miR-223 is upregulated by the transcription factor TAL1 in human T-ALL cells and suppress the FBXW7 tumor suppressor.
Abstract
The oncogenic transcription factor TAL1/SCL is aberrantly expressed in 60% of cases of human T cell acute lymphoblastic leukemia (T-ALL) and initiates T-ALL in mouse models. By performing global microRNA (miRNA) expression profiling after depletion of TAL1, together with genome-wide analysis of TAL1 occupancy by chromatin immunoprecipitation coupled to massively parallel DNA sequencing, we identified the miRNA genes directly controlled by TAL1 and its regulatory partners HEB, E2A, LMO1/2, GATA3, and RUNX1. The most dynamically regulated miRNA was miR-223, which is bound at its promoter and up-regulated by the TAL1 complex. miR-223 expression mirrors TAL1 levels during thymic development, with high expression in early thymocytes and marked down-regulation after the double-negative-2 stage of maturation. We demonstrate that aberrant miR-223 up-regulation by TAL1 is important for optimal growth of TAL1-positive T-ALL cells and that sustained expression of miR-223 partially rescues T-ALL cells after TAL1 knockdown. Overexpression of miR-223 also leads to marked down-regulation of FBXW7 protein expression, whereas knockdown of TAL1 leads to up-regulation of FBXW7 protein levels, with a marked reduction of its substrates MYC, MYB, NOTCH1, and CYCLIN E. We conclude that TAL1-mediated up-regulation of miR-223 promotes the malignant phenotype in T-ALL through repression of the FBXW7 tumor suppressor.
Human T cell acute lymphoblastic leukemia (T-ALL) arises in thymocyte precursors through stepwise alterations in molecular pathways that often include the aberrant expression of intact master developmental regulatory transcription factors (Look, 1997; Ferrando et al., 2002; Armstrong and Look, 2005). One such transcription factor is TAL1/SCL, which is required for definitive hematopoiesis in early hematopoietic stem cells (HSCs), and is expressed by immature thymocytes after they migrate to the thymus, continuing until they reach the double negative 2 (DN2) stage of differentiation, after which its levels are progressively down-regulated (Herblot et al., 2000; Lacombe et al., 2010; Lécuyer and Hoang, 2004; Porcher et al., 1996). Activation of TAL1 expression through intrachromosomal deletion (SIL-TAL) or chromosomal translocation occurs in ∼30% of patients, yet up to 60% of cases of T-ALL overexpress TAL1 through unknown mechanisms (Ferrando et al., 2002; Aifantis et al., 2008). TAL1-positive T-ALL is characterized by an arrest in differentiation at the CD4+CD8+ double-positive (DP) stage of thymocyte development, when the TAL1 gene is normally silent (Larson et al., 1996; Ferrando et al., 2002; Aifantis et al., 2008; Tremblay et al., 2010). Thus, transcriptional activity of TAL1 is clearly aberrant in T-ALL lymphoblasts compared with their normal DP counterparts.
The TAL1 gene encodes a class II basic helix–loop–helix (bHLH) transcription factor that binds E-box motifs only after heterodimerization with one of the class I bHLH E-proteins, such as E2A or HEB (Hsu et al., 1991; Hsu et al., 1994). Several of the core components of the transcriptional complex have now been elucidated and include GATA3, LIM domain only 1/2 (LMO1/2), and runt-related transcription factor 1 (RUNX1; Wadman et al., 1997; Lécuyer et al., 2002; Xu et al., 2003; Palii et al., 2011). Together, members of the core complex differentially interact with co-activators, such as the histone acetyltransferases p300 and p300/CBP-associated factor (PCAF), or with co-repressors such as SIN3A and histone deacetylase 1/2 (HDAC1/2; Huang et al., 1999; Huang and Brandt, 2000). Accordingly, binding of the TAL1 complex to promoter/enhancer regions can exert either a positive or negative influence on target gene expression.
We recently generated high-resolution maps of the genome-wide occupancy of the TAL1 complex in human T-ALL, which includes E2A, HEB, LMO1/2, GATA3, and RUNX1, using chromatin immunoprecipitation coupled to massively parallel DNA sequencing (ChIP-seq; Sanda et al., 2012). This analysis established that TAL1 acts predominantly as a positive regulator of the expression of its direct target genes, forms a positive interconnected auto-regulatory loop involving other members of the TAL1 complex, and activates several important target genes, including the MYB and TRIB2 oncogenes. Despite the emerging regulatory roles of miRNAs in normal and malignant hematopoiesis (Chen et al., 2004; Calin and Croce, 2006; Mavrakis et al., 2011), the underlying transcriptional mechanisms leading to dysregulated miRNA expression in cancer and T-ALL in particular remain poorly understood. Thus, we investigated whether the TAL1 complex might interact with regulatory sequences of one or more of these miRNAs to regulate downstream targets with critical functions in T-ALL.
Here, we report the results of a genome-wide survey of TAL1 binding of miRNA genes by ChIP-seq in both T-ALL cell lines and primary cells, together with an analysis of changes in miRNA gene expression after TAL1 depletion. This strategy has allowed us to identify the miR-223 gene as an important direct transcriptional target of TAL1 in normal and malignant T cells. We show that TAL1 down-regulates the expression of the critical tumor suppressor protein FBXW7 through miR-223, thus promoting the malignant phenotype in T-ALL.
RESULTS
The TAL1 complex regulates a select number of miRNAs in T-ALL
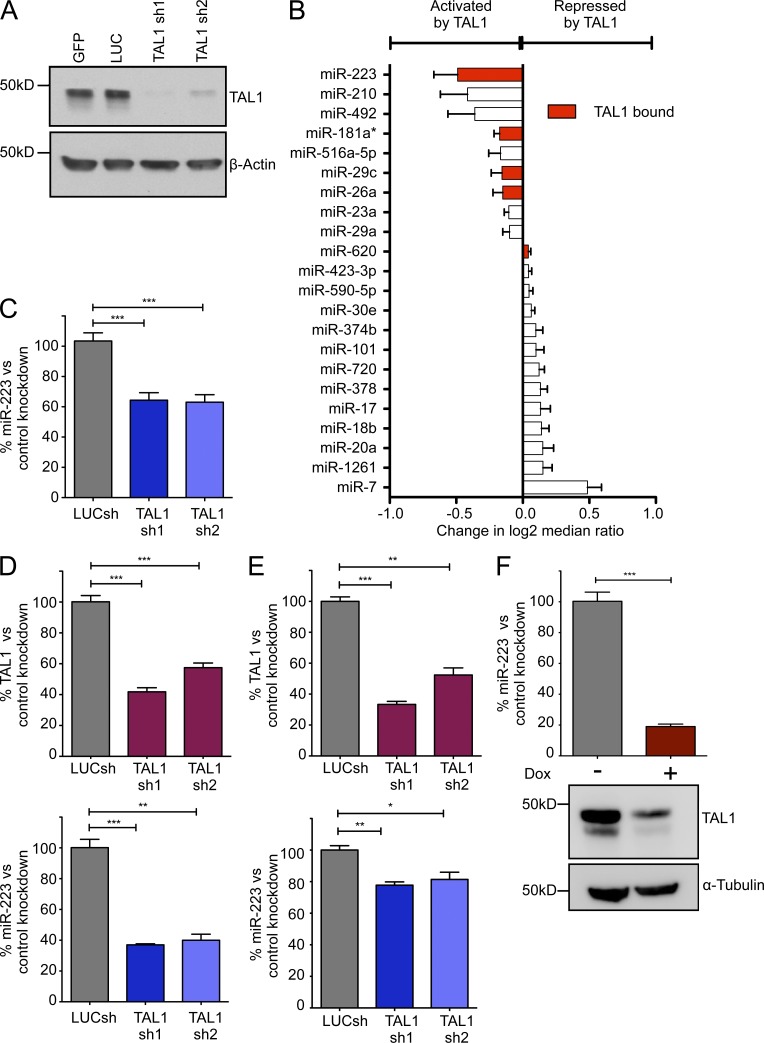
We first sought to determine which miRNAs were either primary or secondary targets of the TAL1 oncoprotein by assessing changes in miRNA expression after knockdown of TAL1 in Jurkat cells, a T-ALL cell line that aberrantly expresses TAL1. By depleting TAL1 using two independent short-hairpin RNAs (shRNAs1 and 2; Fig. 1 A), and assessing global changes in miRNA expression using a locked nucleic acid (LNA)–based profiling platform, we identified significant changes in the expression of 22 miRNAs (Fig. 1 B). 9 of these were down-regulated on TAL1 knockdown (hence positively regulated by TAL1), whereas 13 were up-regulated (hence negatively regulated by TAL1). Because many of these changes were likely to be secondary, we used ChIP-seq analysis of TAL1 binding with its binding partners HEB, E2A, GATA3, and RUNX1 (Sanda et al., 2012) to address this issue. Of the 22 regulated miRNAs, five (miR-223, miR-181a*, miR-29c, miR-26a and miR-620) were judged to be direct targets of the TAL1 complex based on our ChIP-seq data, with evidence of TAL1 occupancy within 12 kb of the candidate miRNA gene or binding a nearby enhancer (Fig. 1 B, red bars).
