SUMMARY
Lipid droplets (LDs) store metabolic energy and membrane lipid precursors. With excess metabolic energy, cells synthesize triacylglycerol (TG) and form LDs that grow dramatically. It is unclear how TG synthesis relates to LD formation and growth. Here, we identify two LD subpopulations: smaller LDs of relatively constant size, and LDs that grow larger. The latter population contains isoenzymes for each step of TG synthesis. Glycerol-3-phosphate acyltransferase 4 (GPAT4), which catalyzes the first and rate-limiting step, relocalizes from the endoplasmic reticulum (ER) to a subset of forming LDs, where it becomes stably associated. ER-to-LD targeting of GPAT4 and other LD-localized TG synthesis isozymes is required for LD growth. Key features of GPAT4 ER-to-LD targeting and function in LD growth are conserved between Drosophila and mammalian cells. Our results explain how TG synthesis is coupled with LD growth and identify two distinct LD subpopulations based on their capacity for localized TG synthesis.
INTRODUCTION
Life is driven by the flow of metabolic energy through biological systems. Variations in energy availability are major challenges for organisms, which consequently have developed storage depots for conserving metabolic energy. These depots are filled in times of excess energy availability and mobilized during times of need. The most widely used molecule for storing metabolic energy is triacylglycerol (TG). Virtually all cells have the capacity to synthesize and store TG in cytosolic lipid droplets (LDs) (Brasaemle and Wolins, 2012; Fujimoto and Parton, 2011; Thiele and Spandl, 2008; Walther and Farese, 2012), and many organisms, particularly mammals, have specialized cells (adipocytes) and tissues for TG storage. Besides their fundamental role in energy metabolism, LDs are linked to numerous other cellular processes, including membrane expansion during cell division and storage of sterols (Bartz et al., 2007; Kurat et al., 2009).
LDs are essentially neutral lipids emulsified within the aqueous cytoplasm. To prevent LD-phase coalescence, the TG core of LDs is coated by a monolayer of phospholipids that act as surfactants. The monolayer contains specific proteins with many biochemical functions. How proteins target to LDs is mostly unknown. Some LD proteins, such as perilipins, are targeted from the cytoplasm (Londos et al., 1999; Wolins et al., 2006). Other proteins, such as the triglyceride lipase adipose triglyceride lipase (ATGL) or methyltransferase-like 7B (METTL7B), have been posited to use hydrophobic domains to relocalize from the endoplasmic reticulum (ER) to LDs (Soni et al., 2009; Zehmer et al., 2008, 2009). The mechanisms for such targeting are unknown. It is also unclear whether all LDs in a cell have similar protein and lipid compositions. Evidence suggests that, at least in some cells, LDs with different protein compositions coexist (Wolins et al., 2005) and that LDs of different lipid composition contain different perilipins (Hsieh et al., 2012).
Little is known about how LDs form and grow. During conditions of excess metabolic energy (e.g., excess fatty acids), cells form many LDs, and some of these subsequently grow dramatically (e.g., up to 30-fold increases in volume within hours; Krahmer et al., 2011). Based on the tight association of LDs with the ER and the ER localization of enzymes involved in synthesizing neutral lipids for LDs, a prominent model posits that LDs form by growing a lipid lens in the ER that subsequently buds off into the cytoplasm (Martin and Parton, 2006). Alternative models have been proposed (Ploegh, 2007; Robenek et al., 2006; Walther and Farese, 2009), and little direct evidence supports any of them.
Large LDs can arise by two mechanisms. One mechanism is fusion or coalescence of LDs, which appears to be relatively rare under normal conditions (Murphy et al., 2010), but occurs frequently with PC deficiency or accumulation of phosphatidic acid (Fei et al., 2011; Guo et al., 2008). Fusion is facilitated in cells expressing FSP-27 (CIDEC; Gong et al., 2011; Jambunathan et al., 2011). The other mechanism for forming large LDs is by growth or expansion. In LD growth, TGs are added to the cores and phospholipids to the surfaces. We recently described a mechanism whereby PC synthesis is activated to maintain sufficient PC at the LD surface (Krahmer et al., 2011). TGs are known to be synthesized in the ER, but how they fill the cores of growing LDs is unclear. DGAT2, one of the two acyl-CoA:diacylglycerol O-acyltransferase (DGAT) enzymes, localizes to LDs when cells are cultured with excess fatty acids, suggesting that TG might be added to growing LDs in situ (Jacquier et al., 2011; Kuerschner et al., 2008; Stone et al., 2009). However, DGAT2 catalyzes only the final step in de novo TG synthesis, and its localization to LDs could be for recycling of hydrolyzed lipids. Thus, its functional role at LDs is unclear.
Here, we investigated the mechanism of LD growth and discovered that relocalization of TG enzymes to a subset of LDs mediates their growth.
RESULTS
Specific Isoenzymes of De Novo TG Synthesis Localize to Mature LDs
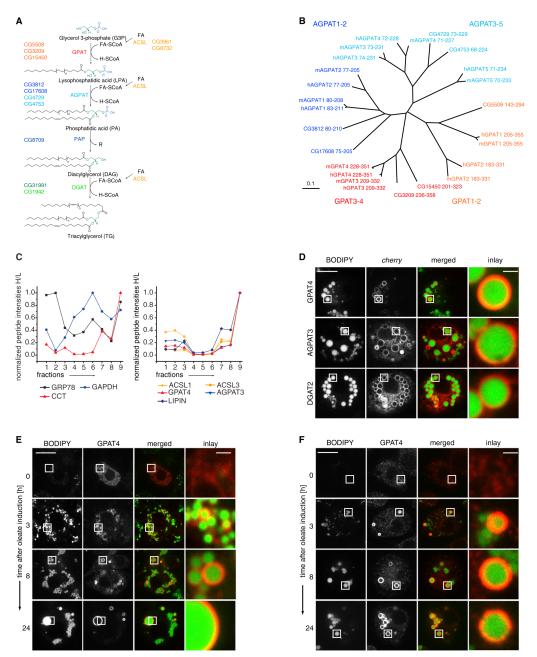
De novo TG synthesis occurs in four sequential steps catalyzed by glycerol-3-phosphate O-acyltransferase (GPAT), 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT), phosphatidic acid phosphatase (PAP), and DGAT enzymes (Figure 1A). In most organisms, multiple isoenzymes, encoded by multiple genes, contribute to the enzymatic activity of each step (Yen et al., 2008). Each step of the pathway is catalyzed by activities contributed by multiple isoenzymes encoded by a family of genes. For example, there are three or four evolutionarily related genes encoding GPAT activity in Drosophila or humans, respectively (Figure 1B; Table S2 available online). Members of the GPAT/AGPAT family of enzymes localize to the ER (Coleman and Lee, 2004). However, several other lipid metabolism enzymes, such as CCT1 or DGAT2, relocalize from the nucleus/cytoplasm or the ER, respectively, to LDs during oleate loading (Krahmer et al., 2011; Kuerschner et al., 2008; Stone et al., 2009).
