca1 mRNA accumulation (Fig. 2 J). Of note, Brca1 expression was down-regulated in COMMA-1D cells treated with Nrf2 siRNA, similarly to Nrf2-target NQO1 (Fig. 2 K). These results further support the notion that BRCA1 may be an Nrf2 transcriptional target gene regulated through an antioxidant response element in the BRCA1 promoter (Wang et al., 2013). Overall, our data suggest the existence of a positive feedback loop regulation between BRCA1 and Nrf2 that may be necessary for an efficient cellular antioxidant response.
BRCA1 interacts with Nrf2 and promotes its stability
Nrf2 protein is tightly regulated by Keap1, a substrate adaptor for Cul3-based E3 ubiquitin ligase (Li and Kong, 2009). Under resting conditions, Keap1 binding triggers Nrf2 ubiquitination and degradation. Under oxidative stress conditions, Keap1 is oxidized on specific cysteine residues, which affect Nrf2 binding, thereby preventing Nrf2 degradation and resulting in transactivation of Nrf2-dependent antioxidant genes.
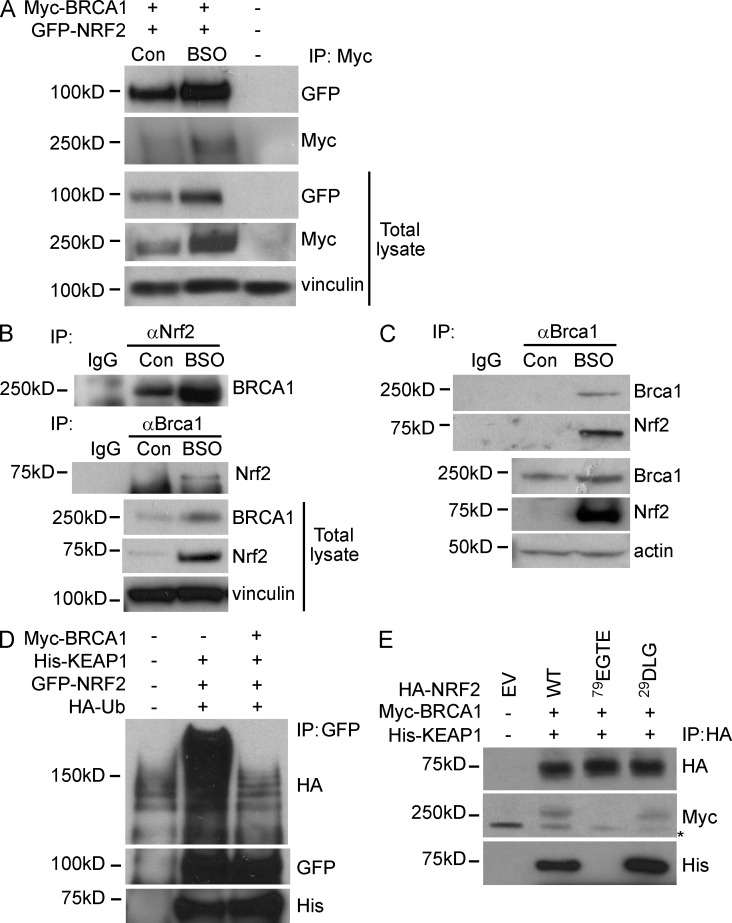
We determined whether down-regulation of Nrf2 protein levels in Brca1-deficient cells was not only caused by low mRNA expression but also a consequence of low protein stability. GFP-tagged NRF2 and Myc-tagged BRCA1 were transiently overexpressed in 293FT cells in the presence or absence of BSO to induce oxidative stress. GFP-NRF2 was detected in Myc-BRCA1 immunoprecipitates, and complex formation was increased by oxidative stress (Fig. 3 A). Similarly to GFP-NRF2, Myc-BRCA1 accumulated upon BSO treatment, suggesting the involvement of posttranslational regulation of exogenously expressed BRCA1 in the presence of oxidative stress.
Figure 3.
BRCA1 physically interacts with Nrf2 and affects Keap1-mediated Nrf2 ubiquitination. (A) 293FT cells were transfected with Myc-BRCA1 and GFP-NRF2 constructs and then left untreated (Con) or treated with BSO. BRCA1 was immunoprecipitated with anti-Myc Ab. The blot was probed with anti-Myc and anti-GFP Abs. “−” indicates untreated EV control. Vinculin was used as a loading control. (B) COMMA-1D cells were left untreated (Con) or treated with BSO and processed for immunoprecipitation (IP) with anti-Nrf2 (H-300) or anti-Brca1 (C20) Abs to detect the endogenous Brca1 and Nrf2 proteins. (C) COMMA-1D cells were treated as in B and subjected to immunoprecipitation with anti–mouse Brca1 Ab to detect endogenous Brca1 and Nrf2 complex. Nrf2 was detected with an affinity-purified Ab as described in Materials and methods. (B and C) IgG served as an isotype control. (D) 293FT cells were transfected with constructs expressing HA-ubiquitin, His-KEAP1, GFP-NRF2, and/or Myc-BRCA1 (as indicated). Immunoprecipitation was performed with anti-GFP Ab followed by Western blot to detect ubiquitinated NRF2 (HA, GFP) and His-KEAP1. (E) 293FT cells were transfected with Myc-BRCA1, His-KEAP1, HA–WT NRF2 (WT), HA–79EGTE NRF2 mutant (79EGTE), and HA–29DLG NRF2 mutant (29DLG). Immunoprecipitation was performed with anti-HA Ab, and Western blot was probed with anti-Myc and anti-His Abs. The asterisk indicates an unspecific band.
Endogenous Brca1–Nrf2 interaction was confirmed by reciprocal Brca1 and Nrf2 immunoprecipitations in control and BSO-treated COMMA-1D cells (Fig. 3 B). The mutual endogenous interaction was also found using an affinity-purified Ab against mouse Brca1 (Fig. 3 C; Bouwman et al., 2010).
To determine whether BRCA1 interfered with Keap1-mediated Nrf2 ubiquitination, we performed an in vivo ubiquitination assay by transfecting 293FT cells with GFP-NRF2, His-KEAP1, and HA-ubiquitin, with or without Myc-BRCA1. As shown in Fig. 3 D, Keap1-mediated ubiquitination of GFP-NRF2 was abrogated by Myc-BRCA1 overexpression. Myc-BRCA1 was unable to dissociate the NRF2–KEAP1 complex because His-KEAP1 equally immunoprecipitated using GFP-NRF2 with or without Myc-BRCA1 (Fig. 3 D). Thus, upon oxidative stress, BRCA1 up-regulation interferes with KEAP1-mediated NRF2 ubiquitination, resulting in NRF2 protein accumulation.
The capacity of BRCA1 to interfere with KEAP1-mediated NRF2 ubiquitination prompted us to evaluate which domain of NRF2 protein mediated the binding to BRCA1. NRF2 and KEAP1 interaction is based on a two-site substrate recognition model at two distinct domains of NRF2, the ETGE and DLG motifs, known as the “hinge and latch mechanism” (Li and Kong, 2009). We determined the possibility that BRCA1 interfered with KEAP1 binding to one of these two domains. HA-tagged WT NRF2 and HA-tagged NRF2 mutants where the 29DLG or 79ETGE motif were replaced with alanine residues (Chen et al., 2009) were expressed in 293FT cells along with Myc-BRCA1 and His-KEAP1. Immunoprecipitation of HA-tagged NRF2 proteins showed that mutation of 79ETGE diminished both KEAP1 and BRCA1 binding to NRF2, whereas replacement of 29DLG did not affect either KEAP1 or BRCA1 affinity for NRF2 (Fig. 3 E).
As previously reported, the ETGE domain is the strongest interacting motif between Nrf2 and Keap1 compared with the DLG motifs (Chen et al., 2009). Our data postulate a scenario in which BRCA1 interferes with the conformational dynamics of NRF2–KEAP1 interaction at the ETGE site, thus affecting NRF2 ubiquitination and stability.
Brca1-deficient pMECs have a limited lifespan in vivo
Antioxidant signaling and efficient DNA repair are essential for hematopoietic stem cell self-renewal (Pang, 2011). Similarly, high oxidative stress caused by ATM deficiency triggers senescence and limits long-term in vivo repopulation of the hematopoietic compartment (Ito et al., 2004). ROS levels have been demonstrated to regulate mammary stem/progenitor cell function (Diehn et al., 2009). To determine in vivo whether MaSC functions were impaired by ROS accumulation associated with BRCA1 deficiency, we subjected pMECs to a mammary fat pad transplantation assay.
