Abstract
Prostate cancer (PCa) is the most commonly diagnosed noncutaneous malignancy and second leading cause of cancer-related deaths in US males. Clinically, locally confined disease is treated surgically and/or with radiation therapy. Invasive disease, however, must be treated with pharmacological inhibitors of androgen receptor (AR) activity, since disease progression is fundamentally reliant on AR activation. However, despite initially effective treatment options, recurrent castration-resistant PCa often occurs due to aberrant reactivation of AR. Additionally, it is appreciated that many other signaling molecules, such as transcription factors, oncogenes, and tumor suppressors, are often perturbed and significantly contribute to PCa initiation and progression to incurable disease. Understanding the interplay between AR signaling and other signaling networks altered in PCa will advance therapeutic approaches. Overall, comprehension of the molecular composition promoting neoplastic growth and formation of CRPC is paramount for developing durable treatment options.
Keywords: prostate cancer, androgen receptor, castration resistance, AR antagonists, TMPRSS2:ERG, Myc, initiation, progression, precision medicine
Prostate cancer (PCa) is the most commonly diagnosed noncutaneous malignancy and second leading cause of cancer-related deaths in US males. The prognosis for locally confined disease is favorable with treatment by surgical resection and/or radiation therapy. PCa at all stages is impingent on activity by the androgen receptor (AR) nuclear receptor, and as such, treatment options for invasive disease relies on targeting this pathway. Androgen deprivation therapy (ADT), often coupled with direct AR antagonists, is the standard of care for disseminated disease, and affords patients with 24–36 months of disease regression. However, despite initially effective treatment options, recurrent PCa (termed castrate-resistant (CRPC)) is often detected due to aberrant reactivation of AR. Progression to CRPC also accumulates perturbations in signaling pathways that propagate cell cycle progression and uncontrolled proliferation. The ability to understand the molecular composition that promotes neoplastic growth, and drives formation of incurable CRPC is paramount for developing durable treatment options. This review will focus on key molecules that have been implicated in PCa initiation and progression including transcription factors, oncogenes, and tumor suppressors.
Prostate Development and Hyperplasia
The developed prostate gland is comprised of luminal and basal epithelial cell layers surrounded by stroma. Luminal epithelial cells are polarized, columnar cells that line prostate lumen, and basal cells are elongated cells that separate lumen from stroma. These cells are pathologically distinguishable based on various cellular markers. For example, luminal epithelia express high levels of AR and are positive for cytokeratin 8 and 18, CD57, and NKX3.1. Basal epithelial cells express low levels of AR, and stain positive for cytokeratin 5 and 14, p63, CD44, and GSTP1. A rare cell type within this compartment are neuroendocrine cells, which are AR-negative, post-mitotic, and secrete neuropeptides and growth factors for luminal cell growth 1. Finally, while not discussed further here, stroma plays a potential role in epithelial cell homeostasis and PCa progression 2, 3. Prostate embryonic and postnatal development depends on multiple critical regulators of differentiation, such as NKX3.1, Forkhead box A1 (FOXA1), and AR. The NKX3.1 transcription factor is a member of the NK subfamily of homeobox genes, which play a role in organogenesis in multiple species. In prostate, NKX3.1 expression at embryonic day 15.5 represents the earliest prostate-specific gene, and is instrumental for development of prostatic bud formation 4. In postnatal mice, NKX3.1 expression in luminal epithelial nuclei corresponds with regions of active morphogenesis within the end of prostate buds, while genetic deletion of nkx3.1 results in decreased ductal branching and altered secretory protein production 4. Interestingly, although nkx3.1 is an androgen-regulated gene and castration of adult mice decreases protein expression, NKX3.1 precedes androgen production, which suggests there are alternative mechanisms to initiate expression during development 5.
Androgens and AR signaling is requisite for prostate cellular functions and architectural maintenance. For example, androgen ablation during early stages of murine embryonic development inhibits adult prostate development, and lack of functional AR results in prostatic absence 6. ADT in developed prostate glands results in luminal epithelial cell apoptosis and subsequent lumen involution, which can be rescued by testosterone supplementation 7. AR signaling within the context of normal tissue promotes a pro-differentiation gene signature and is important for differentiation of prostate terminal end buds. Despite dependence on androgens for normal prostate development, there is limited evidence to suggest that androgens autonomously promote PCa. Testosterone administration does not increase the incidence of PCa, and serum testosterone levels and PCa risk are not correlative 8, 9. These findings illustrate the differences in androgen function in developing and normal prostate versus PCa, wherein androgens are potent mediators of cancer growth and progression. Finally, FOXA1 transcription factor is expressed throughout all stages of prostate development and maturation and regulates expression of AR target genes. Mice with prostate-specific FOXA1 deletion contain improperly formed pubertal ducts composed of immature luminal epithelial cells and an abundance of surrounding stroma. Furthermore, FOXA1 is coexpressed with AR and regulates AR target gene expression. As will be highlighted, deregulation of developmental genes in differentiated cells is a common mechanism to promote aberrant growth and proliferation.
As men age, the incidence of hyperplastic lesions significantly increases, such that 80% of 80-year-old men harbor detectable benign prostate hyperplasia (BPH). However, BPH is not a precursor to PCa and does not require treatment. In contrast, High Grade Prostate Intraepithelial Neoplasia (HGPIN) represents in situ adenocarcinoma and is recognized to progress to PCa. HGPIN is a multifocal lesion identified by dysplasia of prostate luminal epithelial cells and a decrease in basal layer cells. The vast majority of PCa are multifocal adenocarcinoma, which contain invasive epithelia cells that are classified as luminal based on cytokeratin staining. There is a great need to diagnose and treat PCa early while therapies are effective and clinical responses are durable. One potential facet of PCa biology to provide insight into tumor prognosis for therapeutic options is determining the PCa cell of origin capable of tumor initiation.
PCa Cell of Origin
Luminal cells are the purported cell of PCa origin primarily due to luminal cell markers present in PCa and the absence of basal cells. However, several recent studies have provided evidence that basal and/or luminal epithelial cells could be tumor initiating. A prominent mechanism to address questions of prostate gland formation, PCa initiation, and cell of origin is tissue recombination, which involves injecting a combination of prostate epithelial cells and murine urogenital sinus mesenchyme (UGSM) under the kidney capsule. This technique results in development of a fully differentiated and functional prostate gland under physiologic androgen levels, and enables manipulation of molecules required for prostate gland formation. It was hypothesized that initiating cells resided amongst a subset of epithelial cells capable of repopulating prostatic lumen following ADT-mediated involution. Castrate-resistant Nkx3.1-expressing cells (CARNs) were identified as a rare population of epithelial cells that survive androgen ablation and are only found in the luminal compartment 10. CARNs promote a 9-fold growth of glands that contain luminal, basal and neuroendocrine cells when repopulated following androgen supplementation, and when clinically relevant genetic lesions were modeled in CARNs, such as PTEN deletion, there was a rapid development of luminal PCa without the presence of basal cells 10. Using a similar approach, human luminal prostate cells were sorted for basal cells, and found to be sufficient to produce glandular structures containing both luminal and basal cells, which suggested basal cells have a role in PCa initiation 11. In the same study, luminal epithelial cells were not able to produce glands in tissue recombination experiments. Furthermore, primary basal cells, but not luminal cells, transduced with AKT/ERG, or AKT/ERG/AR were capable of promoting abnormal structures or adenocarcinoma 11. Finally, in contrast to the aforementioned studies, basal and luminal cells were tracked using lineage-specific keratin-14 and keratin-8-mediated expression of fluorescent proteins, respectively 12. The authors determined that both cell types are independently capable of giving rise to PCa when combined with PTEN deletion. Based on these studies, there is no definitive answer for which cell type gives rise to PCa, which could be due to differences between human and murine cells employed in tissue recombination, technical differences, or an incomplete comprehension of cellular plasticity under dedifferentiation pressures. It is possible that genes controlling differentiation status are variable between model systems or cell types and, given the importance of these factors on disease state, have influence on the formation and progression of PCa.