Figure 1. A Subset of TG Synthesis Isoenzymes Localizes to LDs in Drosophila S2 Cells.
(A) Overview of de novo TG synthesis. CG numbers identify the Drosophila genes encoding each activity.
(B) Neighbor-joining tree based on the degree of sequence similarity between acyltransferase domains of the GPAT/AGPAT family from Homo sapiens (h), Mus musculus (m), and Drosophila melanogaster (dm). The tree branch lengths are scaled in substitutions per site.
(C) Abundance profiles of proteins throughout the fractions of an LD purification measured by quantitative mass spectrometry. Normalized peptide abundances measured by proteomic analyses of fractions of a purification from cells metabolically labeled with heavy isotope-containing amino acids are shown normalized to an LD protein standard obtained by a similar purification. Fraction 9 is the LD fraction. The left graph indicates profiles representative of cytosolic (GAPDH), ER (GRP78), and LD (CCT1) proteins. The right graph shows profiles of isoenzymes involved in TG synthesis highly enriched in the LD fraction.
(D) GPAT4, AGPAT3, or DGAT2 tagged with fluorescent cherry tag (red) localizes to LDs (BODIPY, green) in oleate-loaded Drosophila S2 cells.
(E) GPAT4 accumulates at LDs during LD formation. Cherry-GPAT4 images during LD formation induced by oleate-containing medium at the indicated time points are shown.
(F) Immunofluorescence staining of GPAT4 (red) on forming LDs (BODIPY, green).
Scale bars, 10 μm (overview) and 1 μm (magnification). See also Figure S1, and Tables S1 and S2.
To determine if TG synthesis enzymes other than DGAT2 localize to LDs, we comprehensively analyzed by quantitative proteomics the abundance of TG enzymes in LD fractions from Drosophila S2 cells incubated for 12 hr with oleate (Krahmer et al., 2013). Unexpectedly, at least one isoenzyme for each step of the TG synthesis pathway (e.g., GPAT4, AGPAT3, PAP [LIPIN]) was strongly enriched in the LD fraction and far less abundant in other fractions of the purification, closely resembling the profile of the LD protein CCT1 (Figure 1C; Table S1; Krahmer et al., 2011). Consistent with previous findings, we found similar LD localization patterns for acyl-CoA synthetase enzymes (ACSL1 and ACSL3), which activate fatty acids and enable them to serve as substrates for TG synthesis enzymes (Figure 1C; Brasaemle et al., 2004; Fujimoto et al., 2007; Poppelreuther et al., 2012). To confirm the localization of TG synthesis isoenzymes to LDs, we expressed each candidate protein fused to the cherry red fluorescent protein and analyzed its localization relative to BODIPY-stained LDs. Each of the proteins localized to LDs (Figures 1D and 1E), whereas other isoenzymes not found in LD fractions by quantitative proteomics did not (Figures S1A and S1B; data not shown). We next determined if the localization of fluorescently tagged GPAT4 reflected the distribution of the endogenous GPAT4 enzyme. Endogenous GPAT4 localized to LDs and prominently stained only a subset of large LDs (Figures 1F, S1C, and S1D).
GPAT4 Relocalizes to Distinct ER Domains during LD Formation
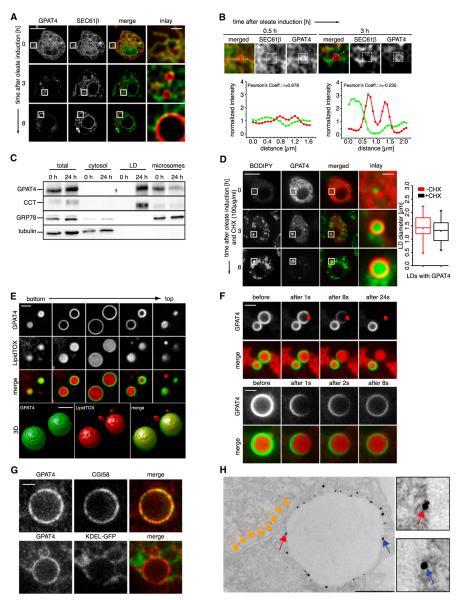
Finding that GPAT4 localizes to LDs raised an apparent contradiction as members of the GPAT/AGPAT enzyme family had been reported as ER enzymes based on their cofractionation with microsomes (Aguado and Campbell, 1998; Coleman and Lee, 2004; Nimmo, 1979). To address this question, we localized GPAT4 and an ER marker (N-terminally GFP-tagged SEC61β). Consistent with the previous reports, both endogenous GPAT4 and fluorescently tagged GPAT4 colocalized with the ER marker in cells that were not treated with oleate and had no or few LDs (Figure 2A). However, within 0.5 hr of inducing LD formation with oleate supplementation, GPAT4 accumulated in ER domains near forming LDs (Figures 2B and S2A). After 3 hr of LD formation, GPAT4 was present in domains continuous with the ER but segregated from the ER marker SEC61β-GFP (Figure 2B) and a luminal ER protein (signal sequence-GFP-KDEL; Figure S2B).
Figure 2. GPAT4 Relocalizes from the ER to the Surface of LDs.
(A) A time course of LD formation induced by oleate-containing medium shows segregation of cherry-tagged GPAT4 (red) from the ER marker (GFP-SEC61β; green). Optical sections focus on LDs, resulting in different sections of the nuclear envelope (NE) at each time point (plane of the NE at 0 hr; cross-section at other time points). Scale bars, 10 μm (overview) and 1 μm (magnification).