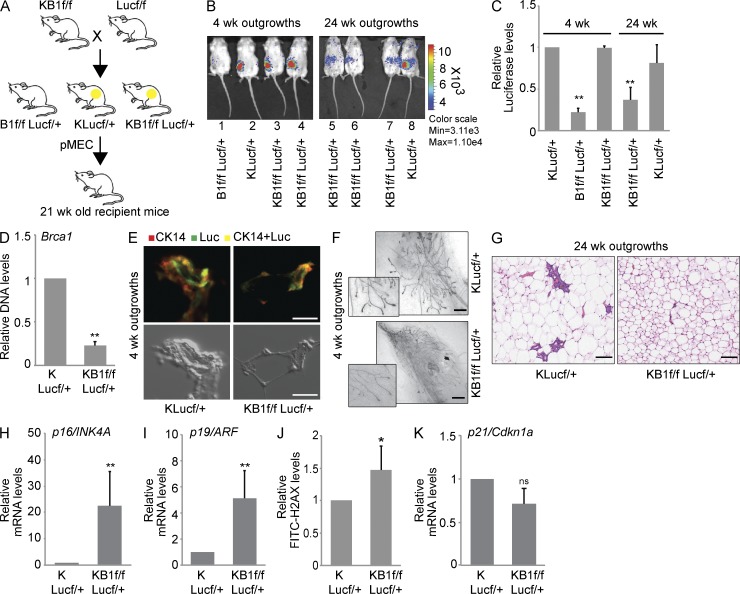
One of the most unique features of mammary gland studies is the ability to transplant pMECs into their normal microenvironment and assess their in vivo growth and differentiation capabilities (Smith and Medina, 2008). To track transplanted pMECs in vivo, we crossed KB1f/f mice with Luc-expressing transgenic mice (Safran et al., 2003) and generated B1f/fLucf/+ (negative control), KLucf/+, and KB1f/fLucf/+ progeny (Fig. 4 A). In KB1f/fLucf/+ mice, K14cre activation controls both Luc expression and Brca1 deletion. pMECs isolated from B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ mice were injected into precleared mammary fat pads of isogenic 21-d-old FVB recipient mice. Anesthetized mice received intraperitoneal luciferin administration, and Luc activity was analyzed in pMEC outgrowths using IVIS imaging. KB1f/fLucf/+ pMECs produced Luc-positive outgrowths by 4 wk after injection, similar to control KLucf/+ pMECs, but by 8 wk, Luc activity in KB1f/fLucf/+ pMECs was progressively lost until barely detectable at 24 wk after injection (Fig. 4, B and C). Genomic DNA qPCR in pMECs isolated from 4-wk-old KB1f/fLucf/+ outgrowths confirmed low levels of Brca1 WT allele compared with KLucf/+ control cells, suggesting that the majority of pMECs lacked Brca1 expression (Fig. 4 D). Furthermore, immunohistochemical analysis confirmed similar CK14 and Luc signals in both KLucf/+ and KB1f/fLucf/+ outgrowths (Fig. 4 E). Whole-mount staining with carmine aluminum of 4-wk-old outgrowths from KLucf/+ and KB1f/fLucf/+ pMECs did not reveal any major morphological differences (Fig. 4 F). However, the repopulation defects reached a dramatic phenotype at 24 wk after transplantation. As shown by hematoxylin-eosin staining at 24 wk after injection, normal ducts were present in outgrowths from KLucf/+ cells, whereas outgrowths from KB1f/fLucf/+ cells showed disorganized ductal structures (Fig. 4 G).
Figure 4.
Brca1-deficient pMECs can regenerate a functional mammary gland but have a limited lifespan in vivo. (A) Breeding strategy used to obtain B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ animals used for in vivo fat pad transplantation assays. pMECs isolated from B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ donor mice were transplanted in 21-wk-old recipient mice to generate outgrowths. (B) In vivo Luc activity in outgrowths derived from B1f/fLucf/+ (mouse 1 as negative control), KLucf/+ (mice 2 and 8 as positive controls), and KB1f/fLucf/+ (mice 3–7) pMECs at 4 and 24 wk after transplantation. (C) Quantitation of Luc activity shown in B using Living Image 3.0 software. (D) qPCR of Brca1 WT allele with genomic DNA from pMECs isolated from KLucf/+ and KB1f/fLucf/+ 4-wk outgrowths. (E) Immunohistochemical staining of 4-wk KLucf/+ and KB1f/fLucf/+ outgrowths with anti-Luc and anti-CK14 Abs. Phase-contrast images of the same glands are shown below. (F) Whole-mount staining of KLucf/+ and KB1f/fLucf/+ 4-wk outgrowths. Terminal end buds are shown at higher magnification. (G) Hematoxylin-eosin staining of KLucf/+ and KB1f/fLucf/+ outgrowths at 24 wk after transplantation. Data are representative of 10 outgrowths examined per genotype. Bars: (E) 16 µm; (F and G) 50 µm. (H and I) RT-PCR analysis of p16/INK4 (H) and p19/ARF (I) mRNA levels in KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths. (J) pMECs from KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths were stained with FITC-γH2AX and analyzed by flow cytometry. (K) p21/Cdkn1a mRNA levels in pMECs from KLucf/+ and KB1f/fLucf/+ 8-wk outgrowths. (C, D, and H–K) Data represent the mean ± SEM of n = 5 outgrowths of each genotype. *, P < 0.05; **, P < 0.01.
Both BRCA1 loss and high ROS levels have been associated with cellular senescence and DNA damage. Interestingly, compared with control KLucf/+ pMECs, KB1f/fLucf/+ pMECs from 8-wk-old outgrowths showed increased mRNA levels of the senescence markers p16/INK4A and p19/ARF (Fig. 4, H and I) and evidence of DNA damage as measured by γH2AX staining (Fig. 4 J). Loss of Brca1 can also lead to up-regulation of the cell cycle arrest gene p21/Cdkn1a (Hakem et al., 1996, 1997). However, KB1f/fLucf/+ pMECs did not show any increase in p21 expression (Fig. 4 K). These data demonstrate that Brca1-deficient pMECs initially survive in vivo transplantation in the clear mammary fat pad but soon lose their repopulation capacity as a result of the onset of senescence, presumably triggered by DNA damage and ROS accumulation.
Nrf2 activation restores ROS levels and survival in Brca1-deficient MECs
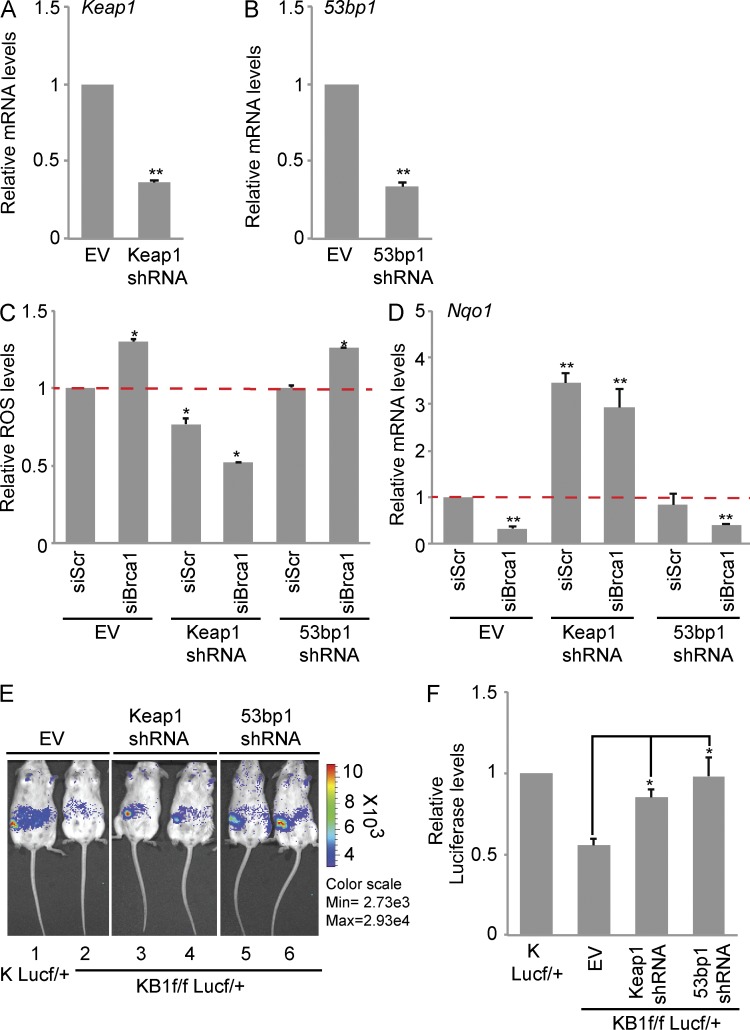
We next determined whether constitutive Nrf2 activation could rescue Brca1-dependent ROS accumulation and in vivo survival defects. COMMA-1D cells were infected with lentiviral shRNA against Keap1 to stabilize and activate Nrf2. As expected, Keap1 shRNA-infected cells (Keap1 shRNA) had low Keap1 expression (Fig. 5 A) and elevated expression of Nqo1 (Fig. 5 D and not depicted) compared with empty vector (EV) control. EV- and Keap1 shRNA–infected cells were subsequently transfected with scrambled siRNAs (siScr) or Brca1 siRNA (siBrca1), and ROS levels were measured. As shown in Fig. 5 D, Keap1 silencing lowered ROS levels in control siScr cells but to a greater extent in siBrca1 cells. To evaluate whether modulation of ROS levels was caused by Nrf2 activation, we measured Nqo1 expression in the same transfected cell lines. Keap1 shRNA significantly increased Nqo1 levels both in siScr and siBrca1 cells (Fig. 5 E).