Figure 1.
TAL1 regulates a select number of miRNAs in T-ALL cells. (A) TAL1 was silenced by lentiviral knockdown in Jurkat cells and protein expression was assessed by Western blot. Two independent shRNAs were compared with control shRNAs targeting GFP or luciferase (LUC). (B) Global miRNA expression profiling with the Exiqon miRCURY LNA platform after shRNA knockdown of TAL1 in Jurkat cells. Horizontal bars show the mean change in log2 median ratios (Hy3/Hy5) ± SD after TAL1 knockdown in duplicate experiments with shRNAs 1 and 2 compared with control shRNAs targeting GFP or luciferase. Only miRNAs with a P value ≤ 0.05 by two-tailed Student’s t test are shown. Red bars represent miRNAs bound by TAL1 in Jurkat cells as determined by ChIP-seq analysis. (C) Quantitative RT-PCR analysis of miR-223 expression normalized to RNU5A in Jurkat cells after TAL1 knockdown with shRNAs 1 and 2 compared with a LUC control shRNA. Bars represent mean ± SEM from three independent experiments performed in triplicate. TAL1 was silenced in RPMI-8402 (D) and CCRF-CEM (E) cells and miR-223 expression was determined by qRT-PCR. (top) TAL1 expression normalized to GAPDH and bottom panel shows miR-223 expression normalized to RNU5A. Bars represent the mean ± SEM of two experiments performed in triplicate. (F) miR-223 expression was assessed by qRT-PCR in a doxycycline-inducible TAL1 knockdown system developed in Jurkat cells (Jurkat S1C1) for 48 h with (+) or without (−) the addition of doxycycline (Dox). Bars represent the mean ± SEM of two experiments performed in triplicate. Western blot (bottom) shows TAL1 protein expression after the addition of doxycycline. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by two-tailed Student’s t test.
miR-223 is directly regulated by the TAL1 complex in TAL1-positive T-ALL
Of the five miRNAs directly regulated by TAL1, we chose to focus on miR-223, because it exhibited the most robust down-regulation after TAL1 knockdown. After using qRT-PCR to confirm the changes we had identified by miRNA expression profiling (Fig. 1 C), we asked whether miR-223 was regulated by TAL1 in TAL1-positive T-ALL cell lines other than Jurkat cells. Knockdown with both TAL1 shRNAs1 and 2 in the TAL1-positive T-ALL cell lines RPMI-8402 and CCRF-CEM (each harboring an activating SIL-TAL1 deletion) resulted in down-regulation of miR-223 as determined by qRT-PCR, similar to our results in Jurkat cells (Fig. 1, D and E). We also examined the changes in miR-223 expression in an inducible TAL1 knockdown model previously established in Jurkat cells that expresses a TAL1 shRNA upon addition of doxycycline (termed Jurkat S1C1 cells; Palii et al., 2011). In agreement with our results from lentiviral knockdown, we found marked down-regulation of miR-223 after TAL1 depletion (Fig. 1 F). Furthermore, transduction of the TAL1-negative cell line KOPT-K1 with TAL1 shRNAs 1 and 2 did not affect miR-223 expression by qRT-PCR (unpublished data), consistent with the interpretation that TAL1 depletion mediates the reduction in miR-223 in TAL1-positive cell lines. Thus, the expression of miR-223 is consistently and directly activated by TAL1 in T-ALL.
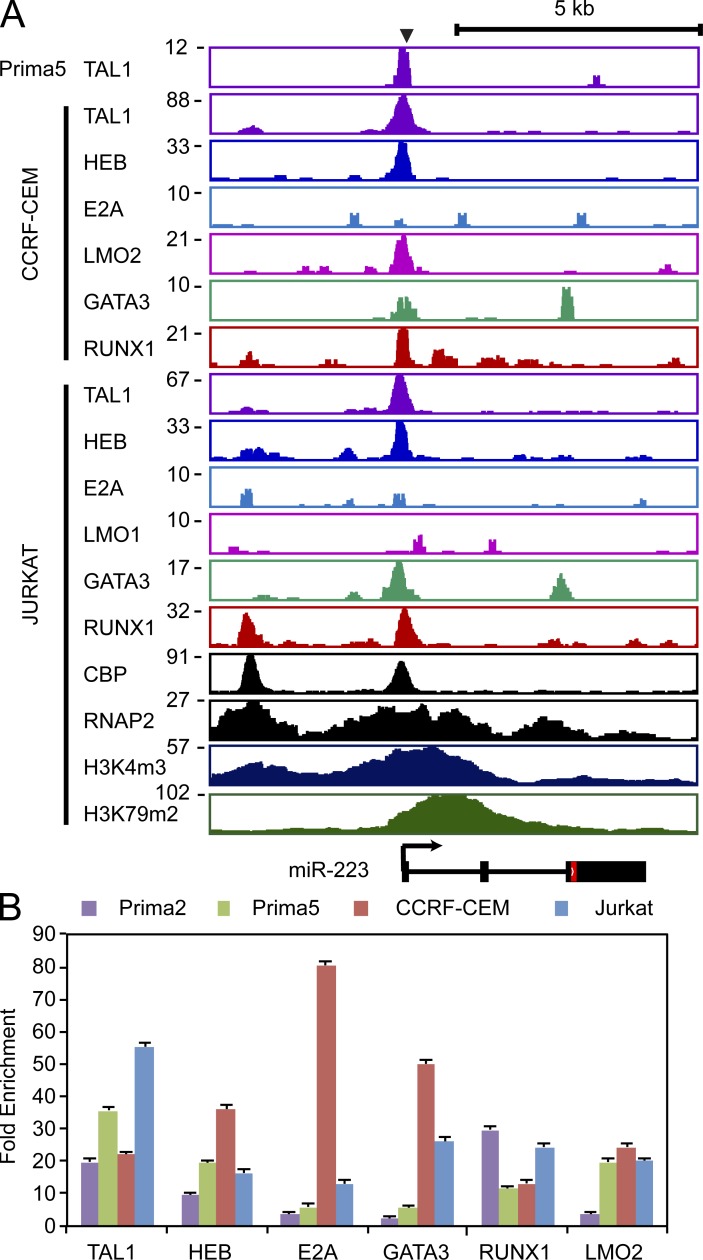
Next, we examined the miR-223 locus in detail in TAL1-positive T-ALLs, including two cell lines (Jurkat and CCRF-CEM) and two “primagraft” samples (designated “Prima 2” and “Prima 5”), which were derived from primary T-ALL cells expanded in immunocompromised mice without prior exposure to in vitro culture. In all four T-ALL samples, a region within 4 kb of the miR-223 transcriptional start site was occupied by TAL1, HEB, E2A, LMO1/2, GATA3 and RUNX1 (Fig. 2 A), a result validated by ChIP-qPCR (Fig. 2 B). Furthermore, this putative regulatory element was co-occupied by the transcriptional co-activator CREB-binding protein (CBP). The promoter and body of the miR-223 gene had high levels of trimethyl histone H3 on lysine 4 (H3K4me3) and dimethyl histone H3 on lysine 79 (H3K79me2) and broad enrichment of RNA polymerase II (RNAPII), indicative of transcriptional initiation and elongation of this locus (Fig. 2 A).
Figure 2.
TAL1 and its partners HEB, E2A, LMO1/2, GATA3, and RUNX1 bind a region 4-kb upstream of the miR-223 transcriptional start site. (A) ChIP coupled to massively parallel DNA sequencing (ChIP-seq) was performed for TAL1, HEB, E2A, and LMO1 or LMO2, GATA3, and RUNX1 (Sanda et al., 2012). Gene tracks represent binding at the miR-223 locus in a human primagraft sample (Prima 5) and the T-ALL cell lines CCRF-CEM (top) and Jurkat (bottom), with the miR-223 locus shown below. Y-axis values represent the number of sequence reads. (B) Enrichment of miR-223 regions indicated in A in four T-ALL cell samples (Jurkat, CCRF-CEM, Prima 2, and Prima 5) was analyzed by ChIP-qPCR. Mean ± SD fold enrichment as normalized to binding at the NANOG promoter (negative control) is shown from two independent experiments performed in triplicate.