(B) GPAT4 tagged with fluorescent cherry tag (red) accumulates in specialized regions of the ER (marked by GFP-SEC61β; green). Scale bars, 10 μm (overview) and 1 μm (magnification). Intensity profiles of the two signals along the dotted line are shown. Pearson correlations for GFP-Sec61β and cherry-GPAT4 signal within the white boxes at 0.5 or 3 hr of oleate are shown.
(C) GPAT4 fractionates with LDs after oleate treatment. Western blots of equal protein amounts of cytosol, microsome, and LD fractions against the LD marker CCT1, the ER marker GRP78, and the cytosolic marker tubulin are shown.
(D) cherry-GPAT4 relocalizes to LDs from the ER during LD formation in cells where the synthesis of new proteins is blocked by cycloheximide added prior to oleate. On the right, sizes of 40 GPAT4-containing LDs from cells cultured 8 hr in control medium or medium containing cycloheximide are shown.
(E) Cherry-GPAT4 signal covers the entire LD. z sections and a 3D reconstruction of cherry-GPAT4 containing LD are shown. Scale bar, 2 μm.
(F) Cherry-GPAT4 decreases uniformly during continuous bleaching of a small LD surface section (upper panels). Cherry-GPAT4 recovers uniformly after photobleaching of a small LD surface section (lower panel). Scale bars, 2 μm.
(G) Endogenous GPAT4 (red) colocalizes with the LD marker CGI58 (green) but segregates from the ER (ss-GFP-KDEL; green). Scale bar, 1 μm.
(H) Silver-enhanced Immunogold staining of endogenous GPAT4 on LDs in thin-section EM. Red and blue arrows show representative gold particles on the surface of LDs. Yellow arrows show close-by ER structures. Scale bar, 500 nm.
See also Figures S2 and S3.
GPAT4 relocalization was reflected in its biochemical purification properties. Before LD induction with oleate, GPAT4 copurified with microsomes as reported by Beigneux et al. (2006), Nimmo (1979), and Saggerson et al. (1980). However, after induction of LDs, most GPAT4 was found in the LD fraction (Figure 2C).
GPAT4 localization to sites of forming LDs could be either due to relocalization of the existing pools of the protein or due to degradation of the ER pool and replenishment of newly synthesized GPAT4 only at LDs. To distinguish between these possibilities, we induced LD formation in GPAT4-cherry-expressing cells pretreated with cycloheximide, which blocks synthesis of GPAT4 (Figure S3A). Under this condition and oleate loading, GPAT4 targeted to LDs efficiently, indicating that new protein synthesis is not required for LD targeting (Figure 2D).
GPAT4 Relocalizes to the Phospholipid Monolayer Delimiting Mature LDs
LDs are bounded by a monolayer of phospholipids and are often found in close proximity to other organelles, including the ER, mitochondria, peroxisomes, and endosomes (Murphy et al., 2009). Most prominently, the ER is often found in extended, intimate contact with LDs (e.g., Blanchette-Mackie et al., 1995;Robenek et al., 2006).
To determine whether GPAT4 localizes to the LD surface or some associated membrane, we recorded optical sections through GPAT4-containing LDs and reconstructed a three-dimensional (3D) LD surface from deconvoluted images. GPAT4 localizes to the entire LD surface in the resulting images, and not a subsection of it, as might be expected if it was embedded in an LD-associated organelle (Figure 2E). Consistently, fluorescence recovery after photobleaching (FRAP) experiments show uniform recovery of bleached cherry-GPAT4 signal, and fluorescence loss in photobleaching (FLIP) experiments show uniform signal depletion. These results suggest that GPAT4 localizes to a uniform surface covering the whole LD (Figures 2F, S3B, and S3C).
Because of the close proximity of LDs to other organelles, it is difficult for conventional microscopy to directly visualize localization to the LD and to distinguish this from localization to closely apposed ER. To overcome this challenge, we employed stimulated emission depletion (STED) super high-resolution microscopy to compare the localization of endogenous GPAT4 with LD or ER markers. STED microscopy showed clear colocalization of GPAT4 with the LD marker CGI58 and separation from the ER marker ss-KDEL-GFP (Figure 2G). Consistently, Immunogold-labeled GPAT4 was specifically detected on the phospholipid monolayer surrounding mature LDs, but not on nearby LD-associated ER membranes in thin-section transmission electron microscopy (EM) (Figures 2H and S2C).
GPAT4 Is an Integral Membrane Protein without Luminal Domains
Analysis of the primary sequence of GPAT4 predicts three membrane-spanning helices (Figure S4A). However, LDs should exclude transmembrane domain-containing proteins with hydrophilic regions of the protein on both sides of the membrane.
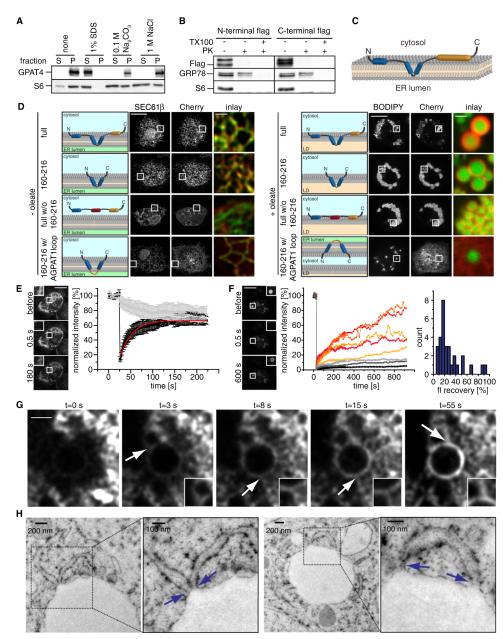
To assess whether GPAT4 is an integral membrane protein, we analyzed its biochemical extraction behavior from membranes of cells not treated with oleate. In contrast to peripheral membrane proteins such as S6 ribosomal protein, GPAT4 could not be extracted from membranes by high pH (Na2CO3 buffer [pH 11.5]) or high salt (1 M NaCl) but could be solubilized by detergents (Figure 3A), indicating that GPAT4 is an integral ER membrane protein.