Figure 5.
Keap1 down-regulation rescues in vivo survival defect and ROS levels in BRCA1-deficient cells. (A) Keap1 mRNA levels in COMMA-1D cells infected with EV or Keap1 shRNA. (B) 53bp1 mRNA levels in COMMA-1D cells infected with EV or 53bp1 shRNA. (C) ROS levels in COMMA-1D cells stably expressing EV or Keap1 and 53bp1 shRNAs after transfection with scrambled siRNA (siScr) or Brca1-specific siRNA (siBrca1). Data are normalized to ROS levels in EV/siScr cells. (D) Nqo1 mRNA levels in cells as described in C. Data are normalized to siScr/EV cells. (A–D) Data represent the mean ± SEM of three biological replicates. (E) Representative images of in vivo Luc activity in outgrowths derived from KLucf/+ pMECs (mouse 1, positive control) and KB1f/fLucf/+ pMECs infected with EV (mouse 2), Keap1 shRNA (mice 3 and 4), or 53bp1 shRNA (mice 5 and 6). (F) Quantitation of Luc activity shown in E using Living Image 3.0 software. Data represent the mean ± SEM from n = 5 outgrowths of each combination. *, P < 0.05; **, P < 0.01.
53BP1 inactivation has been shown to rescue the proliferative defects associated with BRCA1 loss-of-function. In particular, 53BP1 loss alleviates G2/M cell cycle arrest, ATM-dependent checkpoint response, and genomic instability (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010). To investigate the impact of 53BP1 loss on ROS regulation, we used COMMA1-D cells infected with 53bp1 shRNA and cotransfected with siScr or siBrca1 (Fig. 5, B and C). Surprisingly, 53bp1 shRNA did not affect intracellular ROS levels in control cells or alleviate ROS accumulation caused by Brca1 down-regulation (Fig. 5 C). In addition, 53bp1 shRNA did not alter Nqo1 down-regulation caused by Brca1 siRNA (Fig. 5 D).
We then evaluated whether Keap1 or 53bp1 inactivation could rescue the repopulation defects associated with Brca1 loss in vivo by fat pad transplantation assay. Lentiviral-mediated down-regulation of Keap1 and 53bp1 restored cell survival in KB1f/f pMECs into the precleared fat pad of normal recipient mice to a level similar to KLucf/+ pMECs (Fig. 5, E and F).
These data show that suppression of Keap1 rescues cell survival defects associated with Brca1 deficiency through Nrf2-dependent regulation of intracellular ROS. This is in contrast to cell survival associated with suppression of 53bp1, which appears to be independent of Nrf2 transactivation and regulation of ROS.
Estrogen induces an Nrf2-dependent antioxidant response
Tumor development in BRCA1 mutation carriers is largely restricted to hormonally regulated tissues. Although mouse BRCA1 tumor models do not recapitulate the exquisite organ specificity of the human cancer counterpart, some mouse studies suggest that estrogen (E2)-mediated survival of BRCA1-deficient cells is at the basis of BRCA1 tissue–specific tumor suppression (Elledge and Amon, 2002; Li et al., 2007).
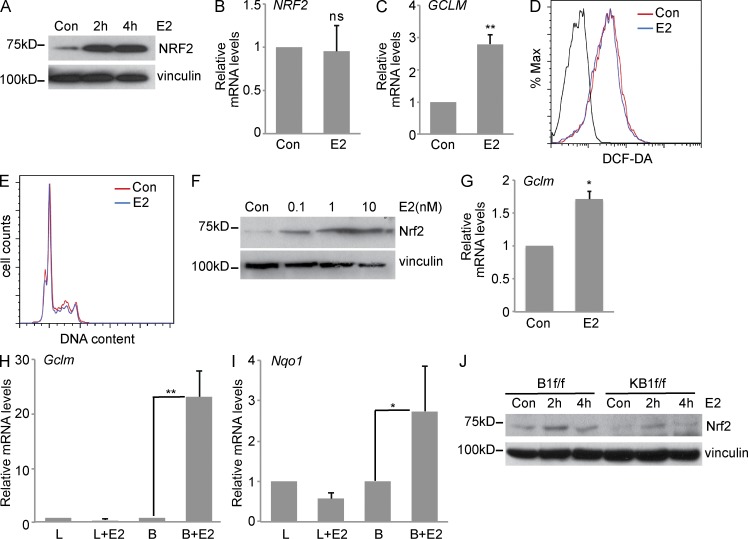
To determine whether E2-regulated signaling can counteract Nrf2 defective response induced by BRCA1 loss, we treated the E2-responsive human breast cancer cell line MCF7 with 17-β estradiol (E2). As previously reported (Dubik and Shiu, 1992), c-MYC mRNA was up-regulated by 10 nM E2 treatment in MCF7 cells (not depicted). Interestingly, E2 stimulation led to significant NRF2 protein increase (Fig. 6 A), with no change in NRF2 mRNA expression (Fig. 6 B). E2 also induced NRF2-driven transactivation, as shown by elevated GCLM and HMOX1 mRNA expression (Fig. 6 C and not depicted).
Figure 6.
Estrogen stimulates an Nrf2-regulated antioxidant response. (A) Representative immunoblot analysis of NRF2 protein in estrogen (E2)-starved MCF7 cells that were treated with 10 nM E2 for 2 or 4 h. (B and C) MCF7 cells that were left untreated (Con) or treated as in A for 4 h were used to detect NRF2 (B) and GCLM (C) mRNA levels by RT-PCR. (D) Representative ROS analysis by FACS in MCF7 cells treated as in B. (E) Representative cell cycle analysis by PI staining of control (Con) or E2-treated MCF7 cells (10 nM for 4 h). (F) HC11 cells were left untreated or stimulated with different doses of E2 as indicated for 4-h Nrf2, and vinculin protein levels were detected. Representative Western blot is shown. (G) Gclm mRNA levels in HC11 treated with 10 nM E2 for 4 h. (B, C, and G) Data represent the mean ± SEM of three biological replicates. (H and I) RT-PCR analysis of Gclm (H) and Nqo1 (I) mRNA levels in luminal (L) and MaSC/basal (B) cells isolated from ovariectomized FVB mice that were treated with vehicle or 0.14 mg E2 for 14 d. Data represent the mean ± SEM of n = 3 independent isolates. L, B = luminal or MaSC/basal cells from vehicle-treated mice; L+E2, B+E2 = luminal or basal cells from E2-treated mice (n = 3/group). (J) Representative Western blot analysis of Nrf2 protein levels in B1f/f and KB1f/f pMECs treated with 10 nM E2 for 2 and 4 h. Vinculin was used as a loading control. *, P < 0.05; **, P < 0.01.
E2 has been shown to elicit pleiotropic effects, including cell cycle regulation (Foster et al., 2001) and ROS generation, through oxidative metabolism or by acting on mitochondrial uncoupling proteins (Fussell et al., 2011). To evaluate whether NRF2 accumulation was caused by E2-induced ROS increase, we measured ROS levels in MCF7 at 4 h after treatment with 10 nM E2, but we did not detect any changes in ROS levels between E2-treated cells and control cells (Fig. 6 D). Similarly, we did not observe any difference in cell cycle profile between E2-starved MCF7 cells before and after E2 stimulation (Fig. 6 E). E2 treatment of the ER-positive mouse MEC line, HC11, also resulted in Nrf2 protein accumulation (Fig. 6 F) and expression of the Nrf2 target gene, Gclm (Fig. 6 G). Collectively, these data suggest that E2 results in Nrf2 accumulation and activation.