Developmental Genes Re-wired
As described above, throughout prostate development and maturation, genes such as nkx3.1, FOXA1 and AR function to mediate gland formation and cellular differentiation. However, aberrant activation of developmental genes within differentiated prostate epithelium can promote hyperplastic proliferation and/or progression of PCa.
A. NKX3.1
Nkx3.1 homeobox gene is one of the earliest expressed prostate-specific proteins that is maintained and necessary at all stages of prostate differentiation and into adulthood. NKX3.1 acts as a transcriptional repressor when bound to consensus DNA sequences by recruiting co-repressor complexes or suppressing transcriptional activators. The gene is located on chromosome 8p21, which is a region displaying loss of heterozygosity in 50% of primary PCa and 80% of metastatic disease. NKX3.1 mutation has likely relevance for human disease, as variants unable to bind DNA are linked to increased risk for PCa development, and functional variants that result in reduced Nkx3.1 stability are a risk factor for sporadic PCa. However, NKX3.1 is not typically considered a classical tumor suppressor since loss is not sufficient for tumorigenesis, both alleles are not deleted in PCa, and NKX3.1 is still significantly expressed in aggressive PCa 13.
Based on mouse models of PCa progression, the stage where NKX3.1 downregulation occurs is variable. Analysis of established mouse models of PCa identified spontaneous NKX3.1 loss coincident with PTEN loss or MYC overexpression-driven tumors. However, in PTEN null tumors, NKX3.1 loss was detected in the earliest hyperplastic stages, while MYC overexpression resulted in NKX3.1 loss during the transition from PIN to invasive adenocarcinoma 14. Furthermore, only the PTEN mouse model recapitulates an “NKX3.1 loss gene signature” suggesting that timing of loss is important for activation of downstream signals driving tumorigenesis. Recently, analysis of NKX3.1 binding sites from wild type (WT), heterozygous, and null mice determined that NKX3.1 repressed and activated target genes 15. Interestingly, one quarter of these targets were also direct MYC targets, which NKX3.1 could antagonize direct interaction at gene promoters with MYC. Biologically, NXK3.1 loss and MYC overexpression cooperate to promote tumorigenesis in mouse models resulting in Low Grade PIN (LGPIN) and microinvasive cancer 15. Frequent deletion of the NKX3.1-containing region in human PCa, and the ability to promote hyperplasia and PIN in murine models strongly suggest that NKX3.1 plays a significant role in the initiation of PCa.
B. AR
Years of work have aimed to understand the basis of AR reactivation and progression toward CRPC. Although CRPC was initially described as “androgen-independent”, more recent investigations demonstrated that recurrent AR activity is the key mediator of progression to castrate-resistant stage. Numerous mechanisms to reestablish AR activity are utilized in CRPC, and sufficient to drive progression. Prevalent means of AR reactivation will be discussed below.
1. AR amplification/overexpression
The most common mechanism to reactivate AR signaling is through increased expression, achieved through gene amplification or upregulated protein levels. While most studies address the impact of increased AR expression in progression to CRPC, mouse models demonstrate the ability of de novo AR upregulation to promote murine PIN or PCa 16, 17. Clinically, however, AR amplification is seen nearly exclusively in tumors that recur after ADT 18, suggesting there is a selection for increased AR as a means to circumvent therapeutic intervention. Mechanistically, increased AR expression sensitizes cells to sub-physiologic concentrations of androgen 19, overrides inhibitory affects of AR antagonists, such as bicalutamide 20, and correlates with shorter recurrence-free survival in patients 21. PCa cells with AR overexpression have an increased number of AR-binding sites on chromatin that promote an altered androgen-dependent gene signature 22. PCa cell xenograft models demonstrate that increased AR expression is the only consistently detected aberration in tumors that are refractory to ADT 23 and, importantly, elevated AR is significantly overrepresented in CRPC tumor samples, compared to benign tissue or untreated primary tumors 24. Given the ability of increased AR in model systems to promote murine PIN and the lack or AR amplification in primary PCa, understanding mechanisms to alter AR protein stability could afford new therapeutic opportunities for treating early stage disease.
2. AR Mutations
AR mutations represent a selective response to ADT, and as such, somatic mutations are uncommon in early-stage, treatment naïve tumors but are routinely detected in CRPC patients. Germline mutations associated with cancer risk are rare in all stages of PCa progression, although at least one missense mutation has been identified and may predict early onset PCa 25. Most AR mutations are gain-of-function, which can be mapped to the ligand-binding domain and result in androgen hypersensitivity or decreased ligand specificity. For example, T877A is the most common point mutation, which is located in helix 11 of ligand binding domain (LBD) and broadens ligand specificity such that the anti-androgen, hydroxyflutamide (flutamide), can have partial agonist activity and also be activated by other steroids (eg. estrogen, cortisol) 26, 27. Furthermore AR-T877A is activated by endocrine disruptor, bisphenol A (BPA), which promotes AR transcriptional activity and cell cycle progression in castrate conditions 28, 29. While the precise incidence of AR mutations is still unclear, ranging from 8 to 25% of metastatic tumors, it is hypothesized that mutations impact progression to CRPC 26. Recently, whole genome sequencing of treatment naïve PCa and heavily treated CRPC tumors revealed that AR was commonly mutated only in CRPC samples 30, 31. Sequencing of AR mRNAs from CRPC metastases identified that, compared to treatment naïve tumors, patients treated with AR antagonists flutamide or bicalutimide specifically contained LBD gain-of-function mutations. Interestingly, mutational variations existed between different antagonist treatments, suggesting that chemical compositions of therapies drive conformational changes in AR that promote altered activity 32. Given the selection of AR mutations in response to various therapeutic pressures, new individualized treatment regimens may be necessary to specifically target mutated AR in CRPC patients.
3. AR Splice Variants
Recently, constitutively active AR splice variants (SVs), created by gene splicing or genomic rearrangement, were detected in PCa cell lines and tumor samples 33–35. For example, AR variant AR3/AR-V7 is the most abundantly detected variant, and is linked to a 35-kb intragenic tandem duplication of AR exon 3 and flanking sequences 34, 36, while ARv567es is produced by intragenic deletion of exons 5, 6, and 7 37. Consequently, these variants remain constitutively active in the absence of the ligand-binding domain, having maintained the elements necessary for their transcriptional regulatory function (N-terminal AF1 and DNA binding domain) 38. Although variants retain the elements necessary for AR function, it is unclear whether the ability to dimerize with full length AR in vivo is affected. Based on siRNA-mediated depletion, both the necessity and expendability of full length AR for SV expression was demonstrated 39–41. Although AR SVs conserve the ability to drive classical AR target gene transcription (like KLK3/PSA), there is interest in determining whether alternative gene signatures in CRPC cells are due to SV expression 36, 40, 42. A SV gene signature, which was independent of FL AR, was found to be unique from parental AR full length-driven transcriptional profile 40. This suggests the ability of SVs to promote CRPC through alternative transcriptional output. However, the impact of AR SVs in the clinical context of CRPC is only beginning to be appreciated. Analysis of CRPC bone metastases detected AR3 variant transcript and protein, which correlated with decreased rate of survival after surgery 43. Based on IHC staining and transcript analysis of metastatic CRPC, compared to primary tumors, a significant increase in SV-positive staining was observed with multiple variants detected per patient, yet there was no correlation between SV expression and proliferation index of tumors 44. As most current AR antagonists target AR LBD, presence of variants renders the cell refractory to effects of ADT. Thus, there is a critical need to elucidate the transcriptional network that is controlled by these variants in order to more effectively combat these clinically challenging tumors.