Figure 3. GPAT4 Is an Integral Membrane Protein Containing a Membrane Hairpin Sequence Localizing to LDs via ER-LD Bridges.
(A) GPAT4 is a membrane protein not extracted from microsomes by buffers containing 1% SDS, 100 mM Na2CO3, or 1 M NaCl.
(B) Similar to cytosolic ribosomal protein S6, but in contrast to GRP78, the N and C termini of GPAT4 are in the cytosol and accessible to Proteinase K in the presence or absence of Triton X-100.
(C) Membrane topology model of GPAT4. Blue regions indicate predicted transmembrane domains and orange regions the catalytic domain.
(D) The membrane hairpin of GPAT4 is necessary and sufficient for ER targeting. GPAT4 constructs containing (i) the entire GPAT4 sequence (ii) only the hairpin loop (iii) having replaced the hairpin loop with a flexible linker (iv) the interhelix segment of the hairpin loop with a linker sequence consisting of loop sequence of AGPAT1 were localized by fluorescence microscopy relative to an ER (GFP-SEC61β; in cells without oleate) or LD marker (BODIPY; in cells with oleate). Scale bars, 10 μm (overview) and 1 μm (magnification).
(E) GPAT4 is mobile within the ER, as indicated by fast recovery of signal into a bleached region (indicated by a box). Representative images are shown. Scale bar, 10 μm. Quantitation shows the recovery of GPAT4-GFP signal from three independent experiments. Error bars indicate SDs.
(F) Fluorescence recovery in a bleached region of GFP-GPAT4 signal on a forming LD (after 2 hr of incubation with oleate-containing medium). Representative images are shown. Single experiments show recovery of GFP-GPAT4 signal with different rates. Scale bar, 10 μm. Histogram of fluorescence recovery levels from 30 FRAP experiments.
(G) Recovery of GPAT4 occurs through ER-LD connections. Representative images from a FRAP movie are shown. Arrowheads highlight bridges between the GPAT4 ER and LD surface signal. Scale bar, 2 μm.
(H) Thin-section EM analysis of high-pressure frozen cells shows membrane continuity between LDs and the ER (highlighted by boxes). Blue arrows indicate ER-LD connections.
See also Figures S4 and S5, and Movie S1.
We next determined if the N and C termini of GPAT4 are localized in the cytoplasm or the ER lumen. FLAG tags fused to either terminus were readily digested by Proteinase K under conditions in which cytosolic proteins, such as ribosomal protein S6, but not luminal proteins, such as GRP78, were digested (Figure 3B). These data indicate that the N and the C termini are localized to the cytosol and that GPAT4 has an even number of membrane-inserted helices.
With this result and predictions from its sequence analysis, GPAT4 topology likely has a region (amino acids 163–207) with two hydrophobic helices that are separated only by a few, relatively hydrophobic residues, and forms a hairpin sequence embedded in lipids (Figure 3C). The three amino acids between the hydrophobic segments could be exposed to the lumen of the ER or contained within the ER membrane. Sequence analysis of Drosophila AGPAT3 and DGAT2, which also localize to LDs, revealed that these proteins have similar hydrophobic domains (Figure S4A). In contrast, the Drosophila isoenzymes of TG synthesis that remain localized within the ER during LD formation (e.g., AGPAT1, AGPAT2, AGPAT4, and DGAT1) have hydrophobic helices that are separated by larger stretches of hydrophilic amino acids, making it more likely that they are membrane-spanning helices.
We tested whether the GPAT4 membrane hairpin is required for its ER and/or LD targeting in cells. Figure 3D shows that expression of the GPAT4 hairpin region alone is sufficient for protein ER targeting and relocalization to LDs during oleate loading (Figure 3D). In contrast, a GPAT4 hairpin in which the sequence between membrane helices was replaced with the sequence from ER-localized AGPAT1 was targeted to the ER but failed to relocalize to LDs. Therefore, the hydrophobic hairpin configuration is sufficient to mediate GPAT4 ER insertion and relocalization from there to LDs. GPAT4, in which the hairpin is replaced by a hydrophilic linker sequence, failed to insert into the ER but was recruited to LDs from the cytoplasm, indicating that the hairpin is required for ER targeting of GPAT4 but that additional sequences in the N and C terminus may contribute to LD binding of GPAT4 (Figure 3D).
GPAT4 Diffuses on Membrane Bridges from the ER to LDs during Their Formation
We next investigated how GPAT4 relocalizes to LDs. We hypothesized that the protein is mobile within the ER, enabling it to move to LD formation sites. Consistent with this model, the GFP-tagged GPAT4 was highly mobile in FRAP experiments in cells that contain no or very few LDs (t1/2 = 54.9 s; Figure 3E). Similarly, FRAP of GPAT4 on mature LDs was mobile on LDs (Figure S3B). The latter finding makes it unlikely that GPAT4 at LDs is stably anchored to a static LD structure.
To assess whether GPAT4 is mobile between the ER and LDs in cells containing mature LDs (e.g., after 24 hr of oleate loading), we repeatedly bleached an ER region (FLIP) and found that GPAT4-GFP signal on LDs did not decrease over time (Figures S4B and S4C). Similarly, GPAT4 signal did not equilibrate between droplets in FRAP experiments bleaching a whole LD (Figure S4D). These data indicate little or no exchange between GPAT4 on different LDs once they are mature (Figure S4D).
If the GPAT4 pool of mature LDs does not exchange with an ER pool, how does the ER-localized GPAT4 access LDs? To address this question, we performed FRAP experiments during an intermediate time point of LD formation (e.g., after 2 hr oleate treatment). Under these conditions, the majority of LDs (60%) showed variable rates of recovery of fluorescence when bleached (Figure 3F). Analysis of GPAT4 localization during its recovery revealed many striking connections of the signal between the LD surface and the surrounding ER (Figure 3G). These structures appeared as flexible tubules or bridges through which the GPAT4 signal reached the LD (Figure 3G; Movie S1). Consistent with ER bridges to LDs during their formation, we observed a variable number of connections between the LD-delimiting monolayer and the membrane tubules by thin-section EM of LDs in cells prepared by high-pressure freezing or chemical fixation (Figures 3H, S4E, and S5).