To explore whether E2 induced an Nrf2-dependent antioxidant program in vivo, E2 was administered to bilaterally ovariectomized FVB mice. After 2 wk of administration, MaSCs/basal and luminal cells were sorted from mammary gland isolates obtained from vehicle- and E2-treated mice. E2 responsiveness was confirmed by up-regulation of the E2-responsive gene Greb1 (Ghosh et al., 2000) in both MaSCs/basal and luminal cells (not depicted). Interestingly, the Nrf2 targets Gclm and Nqo1 were mainly up-regulated by E2 in MaSCs/basal cells (Fig. 6, H and I). Because MaSCs/basal cells lack ER expression, these data suggest that in vivo E2-induced Nrf2 activation occurs through a paracrine mechanism.
The capacity of E2 to induce Nrf2 accumulation was next evaluated in BRCA1-deleted pMECs. We found that in vitro E2 treatment resulted in differential Nrf2 protein accumulation in B1f/f versus KB1f/f pMECs (Fig. 6 J). Untreated KB1f/f cells showed undetectable Nrf2 levels that increased after 2-h treatment with E2 to a level similar to control B1f/f cells but lower than 2-h E2-treated B1f/f cells. This result shows that E2 may rescue compromised Nrf2 stabilization in the absence of BRCA1 to counteract ROS accumulation and promote cell survival.
Nrf2 antioxidant signaling is defective in human BRCA1-mutated breast tumor cells
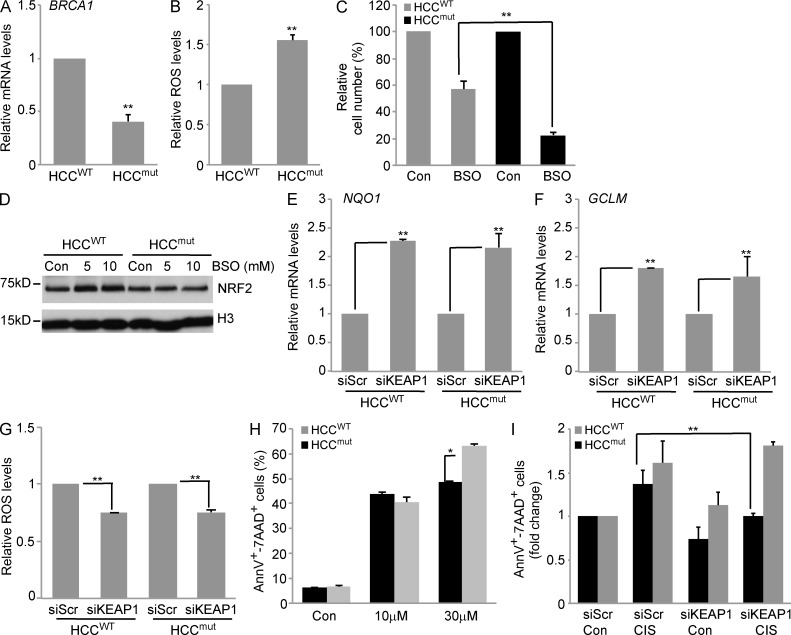
To determine whether human BRCA1-mutated tumor cells retain a defective NRF2 antioxidant response, we analyzed human breast cancer HCC1937 cells (Scully et al., 1999) either carrying a 5382insC BRCA1 mutation (HCCmut) or reconstituted with WT BRCA1 (HCCWT). Reconstitution of HCCmut cells with WT functional BRCA1 increased BRCA1 mRNA levels (Fig. 7 A), reduced intracellular ROS levels (Fig. 7 B), and lowered sensitivity to oxidative stress–induced cell death after treatment with a high dose of BSO (Fig. 7 C).
Figure 7.
Reconstitution of WT BRCA1 in a human BRCA1-mutated breast cancer cell line restores Nrf2 activation. (A) RT-PCR analysis of BRCA1 mRNA levels in HCCWT and HCCmut cells. (B) Quantitation of ROS levels analyzed by FACS in HCCWT and HCCmut cells. (C) Cell viability of HCCWT and HCCmut cells at 4 d after treatment with 5 mM BSO. Data are the mean percentage ± SEM of viable cells compared with untreated controls (n = 3/group). (D) Representative immunoblot of NRF2 protein levels in HCCWT and HCCmut cells that were left untreated (Con) or treated with the indicated doses of BSO. H3 served as a loading control. (E and F) NQO1 (E) and GCLM (F) expression levels in HCCWT and HCCmut cells transfected with scrambled siRNA (siScr) and KEAP1 siRNA (siKEAP1). (G) ROS levels in siScr- and siKeap1-transfected HCCWT and HCCmut cells. Data in siKeap1 cells are normalized to siScr controls for each cell line (n = 3/group). Data represent the mean ± SEM of three biological replicates, normalized to siScr-transfected cells. (H) Analysis of apoptosis by Annexin V/7-AAD staining of HCCWT and HCCmut cells left untreated (Con) or treated with 10 or 30 µM cisplatin for 24 h. (I) Analysis of apoptosis by Annexin V/7-AAD staining in siScr- and siKeap1-transfected HCCWT and HCCmut cells left untreated (Con) or treated with 10 µM cisplatin (CIS) for 24 h. Data are normalized to untreated siScr cells for each cell line. (A, B, E, F, H, and I) Data represent the mean ± SEM of three biological replicates. *, P < 0.05; **, P < 0.01.
We determined whether increased sensitivity of HCCmut cells to BSO was in part caused by compromised NRF2 signaling. Compared with HCCWT cells, HCCmut cells failed to up-regulate NRF2 upon BSO treatment (Fig. 7 D). However, transfection with KEAP1 siRNA increased the expression of the NRF2 target genes NQO1 (Fig. 7 E) and GCLM (Fig. 7 F) and reduced intracellular ROS levels to a similar extent in both HCCmut and HCCWT (Fig. 7 G).
BRCA1-associated tumors are more sensitive to interstrand DNA cross-linking agents, such as cisplatin and carboplatin (D’Andrea, 2010). As platinum-based therapy generates high intracellular ROS levels (Conklin, 2004), we evaluated whether modulating ROS levels may alter platinum sensitivity. HCCWT and HCCmut cells were treated with cisplatin after KEAP1 down-regulation by siRNA. As previously reported (Tassone et al., 2009), HCCmut cells were more sensitive to cisplatin than HCCWT cells but only at higher concentration (30 µM compared with 10 µM; Fig. 7 H). Because 30 µM cisplatin induced high levels of cell death, we determined the role of KEAP1 siRNA in cells treated with low-dose (10 µM) cisplatin. We found that KEAP1 down-regulation preferentially decreased cisplatin-induced cell death in HCCmut cells compared with HCCWT cells (Fig. 7 I). These data suggest that constitutive NRF2 signaling achieved by KEAP1 silencing modulates the sensitivity to cisplatin in BRCA1-mutated tumor cells.
NQO1 expression correlates with tumor-associated BRCA1 loss or mutation
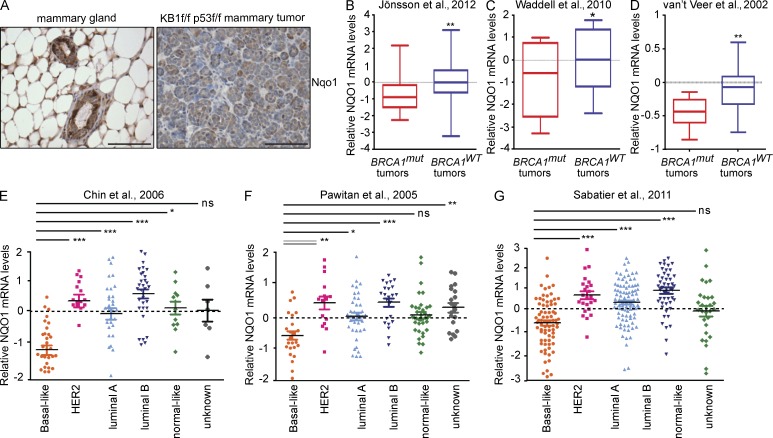
We next evaluated whether mouse and human BRCA1-related tumors show evidence of impaired Nrf2 activation. Mouse mammary tumors were generated from crossing conditional p53f/f with KB1f/f mice as previously described (Liu et al., 2007). Immunohistochemical analysis of KB1f/fp53f/f mammary tumors showed low expression of the Nrf2 target gene Nqo1 compared with normal mammary acini (Fig. 8 A).