3. Posttranslational Modifications of AR
Numerous posttranslational modifications (PTMs) of AR dictate activity, structure, and stability, including: phosphorylation, ubiquitylation, SUMOylation, acetylation and methylation. Overall, the majority of PTMs result in AR activation and are selected for during progression to CRPC. AR is capable of being phosphorylated on sixteen different tyrosine, serine, or threonine residues, which represents the most abundant PTM. For example, Ser81 is the most highly phosphorylated residue in response to androgens and plays a role in AR-mediated cell cycle progression manifest through AR binding to chromatin and transcriptional activity 45, 46. One potential mechanism of AR protein upregulation, which is implicated in CRPC, is altered protein stability mediated by ubiquitylation (Ub) and proteasomal degradation pathways. Ubiquitylation of AR by CHIP and MDM2 E3 ligases promote protein degradation 47, 48, while deubiquitylases cleave ubiquitin moieties from AR and increase protein expression due to degradation bypass 49. AR Ub is also implicated in transcriptional activation since RNF6-mediated Ub increases AR activity without altering protein stability 50, and MDM2 Ub of AR at gene promoters is necessary for AR turnover and effective transcription 51. Small Ubiquitin-like MOdifier (SUMO) addition is similar in structure to Ub, although it reduces transcriptional activity without altering protein stability 52. Therefore, Ub and SUMOylation are two PTMs that inhibit AR activity. AR acetylation by several acetyltransferases occurs primarily in the hinge region and increases transactivation, and reversal of this process by HDAC/SIRT1 is inhibitory 53. Acetylation and phosphorylation appear to be integrated since acetylation-dead AR mutants are not as efficiently phosphorylated 54. Finally, AR lysine methylation by SET9 increases activity by regulating N- and C-terminal interactions and recruitment to target genes 55, 56.
Although AR PTMs are all capable of modulating AR activity and influencing PCa progression in vitro, there is limited evidence of clinical implication. In one study, AR serine 210 phosphorylation, mediated by Akt, was identified to correlate with decreased survival in hormone-therapy refractory patient samples 57. Together with in vitro data demonstrating AR pSer-210 activates AR under low androgen conditions 58, it is proposed that AR phosphorylation mediates progression to CRPC. While biochemically intriguing, most PTMs have not been studied within the context of clinical disease and need to be translated to the patient setting to understand clinical utility.
4. Intratumoral androgen synthesis
An alternative mechanism to reactive AR signaling in absence of circulating androgens is autocrine androgen production by prostate tumor cells. Multiple studies examining the enzymes necessary for androgen synthesis in PCa specimens demonstrated that all components are present and many are elevated in metastatic or recurrent tumors 26. Key enzymes responsible for conversion of cholesterol to androgen precursors (CYP17A1 & HSDD3B2) were shown to be nearly 10-fold higher in metastatic PCa, while reduction enzymes, AKR1C3 and SRD5A1/2, responsible for conversion of precursors to androgen, were elevated 8 and 9.2 fold respectively 59. Such a robust increase in androgen production machinery likely increases local concentrations of available androgens during CRPC, reactivating AR signaling cascade. It is possible that tumors with intratumoral androgen synthesis are not truly castration resistant and could represent a treatable patient population with combinational therapy targeting localized androgen synthesis and AR activity.
Inhibition of cholesterol metabolism to testosterone has led to clinically viable options for treating CRPC tumors. Abiraterone acetate (Abiraterone) inhibits CYP17A1 enzymatic activity, which converts cholesterol into androgen precursors. Abiraterone treatment effectively decreases growth of PCa bone metastases 60 and is FDA approved for treatment of metastatic PCa with prior docetaxel treatment 61. However, Abiraterone treatment is capable of selecting for resistance due to increasing intratumoral levels of CYP17A1 62, and promoting accumulation of androgen precursors capable of activating AR 63. Therefore, it may be necessary and advantageous to combinatorially treat PCa patients with CYP17A1 inhibitors and AR antagonists to abrogate resistance.
5. New Approaches to Target AR Signaling
AR antagonists are initially effective against invasive PCa, but AR perturbations are selected that diminish overall efficacy of prolonged treatment and give rise to CRPC. Therefore, alternative strategies of inhibiting AR are imperative for durable treatment of aggressive PCa. Second generation AR antagonists, such as MDV3100 and ARN-509, are currently in clinical trials and have greater preclinical efficacy than Casodex 20, 64. Casodex acts as a partial antagonist by binding AR in place of androgen, resulting in nuclear translocation, binding to DNA, and recruiting corepressors to inhibit transcription. In comparison, MDV3100 impairs AR translocation and blocks recruitment to DNA, which functions to more profoundly inhibit AR transactivation 20. Based on phase I/II clinical data, 22/30 men with CRPC had steady PSA level declines, with nearly half obtaining greater than 50% reduction 65. Early AFFIRM phase III trial results comparing MDV3100 with placebo in over 1000 patients demonstrated 37% reduction in the risk of death 66. ARN-509 is earlier in phase I/II trials, but preclinical experiments demonstrated greater efficacy in xenograft models and improved bioavailabilty, compared to MDV3100 64. Alternatively, targeting the unstructured AR N-terminal domain, rather than the LBD like other antagonists has made EPI-001 a novel AR antagonist currently in preclinical testing. EPI-001 reduces AR transcriptional activity by disrupting coactivator recruitment independent of ligand binding and effectively inhibits xenograft tumor growth in mice 67. Therefore, if clinical testing validates preclinical findings, EPI-001 should potently inhibit constitutively active AR SVs that are not affected by traditional LBD-targeting antagonists. Finally, AR antisense oligonucleotides, such as the third generation EZN-4176, have shown promising preclinical results due to downregulation of AR expression 68, and are in phase I/II clinical trial. Although novel AR antagonists are still awaiting approval for clinical applications, they will greatly enhance first line treatment options for patients and potentially give physicians a mechanism to target sustained AR activity in CRPC.