miR-223 and TAL1 expression are closely correlated during normal thymic development and in T-ALL
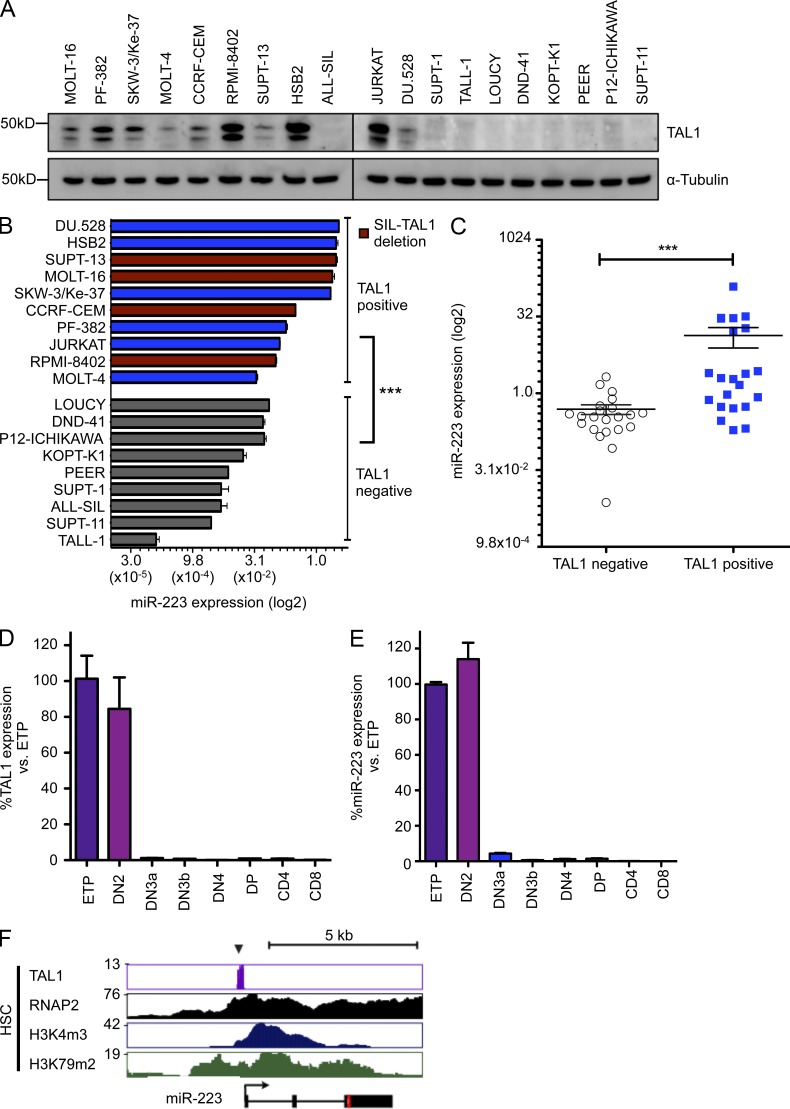
Based on our evidence that the TAL1 complex directly and positively regulates miR-223, we investigated whether this miRNA is selectively up-regulated in the subset of T-ALLs transformed by the TAL1 complex. Of the 19 T-ALL cell lines interrogated, 10 overexpressed TAL1, of which 4 harbored the SIL-TAL1 deletion (Fig. 3 A). Expression of miR-223 was significantly higher in the TAL1-positive cell lines as compared with the TAL1-negative cells (Fig. 3 B; P < 0.0001). To assess whether TAL1 regulates miR-223 in primary T-ALL, we also analyzed RNA from primary pediatric T-ALL samples collected at presentation (Ferrando et al., 2002). Consistent with our findings in T-ALL cell lines, TAL1-positive patients exhibited significantly higher expression of miR-223 than TAL1-negative patients (P < 0.001), indicating that TAL1 regulates the expression level of miR-223 in T-ALL cells as they exist in patients and not only in T-ALL cell lines growing in tissue culture (Fig. 3 C).
Figure 3.
TAL1 expression correlates with miR-223 expression in human T-ALL and during normal thymic development. (A) TAL1 protein expression as determined by Western blot in T-ALL cell lines. The double band is likely to reflect differing TAL1 isoforms. A representative blot from two independent experiments is shown. (B) Comparison of miR-223 expression as determined by qRT-PCR in T-ALL cells positive or negative for TAL1. Red bars indicate presence of an activating SIL-TAL1 deletion. Bars represent mean ± SEM from two independent experiments performed in triplicate. miR-223 expression is given as the percentage of RNU5A control. ***, P < 0.001 by two-tailed Student’s t test. (C) miR-223 expression as determined by qRT-PCR in 44 pediatric T-ALL patients according to TAL1 expression (n = 22 TAL1-positive; n = 22 TAL1-negative). Horizontal bars represent mean with SEM. TAL1 status was determined by qRT-PCR. miR-223 expression is given as the percentage of RNU5A control. ***, P < 0.001 by Mann-Whitney-Wilcoxon test. Expression levels of TAL1 (D) and miR-223 (E) as determined by qRT-PCR from thymic subsets sorted from mice (Pearson’s correlation, r = 0.89; P < 0.001). Bars represent the mean ± SEM of two experiments performed in triplicate. (F) Gene tracks showing binding of TAL1 in relation to the miR-223 locus in highly purified human hematopoietic stem cells. Binding of RNA polymerase II (RNAP2), together with H3K4m3 (marking transcriptional initiation) and H3K79m2 (marking transcriptional elongation) are also shown. Data are from Cui et al. (2009) and Novershtern et al. (2011).
In normal murine thymocytes, Tal1 expression is highest in early thymic development and then is rapidly silenced as the cells differentiate beyond the DN2 stage (Herblot et al., 2000). To explore whether miR-223 expression is a physiological or promiscuous target of TAL1, we isolated thymic subsets from mice and analyzed them for Tal1 and miR-223 expression by qRT-PCR. In accord with published studies, Tal1 expression was highest in early T cell progenitor and DN2 cells and was dramatically down-regulated in more differentiated cells (Fig. 3 D). Interestingly, miR-223 expression closely mirrored Tal1 expression (Pearson’s correlation r = 0.89; P < 0.001), suggesting that TAL1 regulates miR-223 physiologically during normal thymocyte development (Fig. 3 E). Consistent with this finding, TAL1, RUNX1, and LMO2 bind the miR-223 promoter in murine hematopoietic precursor-7 (HPC7) cells, and Gata3 binds the miR-223 promoter in murine DN1 and DN2 cells, but not in DP cells that do not express Tal1 (not depicted; Wilson et al., 2010; Zhang et al., 2012).
Because TAL1 is expressed during early hematopoietic development, we also analyzed TAL1 binding at the miR-223 locus in highly enriched human HSCs (Cui et al., 2009; Novershtern et al., 2011). There was significant enrichment of TAL1 binding at the miR-223 locus, together with chromatin marks consistent with transcriptional activation (Fig. 3 E). Thus, in normal hematopoiesis, TAL1 is likely to regulate miR-223 as a canonical target in HSCs and in T lymphoid progenitors up to the DN2 stage of development, whereas the sustained high expression of miR-223 in TAL1-positive T-ALL cells, arrested at the DP stage, is clearly aberrant as compared with their normal thymocyte counterparts.
TAL1-positive T-ALL cells require sustained miR-223 expression
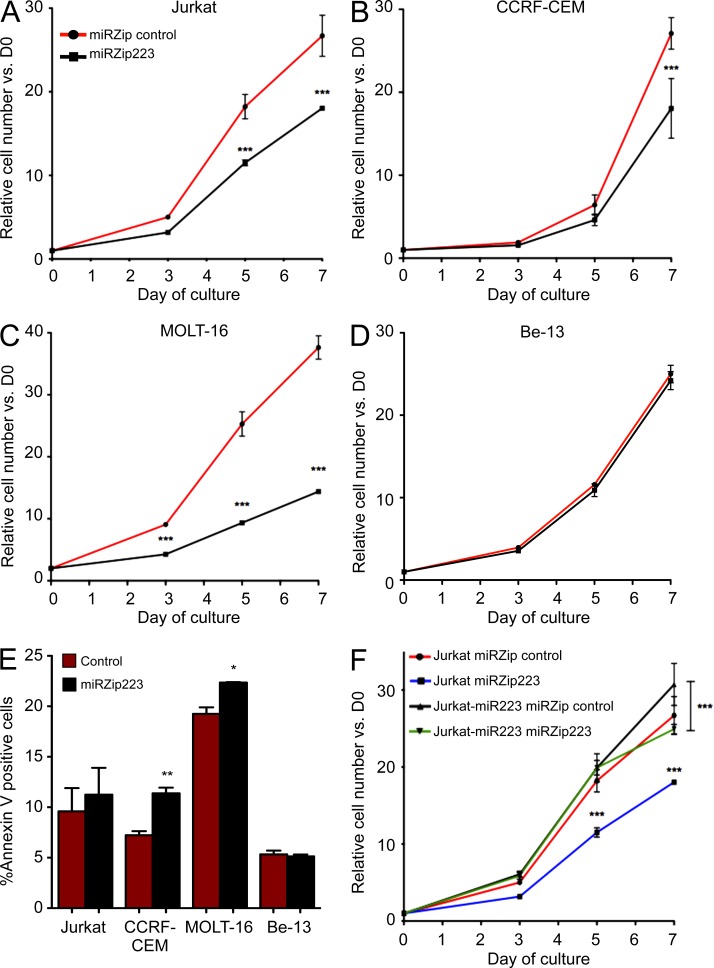
Considering T-ALL cells transformed by the TAL1 oncoprotein are dependent on its maintained expression for optimal growth, we assessed the contribution of its target, miR-223, to this dependency by stably inhibiting mature miR-223 by lentiviral transduction of a miR-223 shRNA construct (termed miRZip-223) in T-ALL cell lines. All three T-ALL cell lines tested demonstrated significant reductions in growth compared with controls (Fig. 4, A–C), an effect that was mediated through an increase in apoptotic cells as determined by Annexin V staining (Fig. 4 E). The most dramatic inhibition of cell growth was observed in MOLT-16 cells that express the highest levels of miR-223 and are FBXW7 wild-type, whereas miR-223 inhibition had no effect on the cell growth or survival of Be-13 cells that harbor a biallelic deletion of FBXW7 (Fig. 4, D and E), indicating that miRZip-223 does not have antiproliferative effects other than those caused by FBXW7 reexpression (O’Neil et al., 2007). When we retrovirally overexpressed miR-223 in Jurkat cells, it rescued most of the phenotype induced by miRZIP-223, arguing against an off-target effect by the construct (Fig. 4 F).