Figure 8.
NQO1 is down-regulated in Brca1-null mouse mammary tumors and BRCA1-mutated human breast cancers. (A) Immunohistochemical analysis of Nqo1 protein in normal mouse mammary gland (left) and in a mammary tumor arising in a K14cre Brca1f/fp53f/f mouse (KB1f/fp53f/f; right). Results are representative of five glands from normal FVB mice and five tumors from KB1f/fp53f/f mice. Bars, 50 µm. (B–D) Box plots representing NQO1 mRNA levels in human primary BRCA1-mutated breast cancers (BRCA1mut) and BRCA1-proficient breast cancers (BRCA1WT) from three different datasets as indicated in each panel and described in Materials and methods. (E–G) Dot plots showing NQO1 mRNA levels in the indicated human primary breast tumor subtypes from three different datasets as indicated in each panel and described in Materials and methods. Vertical lines indicate the variability outside the upper and lower quartiles represented in the box plots. P-values were determined by Kruskal–Wallis analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
To translate our experimental findings to primary human breast tumors, we examined the expression levels of NQO1 in publicly available gene expression datasets from three cohorts of, respectively, 577, 117, and 75 primary breast cancer tumors with known BRCA1 mutational status (Jönsson et al., 2012; van ’t Veer et al., 2002; Waddell et al., 2010). All three analyses showed that NQO1 expression levels were significantly lower in tumors from BRCA1 mutation carriers (Fig. 8, B–D).
The strong association between BRCA1 mutations and BRCA1 promoter hypermethylation within the basal-like subtype (Jönsson et al., 2012) prompted us to determine NQO1 expression levels in primary sporadic breast tumors. We analyzed 555 breast tumors from three independent gene expression datasets (Pawitan et al., 2005; Chin et al., 2006; Sabatier et al., 2011). Tumor subtype classification was determined as described in each study. We found that the lowest NQO1 expression levels were characteristic of basal-like tumors (Fig. 8, E–G), suggesting that NQO1 deregulation may be involved in both BRCA1-mutated tumors and sporadic basal-like tumors.
DISCUSSION
Our current understanding of BRCA1-related breast carcinogenesis posits that, in carriers of BRCA1 mutations, loss of heterozygosity of the remaining WT BRCA1 allele must be accompanied by a secondary event such as p53 inactivation to achieve full transformation (Foulkes, 2008). Cells lacking both p53 and BRCA1 are genomically unstable and acquire additional mutations that drive tumor development such as loss of PTEN (Saal et al., 2008). Recent data suggest that 53BP1 inactivation also alleviates the proliferation defects, DNA damage hypersensitivity, and genomic instability induced by BRCA1 deficiency (Cao et al., 2009; Bouwman et al., 2010; Bunting et al., 2010).
Our work provides new insights in BRCA1-associated tumorigenesis through the regulation of oxidative stress. We showed that BRCA1-deficient cells have increased ROS levels (Fig. 1) caused by impaired Nrf2-mediated antioxidant signaling (Fig. 2) and BRCA1 regulates Nrf2 transcription and protein stability by interacting with Nrf2 (Figs. 2 and 3). Our data also postulate the existence of a positive regulatory feedback loop between BRCA1 and Nrf2 that is sustained by other recent studies (Kang et al., 2011; Wang et al., 2013).
We then translated our in vitro findings to the in vivo fat pad transplantation assay. In the absence of additional events, we demonstrated that BRCA1-related oxidative stress is, at least partially, responsible for the impaired repopulation defects of Brca1-null pMECs in the mammary gland (Figs. 4 and 5). As a proof, Nrf2 constitutive activation by Keap1 silencing is able to rescue ROS levels, Nqo1 down-regulation, and in vivo survival caused by Brca1 deficiency (Fig. 5).
The role of BRCA1 as sensor and regulator of ROS has important implications in BRCA1-associated tumor formation. Among the previously characterized cellular defects induced by BRCA1 deficiency such as lack of DNA repair and cell cycle abnormalities, high oxidative stress may represent another barrier that prevents transformation of BRCA1-deficient cells.
From this study, it appears that the control of DNA damage signaling through 53BP1 and the regulation of ROS homeostasis through Nrf2 are two parallel and independent pathways downstream of BRCA1. However, both pathways are required to alleviate the survival defect in BRCA1-null cells.
The tissue specificity of the tumors that develop in BRCA1 mutation carriers invokes a critical role for estrogen signaling. Our findings suggest that estrogen plays a role in regulating Nrf2 activation and in particular may counteract down-regulation of Nrf2-dependent signaling induced by BRCA1 loss to promote cell survival (Fig. 6). Increased survival of BRCA1-deficient cells permits proliferation and increases genomic instability and acquisition of p53, 53BP1, and/or PTEN mutations that promote malignant transformation.
Current efforts are devoted to the discovery of novel and more efficacious therapies to treat BRCA1-mutated breast cancers (Drost and Jonkers, 2009; Foulkes et al., 2010). In this context, our study suggests that deregulated Nrf2-dependent antioxidant signaling sensitizes human BRCA1-mutated tumors to chemotherapy-induced oxidative stress and may provide new avenues for therapeutic intervention.
MATERIALS AND METHODS
Mice.
K14cre transgenic mice and p53 conditional knockout mice were obtained from A. Berns (Netherlands Cancer Institute, Amsterdam, Netherlands; Jonkers et al., 2001). Brca1 conditional knockout mice were provided by J. Jonkers (Liu et al., 2007). Luc-expressing transgenic mice were purchased from the Jackson Laboratory (Safran et al., 2003). All of the strains were in the FVB background. TdTomato mice in the 129/B6 background were purchased from the Jackson Laboratory (Madisen et al., 2010). Mice were maintained and handled according to protocols approved by the Animal Care and Use Committee of the University Health Network, Toronto, Ontario, Canada. Before mammary gland isolation, 8–10-wk-old females were staged by vaginal smear cytology as described previously (Joshi et al., 2010). For in vivo estrogen experiments, mice were ovariectomized and treated as described previously (Joshi et al., 2010).
Cell lines and culture conditions.
The following cell lines were used: COMMA-1D cells (provided by S. Muthuswamy, Ontario Cancer Institute, Toronto, Ontario, Canada) in DMEM/F12 medium containing 10% FBS, l-glutamine, 1 µg/ml hydrocortisone (Sigma-Aldrich), 5 µg/ml insulin (Sigma-Aldrich), and 5 ng/ml epidermal growth factor (EGF; Sigma-Aldrich); mouse HC11 cells (provided by M. Jason, University of Toronto, Toronto, Ontario, Canada) in RPMI 1640 medium containing 10% FBS, l-glutamine, 5 µg/ml insulin, and 5 ng/ml EGF; MCF7 cells (provided by H. Okada, The Campbell Family Cancer Research Institute, Toronto, Ontario, Canada) in DMEM with 10% FBS; and HCC1937 BRCA1 mutant breast cancer cells (HCCmut) and HCC1937 reconstituted with WT BRCA1 (HCCWT; provided by R. Scully, Harvard Medical School and Beth Israel Deaconess Medical Center, Boston, MA) in RPMI 1640 with 10% FBS (Scully et al., 1999).
Oxidative stress was induced for 48 h in medium containing 2 mM BSO (Sigma-Aldrich). In the experiments with antioxidants, cells were cotreated with both 250 µM Trolox and BSO. For estrogen treatment, MCF7, HC11, and pMECs were maintained in their respective E2-stripped media (Phenol red–free media and with 10% charcoal-filtered FBS) for 3 d before addition of 10 nM 17-β estradiol (E2; Sigma-Aldrich) for 2 or 4 h. HCC cells were exposed to 10 or 30 µM cisplatin for 24-h treatment.
Mouse and HMEC isolation, flow cytometric analysis, and cell sorting.
Mouse pMECs were obtained through dissociation of #4 mammary glands and processed as described previously (Joshi et al., 2010). All reagents were obtained from STEMCELL Technologies. For flow cytometry, cells were stained with the Abs PE–anti-CD24 (STEMCELL Technologies) and APC–anti-CD49f (R&D Systems). pMECs were either sorted to >96% purity using a FACSAria instrument (BD) or cultured in serum-free medium for 3 d to kill stromal fibroblasts. Flow cytometry for H2AX was determined using FITC–anti-H2AX Ab (EMD Millipore) after intracellular fixation and permeabilization with buffers from eBioscience according to the manufacturers’ instructions.