GPAT4 Localizes to Growing LDs
LDs grow dramatically after their initial formation (Krahmer et al., 2011). We hypothesized that the LD-localized TG synthesis enzymes, such as GPAT4, mediate growth by synthesizing TG on LDs.
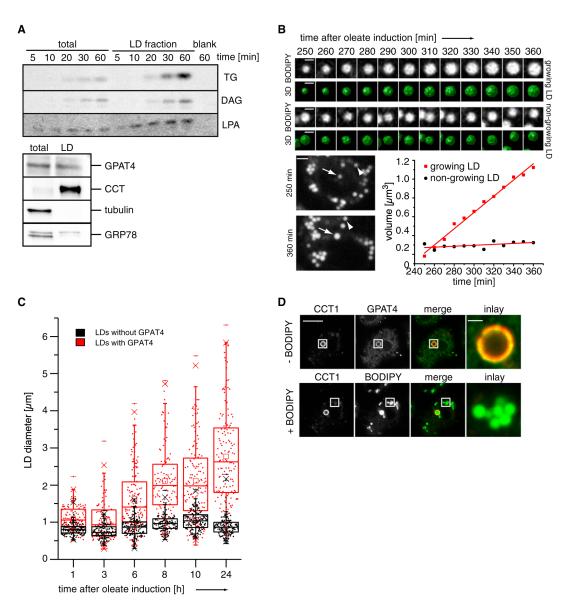
This model implies several strong predictions. First, LD fractions should contain all activities required for de novo TG synthesis. Incubating radiolabeled fatty acyl-CoA as a substrate with LD fractions led to the synthesis of LPA, DG, and TG, showing presence of GPAT, AGPAT, PAP, and DGAT activities. Since the activity of the LD fraction was greater than the total, whereas ER proteins were deriched, this activity is very likely due to enzymes directly associated with LDs.
Second, if LDs in our experimental system grow by addition of TG rather than by fusing existing LDs, live-cell imaging should reveal gradual growth of LDs. Consistent with this notion, 3D time-lapse imaging revealed that some, but not all LDs, grow gradually during oleate loading of cells in the absence of nearby LDs that could fuse to the growing LD (Figure 4B).
Figure 4. GPAT4 Localizes to Growing LDs.
(A) LDs are able to in vitro generate TG, DG, and LPA from radiolabeled oleoyl-CoA, resolved by TLC. Western blot analysis of the LD fractions (lower panel).
(B) A subset of LDs grows as shown by 3D time-lapse live-cell imaging from 0 to 8 hr of oleate loading. Representative optical midsections and reconstructions for a growing (top) and nongrowing (bottom) LD are shown from 240 to 360 min. Scale bars, 1 μm (BODIPY and 3D) and 2 μm (whole cell). The graph shows LD volume over time.
(C) Only LDs with GPAT4 expand over time. Scatterplots and box plots (median, lower and upper quartile, 1.5 interquartile range for whiskers) of diameters of LDs with (red) or without (black) GPAT4 are shown during their formation induced by oleate-containing medium.
(D) GPAT4 (green) localizes to expanding LDs marked by CCT1 (red) by immunofluorescence. After imaging the immunofluorescence signal, cells were stained with BODIPY to visualize all LDs (green channel). Scale bars, 10 μm (overview) and 1 μm (magnification).
Third, among LDs, only those containing GPAT4 should be able to grow. Consistent with live-cell imaging revealing two classes of LDs, we found that the diameters of LDs containing GPAT4 increased over time, whereas the diameters of LDs lacking GPAT4 remained relatively constant (Figure 4C). We never observed large LDs without GPAT4 signal.
Finally, only GPAT4-containing LDs that are growing should show evidence of surface expansion. Previously, we found that CCT1 localizes specifically to expanding LDs, where it apparently detects deficiency in PC as LD surface increases (Krahmer et al., 2011). Thus, CCT1 acts as a marker for expanding LDs. Strikingly, CCT1 targeted the same subset of growing LDs that contained endogenous GPAT4 (Figure 4D).
LD-Localized TG Synthesis Enzymes Are Required for LD Growth
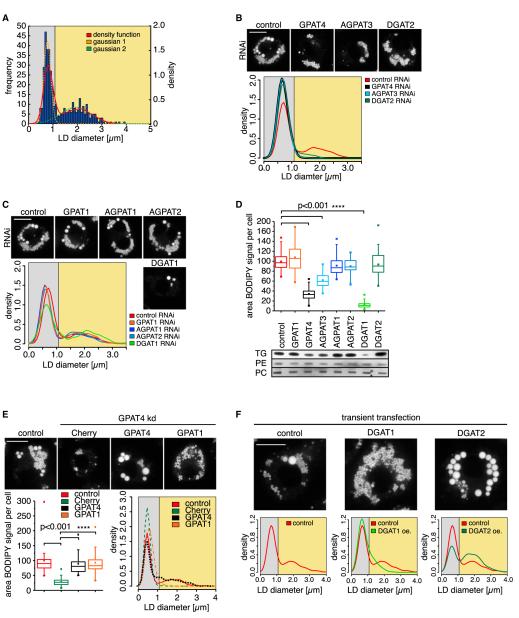
If GPAT4 and other LD-localized enzymes of the TG synthesis pathway mediate local TG synthesis, they should be specifically required to form large LDs. To test this prediction, we measured LD sizes in oleate-loaded cells depleted for each of the enzymes with RNAi. In Figure 5A, we show sizes of the largest LDs per cell, which reveals many small LDs and fewer larger ones in control cells (red line). Each of these populations can be fitted well to a Gaussian distribution. Assuming that all LDs are round, we estimate that Drosophila cells store roughly 80% of TG in large LDs and the remainder in smaller LDs after 12 hr of oleate treatment (F.W., R.V.F., and T.C.W., unpublished data). Depletion of the LD-localized TG synthesis enzymes GPAT4, AGPAT3, and DGAT2 resulted in major decreases of the large LD population. Consequently, the LDs in these cells were of the same size as the smaller LDs in wild-type cells (Figure 5B). In contrast, depletion of GPAT1, or AGPAT1/AGPAT2, which do not relocalize to LDs, had little effect on the number of large LDs. Similarly, depletion of ER-resident DGAT1 had no effect on large LDs but decreased the number of small LDs (Figure 5C). Depletion of GPAT, AGPAT, and DGAT enzymes had differential effects on TG amounts in cells as detected by quantitating BODIPY staining or TLC (Figure 5D). Notably, the effect of depleting LD-localized TG synthesis enzymes on LD sizes was not due to just decreased overall TG levels, as those phenotypes did not always correlate. Depletion of DGAT1, for example, had a strong effect on TG levels but did not abolish formation of large LDs.