C. FOXA1
AR activation results in binding to enhancer and promoter elements on DNA and culminates in transcription of target genes. ‘Pioneer’ transcription factors were identified to rearrange chromatin structure from a closed to open state to enable binding of transcriptional regulators, such as AR. FOXA1 binding facilitates AR accessibility to chromatin and has been shown to coincide with the majority of AR-binding sites 69. Despite a high degree of AR and FOXA1 overlap, depletion of FOXA1 does not completely abrogate AR recruitment to chromatin but redistributes AR binding and promotes a new gene profile 69, 70. Overall, FOXA1 in PCa appears to both inhibit and promote AR binding to regulate cell-specific transcriptional programs 71. Interestingly, FOXA1 has a similar influence on AR chromatin accessibility in apocrine breast cancer cells, which lack estrogen receptor expression but are AR positive and sensitive to AR antagonism 72, 73. Analysis of PCa samples indicates that FOXA1 is elevated in primary PCa, independently predicts for shorter time to PCa death 69, and is highly expressed in CRPC metastases 74, 75. Non-silent mutations in FOXA1 DNA-binding domain were identified in primary PCa, which could influence interaction between FOXA1 and target genes. Overall, FOXA1-mediated regulation of AR binding drives a specific gene profile that appears to govern primary PCa, CRPC, and a subtype of breast cancer. Based on in vitro FOXA1 depletion studies, it is unclear whether targeting FOXA1 would attenuate AR activity and tumor growth, or if new AR binding sites would promote a similar tumorigenic signaling network.
Oncogenes and Tumor Suppressors in Prostate Cancer
Oncogene activation and tumor suppressor abrogation are common events in initiation and progression of most cancers, though relatively few are implicated in PCa. As whole-genome sequencing becomes commonplace, our understanding of oncogene drivers and tumor suppressor gatekeepers will expand. Currently, there are several molecules altered in PCa initiation and progression. The prototypical oncogene Myc and the product of TMPRSS2:ETS gene fusions are often elevated throughout the course of disease progression. Additionally, the PI3K/AKT/mTOR pathway is regularly activated in PCa due to loss of tumor suppressor PTEN, while RB activity and/or expression loss promotes CRPC.
A. Myc
Oncogenic transformation of prostate epithelial cells differs from most solid tumors because there are few bona fide oncogenes implicated in PCa initiation and progression. The proto-oncogene, Myc, is a transcription factor that regulates expression of genes involved in nearly every biological process, and is often upregulated in PCa. In non-transformed cells, Myc expression is tightly controlled because while increased expression can promote growth and proliferation, it can also drive cell senescence or apoptosis. myc is located at chromosome 8q24, which is a locus often found amplified in PCa. As such, myc amplification is detected in roughly 30% of PCa patients but there remains some discrepancy as to whether Myc is overexpressed or amplified in primary tumors 76, 77. Myc overexpression is sufficient for immortalization and transformation of human prostate epithelial cells 78, and Myc alone is capable of promoting tumor formation in tissue recombination experiments 79. Similarly, mice engineered to maintain ‘Super-Lo’, ‘Lo’, or ‘Hi’ levels of Myc expression 80 all developed PIN lesions spontaneously, and Lo and Hi models progressed to invasive disease 81. Conversely, Myc was shown to initiate progression of epithelial cells to PIN and invasive PCa only when combined with other oncogenic lesions, such as PTEN/p53 heterozygosity 82, although Myc expression levels or mouse genetics could account for discrepancies.
In addition to the role of Myc for PCa initiation, the relationship between Myc expression and PCa progression has also been assessed. myc amplification is significantly higher in metastatic tumors, compared to primary, and Myc protein levels correlate with disease progression 83, 84. Although it is not clear if Myc amplification causes CRPC or is selected for during ADT, one study concluded that amplification was observed as a consequence of endocrine therapy 85. In addition to expression levels, intracellular localization of Myc may be a prognostic indicator. Under conditions of elevated Myc expression, nuclear Myc protein was upregulated in PIN and prostate adenocarcinoma in the absence of amplification 86. Additionally, high Myc expression in primary PCa predicted recurrence, aggressive disease, and poor prognosis 83, 87, 88.
These data suggest that mechanisms distinct from gene amplification can influence PCa progression. Despite data implicating Myc in disease progression, poor prognosis and metastatic phenotype 89, this pathway cannot currently be targeted pharmacologically. Myc amplification and/or expression status may be most useful to stratify patients for optimizing therapeutic outcome.
B. TMPRSS2:ERG Fusion
Chromosomal rearrangements in leukemias and sarcomas are known to play a role in tumorigenesis, but are rarely found in cancers of epithelial origin 90. Recently, a fusion between the 5′ untranslated region of transmembrane protease serine 2 (TMPRSS2), an androgen-regulated gene, and the 3′ exon of ERG, an erythroblast transformation specific (ETS) transcription factor family member, was discovered under conditions of ERG overexpression. While there are several ETS family members capable of rearranging with TMPRSS2, ERG is most commonly detected. Fusion is created through interstitial deletion, which excises intragenic regions on chromosome 21 91, 92, or by interchromosomal rearrangements between two independent chromosomes 21. Concordant with TMPRSS2 as an AR target, androgen-mediated AR activation regulates TMPRSS2:ERG expression 93–95, while fusion prevalence decreases upon castration of mice with TMPRSS2:ERG fusion-positive xenografts 96. As a potential mechanism for AR-mediated fusion formation, acute DHT stimulation was described to shorten distances between TMPRSS2 and ERG in androgen-responsive cell lines, and AR mediated chromosome proximity and double strand breaks 97, 98.
Given dependence on AR signaling, TMPRSS2:ERG fusions were initially hypothesized to be critical for PCa initiation. However, numerous in vitro and clinical studies analyzing the incidence of ETS fusions in PCa implicate invasion and metastasis functionality. In mouse models, ERG upregulation was not sufficient for epithelial cell transformation, but did corroborate with enhanced AR signaling and AKT/PI3K perturbation to promote invasive PCa 99. ERG modulation did not correspond with changes in cell proliferation, but overexpression induced a gene signature profile similar to networks associated with invasion while ERG depletion increased expression of genes related to differentiated luminal prostate epithelial cells 100. Despite AR-mediated regulation of TMPRSS2:ERG fusions, ERG can negatively regulate AR by binding to the AR gene and promoting a dedifferentiated ‘polycomb gene signature’ via EZH2 101, 102. These data suggest that TMPRSS2:ERG may maintain or promote PCa into a de-differentiated state that could contribute to a migratory, mesenchymal cell phenotype.
Overall, approximately 50% of primary and metastatic PCa contain a variation of the fusion, which correlates with poor prognosis 103, 104. Compared to fusion-negative patients, those who presented with TMPRSS2:ERG-positive disease and participated in ‘watchful waiting’ treatment regimen had more aggressive disease and a poorer prognosis 105. The contribution of TMPRSS2:ERG fusions in metastatic disease has been aided by rapid autopsy biopsies. By comparing primary tumor and metastasis, it was concluded that the mechanism of fusion rearrangement was conserved, and this suggested tumor aggressiveness could be predicted based on rearrangements found in primary disease 91. Furthermore, analysis of hormonally naïve PCa and CRPC demonstrated that selection for fusion-positive cells is not occurring over course of tumor progression based on equivalent levels of TMPRSS2:ERG fusions 91. TMPRSS2:ERG fusions gained significant attention because of the high incidence in both primary and metastatic tumors, however, the impact on clinical treatment regimen is questionable. Recently, TMPRSS2:ERG was identified to interact with poly(ADP-ribose) polymerase 1 (PARP1) and DNAPKcs, which are both required for ERG-mediated transactivation and invasion 106. Additionally, PARP1 inhibition significantly attenuated growth of PCa cell xenografts, demonstrating a potential approach to treat ETS-fusion positive tumors. TMPRSS2:ETS fusions have also increased interest in mining for additional fusions that may be playing a role in PCa. To that end, SLC45A3-BRAF, ESRP1-RAF1, RAF1-ESRP1, and C15orf21:MYC fusions were described in PCa107, 108. However, additional study is needed to rigorously determine if the fusion is a consequence and passenger of PCa progression or it actively promotes CRPC.