Figure 4.
Inhibition of miR-223 impairs growth and induces apoptosis in TAL1-positive T-ALL cell lines. Four cell lines were transduced with a control miRZip shRNA or an shRNA targeting miR-223 (miRZip-223) and growth kinetics were analyzed. (A) Jurkat cells, harboring a heterozygous FBXW7 R505C mutation (B) CCRF-CEM, harboring a heterozygous FBXW7 R465C mutation (C) MOLT-16, wild-type for FBXW7 and (D) Be-13, which have biallelic deletion of the FBXW7 locus. (E) Annexin V expression was determined by flow cytometry in T-ALL cells 72 h after transduction with control miRZip versus miRZip-223. (F) Jurkat cells were transduced with a retrovirus to overexpress miR-223 and the growth kinetics analyzed after transduction with miRZip-control or miRZip-223 shRNA constructs. All data points represent mean ± SEM for triplicate experiments. P values are for *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by two tailed t test for each time point.
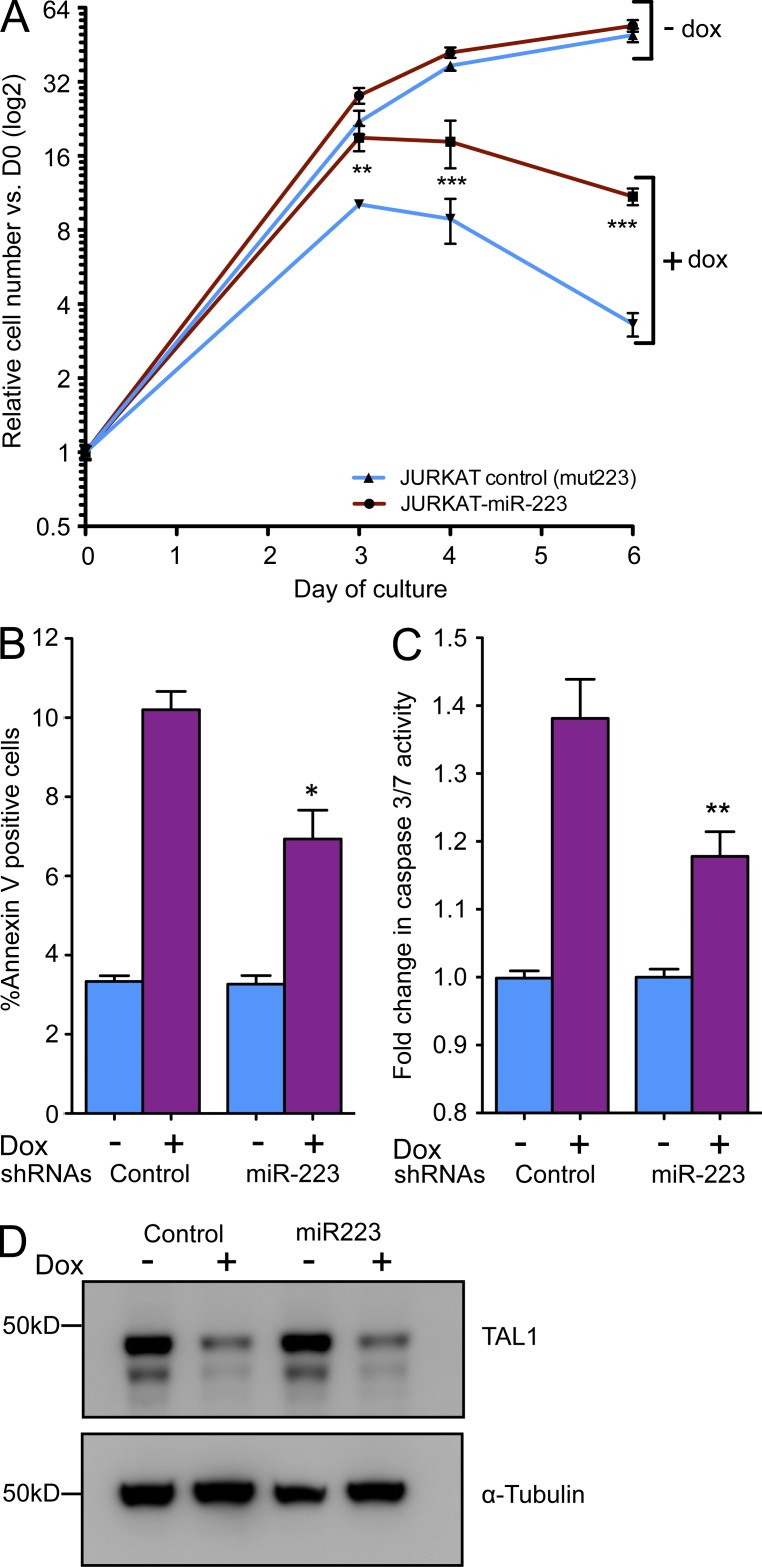
We and others have shown that knockdown of TAL1 slows the growth of TAL1-positive T-ALL cell lines through the induction of apoptosis without affecting the cell cycle (Palii et al., 2011; Sanda et al., 2012). We thus tested whether the decrease in miR-223 expression observed after TAL1 knockdown was responsible, at least in part, for growth inhibition and apoptosis. We therefore infected the doxycycline-inducible TAL1 knockdown Jurkat cells with a retroviral vector expressing miR-223, such that miR-223 expression was no longer under the control of TAL1 in these cells (termed Jurkat-223 cells). Remarkably, retroviral expression of miR-223 partially rescued the growth inhibitory effects induced by doxycycline-induced TAL1 knockdown, whereas a control miR-223 construct in which four bases of the miR-223 seed sequence had been mutated (termed Jurkat-mut223) was unable to rescue cell growth (Fig. 5 A). Accordingly, levels of Annexin V staining and activation of caspases 3 and 7 were significantly lower in Jurkat-223 cells after TAL1 knockdown as compared with control cells (Fig. 5, B and C). Importantly, the degree of TAL1 knockdown was equivalent between Jurkat-223 and Jurkat-mut223 cells (Fig. 5 D). Thus, as a key downstream target of TAL1, miR-223 is essential for the survival and optimal growth of TAL1-positive T-ALL cells.
Figure 5.
miR-223 can partially rescue TAL1-positive cells from apoptosis induced by TAL1 knockdown. (A) Doxycycline-inducible TAL1-knockdown Jurkat S1C1 cells were retrovirally transduced to express miR-223, such that its expression was no longer under the control of TAL1. Growth kinetics were assessed with and without the addition of doxycycline, compared with control Jurkat S1C1 cells expressing a mutant miR-223 in which four bases of the seed sequence had been mutated (mut223). Values represent mean ± SD from two experiments performed in triplicate. (B) Percentage of Annexin V–positive cells as assayed by flow cytometry at 48 h after the addition of doxycycline. (C) Change in cleaved caspase-3 and -7 activity as determined by luminescence in the Caspase-Glo 3/7 assay. Values represent mean ± SEM fold changes from two independent experiments performed in triplicate. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 by two-tailed Student’s t test. (D) Western blot analysis showing equivalent TAL1 knockdown between samples.