HMECs were isolated from fresh human mammary tissue as described previously (Gauthier et al., 2007). Human tissue was obtained with informed patient consent and Institutional Research Ethics Board approval from St. Michael’s Hospital and the University Health Network.
Mammary fat pad transplantation.
pMECs (2 × 105/genotype) from B1f/fLucf/+, KLucf/+, and KB1f/fLucf/+ mice were resuspended 1:1 in 1× PBS/Matrigel (Matrigel Matrix High Concentration; BD) and injected into precleared fat pads of 21-d-old FVB recipient mice. Fat pad clearance of preexisting epithelium was performed as previously described (Joshi et al., 2010). For in vivo tracking of engrafted pMECs, 150 mg d-luciferin (Caliper)/kg mouse body weight was injected into the intraperitoneal cavity of anesthetized mice. Outgrowths were analyzed by in vivo imaging (IVIS Spectrum; PerkinElmer), and Luc activity was measured using Living Image Software 4.3.
Genomic DNA PCR.
Genomic DNA was extracted from pMECs using the NucleoSpin Tissue kit (Macherey-Nagel) according to the manufacturer’s protocol. For qualitative PCR, P1, P2, P3, and P4 DNA primers were used as described in Liu et al. (2007). For qPCR, specific primers that detect a DNA region comprised within exon 5–13 that is missing in KB1f/f mice were used (gBrca1_F/R) along with an internal control (gIC_F/R). See Table S1 for primer sequences.
RT-PCR.
RNA was isolated using the NucleoSpin RNA II kit (Macherey-Nagel) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturers’ instructions. qRT-PCR was performed using SYBR green primers (Applied Biosystems). Samples from ovariectomized mice were processed for RNA isolation and cDNA preparation as previously described (Joshi et al., 2010). qRT-PCR was performed using SYBR green primers. All data were normalized to values obtained for luminal and MaSCs/basal cells isolated from their respective vehicle-treated mice.
Mouse β-actin, mouse ribosomal protein S9 (rps9), and human ribosomal protein S18 (rps18) were used as housekeeping genes to determine relative mRNA expression. All RT-PCR primer sequences are described in Table S2.
Cell cycle analysis.
Cell cycle analysis was performed by propidium iodide (PI) staining. In brief, cells were fixed overnight in cold 70% ethanol. After washes with 1× PBS, cells were treated with 50 µl of 100 µg/ml stock of RNase and stained with 200 µl PI (from 50 µg/ml stock solution). Cells were analyzed by a FACSCalibur flow cytometer (BD) immediately after staining. Cell cycle distribution was analyzed as typical DNA content histogram.
Apoptosis measurement.
Apoptosis was evaluated by Annexin V/7-AAD staining. In brief, cells were collected and stained with PE-conjugated Annexin V and 7-AAD for 15 min at room temperature in 10× binding buffer. All reagents were purchased from BD. Cells were analyzed by a FACSCalibur flow cytometer immediately after staining.
ROS measurement.
To measure intracellular ROS in pMECs, surface-stained cells were incubated with 300 nM CM-H2DCFDA (DCF-DA; Invitrogen) for 10 min at 37°C. DCF-DA fluorescence was analyzed by flow cytometry using a FACSCanto instrument (BD) and FlowJo software (Tree Star). Because HCCmut or HCCWT cells express GFP, ROS measurements were performed in these cells using dihydroethidium (DHE; Sigma-Aldrich). Mean fluorescence values (FL-1 for DCF-DA or FL-2 for DHE) were displayed as bar graphs. In case of sorted luminal and MaSCs/basal pMECs, data were shown in the form of histogram overlays using the %Max option, which scaled each population curve to mode = 100% on the y axis and log10 FL-1 (DCF-DA) fluorescence intensity on the x axis.
Cell viability measurement.
Cell viability was measured by Trypan blue exclusion assay. In brief, 105 HCC1937 cells were seeded in 6-well plates 24 h before being treated with 5 mM BSO for 4 d. Trypan blue–negative viable cells were scored each day after staining with 0.4% Trypan blue solution.
Gene silencing by siRNA oligonucleotides and lentiviral shRNAs.
For gene silencing by siRNA, the following Thermo Fisher Scientific siRNAs were used: nontargeting scrambled siRNA, mouse Brca1 siRNA, mouse Nfe2l2 siRNA, human NFE2L2 siRNA, and human KEAP1 siRNA. For siRNA experiments, cells were transfected with 25–100 pmol siRNA using Lipofectamine 2000 in a 6-well plate. Transfection media was removed after 5 h. Transfected cells were cultured for 24–48 h before experiments. For shRNA experiments, the following lentiviral shRNA constructs were used: dox-inducible lentiviral mouse Brca1 shRNA from Thermo Fisher Scientific (pTRIPZ) and lentiviral pLKO shRNAs against human Luc, mouse Trp53bp1, and mouse Keap1 (a gift of T. Ketela, University of Toronto). The human BRCA1 shRNA target sequence, 5′-AAGCTCCTCTCACTCTTCAGT-3′ (Sankaran et al., 2005), was subcloned into pLKO.1puro lentiviral vector (Addgene). Lentiviruses were produced in 293FT packaging cells transfected with Lipofectamine 2000. Cells were infected with lentiviruses in the presence of 8 µg/ml Polybrene (Sigma-Aldrich) for 4 (HMECs) and 18 h (COMMA-1D; pMECs). Cells were cultured for 24 h after infection before antibiotic selection with 2–4 µg/ml puromycin (Sigma-Aldrich). pMECs were in vivo injected the day after infection without antibiotic selection.
Immunoprecipitation and immunoblotting.
Immunoprecipitations were performed in 293FT cells using Lipofectamine 2000 with the following overexpression constructs: Myc-BRCA1 (provided by R. Hakem, Ontario Cancer Institute; Wilson et al., 1997); HA-ubiquitin (gift of E. Guccione, A*Star, Singapore); His-Keap1 (gift of J.P. Ting, University of North Carolina at Chapel Hill, Chapel Hill, NC); GFP-Nrf2 (Addgene; Invitrogen); and HA-Nrf2, HA-29DLG Nrf2, and HA-79ETGE Nrf2 (gifts of D.D. Zhang, University of Arizona, Tucson, AZ). Cells were collected at 36 h after transfection and lysed in RIPA buffer. Lysates (>1 mg) were incubated at 4°C first with 2 µg of primary Ab for 2 h followed by incubation with Protein A–agarose beads (GE Healthcare) for 1 h. Beads were washed three times with RIPA buffer, resuspended in NuPAGE LDS sample buffer (Invitrogen), and incubated at 70°C for 5 min before loading on precast SDS-PAGE protein gels (Invitrogen). For immunoprecipitation of ubiquitinated GFP-Nrf2, 25 µM MG132 (Sigma-Aldrich) was added 4 h before cell lysis.
For endogenous immunoprecipitations, we modified the protocol described above as follows. Cells lysed in RIPA buffer and lysates (>1 mg) were incubated at 4°C with 2 µg of primary Ab for 16 h followed by incubation with Protein A–agarose beads (GE Healthcare) for 1 h.
Immunoblotting was performed using standard protocols with the following primary Abs: Nrf2 (Santa Cruz Biotechnology, Inc.), mouse Nrf2 (gift from D. Tuveson [Cold Spring Harbor Laboratory, Cold Spring, New York] and E. Schmidt [Montana State University, Bozeman, MT]), BRCA1 (Santa Cruz Biotechnology, Inc.), mouse BRCA1 (GH118; gift from J. Jonkers), vinculin (Abcam), β-actin (Sigma-Aldrich), H3 (Genetex), Myc (Cell Signaling Technology), GFP (Novus Biologicals), and His (Invitrogen). Anti–mouse and anti–rabbit HRP-conjugated secondary Abs were obtained from Thermo Fisher Scientific. When required, immunoblots were quantified by ImageJ 1.45 software (National Institutes of Health).
Whole-mount mammary gland staining.
#4 mammary glands were dissected and placed on glass slides and fixed overnight with Carnoy’s solution (1:3:6 glacial acetic/chloroform/ethanol). The glands were rehydrated before staining with aluminum carmine overnight. After dehydration, glands were cleared in toluene and mounted. Imaging was performed on a stereo microscope (MZ16F; Leica) in conjunction with Volocity acquisition software (version 4.0; PerkinElmer).
Tissue preparation and immunohistochemistry.
Mammary glands from normal FVB, KLucf/+, and KB1f/fLucf/+ mice and mammary tumors from KB1f/fp53f/f mice were fixed in formalin, processed, and embedded in paraffin for sectioning. 5-µm paraffin sections were dewaxed and rehydrated according to standard procedures.