C. PI3K/PTEN/AKT
Phosphatidylinositol 3-kinase (PI3K) functions by transducing signals generated by receptor and non-receptor tyrosine kinases that govern cell growth, proliferation, survival and motility. PI3K can drive cell proliferation through phosphorylation of membrane-bound phospholipids, which recruits and activates AKT, and subsequently transduces a signal transduction cascade through mammalian Target of Rapamycin (mTOR) 109. Conversely, tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) opposes PI3K activity by PIP3 dephosphorylation, which abrogates PI3K-mediated downstream signaling, including AKT and mTOR activation. Given the complex regulation of PI3K activation it is not surprising that AKT, PTEN, and mTOR are routinely altered in PCa 110, 111. For example, 70% of primary PCa are PTEN haploinsufficient, while complete loss is often described in metastatic disease. pAKT is detected most often in high grade (≥Gleason 8) PCa, compared to low grade (≤Gleason 7) PCa 112, which also correlates with elevated mTOR activity.
Models of Pten loss in mice closely mimic human PCa based on timing of disease progression 113, expression of molecular markers, such as downregulation of NKX3.1, and similarities in gene expression profiles 114. Furthermore, these models provide evidence that PTEN loss is involved in PCa initiation. For examples, heterozygous deletion of PTEN in murine embryo fibroblasts (MEFs) significantly increases cell proliferation, whereas homozygous deletion renders cells senescent via a p53-mediated mechanism 115, 116. Overall, loss of a single PTEN allele is principal for initiation of early stage PCa, as this is often detected in primary PCa, and loss of the second allele occurs in combination with other aberrations to promote invasive cancer.
In addition to initiation, PTEN loss alters AR signaling and the cellular response to ADT, and is a key determinant for CRPC formation. In mouse models of homozygous PTEN loss, animals castrated as early as 2 weeks still developed PIN lesions and gene profiling demonstrate that PTEN loss suppresses androgen-responsive gene expression by modulating AR activity. Thus, PI3K and AR are involved in a reciprocal feedback loop under conditions of PTEN deletion, such that PI3K inhibition activates AR and vice versa 117. Interestingly, castrated PTEN WT mice and intact PTEN null mice express a similar AR gene signature, suggesting that PTEN-mediated PIN is independent of AR signaling. In fact, PTEN epithelial loss represses AR signaling in concert with increased EZH2 118. In human tumor biopsies, AR staining was heterogeneous in PTEN null regions, but adjacent PTEN positive regions stained uniformly for AR. Also, tumors were reliant on PI3K/PTEN/mTOR pathway and showed additive benefit of combining ADT and mTOR, demonstrating utility of combinatorial treatment. 118
PTEN loss or PI3K/AKT pathway activation is often found in invasive and metastatic human PCa, however, PTEN deletion in mice fails to recapitulate fatal metastases 119. One explanation of this observation is that PTEN loss alone can promote cellular senescence. When PTEN loss is combined with other prominently altered pathways, which alone have little influence on tumor initiation or disease aggressiveness, a cooperative effect can be achieved. For example, loss of the tumor suppressor, p53, in concert with PTEN loss increases tumor progression rate and drives lethal PCa progression 120. Additionally, overexpression of human epidermal growth factor 2 and 3 (HER2/3) receptor tyrosine kinases confer poor prognosis in PCa patients whose tumors have low PTEN expression, and is sufficient to overcome PTEN-induced cellular senescence 121. Finally, BMI1 inhibition slows growth of PTEN-deletion-induced PCa, suggesting that BMI1 deregulation can contribute to PTEN-mediated PCa progression 122.
PTEN loss is a common event in PCa that appears to play a role in initial hyperplastic events and can promote progression to invasive PCa and CRPC when combined with other genetic insults, such as p53 loss, and overexpression of BMI1 and HER2/3. There are currently numerous approaches being investigated to inhibit this pathway. mTor inhibitors have shown some efficacy as single agents in vitro 123, however clinical trials have yielded less promising results. Given that PI3K and AR reciprocally regulate functions 117, combinatorial PI3K and AR inhibition is currently being pursued. Preclinical assessment of this strategy identified a profound antiproliferative effect of combining PI3K inhibition and AR antagonism, compared to single treatments, in the absence of PTEN 117. This combination therapy represents the future of PCa treatment, whereby dual modulation of dominant signaling nodes dramatically increases therapeutic efficacy.
D. RB
p130 retinoblastoma (RB) protein is a tumor suppressor inactivated in the majority of epithelial cancers. RB functions to regulate cell cycle progression via binding to E2F family members and suppressing E2F-mediated gene transcription. Phosphorylation of RB by cyclin-dependent kinases (CDKs) results in inactivation and subsequent E2F-mediated cell cycle progression 124. Within the context of cancer, absence of RB signaling allows cells to improperly cycle, leading to aberrant proliferation.
Recently, RB loss was identified as being overrepresented in PCa metastases and CRPC 125. When non-neoplastic tissue, PCa, and CRPC were applied to a previously defined RB loss signature, only CRPC specimens clustered with the signature. Overall, greater than 70% of metastatic CRPC specimens were RB negative, based on immunohistochemistry, and an RB loss signature was associated with poor patient prognosis. Furthermore, modeling RB loss in ADT-sensitive PCa cells was sufficient to confer castration resistant tumor growth. The mechanism for this CRPC phenotype was identified to be E2F1-mediated activation of AR gene transcription, which resulted in elevated AR protein expression 125. In patient samples, AR inversely correlated with RB status, but directly correlated with E2F1. These data demonstrate the ability of RB loss to impinge on AR signaling and attenuate ADT, which represents a clinical biomarker for stratification of treatment strategies. For example, patients with RB-negative tumors have been shown to respond to chemotherapy, while RB-proficient tumors that have been inactivated through CDK-mediated phosphorylation may demonstrate benefit from treatment with CDK inhibitors.
Polycomb Repression
Polycomb group (PcG) proteins were originally characterized in drosophila to regulate expression of genes critical for development. Two main complexes, Polycomb Repressive Complex (PRC) 1 and 2, silence genes by enzymatically modifying histones and altering chromatin structure, and upregulation of PRC1/2 in cancers can block cell differentiation and promote a stem cell phenotype 126. PRC1 contains BMI1 and RING1B, which are ring domain-containing enzymes that add monoubiquitin moieties to lysine 119 of histone H2A (H2AK119ub1) and perturb chromatin compaction. In comparison, PRC2 represses gene expression by promoting di- and tri-methylation of lysine 79 on histone H3 (H3K79me2/3) through EZH2 127. In particular, EZH2 is the most extensively studied enzyme of the PcG proteins and is responsible for H3K79me3-mediated gene silencing. As these complexes control expression of genes critical for development, BMI1 and EZH2 are often deregulated in cancers 128 and promote initiation and progression of Pca 129. EZH2 regulation in PCa can be controlled through gene amplification 130, microRNAs 131, ERG-mediated activation 132, and Myc 133. Recently, EZH2 was identified to promote BMI1 expression by repressing miRNAs that directly inhibited BMI1 expression 134, demonstrating a complex interplay between EZH2 and BMI1.