TAL1 regulates the FBXW7 tumor suppressor through miR-223
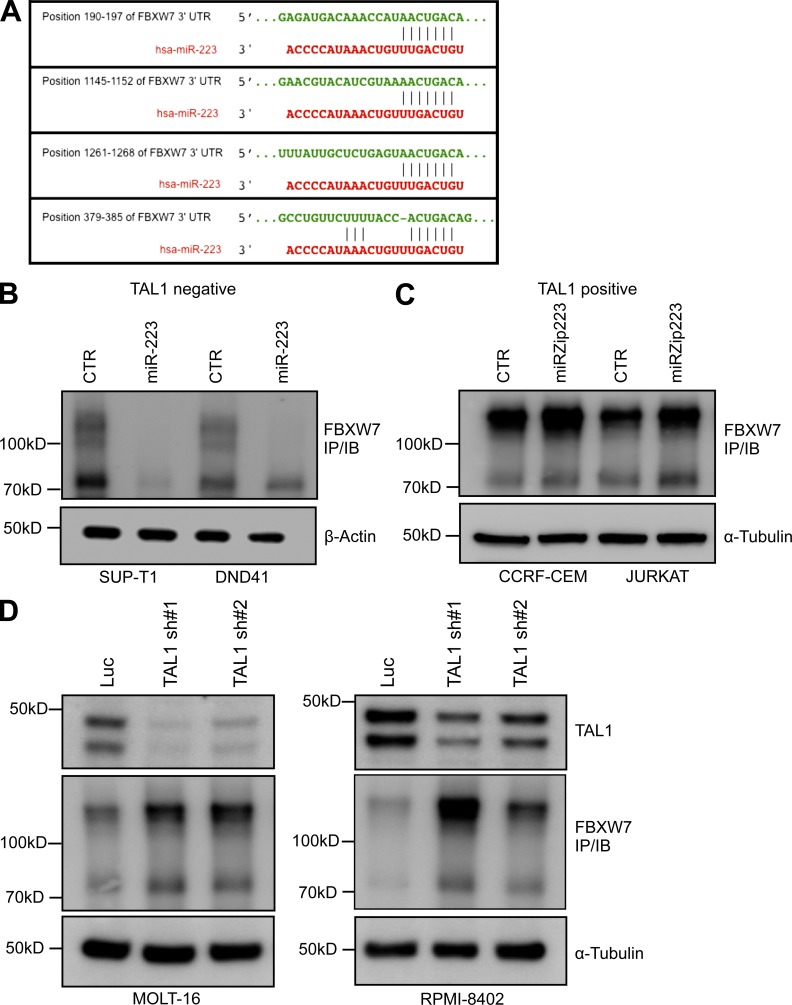
To identify candidate mRNA targets that could mediate the effects of miR-223 on cell survival and growth, we interrogated computational databases (TargetScan and microRNA.org; John et al., 2004; Lewis et al., 2005) and published studies (Xu et al., 2010; Lerner et al., 2011; Mavrakis et al., 2011). The highest ranked target of miR-223 predicted by TargetScan is the FBXW7 tumor suppressor, by virtue of three highly conserved and one poorly conserved binding site in its 3′UTR (Fig. 6 A). Notably, the colorectal carcinoma line HCT116Dicer1−/− shows marked up-regulation of FBXW7 protein expression, consistent with its regulation by miRNAs (Xu et al., 2010; Lerner et al., 2011). Furthermore, in a miRNA library screen to identify miRNAs that can regulate a luciferase reporter containing the FBXW7 3′UTR, miR-223 exhibited the most potent suppression of luciferase activity, a finding consistent with other studies (Lerner et al., 2011; Xu et al., 2010). To demonstrate that this interaction is functional in human T-ALL, we stably overexpressed miR-223 in SUP-T1 and DND41 T-ALL cells, which lack TAL1 and have low miR-223 levels, and then analyzed endogenous FBXW7 protein expression by immunoprecipitation/immunoblot. As expected, FBXW7 expression was markedly down-regulated by miR-223 overexpression in both cell lines, confirming FBXW7 as a target of miR-223 in human T-ALL cells (Fig. 6 B). Additionally, inhibition of miR-223 in TAL1-positive cells with the miRZip-223 lentivirus resulted in reexpression of FBXW7, indicating its protein levels are repressed by miR-223 in these cells (Fig. 6 C).
Figure 6.
TAL1 represses FBXW7 via miR-223 in TAL1-positive T-ALL. (A) Binding sites for miR-223 and the FBXW7 3′UTR are shown for three conserved and one poorly conserved site (bottom), as determined by the TargetScan algorithm. Position numbers refer to distance from the FBXW7 stop codon. (B) TAL1-negative miR-223 low T-ALL cell lines, SUP-T1 and DND-41, were retrovirally transduced to overexpress miR-223, and FBXW7 protein expression determined by immunoprecipitation/immunoblot. Loading controls are 10% of immunoprecipitation input. A representative blot from two independent experiments is shown. (C) FBXW7 protein expression was determined after transduction of TAL1-positive CCRF-CEM and Jurkat cells with miRZip-223 compared with a miRZip control. A representative blot from two independent experiments is shown. (D) TAL1 was depleted in TAL1-positive MOLT-16 and RPMI-8402 cells by lentiviral knockdown with two independent shRNAs, and FBXW7 protein expression was determined by immunoprecipitation/immunoblot. A representative blot from two independent experiments is shown.
FBXW7, an E3 ubiquitin ligase, targets a host of oncoproteins, including MYC, MYB, CYCLIN E, MCL-1, MTOR, and NOTCH-1 for proteasomal degradation, and is an important tumor-suppressor in T-ALL, with loss-of-function mutations present in 20% of patients (Maser et al., 2007; O’Neil et al., 2007; Thompson et al., 2007; Welcker and Clurman, 2008). Furthermore, Fbxw7 knockout mice develop T-ALL (Onoyama et al., 2007). Given the dynamic regulation of miR-223 by TAL1, we hypothesized that TAL1 represses FBXW7 by controlling miR-223 expression. Indeed, knockdown of TAL1 with two independent shRNAs in two TAL1-positive cell lines led to a significant increase in FBXW7 protein expression (Fig. 6 D). This effect was not mediated by direct regulation of FBXW7 transcription by TAL1, as we did not detect changes in FBXW7 mRNA levels after TAL1 knockdown (Sanda et al., 2012).
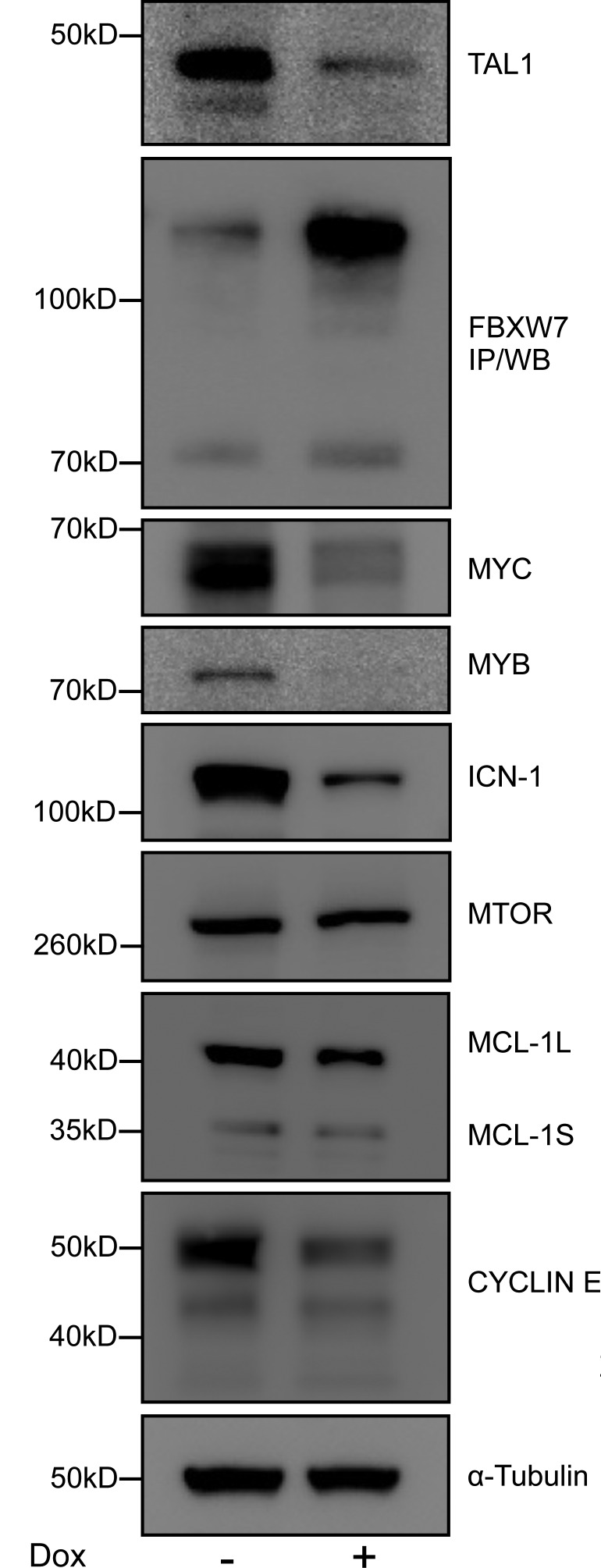
We also examined known oncogenic substrates of FBXW7, to further strengthen the evidence that FBXW7 is the primary mediator of cell growth inhibition downstream of TAL1 and its direct target miR-223. After the addition of doxycycline to Jurkat cells harboring a doxycycline-inducible TAL1-specific shRNA, the reduction in TAL1 was associated with re-expression of FBXW7 and a striking reduction in the protein levels of MYC, intracellular NOTCH1 (ICN-1), MYB, and CYCLIN E (CCNE1), consistent with reactivation of FBXW7 activity (Fig. 7 A). However, we observed only marginal reductions in MCL-1 protein and no changes in MTOR levels, which may relate to cell type–specific aspects of the control of degradation of these FBXW7 substrates in T-ALL.