In case of staining with anti-Luc (Sigma-Aldrich) and anti-CK14 (Covance), microwave antigen retrieval using 10 mM Tris EDTA, pH 9, was used. Primary Abs were incubated overnight at 4°C. Secondary Abs were applied after washing using 1× PBS (Cy2-conjugated anti–rabbit and Cy3-conjugated anti–mouse; Jackson ImmunoResearch Laboratories, Inc.) and were incubated at room temperature for 45 min. Slides were washed with PBS and mounted using Entellan. Imaging was performed on a DM6000 fluorescent microscope (Leica) with appropriate filter sets and high-resolution camera (C4742-80; Hamamatsu Photonics) in conjunction with Volocity acquisition software (version 4.0).
In case of staining with anti-NQO1 Ab (Abcam), antigen retrieval was performed in 1 µM EDTA, pH 9.0, and sections were incubated overnight at 4°C with the primary Ab. Antigen–Ab complexes were visualized using the VECTASTAIN Elite ABC kit followed by 3,3′-diaminobenzidine staining (DAB; Vector Laboratories) and counterstained with Mayer’s hematoxylin (Thermo Fisher Scientific). Specimens were viewed with a brightfield and epifluorescence microscope (Leica DM2500 equipped with Micropublisher 3.3-QI imaging camera) using Q-Capture Pro software (QImaging) and processed with Photoshop CS5 (Adobe).
Statistical analyses.
Results were reported as the mean ± SEM. Two-sided independent Student’s t test without equal variance assumption was used to determine p-values (*, P < 0.05; **, P < 0.01). For analysis of gene expression microarray in primary human breast tumors, box plots and dot plots were performed on three publicly available datasets of BRCA1 mutated breast tumors (van ’t Veer et al., 2002; Jönsson et al., 2010; Waddell et al., 2010) and on three sporadic breast tumor datasets (Pawitan et al., 2005; Chin et al., 2006; Sabatier et al., 2011), respectively. Tumor subtype classification was determined as described in each study. Values indicated in the plots are median-centered levels of NQO1 gene expression as assessed from normalized microarray data. Differences in NQO1 (201468_s_at) mRNA levels among breast tumor subtypes were evaluated using Man–Whitney analysis.
Online supplemental material.
Table S1 includes the list of primers used for PCR with genomic DNA. Table S2 includes the list of primers used for RT-PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20121337/DC1.
Supplementary Material
Acknowledgments
We thank D.S. Park (Ottawa Hospital Research Institute, Ottawa, Ontario, Canada) and J.P. Ting for valuable insights; R. Hakem for providing DNA constructs; T. Deng, J.C. Liu, and E. Zacksenhaus (all from Toronto General Research Institute, Toronto, Ontario, Canada) for insightful comments and assistance with mammosphere assays and fat pad transplantation; D.D. Zhang, J. Parvin (The Ohio State University, Columbus, OH), D. Tuveson, and E. Schmidt for providing valuable tools; and M. Saunders for scientific editing.
This research was partially supported by a European Molecular Biology Organization Long-term Fellowship to C. Gorrini and by grants from the Canadian Institutes of Health Research to M.L. Gauthier and T.W. Mak and from the Ontario Ministry of Health and Long-Term Care (OMOHLTC) to T.W. Mak. Please note that the views expressed do not necessarily reflect those of the OMOHLTC.
The authors have no conflicts of interest to disclose.
Footnotes
Abbreviations used:
- Ab
- antibody
- BSO
- l-buthionine-sulfoximine
- dox
- doxycycline
- ER
- estrogen receptor
- EV
- empty vector
- HMEC
- human MEC
- Luc
- luciferase
- MaSC
- mammary stem cell
- MEC
- mammary epithelial cell
- PI
- propidium iodide
- pMEC
- primary MEC
- ROS
- reactive oxygen species
References
- Bae I., Fan S., Meng Q., Rih J.K., Kim H.J., Kang H.J., Xu J., Goldberg I.D., Jaiswal A.K., Rosen E.M. 2004. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 64:7893–7909 10.1158/0008-5472.CAN-04-1119 [DOI] [PubMed] [Google Scholar]
- Barbano R., Muscarella L.A., Pasculli B., Valori V.M., Fontana A., Coco M., la Torre A., Balsamo T., Poeta M.L., Marangi G.F., et al. 2013. Aberrant Keap1 methylation in breast cancer and association with clinicopathological features. Epigenetics. 8:105–112 10.4161/epi.23319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns R.A., Harris I.S., Mak T.W. 2011. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 11:85–95 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- Cao L., Xu X., Cao L.L., Wang R.H., Coumoul X., Kim S.S., Deng C.X. 2007. Absence of full-length Brca1 sensitizes mice to oxidative stress and carcinogen-induced tumorigenesis in the esophagus and forestomach. Carcinogenesis. 28:1401–1407 10.1093/carcin/bgm060 [DOI] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S.F., Liu J., Wang R.H., Cao L.L., Wu J.J., Peng T.N., Chen J., Nussenzweig A., et al. 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell. 35:534–541 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Wang R.H., Akagi K., Kim K.A., Martin B.K., Cavallone L., Haines D.C., Basik M., Mai P., Poggi E., et al. ; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) 2011. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 17:1275–1282 10.1038/nm.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. 2009. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 34:663–673 10.1016/j.molcel.2009.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Farmer A.A., Chen C.F., Jones D.C., Chen P.L., Lee W.H. 1996. BRCA1 is a 220-kDa nuclear phosphoprotein that is expressed and phosphorylated in a cell cycle-dependent manner. Cancer Res. 56:3168–3172 [PubMed] [Google Scholar]
- Chin K., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., et al. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 10:529–541 10.1016/j.ccr.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Conklin K.A. 2004. Chemotherapy-associated oxidative stress: impact on chemotherapeutic effectiveness. Integr. Cancer Ther. 3:294–300 10.1177/1534735404270335 [DOI] [PubMed] [Google Scholar]
- D’Andrea A.D. 2010. Susceptibility pathways in Fanconi’s anemia and breast cancer. N. Engl. J. Med. 362:1909–1919 10.1056/NEJMra0809889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M.J., Forni L.G., Willson R.L. 1988. Vitamin E analogue Trolox C. E.s.r. and pulse-radiolysis studies of free-radical reactions. Biochem. J. 255:513–522 [PMC free article] [PubMed] [Google Scholar]
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. 2011. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 475:106–109 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. 2009. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 458:780–783 10.1038/nature07733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost R.M., Jonkers J. 2009. Preclinical mouse models for BRCA1-associated breast cancer. Br. J. Cancer. 101:1651–1657 10.1038/sj.bjc.6605350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubik D., Shiu R.P. 1992. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene. 7:1587–1594 [PubMed] [Google Scholar]
- Elledge S.J., Amon A. 2002. The BRCA1 suppressor hypothesis: an explanation for the tissue-specific tumor development in BRCA1 patients. Cancer Cell. 1:129–132 10.1016/S1535-6108(02)00041-7 [DOI] [PubMed] [Google Scholar]
- Foster J.S., Henley D.C., Ahamed S., Wimalasena J. 2001. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol. Metab. 12:320–327 10.1016/S1043-2760(01)00436-2 [DOI] [PubMed] [Google Scholar]
- Foulkes W.D. 2008. BRCA1—sowing the seeds crooked in the furrow. Nat. Genet. 40:8–9 10.1038/ng0108-8 [DOI] [PubMed] [Google Scholar]
- Foulkes W.D., Smith I.E., Reis-Filho J.S. 2010. Triple-negative breast cancer. N. Engl. J. Med. 363:1938–1948 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- Fussell K.C., Udasin R.G., Smith P.J., Gallo M.A., Laskin J.D. 2011. Catechol metabolites of endogenous estrogens induce redox cycling and generate reactive oxygen species in breast epithelial cells. Carcinogenesis. 32:1285–1293 10.1093/carcin/bgr109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier M.L., Berman H.K., Miller C., Kozakeiwicz K., Chew K., Moore D., Rabban J., Chen Y.Y., Kerlikowske K., Tlsty T.D. 2007. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 12:479–491 10.1016/j.ccr.2007.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M.G., Thompson D.A., Weigel R.J. 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60:6367–6375 [PubMed] [Google Scholar]
- Griffith O.W. 1999. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 27:922–935 10.1016/S0891-5849(99)00176-8 [DOI] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Sirard C., Mo R., Woo M., Hakem A., Wakeham A., Potter J., Reitmair A., Billia F., et al. 1996. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 85:1009–1023 10.1016/S0092-8674(00)81302-1 [DOI] [PubMed] [Google Scholar]