Figure 5. LD-Localized TG Synthesis Enzymes Are Required for the Formation of Large LDs.
(A) Histogram of LD sizes from control RNAi-treated cells shows a bimodal distribution of LD diameters (red curve), modeled by two normal distributions (; orange and green dashed lines; a1, 42.25; b1, 0.7672; c1, 0.2; a2, 8.377; b2, 1.936; c2, 0.8925) for small (gray area) and large LDs (yellow area).
(B) Depletion of LD-localized TG synthesis enzymes (GPAT4, AGPAT3, DGAT2; black, light-blue, dark-green density curves) diminishes the abundance of large LDs present in the control (red line). Scale bar, 10 μm.
(C) Depletion of ER-localized TG synthesis enzymes (GPAT1, AGPAT1, AGPAT2, DGAT1; orange, blue, dark-blue, light-green density curves) has little effect on the size distribution of large LDs. Representative images are shown. Scale bar, 10 μm.
(D) Effect of TG synthesis enzyme depletion by RNAi on TG levels. A box plot quantitation of BODIPY signal area per cell (median, lower, and upper quartile, 1.5 interquartile range for whiskers) is shown on top. Significance was tested by ANOVA. Bottom panels show a TLC of TG, PC, and PE levels in cells depleted for the indicated enzymes.
(E) Although both GPAT4 and GPAT1 transfection rescue total TG levels in GPAT4-depleted cells, only GPAT4 transfection leads to formation of large LDs. Scale bar, 10 μm. Significance was tested by ANOVA.
(F) Overexpression of DGAT2 and DGAT1 leads to large or small LDs, respectively. Scale bar, 10 μm.
See also Figure S6.
To further test whether the phenotype of GPAT4 depletion on LD size is specifically due to its localization on LDs, rather than reduction of cellular GPAT activity, we restored GPAT activity in GPAT4-depleted cells by transfecting either with mitochondrial GPAT1 or LD-localized GPAT4. Since differences in transfection rates might mask changes in TG synthesis, we assayed TG synthesis at cell level by BODIPY staining. This analysis showed that transfection of either GPAT restored roughly wild-type TG levels in transfected cells (Figure 5E). However, either enzyme promoted the formation of different LDs: GPAT4 transfection led to formation of relatively few, large LDs, whereas GPAT1 promoted the formation of many small LDs (Figure 5E).
If LD and ER-localized TG synthesis enzymes contribute primarily to the formation of large and small LDs, respectively, we hypothesize that overexpression of either enzyme will push the distribution of LD sizes in different directions. Indeed, overexpression of LD-localized DGAT2 led to the formation of very large LDs, and overexpression of ER-localized DGAT1 resulted in small LDs (Figures 5F and S6A).
Our data suggest that two different LD populations are generated by different sets of enzymes. One LD population is uniformly smaller and derived exclusively from ER-localized TG synthesis, whereas the other population is larger due to addition of TGs locally on LDs.
TG Synthesis Enzymes Localize to the Same LDs
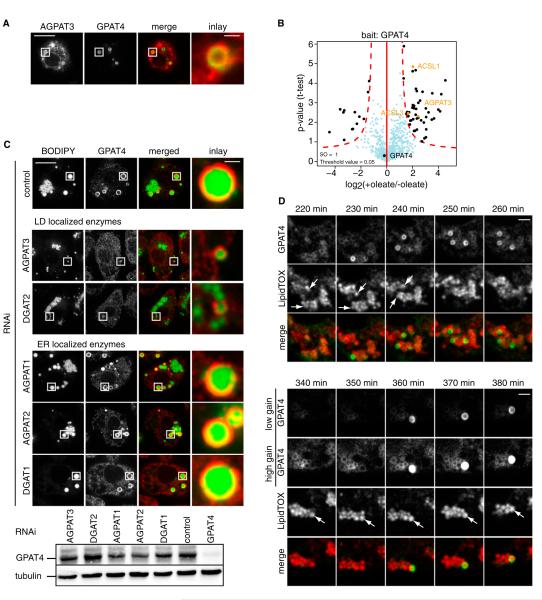
To mediate LD growth, TG enzymes localizing to LDs should target the same LD. Consistent with this notion, cotransfected GFP-GPAT4 and cherry-AGPAT3 localized to the same set of LDs by fluorescence microscopy (Figure 6A). To independently, biochemically test the colocalization of GPAT4 with other TG synthesis enzymes, we affinity purified endogenous GPAT4 from cells before and after incubation with oleate and identified copurifying proteins by quantitative, label-free mass spectrometry (Hubner et al., 2010). Intriguingly, these experiments showed that GPAT4 on LDs specifically interacts with ACSL1 and ACSL3, catalyzing the enzymatic step providing the substrate for the GPAT reaction, as well as AGPAT3, metabolizing the product of the GPAT4 reaction (Figure 6B and Table S3). Minimally, these data show that these enzymes are located on the same LDs, but it is also possible that they form a physical complex specifically on LDs.
Figure 6. A Subset of TG Synthesis Enzymes Localizes to the Same LDs.
(A) GFP-GPAT4 and cherry-AGPAT3 colocalize to the same LD by fluorescence microscopy in cotransfected cells. Scale bars, 10 μm (overview) and 1 μm (magnification).
(B) Volcano plot of proteins associating with GPAT4 in the presence or absence of oleate, analyzed by label-free proteomics. The logarithmic ratios of protein intensities are plotted against negative logarithmic p values of the t test performed from three independent experiments. The red dotted line (significance, 0.05) separates specifically interacting proteins marked in black from background (blue dots). All specific interactors are reported in Table S3.