Figure 7.
TAL1 stabilizes a host of oncogenic substrates through the repression of FBXW7. Western blots for TAL1, FBXW7, and its substrates MYC, MYB, intracellular NOTCH1 (ICN-1), MTOR, MCL-1 (L, long isoform; S, short isoform), and CYCLIN E 48 h after the addition of doxycycline in doxycycline-inducible TAL1-knockdown Jurkat S1C1 cells. Representative blots from three independent experiments are shown.
Together, our results suggest a model in which the TAL1 complex directly regulates the expression of miR-223, leading to its aberrant expression in transformed DP thymocytes, where it represses the ubiquitin ligase FBXW7, a tumor suppressor that can degrade oncoproteins with critical functions in the pathogenesis of T-ALL (Fig. 7 B).
DISCUSSION
The t(1;14)(p34;q11) translocation involving TAL1 in T-ALL was first described over 20 yr ago (Finger et al., 1989; Bernard et al., 1990; Xia et al., 1992), yet the mechanisms through which TAL1 is able to transform thymocytes are only beginning to be dissected. Studies from our laboratory and others using ChIP-seq analysis have shed light on the regulatory networks activated during malignant transformation by TAL1 (Wilson et al., 2010; Palii et al., 2011; Sanda et al., 2012). In the T cell lineage, TAL1 has preferential binding to E-boxes with adjacent GATA, ETS, and RUNX motifs and regulates genes involved in T cell differentiation (Palii et al., 2011). It also targets several downstream oncogenes, including MYB, TRIB2, and STAT5A (Sanda et al., 2012). Although several other genes have been identified as direct targets of TAL1 and its regulatory partners in human T-ALL, such as pre–T cell receptor α (pTα), aldehyde dehydrogenase 1 member A2 (ALDH1A2), NK3 homeobox 1 (NKX3-1), and cyclin-dependent kinase 6 (CDK6), the relative contributions of these targets to leukemogenesis remain unclear (Bernard et al., 1998; Ono et al., 1998; Herblot et al., 2000; Palomero et al., 2006; Kusy et al., 2010; Palii et al., 2011; Sanda et al., 2012).
Genome-wide analysis of transcription factor binding using ChIP-seq has made it possible to assess the direct transcriptional regulation of miRNAs within the genome as a contributing factor to carcinogenesis. Here, we used this approach to address the possibility that TAL1-regulated miRNAs contribute to the pathogenesis of T-ALL driven by this oncoprotein. Of 22 miRNAs that changed expression on TAL1 knockdown, we identified five as direct targets of the TAL1 complex, including miR-26a, miR-29c, miR-181a*, miR-620, and miR-223. Of these, miR-223 was selected for further study because it was most highly down-regulated upon TAL1 knockdown in several different T-ALL cell lines.
Our results are in accord with the notion that TAL1-mediated oncogenesis occurs in an anachronistic manner, i.e., the physiological target genes of TAL1 are activated at a stage of thymocyte differentiation in which they are normally silent. Thus, we found evidence of TAL1-binding at the miR-223 locus in HSCs, and there was a close correlation between TAL1 and miR-223 expression throughout normal thymocyte maturation, suggesting that the regulation of miR-223 by TAL1 is part of a conserved developmental pathway. These findings may explain the high levels of miR-223 that have been described in T-ALL cases with features characteristic of an immature cell of origin (Chiaretti et al., 2010; Van Vlierberghe et al., 2011). Considering that miRNA expression during T cell development is tightly regulated (Neilson et al., 2007; Ghisi et al., 2011; Kirigin et al., 2012), the expression of miR-223 in DP thymocytes would be expected to aberrantly down-regulate the expression of several key proteins, including FBXW7, that are not normally regulated by this miRNA at such a developmental time point.
miR-223 was one of the first miRNAs shown to be expressed by hematopoietic cells and until recently was considered a myeloid-specific miRNA (Chen et al., 2004). In acute promyelocytic leukemia, ATRA induces the expression of CCAAT/enhancer binding protein-α (C/EBPA), which up-regulates miR-223 through binding to its promoter (Fazi et al., 2005). miR-223 then acts to repress nuclear factor-IA (NFI-A), thereby contributing to myeloid differentiation (Fazi et al., 2005). Whether the regulation of miR-223 by TAL1 contributes to early myeloid lineage commitment would be an interesting area for future study because the depletion of TAL1 in HSCs leads to a reduction in myeloid cells through an unknown mechanism (Brunet de la Grange et al., 2006; Zardo et al., 2012).
Using a combination of miRNA expression profiling of primary T-ALL patient samples and an unbiased library screen of miRNAs that were able to rescue lymphocytes from cytokine withdrawal, Mavrakis et al. (2011) recently identified several oncogenic miRNAs in T-ALL. Of the miRNAs that scored on their screen, miR-223 was the most abundantly expressed in T-ALL patients and accelerated Notch-induced leukemia in a mouse model, although the mechanism leading to its dysregulation in human T-ALL was not established (Mavrakis et al., 2011). It is notable that in this study miR-223 had a pro-survival role in lymphocytes. Similarly, we found that inhibition of miR-223 in T-ALL induced apoptosis, suggesting T-ALL cells are dependent on its sustained expression. Moreover, in an inducible TAL1 knockdown system, we found that miR-223 undergoes significant down-regulation when TAL1 expression is blocked, and that retroviral transduction of miR-223 significantly rescued cell growth on TAL1 knockdown. Together these results identify miR-223 as a vital TAL1 target gene whose expression is essential for the optimal growth of TAL1-positive leukemic cells.
By virtue of possessing three conserved binding sites in its 3′UTR, the FBXW7 tumor suppressor gained the highest rank among the miR-223 targets predicted by TargetScan, and several groups have shown that miR-223 regulates FBXW7 expression in a variety of cellular contexts (Lerner et al., 2011; Mavrakis et al., 2011; Xu et al., 2010). Here, we show miR-223 can also regulate FBXW7 protein expression in human T-ALL cells. The reactivation of FBXW7 protein expression observed after TAL1 knockdown indicates it is actively suppressed by TAL1 through its regulation of miR-223, although we cannot rule out the presence of other miR-223 targets important to the malignant phenotype. Regulation of FBXW7 positions TAL1 at the top of a genetic network that directly stabilizes oncoproteins such as c-MYC, MYB, and NOTCH that have vital roles in T-ALL pathogenesis. FBXW7 is known to play a prominent role as a tumor suppressor in T-ALL, and our paper firmly establishes a major mechanism leading to its down-regulation by TAL1 in the majority of T-ALL cases that lack gene-specific FBXW7 inactivating mutations or deletions. Our paper implicates a carcinogenic pathway involving overexpression of an oncogenic transcription factor in the coordinate down-regulation of a key tumor suppressor through a miRNA intermediate, and further study will establish whether this mechanism is more broadly used in cancer biology.
MATERIALS AND METHODS
T-ALL cell samples.
Human T-ALL cell lines were maintained as described previously, and the identity of all cell lines was confirmed by short-tandem repeat analysis (Sanda et al., 2010). Diagnostic T-ALL samples from TAL1-positive cases were obtained with informed consent and Institutional Review Board approval from children treated on Dana Farber Cancer Institute study 05–01. The development of primagraft samples for ChIP-seq analysis has been previously described (Sanda et al., 2012). RNA was obtained at presentation from 44 pediatric T-ALL patients as previously reported (Ferrando et al., 2002).
Microarray expression analysis for mRNA or miRNA.
For global miRNA expression analysis, Jurkat cells were harvested at 48 h after lentiviral transduction with TAL1 shRNAs 1 and 2, or control shRNAs targeting GFP and luciferase, with each shRNA performed in duplicate. Efficient TAL1 knockdown was assessed by Western blotting. miRNAs were isolated and purified with the High Pure miRNA isolation kit (Roche). Global miRNA expression profiling was performed with the Exiqon miRCURY LNA fifth generation platform (miRBase version 14.0). The difference in log2 median ratios (Hy3/Hy5) between sample groups was calculated and significance was determined by two-tailed Student’s t test. P values of ≤0.05 were considered to indicate statistical significance. For mRNA expression analysis, total RNA samples were extracted by TRIzol (Invitrogen) from T-ALL cell lines grown in parallel under identical conditions and purified by RNeasy Mini kit (QIAGEN). Gene expression was analyzed by Affymetrix HG U133 2.0 plus microarrays.
ChIP.
ChIP was performed according to previously described methods (Lee et al., 2006). The antibodies and detailed ChIP conditions and analysis parameters have been described in detail (Sanda et al., 2012). Data showing TAL1 binding and histone modifications in human HSCs were from previous studies (Cui et al., 2009; Novershtern et al., 2011). Relative enrichment of the miR-223 promoter was determined by ChIP quantitative PCR as described Sanda et al. (2012) using the primers miR-223-F1 5′-CAGAAAGCCCAATTCCATCT-3′ and miR-223-R1 5′-GGGCAAATGGATACCATACC-3′ normalized to the negative control region near the NANOG promoter using primers NANOGpr-F1 5′-TCACAAGGGTGGGTCAGTAG-3′ and NANOGpr-R1 5′-CCAGCAGAACGTTAAAATCC-3′.