Although AR is known to drive transcription of a large number of genes, there is also a large cohort of understudied genes bound by AR and repressed. AR-repressed genes are enriched for H3K79me3, and EZH2 occupancy is increased following AR activation. Interestingly, many AR-repressed genes are developmental regulators that are equally suppressed in embryonic stem cells. In support of the hypothesis that PRC1/2 perturbation alters differentiation status in cancer, CRPC cells have basal repression of pro-differentiation genes 135. Furthermore, EZH2 was identified in a PCa-specific repressive co-regulatory network with histone deacetylases and ERG. EZH2 was capable of mediating repression of AR-induced transcription. In particular, EZH2 has been shown to repress cytoskeletal proteins, vinculin and E-cadherin, which increases metastasis and contributes to epithelial dedifferentiation 136, 137. Clinically, PRC2 genomic sites and H3K79me3 marks in late-stage aggressive PCa confirmed data from cell lines, indicating that developmental regulators were repressed by EZH2 in PCa. Furthermore, similarly repressed genes were identified in multiple metastatic tumors, and a ‘polycomb repressed gene signature’ derived from these samples could predict for patient outcome 138.
BMI1 mediates epigenetic silencing by H2A ubiquitylation and is involved in DNA damage and repair. However, AKT-mediated phosphorylation of BMI1 is critical for oncogenic potential, such that BMI1 overexpression alone in a transgenic mouse model resulted in PIN lesions, but in concert with PTEN haploinsufficiency (and increased AKT activity), invasive PCa that recapitulated human disease was observed 112. Based on human samples, BMI1 expression is detectable in low grade PCa, BMI1 and pAKT correlate with an increased pathological grade, and expression is maintained throughout disease progression 112. Overall, altered EZH2 and BMI1 expression, as well as resultant genes alterations, represent exciting therapeutic targets that control a tumor-specific transcriptional program not present in normal tissue.
Genome-wide Analysis of PCa Genetic Alterations
The ability to assess all genetic gains, losses, and mutations has been applied to PCa and has given great insight into the genetic landscape of this disease 139. The first reported study sequenced the complete genome of 7 high grade primary PCa, along with matched normal tissue 140. Overall, just under 1 somatic mutation per megabase (Mb) was detected, which is comparable to breast cancer. Surprisingly, each sequenced genome contained an average of 100 inter- or intra-chromosomal rearrangements, such as TMPRSS2:ERG 139, 140.
Since that initial report, two additional studies have analyzed larger cohorts of patients to understand the genetic alterations in an unbiased approach. Barbieri et al. sequenced 112 primary PCa tumors that were treatment naïve 30 while Grasso et al sequenced 11 localized treatment naïve PCa tumors and 50 lethal, heavily pretreated CRPC tumors 31. In both studies, rate of mutation was slightly higher than the initial study at 1.4 mutations per Mb and 2.0 mutations per Mb (among CRPC tumors), respectively. These studies are of diagnostic and therapeutic value because they identified new classifications of mutations and potential targets. For example, SPOP was identified as a highly mutated protein in PCa. When SPOP mutation-positive tumors were stratified for other mutation occurrences, it was discovered that TMPRSS2:ERG fusions were never detected, and p53/PI3K/RB mutations were significantly decreased 30. This could represent a new treatment option for patients with specific SPOP mutations. A novel mutation in the pioneering factor FOXA1 was also identified and shown to inhibit AR signaling and promote tumor growth 31. Given the infancy of these publications, many of the detected mutation still require validation to determine if there is any functional consequence. However, it represents the current paradigm shift from single molecule research to genome-wide analysis.
Conclusion
To date, there have been numerous molecules implicated in PCa initiation or progression to CRPC, many of which can be inhibited in vivo (Figure 1). Modeling PCa in murine models has provided extensive understanding of the phenotypic progression from PIN to PCa to CRPC. However, comprehensive understanding of PCa initiation has remained elusive, which is necessary for targeting pathways implicit in early stage disease and for abrogating cancer growth prior to invasion and metastasis. Currently, assessment of molecules implicated in initiation, such as NKX3.1, FOXA1, and Myc is based primarily on genetic mouse models or correlative analysis from human tumor samples. Incomplete insight in this process is further compounded by the inability to effectively target most molecules associated with initiation. For example, Myc is historically difficult to inhibit pharmacologically and is suggested to be ‘undruggable’. However, it was identified that inhibition of molecules that support Myc oncogenic programs is a therapeutic option for Myc inhibition 141. Similarly, understanding that cell-specific Myc targets are not all equal could result in the targeting of targets to effectively inhibit Myc activity in PCa initiation 142. Overall, our understanding of PCa progression to CRPC vastly outweighs what is known regarding disease formation.
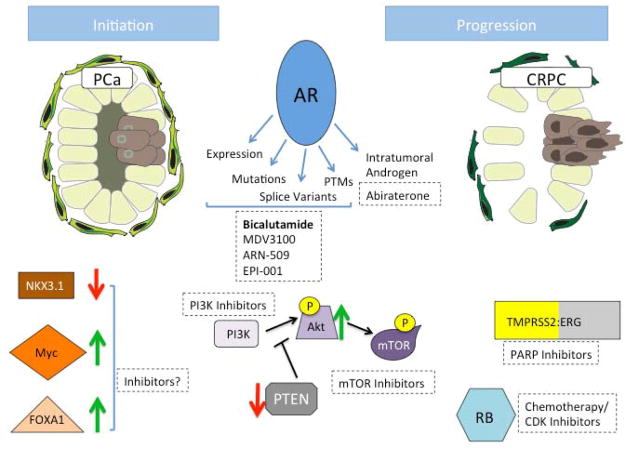
Figure 1. Molecules implicated in PCa initiation and progression.
Initiation of PCa and progression to CRPC involves deregulation of numerous signaling pathways. Expression of NKX3.1, FOXA1, and Myc is altered in early stage PCa, which plays a role in disease initiation. However, there are currently no effective means to target these aberrations. Additional molecules are perturbed in transition to CRPC and are capable of being pharmacologically inhibited. TMPRSS2:ERG fusion expression promotes invasive phenotypes and can be inhibited by PARP1 inhibitors, RB status (loss of expression or function) can stratify patients for responsiveness to chemotherapy or CDK inhibitors, and loss of PTEN expression increases PI3K/AKT/mTOR signaling, which can be inhibited with targeted inhibitors. AR signaling is central to PCa and progression to CRPC. Prior to CRPC, invasive PCa is treated with AR antagonist bicalutamide, although novel inhibitors MDV3100, ARN-509, and EPI-001 are in clinical trials and may provide more effective treatment options. In response to antagonists, AR activity is regained through increased expression, gain-of-function mutations, constitutively active splice variants, posttranslational modifications (PTMs), and intratumoral androgen synthesis. Abiraterone inhibits autocrine androgen synthesis and is approved for CRPC treatment. Red and green arrows indicate decreased or increased protein expression/activity detected in PCa, respectively.