(C) Depletion of LD-localized AGPAT4 or DGAT2, but not ER-localized AGPAT1, AGPAT2, or DGAT1, abolishes GPAT4 localization to LDs. Scale bars, 10 μm (overview) and 1 μm (magnification). Western blot analysis shows expression of GPAT4.
(D) LDs can acquire GPAT4 after their initial formation, as observed by live-cell 3D time-lapse imaging from 0 to 8 hr of oleate. Scale bars, 1 μm. See also Table S3.
Based on the colocalization of some TG enzymes on an LD subset and their requirement to expand LDs, we predict that absence of any of the enzymes required to form expandable LDs leads to lack of this class of LDs. Consequently, targeting of proteins, such as GPAT4, which are specific for expandable LDs, should be abolished when these LDs are absent. To test this model, we depleted either LD-localized AGPAT3 or DGAT2, or as controls the ER-localized AGPAT1/AGPAT2 or DGAT1, and determined the localization of endogenous GPAT4. Depletion of LD-targeted enzymes abolished GPAT4-containing LDs (but not GPAT4 expression; Figure 6C), whereas the depletion of enzymes that are ER localized did not affect LD localization of GPAT4. If anything, depletion of the ER-localized TG synthesis enzyme DGAT1 increased the proportion of LDs containing GPAT4, likely due to the decreased number of small LDs (Figure 6C).
LDs Can Acquire TG Synthesis Enzymes after Their Initial Formation
Our data show that two LD populations exist: those that are expanding and containing TG synthesis enzymes, or those that are smaller and lack TG synthesis enzymes. To determine whether these LDs are part of different LD-independent lineages or alternatively can be interconverted, we followed GPAT4-GFP localization by time-lapse confocal 3D imaging in respect to LDs. Focusing on LDs that were formed initially without containing any GPAT4-GFP signal, we found many examples of single LDs rapidly acquiring GPAT4-GFP signal (Figures 6D and S6B). In each case, GPAT4 accumulated within 10 min on LDs. Thus, previously formed LDs can acquire the TG synthesis machinery and the capacity to expand.
GPAT4 Localization and Function in LD Growth Are Evolutionarily Conserved
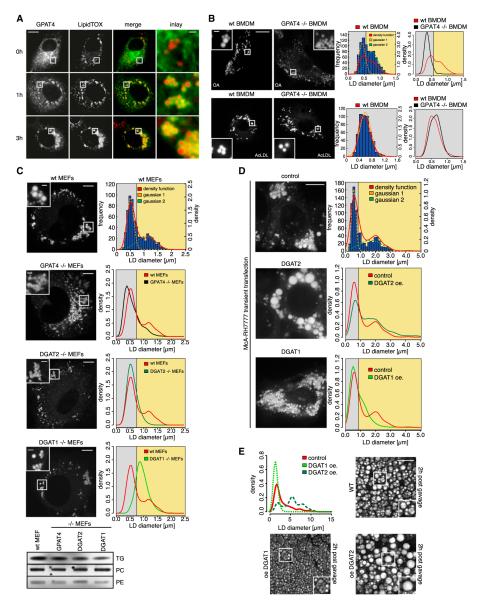
De novo TG synthesis enzymes are highly conserved across species. To test whether our findings in Drosophila cells translate to mammalian cells, we assessed the subcellular localization, relocalization, and function of human GPAT4. GPAT4 relocalized from the ER to LDs in mammalian COS7 cells, similar to what occurred with Drosophila GPAT4 (Figure 7A).
Figure 7. Murine GPAT4 Localizes to LDs and Is Required for the Formation of Large LDs.
(A) Murine GPAT4-GFP (green) expressed in COS7 cells relocalizes from the ER to LDs (red) during incubation in oleate-containing medium. Scale bars, 10 μm (overview) and 1 μm (magnification).
(B) GPAT4 is required for formation of large LDs specifically under conditions of de novo TG synthesis. Fluorescence images of BODIPY-stained LDs in control of Gpat4−/− BMDMs incubated with oleate or AcLDL-containing medium for 12 hr. Scale bars 10 μm (overview) and 1 μm (magnification). The distribution of diameters for 500 measured LDs from 25 different cells for control (red) or Gpat4−/− (black) is shown. Histogram of LD sizes as in Figure 5A (parameters of Gaussians: a1, 109; b1, 0.4387; c1, 0.1438; a2, 79.14; b2, 0.7446; c2, 0.2606).
(C) GPAT4 and DGAT2 are required for large LDs in MEFs. Fluorescence images of BODIPY-stained LDs in control, Gpat4−/−, Dgat2−/−, and Dgat1−/− MEFs incubated with oleate-containing medium for 12 hr. Scale bars, 10 μm (overview) and 2 μm (magnification). The distribution of diameters for 600 measured LDs from 30 different cells for control (red), Gpat4−/− (black), Dgat2−/− (dark green), or Dgat1−/− (light green) is shown (parameters of Gaussians: a1, 85.24; b1, 0.5043; c1, 0.1654; a2, 21.95; b2, 1.067; c2, 0.4563). A TLC showing TG levels in MEFs of the different genotypes is shown.
(D) Overexpression of DGAT2 or DGAT1 in MEFs leads to large or small LDs, respectively. Scale bar, 10 μm. (parameters of Gaussians: a1, 161.2; b1, 0.5535; c1, 0.2082; a2, 28.87; b2, 1.654; c2, 1.073).
(E) Overexpression of DGAT2 or DGAT1 in murine intestine leads to accumulation of large or small LDs visualized by CARS microscopy, respectively, after an acute dietary lipid challenge (2 hr postgavage with 200 μl oil). Scale bar, 10 μm.
See also Movie S2.
To determine if GPAT4 function in LD growth was conserved in mammals, we analyzed the LD phenotype of cells from Gpat4−/− mice. As with fly cells, bone marrow-derived macrophages (BMDMs) from wild-type cells cultured with oleate contain LDs of considerable size variation. Gpat4−/− macrophages had much smaller average diameters of LDs than those of control macrophages (Figure 7B). The effect of GPAT4 on LD size was specific to conditions of TG synthesis, since LDs were similar in size when macrophages were cultured with acetylated low-density lipoproteins (AcLDL), which contain mostly cholesterol esters (Figure 7B). Similar results were obtained for mouse embryonic fibroblasts (MEFs), which showed consistently smaller LDs in Gpat4−/− MEFs compared with controls (Figure 7C). TG levels and, consequently, overall BODIPY staining were only mildly reduced in Gpat4−/− MEFs (Figure 7C; data not shown). This phenotype, as well as the sparse large LDs in Gpat4−/− MEFs, could be due to the highly homologous GPAT3 providing functional redundancy to GPAT4.