Isolation of murine T cell subsets.
For RNA isolation from DP and SP thymocyte subpopulations, cells from 6-wk-old C57BL/6 mice were stained for CD4 (RM4-5), CD8 (53–6.7), and TCRβ (H57-597). Thymocytes with CD4+CD8+ (DP), TCRβ+CD4+CD8− (CD4 single-positive, CD4SP) and TCRβ+CD4−CD8+ (CD8 single-positive, CD8SP) phenotypes were sorted. For CD4/CD8 double-negative (DN) thymocyte subpopulation isolation, cells were first stained with biotinylated antibodies to CD4 and CD8, before CD4+CD8+ depletion using streptavidin-conjugated magnetic beads (Invitrogen). Cells were then stained with fluorochrome-conjugated antibodies against CD25 (PC61), CD44 (IM7), CD28 (E18), and cKit (2B8). Thymocytes with CD4−CD8−cKit+CD44+CD25− (early T cell progenitor), CD4−CD8−cKit+CD44+CD25+ (DN2), CD4−CD8−CD25+CD44−cKit−CD28− (DN3a), CD4−CD8−CD25+CD44−cKit−CD28+ (DN3b) phenotype were sorted. All cell sorts were performed with a FACSAria instrument (BD).
Generation of plasmids.
The pMX-miR-GFP/PURO retroviral expression vector was from Cell Biolabs Inc. (RTV-017). A 310-bp fragment including premiR-223 was amplified by PCR from MOLT-16 DNA using the primers XhoI-miR-223-F 5′-TCGACTCGAGTCCCCGGGGCTAAGGGTGTGA-3′ and XhoI-miR-223-R 5′-TCGACTCGAGAAAGTCACATAAAATAACTTGTGAG-3′. PCR products were cloned into the XhoI sites of the pMX-miR-GFP/PURO vector to generate pMX-miR-223-GFP/PURO. To construct a mutant miR-223 vector, the overlapping primer technique was used by first amplifying MOLT-16 DNA with XhoI-miR-223-F and MUT-miR223-R (5′-ATTTGACAAACGCGGACTCTACCACATGGAGTG-3′), a separate PCR using MUT-miR223-F (5′-GAGTCCGCGTTTGTCAAATACCCCAAGTGC-3′) and XhoI-miR-223-R, followed by a third PCR to anneal the first two fragments using primers XhoI-miR-223-F and XhoI-miR-223-R. The 310-bp fragment was then cloned into the XhoI sites of pMX-miR-GFP/PURO vector to generate pMX-miR-MUT223-GFP/PURO. The mature miRNA of this construct (5′-TCCGCGTTTGTCAAATACCCCA-3′) has no predicted targets by TargetScan. The proof-reading enzyme Phusion (New England Biolabs) was used for all reactions and all plasmids were verified by sequencing.
Western blots and immunoprecipitation.
Whole-cell lysates were prepared in RIPA buffer. Immunoblotting was performed with the following antibodies: mouse monoclonal anti-TAL1 antibody diluted 1 in 1,000 (clone BTL73; Millipore), mouse monoclonal anti–α-tubulin antibody diluted 1 in 10,000 (clone B-5-1-2; Sigma-Aldrich), mouse monoclonal anti–β-actin 1 in 2,000 (Sigma-Aldrich), rabbit monoclonal anti–c-MYC (D84C12; Cell Signaling Technology), mouse anti-MYB (05–175, Millipore), rabbit anti-cleaved NOTCH1 (Val1744; Cell Signaling Technology), rabbit anti-MTOR (7C10; Cell Signaling Technology), rabbit anti-MCL-1 (S-19; Santa Cruz Biotechnology, Inc.), mouse anti-CCNE1 (HE12; Santa Cruz Biotechnology). For immunoprecipitation experiments, 750 ng rabbit FBXW7 antibody (A301-721A; Bethyl Laboratories Inc.) covalently coupled to Dynabeads (Invitrogen) was incubated with 100 µg whole-cell lysate on a rotator overnight at 4°C. Beads were washed five times on a magnet and eluted according to the manufacturer’s instructions (Dynabeads Co-immunoprecipitation kit; Invitrogen). Proteins were immunoblotted with FBXW7 antibody (A301-720A; Bethyl Laboratories Inc.) diluted 1 in 1,000. As a loading control, 10% of coimmunoprecipitation input was immunoblotted with anti–β-actin or anti–α-tubulin antibodies. Secondary horseradish peroxidase–linked IgG antibodies to mouse or rabbit diluted 1 in 10,000 were obtained from Cell Signaling Technology.
shRNA knockdown and doxycycline-inducible TAL1 knockdown cells.
The shRNA sequences directed against TAL1, expressed in the pLKO.1-puro vector, and production of lentivirus have been previously described (Sanda et al., 2012), as have the doxycycline-inducible TAL1 knockdown Jurkat cells (S1C1 clone; Palii et al., 2011). Jurkat S1C1 cells were maintained under selection in blasticidin and G418. After retroviral transduction with pMX-miR-GFP/PURO, cells were also maintained in puromycin. Doxycycline was used at 6.25 µg/ml. The level of knockdown was verified by qRT-PCR for RNA or by Western blotting.
RNA extraction, cDNA, and expression analysis.
miRNAs were isolated and purified with the High Pure miRNA isolation kit (Roche) and reverse transcribed with the miScript kit (QIAGEN), and their expression was quantitated with the miScript SyberGreen kit using a miScript universal primer and miR-223–specific primer (MS00003871; QIAGEN). Expression was normalized to expression of RNU5A (MS00013993; QIAGEN). Both the miR-223 and RNU5A primers are compatible with human and mouse gene orthologues. To assess mRNA knockdown, a small fraction of cells were collected and mRNA was extracted by TRIzol (Invitrogen), followed by column purification using the RNeasy Mini kit (QIAGEN). Purified RNA was reverse-transcribed with the miScript kit (QIAGEN). Quantitative real-time PCR was performed with the AB7300 Detection System (Applied Biosystems) using gene-specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences for qRT-PCR were as follows: human TAL1-F, 5′-AGGGCCTGGTTGAAGAAGAT-3′; human TAL1-R, 5′-AAGTAAGGGCGACTGGGTTT-3′; murine Tal1-F, 5′-CCCCATGTTCACCAACAAC-3′; murine Tal1-R, 5′-CCGCACTACTTTGGTGTGAG-3′; GAPDH-F, 5′-TGCACCACCAACTGCTTAGC-3′; GAPDH-R, 5′GGCATGGACTGTGGTCATGAG-3′. Among T-ALL cases, TAL1 expression exhibited a bimodal distribution, and patients were determined as TAL1-positive if TAL1 transcripts were >3.7% of GAPDH.
Apoptosis assays.
Annexin V staining was assessed by flow cytometry using Annexin V-APC antibody (BD) according to the manufacturer’s recommendations. Cleaved caspase-3 and -7 activity was determined by luminescence using the Caspase-Glo 3/7 assay from Promega after normalization for cell number.
Accession nos.
Expression data and ChIP-seq can be found in the Gene Expression Omnibus under superseries accession no. GSE29181, and chromatin marks and RNA Pol II ChIP-seq data from Jurkat are found under accession no. GSE33850. Chromatin marks and RNA Pol II in human hematopoietic stem and progenitor cells are found under accession no. GSE26015. miRNA expression data are available under accession no. GSE46957.
Acknowledgments
We would like to thank Dr. Alex Kentsis for helpful discussions and critical review. We acknowledge the RNAi Consortium for providing lentivirus shRNA constructs and thank John R. Gilbert for critical review of the manuscript.
This research was supported by the Kay Kendall Leukaemia Fund of the UK (M.R. Mansour), Team Path To the Cure grant (M.R. Mansour and A.T. Look), grants from the National Cancer Institute (5P01CA109901, 5P01CA68484, and 1K99CA157951), and by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. T. Sanda is supported by grants from the William Lawrence and Blanche Hughes Foundation, the Children’s Leukemia Research Association and the Japan Society for the Promotion of Science. A. Gutierrez is supported by NIH grant 1K08CA133103 and is a Scholar of the American Society of Hematology-Amos Faculty Development Program.
The authors declare no financial conflict of interest.