With the increased utility of whole genome sequencing, it is apparent that PCa treatments will increasingly move toward precision medicine. Possessing the capability to determine precisely what mutations and genetic aberrations are present in the tumor of a patient will potentially propel the ability to effectively treat CRPC. However, as new technological advances are attained, it will be necessary to determine how to understand the increasing volumes of data. For example, despite the capability to determine every mutation in a tumor, comprehensive appreciation of biological contexts or mutational crosstalk is not manageable. Therefore, extensive validation of genetic alterations is crucial for translation to therapeutic interventions. Overall, whole genome analysis has the potential to significantly expand the landscape of molecules implicit to PCa.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Knudsen BS, Vasioukhin V. Mechanisms of prostate cancer initiation and progression. Advances in cancer research. 2010;109:1–50. doi: 10.1016/B978-0-12-380890-5.00001-6. [DOI] [PubMed] [Google Scholar]
- 2.Niu YN, Xia SJ. Stroma-epithelium crosstalk in prostate cancer. Asian journal of andrology. 2009;11:28–35. doi: 10.1038/aja.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Josson S, Matsuoka Y, Chung LW, Zhau HE, Wang R. Tumor-stroma co-evolution in prostate cancer progression and metastasis. Seminars in cell & developmental biology. 2010;21:26–32. doi: 10.1016/j.semcdb.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes & development. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation; research in biological diversity. 2008;76:717–727. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.English HF, Santen RJ, Isaacs JT. Response of glandular versus basal rat ventral prostatic epithelial cells to androgen withdrawal and replacement. The Prostate. 1987;11:229–242. doi: 10.1002/pros.2990110304. [DOI] [PubMed] [Google Scholar]
- 7.Sandford NL, Searle JW, Kerr JF. Successive waves of apoptosis in the rat prostate after repeated withdrawal of testosterone stimulation. Pathology. 1984;16:406–410. doi: 10.3109/00313028409084731. [DOI] [PubMed] [Google Scholar]
- 8.Isbarn H, et al. Testosterone and prostate cancer: revisiting old paradigms. European urology. 2009;56:48–56. doi: 10.1016/j.eururo.2009.03.088. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Tindall DJ. Androgen action during prostate carcinogenesis. Methods Mol Biol. 2011;776:25–44. doi: 10.1007/978-1-61779-243-4_2. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang AY, et al. Immunohistochemical differentiation of high-grade prostate carcinoma from urothelial carcinoma. The American journal of surgical pathology. 2007;31:1246–1255. doi: 10.1097/PAS.0b013e31802f5d33. [DOI] [PubMed] [Google Scholar]
- 14.Song H, et al. Loss of Nkx3.1 leads to the activation of discrete downstream target genes during prostate tumorigenesis. Oncogene. 2009;28:3307–3319. doi: 10.1038/onc.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson PD, et al. Nkx3.1 and Myc crossregulate shared target genes in mouse and human prostate tumorigenesis. The Journal of clinical investigation. 2012;122:1907–1919. doi: 10.1172/JCI58540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanbrough M, Leav I, Kwan PW, Bubley GJ, Balk SP. Prostatic intraepithelial neoplasia in mice expressing an androgen receptor transgene in prostate epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10823–10828. doi: 10.1073/pnas.191235898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu C, et al. Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. The Journal of biological chemistry. 2011;286:33478–33488. doi: 10.1074/jbc.M111.269894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bubendorf L, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer research. 1999;59:803–806. [PubMed] [Google Scholar]
- 19.Waltering KK, et al. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer research. 2009;69:8141–8149. doi: 10.1158/0008-5472.CAN-09-0919. [DOI] [PubMed] [Google Scholar]
- 20.Tran C, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donovan MJ, et al. Androgen receptor expression is associated with prostate cancer-specific survival in castrate patients with metastatic disease. BJU international. 2010;105:462–467. doi: 10.1111/j.1464-410X.2009.08747.x. [DOI] [PubMed] [Google Scholar]
- 22.Linja MJ, et al. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer research. 2001;61:3550–3555. [PubMed] [Google Scholar]
- 23.Chen CD, et al. Molecular determinants of resistance to antiandrogen therapy. Nature medicine. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 24.Holzbeierlein J, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. The American journal of pathology. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu SY, et al. Identification of a novel germline missense mutation of the androgen receptor in African American men with familial prostate cancer. Asian journal of andrology. 2010;12:336–343. doi: 10.1038/aja.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends in endocrinology and metabolism: TEM. 2010;21:315–324. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleman MN, Lush RM, Figg WD. The mutated androgen receptor and its implications for the treatment of metastatic carcinoma of the prostate. Pharmacotherapy. 1996;16:376–381. [PubMed] [Google Scholar]
- 28.Wetherill YB, et al. Xenoestrogen action in prostate cancer: pleiotropic effects dependent on androgen receptor status. Cancer research. 2005;65:54–65. [PubMed] [Google Scholar]
- 29.Wetherill YB, et al. Bisphenol A facilitates bypass of androgen ablation therapy in prostate cancer. Molecular cancer therapeutics. 2006;5:3181–3190. doi: 10.1158/1535-7163.MCT-06-0272. [DOI] [PubMed] [Google Scholar]
- 30.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinkamp MP, et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer research. 2009;69:4434–4442. doi: 10.1158/0008-5472.CAN-08-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu R, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer research. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo Z, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer research. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012 doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenster G, van der Korput HA, Trapman J, Brinkmann AO. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 39.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. The Journal of biological chemistry. 2012;287:19736–19749. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu R, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer research. 2012 doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson PA, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16759–16765. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun S, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. The Journal of clinical investigation. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornberg E, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PloS one. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, et al. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PloS one. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, Gulla S, Cai C, Balk SP. Androgen receptor serine 81 phosphorylation mediates chromatin binding and transcriptional activation. The Journal of biological chemistry. 2012;287:8571–8583. doi: 10.1074/jbc.M111.325290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon V, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–2280. doi: 10.1210/me.2010-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic acids research. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gioeli D, Paschal BM. Post-translational modification of the androgen receptor. Molecular and cellular endocrinology. 2012;352:70–78. doi: 10.1016/j.mce.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Dirac AM, Bernards R. The deubiquitinating enzyme USP26 is a regulator of androgen receptor signaling. Molecular cancer research : MCR. 2010;8:844–854. doi: 10.1158/1541-7786.MCR-09-0424. [DOI] [PubMed] [Google Scholar]
- 50.Xu K, et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer cell. 2009;15:270–282. doi: 10.1016/j.ccr.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. The EMBO journal. 2011;30:468–479. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaikkonen S, et al. SUMO-specific protease 1 (SENP1) reverses the hormone-augmented SUMOylation of androgen receptor and modulates gene responses in prostate cancer cells. Mol Endocrinol. 2009;23:292–307. doi: 10.1210/me.2008-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, et al. Inactivation of androgen-induced regulator ARD1 inhibits androgen receptor acetylation and prostate tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3053–3058. doi: 10.1073/pnas.1113356109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu M, et al. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. The Journal of biological chemistry. 2004;279:29436–29449. doi: 10.1074/jbc.M313466200. [DOI] [PubMed] [Google Scholar]
- 55.Gaughan L, et al. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic acids research. 2011;39:1266–1279. doi: 10.1093/nar/gkq861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko S, et al. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol. 2011;25:433–444. doi: 10.1210/me.2010-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCall P, Gemmell LK, Mukherjee R, Bartlett JM, Edwards J. Phosphorylation of the androgen receptor is associated with reduced survival in hormone-refractory prostate cancer patients. British journal of cancer. 2008;98:1094–1101. doi: 10.1038/sj.bjc.6604152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manin M, et al. Androgen receptor expression is regulated by the phosphoinositide 3-kinase/Akt pathway in normal and tumoral epithelial cells. The Biochemical journal. 2002;366:729–736. doi: 10.1042/BJ20020585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Montgomery RB, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer research. 2008;68:4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Efstathiou E, et al. Effects of abiraterone acetate on androgen signaling in castrate-resistant prostate cancer in bone. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:637–643. doi: 10.1200/JCO.2010.33.7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. The New England journal of medicine. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cai C, et al. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer research. 2011;71:6503–6513. doi: 10.1158/0008-5472.CAN-11-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richards J, et al. Interactions of abiraterone, eplerenone, and prednisolone with wild-type and mutant androgen receptor: a rationale for increasing abiraterone exposure or combining with MDV3100. Cancer research. 2012;72:2176–2182. doi: 10.1158/0008-5472.CAN-11-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clegg NJ, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer research. 2012;72:1494–1503. doi: 10.1158/0008-5472.CAN-11-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scher HI, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–1446. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Bono JS, et al. Primary, secondary, and quality-of-life endpoint results from the phase III AFFIRM study of MDV3100, an androgen receptor signaling inhibitor. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:4519. [Google Scholar]