- Hakem R., de la Pompa J.L., Elia A., Potter J., Mak T.W. 1997. Partial rescue of Brca1 (5-6) early embryonic lethality by p53 or p21 null mutation. Nat. Genet. 16:298–302 10.1038/ng0797-298 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., et al. 2004. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 431:997–1002 10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29:418–425 10.1038/ng747 [DOI] [PubMed] [Google Scholar]
- Jönsson G., Staaf J., Vallon-Christersson J., Ringnér M., Holm K., Hegardt C., Gunnarsson H., Fagerholm R., Strand C., Agnarsson B.A., et al. 2010. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Res. 12:R42 10.1186/bcr2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson G., Staaf J., Vallon-Christersson J., Ringnér M., Gruvberger-Saal S.K., Saal L.H., Holm K., Hegardt C., Arason A., Fagerholm R., et al. 2012. The retinoblastoma gene undergoes rearrangements in BRCA1-deficient basal-like breast cancer. Cancer Res. 72:4028–4036 10.1158/0008-5472.CAN-12-0097 [DOI] [PubMed] [Google Scholar]
- Joshi P.A., Jackson H.W., Beristain A.G., Di Grappa M.A., Mote P.A., Clarke C.L., Stingl J., Waterhouse P.D., Khokha R. 2010. Progesterone induces adult mammary stem cell expansion. Nature. 465:803–807 10.1038/nature09091 [DOI] [PubMed] [Google Scholar]
- Kang H.J., Hong Y.B., Kim H.J., Rodriguez O.C., Nath R.G., Tilli E.M., Albanese C., Chung F.L., Kwon S.H., Bae I. 2011. Detoxification: a novel function of BRCA1 in tumor suppression? Toxicol. Sci. 122:26–37 10.1093/toxsci/kfr089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler T.W., Wakabayashi N. 2010. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 31:90–99 10.1093/carcin/bgp231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130–7139 10.1128/MCB.24.16.7130-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M.K., Itoh K., Yamamoto M., Kensler T.W. 2002. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 22:2883–2892 10.1128/MCB.22.9.2883-2892.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kong A.N. 2009. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 48:91–104 10.1002/mc.20465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xiao C., Vonderhaar B.K., Deng C.X. 2007. A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene. 26:7204–7212 10.1038/sj.onc.1210527 [DOI] [PubMed] [Google Scholar]
- Liu X., Holstege H., van der Gulden H., Treur-Mulder M., Zevenhoven J., Velds A., Kerkhoven R.M., van Vliet M.H., Wessels L.F., Peterse J.L., et al. 2007. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. USA. 104:12111–12116 10.1073/pnas.0702969104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R., Brown K.R., Suarez F., Sayad A., Karamboulas K., Krzyzanowski P.M., Sircoulomb F., Medrano M., Fedyshyn Y., Koh J.L., et al. 2012. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2:172–189 10.1158/2159-8290.CD-11-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marí M., Morales A., Colell A., García-Ruiz C., Fernández-Checa J.C. 2009. Mitochondrial glutathione, a key survival antioxidant. Antioxid. Redox Signal. 11:2685–2700 10.1089/ars.2009.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 22:66–79 10.1016/j.ccr.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Narod S.A., Foulkes W.D. 2004. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 4:665–676 10.1038/nrc1431 [DOI] [PubMed] [Google Scholar]
- Nioi P., Nguyen T. 2007. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun. 362:816–821 10.1016/j.bbrc.2007.08.051 [DOI] [PubMed] [Google Scholar]
- Pang Q. 2011. HSCs: stressing out over ROS. Blood. 118:2932–2934 10.1182/blood-2011-07-367755 [DOI] [PubMed] [Google Scholar]
- Pawitan Y., Bjöhle J., Amler L., Borg A.L., Egyhazi S., Hall P., Han X., Holmberg L., Huang F., Klaar S., et al. 2005. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 7:R953–R964 10.1186/bcr1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K., et al. 2011. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 476:346–350 10.1038/nature10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L.H., Gruvberger-Saal S.K., Persson C., Lövgren K., Jumppanen M., Staaf J., Jönsson G., Pires M.M., Maurer M., Holm K., et al. 2008. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat. Genet. 40:102–107 10.1038/ng.2007.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier R., Finetti P., Cervera N., Tallet A., Benchalal M., Houvenaeghel G., Jacquemier J., Birnbaum D., Bertucci F. 2011. Gene expression profiling and its utility in prediction of local relapse after breast-conserving therapy in early breast cancer. Cancer Genomics Proteomics. 8:199–209 [PubMed] [Google Scholar]
- Safran M., Kim W.Y., Kung A.L., Horner J.W., DePinho R.A., Kaelin W.G., Jr 2003. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol. Imaging. 2:297–302 10.1162/153535003322750637 [DOI] [PubMed] [Google Scholar]
- Saha T., Rih J.K., Rosen E.M. 2009. BRCA1 down-regulates cellular levels of reactive oxygen species. FEBS Lett. 583:1535–1543 10.1016/j.febslet.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran S., Starita L.M., Groen A.C., Ko M.J., Parvin J.D. 2005. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol. Cell. Biol. 25:8656–8668 10.1128/MCB.25.19.8656-8668.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer Z.T., Grassian A.R., Song L., Jiang Z., Gerhart-Hines Z., Irie H.Y., Gao S., Puigserver P., Brugge J.S. 2009. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 461:109–113 10.1038/nature08268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D.M. 1999. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 4:1093–1099 10.1016/S1097-2765(00)80238-5 [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. 2006. Generation of a functional mammary gland from a single stem cell. Nature. 439:84–88 10.1038/nature04372 [DOI] [PubMed] [Google Scholar]
- Shibutani S., Takeshita M., Grollman A.P. 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 349:431–434 10.1038/349431a0 [DOI] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Smart C.E., Wronski A., French J.D., Edwards S.L., Asselin-Labat M.L., Waddell N., Peters K., Brewster B.L., Brooks K., Simpson K., et al. 2011. Analysis of Brca1-deficient mouse mammary glands reveals reciprocal regulation of Brca1 and c-kit. Oncogene. 30:1597–1607 10.1038/onc.2010.538 [DOI] [PubMed] [Google Scholar]
- Smith G.H., Medina D. 2008. Re-evaluation of mammary stem cell biology based on in vivo transplantation. Breast Cancer Res. 10:203 10.1186/bcr1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J. 2006. Purification and unique properties of mammary epithelial stem cells. Nature. 439:993–997 [DOI] [PubMed] [Google Scholar]
- Tassone P., Di Martino M.T., Ventura M., Pietragalla A., Cucinotto I., Calimeri T., Bulotta A., Neri P., Caraglia M., Tagliaferri P. 2009. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol. Ther. 8:648–653 [DOI] [PubMed] [Google Scholar]
- van ’t Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415:530–536 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- Vurusaner B., Poli G., Basaga H. 2012. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic. Biol. Med. 52:7–18 10.1016/j.freeradbiomed.2011.09.035 [DOI] [PubMed] [Google Scholar]
- Waddell N., Arnold J., Cocciardi S., da Silva L., Marsh A., Riley J., Johnstone C.N., Orloff M., Assie G., Eng C., et al. ; kConFab Investigators 2010. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res. Treat. 123:661–677 10.1007/s10549-009-0653-1 [DOI] [PubMed] [Google Scholar]
- Wang Q., Li J., Yang X., Sun H., Gao S., Zhu H., Wu J., Jin W. 2013. Nrf2 is associated with the regulation of basal transcription activity of the BRCA1 gene. Acta Biochim. Biophys. Sin. (Shanghai). 45:179–187 10.1093/abbs/gmt001 [DOI] [PubMed] [Google Scholar]
- Wilson C.A., Payton M.N., Elliott G.S., Buaas F.W., Cajulis E.E., Grosshans D., Ramos L., Reese D.M., Slamon D.J., Calzone F.J. 1997. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene. 14:1–16 10.1038/sj.onc.1200924 [DOI] [PubMed] [Google Scholar]
- Yang M., Yao Y., Eades G., Zhang Y., Zhou Q. 2011. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res. Treat. 129:983–991 10.1007/s10549-011-1604-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Pao G.M., Huynh A.M., Suh H., Tonnu N., Nederlof P.M., Gage F.H., Verma I.M. 2011. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 477:179–184 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.