Footnotes
Abbreviations used:
- ChIP-seq
- chromatin immunoprecipitation coupled to massively parallel DNA sequencing
- DN2
- double negative 2
- DP
- CD4+CD8+ double-positive
- HSC
- hematopoietic stem cell
- LNA
- locked nucleic acid
- miRNA
- microRNA
- T-ALL
- T cell acute lymphoblastic leukemia
References
- Aifantis I., Raetz E., Buonamici S. 2008. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 8:380–390 10.1038/nri2304 [DOI] [PubMed] [Google Scholar]
- Armstrong S.A., Look A.T. 2005. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 23:6306–6315 10.1200/JCO.2005.05.047 [DOI] [PubMed] [Google Scholar]
- Bernard O., Guglielmi P., Jonveaux P., Cherif D., Gisselbrecht S., Mauchauffe M., Berger R., Larsen C.J., Mathieu-Mahul D. 1990. Two distinct mechanisms for the SCL gene activation in the t(1;14) translocation of T-cell leukemias. Genes Chromosomes Cancer. 1:194–208 10.1002/gcc.2870010303 [DOI] [PubMed] [Google Scholar]
- Bernard M., Delabesse E., Smit L., Millien C., Kirsch I.R., Strominger J.L., Macintyre E.A. 1998. Helix-loop-helix (E2-5, HEB, TAL1 and Id1) protein interaction with the TCRalphadelta enhancers. Int. Immunol. 10:1539–1549 10.1093/intimm/10.10.1539 [DOI] [PubMed] [Google Scholar]
- Brunet de la Grange P., Armstrong F., Duval V., Rouyez M.C., Goardon N., Romeo P.H., Pflumio F. 2006. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood. 108:2998–3004 10.1182/blood-2006-05-022988 [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6:857–866 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science. 303:83–86 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- Chiaretti S., Messina M., Tavolaro S., Zardo G., Elia L., Vitale A., Fatica A., Gorello P., Piciocchi A., Scappucci G., et al. 2010. Gene expression profiling identifies a subset of adult T-cell acute lymphoblastic leukemia with myeloid-like gene features and over-expression of miR-223. Haematologica. 95:1114–1121 10.3324/haematol.2009.015099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 4:80–93 10.1016/j.stem.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 123:819–831 10.1016/j.cell.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87 10.1016/S1535-6108(02)00018-1 [DOI] [PubMed] [Google Scholar]
- Finger L.R., Kagan J., Christopher G., Kurtzberg J., Hershfield M.S., Nowell P.C., Croce C.M. 1989. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc. Natl. Acad. Sci. USA. 86:5039–5043 10.1073/pnas.86.13.5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisi M., Corradin A., Basso K., Frasson C., Serafin V., Mukherjee S., Mussolin L., Ruggero K., Bonanno L., Guffanti A., et al. 2011. Modulation of microRNA expression in human T-cell development: targeting of NOTCH3 by miR-150. Blood. 117:7053–7062 10.1182/blood-2010-12-326629 [DOI] [PubMed] [Google Scholar]
- Herblot S., Steff A.M., Hugo P., Aplan P.D., Hoang T. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat. Immunol. 1:138–144 10.1038/77819 [DOI] [PubMed] [Google Scholar]
- Hsu H.L., Cheng J.T., Chen Q., Baer R. 1991. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol. Cell. Biol. 11:3037–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.L., Wadman I., Baer R. 1994. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc. Natl. Acad. Sci. USA. 91:3181–3185 10.1073/pnas.91.8.3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Brandt S.J. 2000. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20:2248–2259 10.1128/MCB.20.6.2248-2259.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Qiu Y., Stein R.W., Brandt S.J. 1999. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 18:4958–4967 10.1038/sj.onc.1202889 [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. 2004. Human MicroRNA targets. PLoS Biol. 2:e363 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirigin F.F., Lindstedt K., Sellars M., Ciofani M., Low S.L., Jones L., Bell F., Pauli F., Bonneau R., Myers R.M., et al. 2012. Dynamic microRNA gene transcription and processing during T cell development. J. Immunol. 188:3257–3267 10.4049/jimmunol.1103175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusy S., Gerby B., Goardon N., Gault N., Ferri F., Gérard D., Armstrong F., Ballerini P., Cayuela J.M., Baruchel A., et al. 2010. NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia. J. Exp. Med. 207:2141–2156 10.1084/jem.20100745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J., Herblot S., Rojas-Sutterlin S., Haman A., Barakat S., Iscove N.N., Sauvageau G., Hoang T. 2010. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 115:792–803 10.1182/blood-2009-01-201384 [DOI] [PubMed] [Google Scholar]
- Larson R.C., Lavenir I., Larson T.A., Baer R., Warren A.J., Wadman I., Nottage K., Rabbitts T.H. 1996. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 15:1021–1027 [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Hoang T. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32:11–24 10.1016/j.exphem.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Herblot S., Saint-Denis M., Martin R., Begley C.G., Porcher C., Orkin S.H., Hoang T. 2002. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 100:2430–2440 10.1182/blood-2002-02-0568 [DOI] [PubMed] [Google Scholar]
- Lee T.I., Johnstone S.E., Young R.A. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1:729–748 10.1038/nprot.2006.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M., Lundgren J., Akhoondi S., Jahn A., Ng H.F., Akbari Moqadam F., Oude Vrielink J.A., Agami R., Den Boer M.L., Grandér D., Sangfelt O. 2011. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle. 10:2172–2183 10.4161/cc.10.13.16248 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Look A.T. 1997. Oncogenic transcription factors in the human acute leukemias. Science. 278:1059–1064 10.1126/science.278.5340.1059 [DOI] [PubMed] [Google Scholar]
- Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I., et al. 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 447:966–971 10.1038/nature05886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis K.J., Van Der Meulen J., Wolfe A.L., Liu X., Mets E., Taghon T., Khan A.A., Setty M., Rondou P., Vandenberghe P., et al. 2011. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 43:673–678 10.1038/ng.858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson J.R., Zheng G.X., Burge C.B., Sharp P.A. 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21:578–589 10.1101/gad.1522907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., et al. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 144:296–309 10.1016/j.cell.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J.P., Pieters R., et al. 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204:1813–1824 10.1084/jem.20070876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Fukuhara N., Yoshie O. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18:6939–6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyama I., Tsunematsu R., Matsumoto A., Kimura T., de Alborán I.M., Nakayama K., Nakayama K.I. 2007. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 204:2875–2888 10.1084/jem.20062299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii C.G., Perez-Iratxeta C., Yao Z., Cao Y., Dai F., Davison J., Atkins H., Allan D., Dilworth F.J., Gentleman R., et al. 2011. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 30:494–509 10.1038/emboj.2010.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Odom D.T., O’Neil J., Ferrando A.A., Margolin A., Neuberg D.S., Winter S.S., Larson R.S., Li W., Liu X.S., et al. 2006. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 108:986–992 10.1182/blood-2005-08-3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C., Swat W., Rockwell K., Fujiwara Y., Alt F.W., Orkin S.H. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 86:47–57 10.1016/S0092-8674(00)80076-8 [DOI] [PubMed] [Google Scholar]
- Sanda T., Li X., Gutierrez A., Ahn Y., Neuberg D.S., O’Neil J., Strack P.R., Winter C.G., Winter S.S., Larson R.S., et al. 2010. Interconnecting molecular pathways in the pathogenesis and drug sensitivity of T-cell acute lymphoblastic leukemia. Blood. 115:1735–1745 10.1182/blood-2009-07-235143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda T., Lawton L.N., Barrasa M.I., Fan Z.P., Kohlhammer H., Gutierrez A., Ma W., Tatarek J., Ahn Y., Kelliher M.A., et al. 2012. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 22:209–221 10.1016/j.ccr.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B.J., Buonamici S., Sulis M.L., Palomero T., Vilimas T., Basso G., Ferrando A., Aifantis I. 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204:1825–1835 10.1084/jem.20070872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M., Tremblay C.S., Herblot S., Aplan P.D., Hébert J., Perreault C., Hoang T. 2010. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 24:1093–1105 10.1101/gad.1897910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P., Ambesi-Impiombato A., Perez-Garcia A., Haydu J.E., Rigo I., Hadler M., Tosello V., Della Gatta G., Paietta E., Racevskis J., et al. 2011. ETV6 mutations in early immature human T cell leukemias. J. Exp. Med. 208:2571–2579 10.1084/jem.20112239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grütz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Clurman B.E. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 8:83–93 10.1038/nrc2290 [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 7:532–544 10.1016/j.stem.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Xia Y., Brown L., Tsan J.T., Yang C.Y., Siciliano M.J., Crist W.M., Carroll A.J., Baer R. 1992. The translocation (1;14)(p34;q11) in human T-cell leukemia: chromosome breakage 25 kilobase pairs downstream of the TAL1 protooncogene. Genes Chromosomes Cancer. 4:211–216 10.1002/gcc.2870040304 [DOI] [PubMed] [Google Scholar]
- Xu Z., Huang S., Chang L.S., Agulnick A.D., Brandt S.J. 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23:7585–7599 10.1128/MCB.23.21.7585-7599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Sengupta T., Kukreja L., Minella A.C. 2010. MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. J. Biol. Chem. 285:34439–34446 10.1074/jbc.M110.152306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zardo G., Ciolfi A., Vian L., Starnes L.M., Billi M., Racanicchi S., Maresca C., Fazi F., Travaglini L., Noguera N., et al. 2012. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 119:4034–4046 10.1182/blood-2011-08-371344 [DOI] [PubMed] [Google Scholar]
- Zhang J.A., Mortazavi A., Williams B.A., Wold B.J., Rothenberg E.V. 2012. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 149:467–482 10.1016/j.cell.2012.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]