- 67.Andersen RJ, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, et al. Reduced expression of the androgen receptor by third generation of antisense shows antitumor activity in models of prostate cancer. Molecular cancer therapeutics. 2011;10:2309–2319. doi: 10.1158/1535-7163.MCT-11-0329. [DOI] [PubMed] [Google Scholar]
- 69.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. The EMBO journal. 2011;30:3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. The EMBO journal. 2011;30:3885–3894. doi: 10.1038/emboj.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ni M, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson JL, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. The EMBO journal. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerhardt J, et al. FOXA1 promotes tumor progression in prostate cancer and represents a novel hallmark of castration-resistant prostate cancer. The American journal of pathology. 2012;180:848–861. doi: 10.1016/j.ajpath.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 75.Jain RK, Mehta RJ, Nakshatri H, Idrees MT, Badve SS. High-level expression of forkhead-box protein A1 in metastatic prostate cancer. Histopathology. 2011;58:766–772. doi: 10.1111/j.1365-2559.2011.03796.x. [DOI] [PubMed] [Google Scholar]
- 76.Gallucci M, et al. Cytogenetic profiles as additional markers to pathological features in clinically localized prostate carcinoma. Cancer letters. 2006;237:76–82. doi: 10.1016/j.canlet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 77.Gallucci M, et al. Genetic profile identification in clinically localized prostate carcinoma. Urologic oncology. 2009;27:502–508. doi: 10.1016/j.urolonc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 78.Gil J, et al. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer research. 2005;65:2179–2185. doi: 10.1158/0008-5472.CAN-03-4030. [DOI] [PubMed] [Google Scholar]
- 79.Williams K, et al. Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. The Prostate. 2005;63:369–384. doi: 10.1002/pros.20200. [DOI] [PubMed] [Google Scholar]
- 80.Iwata T, et al. MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PloS one. 2010;5:e9427. doi: 10.1371/journal.pone.0009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ellwood-Yen K, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 82.Kim J, et al. A mouse model of heterogeneous, c-MYC-initiated prostate cancer with loss of Pten and p53. Oncogene. 2012;31:322–332. doi: 10.1038/onc.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jenkins RB, Qian J, Lieber MM, Bostwick DG. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer research. 1997;57:524–531. [PubMed] [Google Scholar]
- 84.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T. Genetic alterations in hormone-refractory recurrent prostate carcinomas. The American journal of pathology. 1998;153:141–148. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaltz-Wittmer C, et al. FISH analysis of gene aberrations (MYC, CCND1, ERBB2, RB, and AR) in advanced prostatic carcinomas before and after androgen deprivation therapy. Laboratory investigation; a journal of technical methods and pathology. 2000;80:1455–1464. doi: 10.1038/labinvest.3780152. [DOI] [PubMed] [Google Scholar]
- 86.Gurel B, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21:1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hawksworth D, et al. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate cancer and prostatic diseases. 2010;13:311–315. doi: 10.1038/pcan.2010.31. [DOI] [PubMed] [Google Scholar]
- 88.Sato K, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. Journal of the National Cancer Institute. 1999;91:1574–1580. doi: 10.1093/jnci/91.18.1574. [DOI] [PubMed] [Google Scholar]
- 89.Wolfer A, et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc Natl Acad Sci U S A. 107:3698–3703. doi: 10.1073/pnas.0914203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mehra R, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer research. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iljin K, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer research. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]
- 93.Bastus NC, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer research. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mertz KD, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200–206. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 96.Hendriksen PJ, et al. Evolution of the androgen receptor pathway during progression of prostate cancer. Cancer research. 2006;66:5012–5020. doi: 10.1158/0008-5472.CAN-05-3082. [DOI] [PubMed] [Google Scholar]
- 97.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zong Y, et al. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tomlins SA, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun C, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu J, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mehra R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 104.Mosquera JM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Demichelis F, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 106.Brenner JC, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu C, et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. The Journal of pathology. 2012;227:53–61. doi: 10.1002/path.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Current opinion in genetics & development. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lonigro RJ, et al. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia. 2011;13:1019–1025. doi: 10.1593/neo.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nacerddine K, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. The Journal of clinical investigation. 2012;122:1920–1932. doi: 10.1172/JCI57477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang S, et al. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang S, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 115.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Narita M, Lowe SW. Senescence comes of age. Nature medicine. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- 117.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mulholland DJ, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mulholland DJ, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer research. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martin P, et al. Prostate epithelial Pten/TP53 loss leads to transformation of multipotential progenitors and epithelial to mesenchymal transition. The American journal of pathology. 2011;179:422–435. doi: 10.1016/j.ajpath.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ahmad I, et al. HER2 overcomes PTEN (loss)-induced senescence to cause aggressive prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16392–16397. doi: 10.1073/pnas.1101263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lukacs RU, Memarzadeh S, Wu H, Witte ON. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell stem cell. 2010;7:682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schiewer MJ, et al. mTOR is a selective effector of the radiation therapy response in androgen receptor-positive prostate cancer. Endocrinerelated cancer. 2012;19:1–12. doi: 10.1530/ERC-11-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Udayakumar T, Shareef MM, Diaz DA, Ahmed MM, Pollack A. The E2F1/Rb and p53/MDM2 pathways in DNA repair and apoptosis: understanding the crosstalk to develop novel strategies for prostate cancer radiotherapy. Seminars in radiation oncology. 2010;20:258–266. doi: 10.1016/j.semradonc.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 125.Sharma A, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. The Journal of clinical investigation. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews. Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 127.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell death & disease. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Leenders GJ, et al. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. European urology. 2007;52:455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 130.Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes, chromosomes & cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 131.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kunderfranco P, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PloS one. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Koh CM, et al. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–683. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cao Q, et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer cell. 2011;20:187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhao JC, et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome research. 2012;22:322–331. doi: 10.1101/gr.131508.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chng KR, et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. The EMBO journal. 2012;31:2810–2823. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu J, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer research. 2007;67:10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 139.Kumar A, Shendure J, Nelson PS. Genome interrupted: sequencing of prostate cancer reveals the importance of chromosomal rearrangements. Genome medicine. 2011;3:23. doi: 10.1186/gm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Van Dang C, McMahon SB. Emerging Concepts in the Analysis of Transcriptional Targets of the MYC Oncoprotein: Are the Targets Targetable? Genes & cancer. 2010;1:560–567. doi: 10.1177/1947601910379011. [DOI] [PMC free article] [PubMed] [Google Scholar]