To test whether our findings were common to deletion of LD-localized TG synthesis enzymes in mammalian cells, we also analyzed Dgat2−/− MEFs and found an even more striking reduction in the number of large LDs (Figure 7C). This effect was not found for MEFs lacking DGAT1, which instead yielded consistently larger LDs.
To further test whether the ability of different TG synthesis enzymes to drive formation of small or large LDs is conserved in mammals, we overexpressed DGAT1 or DGAT2 in McA-RH7777 rat hepatoma cells and cultured them with oleate. Consistent with our findings in Drosophila cells, wild-type hepatoma cells had two populations of LDs: one containing large LDs, and the other containing small ones. With DGAT1 overexpression, cells formed predominantly small LDs, whereas DGAT2 overexpression dramatically increased the proportion of large LDs (Figure 7D). To test the role of LD-localized TG synthesis enzymes in physiological context, we analyzed LDs by coherent anti-Stokes Raman scattering (CARS) microscopy in the small intestine of transgenic mice overexpressing either DGAT1 or DGAT2 after an acute dietary fat challenge. Figure 7E shows both large and small LDs in enterocytes of wild-type mice 2 hr after an oil gavage. The size distribution of LDs in enterocytes was greatly shifted toward large or small LDs by overexpressing either DGAT2 or DGAT1, respectively (Figure 7E; Movie S2).
DISCUSSION
We investigated the role of TG synthesis enzymes in LD formation and growth. Our findings show that cells form two classes of LDs during oleate loading. Large LDs grow by addition of TG, synthesized locally by moving de novo TG synthesis enzymes along membrane bridges to the LD surface. In contrast, LDs of a second class that do not acquire these TG enzymes remain small after their initial formation.
We believe that relocalization of GPAT4 to expanding LDs occurs through bridges between ER and LD, which were observed by EM and likely correspond to the tubular GPAT4 signal found during FRAP experiments. Most likely, these bridges are continuities of the outer ER membrane leaflet with the surface of the LD monolayer. Of note, such connections between the ER and LDs have been seen occasionally in other organisms by EM and, thus, are likely a general feature of LDs (Ohsaki et al., 2008; Wanner et al., 1981). Some LDs acquire GPAT4 (and likely other TG synthesis enzymes) after their initial formation as small LDs. The rapid acquisition of GPAT4 signal of these LDs that were size stable for a long time suggests that relocalization is triggered by establishing a path for the enzymes, e.g., a membrane connection, at this time point.
Once GPAT4 is localized to LDs, it does not leave this compartment under our experimental paradigm. The mechanism mediating unidirectionality of LD targeting is unknown. One possibility is a unidirectional “gating” mechanism, perhaps mediated by protein machinery. Another option is that GPAT4 is under a structural strain as long as it is confined to a bilayer membrane of defined thickness and that this strain is released once the protein localizes to the LD monolayer and underlying hydrophobic core. The resulting conformational change could serve to trap the protein at the LD, preventing its diffusion back into the bilayer membrane. Although membrane topology predictions of GPAT4 based on its primary sequence suggest that the enzyme harbors several transmembrane domains (Figure S4A), our data indicate that it rather forms a hairpin loop within the membrane or monolayer, with the N and C termini being cytosolic, that is necessary and sufficient for ER-to-LD relocalization. Both segments of the hairpin loop are unusually long for an ER-spanning a helix (21 and 22 amino acids), supporting the hypothesis that the protein is embedded under strain in a bilayer membrane. This topology is highly similar to that of oleosins, the major LD proteins of plants (Abell et al., 1997). Topologies similar to GPAT4 are plausible for other enzymes of the de novo synthesis pathway, including, for example, DGAT2 (Stone et al., 2006; Figure S4A). Moreover, caveolin localizes to LDs under some conditions and also contains an intramembrane hairpin, which likely mediates its LD targeting, indicating that hydrophobic hairpins might be common LD-targeting motifs (Thiele and Spandl, 2008). Other proteins, such as ATGL or METTL7B, initially localize to the ER and then concentrate on LDs (Soni et al., 2009; Turró et al., 2006; Zehmer et al., 2009), suggesting that they may target LDs in the same way as GPAT4. Evidence suggests that ER-to-LD relocalization by membrane bridges is not limited to Drosophila. For example, the yeast homologs of GPAT4, Gat1, and Gat2, were found previously in microsomal ER and LD fractions (Athenstaedt and Daum, 1997). Furthermore, Dga1, the yeast homolog of DGAT2, can migrate to LDs via the ER (Jacquier et al., 2011). Additionally, the previous reports of mammalian DGAT2 localizing to LDs upon oleate loading of cells (Kuerschner et al., 2008; Stone et al., 2009) likely represent examples of ER-to-LD relocalization. Thus, this pathway is likely evolutionarily conserved and fundamental for targeting ER proteins to LDs.
Segregation of TG synthesis to LDs and away from the ER may have several important physiological functions. Colocalizing TG synthesis pathway enzymes on LDs might facilitate the efficient handover of substrates from one enzyme to the next in a localized multienzyme complex. Consistent with this possibility, biochemical fractionations (e.g., of intestinal microsomes possibly containing LDs) demonstrated a complex containing several of the activities needed for TG synthesis (Lehner and Kuksis, 1995; Rao and Johnston, 1966), and recent data in C. elegans provide evidence for an interaction of DGAT2 and an ACSL (Xu et al., 2012). In further support of this notion, we find ACSL1, ACSL3, GPAT4, and AGPAT3 physically associated when LDs are present and AGPAT3 and GPAT4 colocalizing together to a subset of LDs. Moreover, depletion of a single LD-localized enzyme impaired localization of other enzymes and TG synthesis. Segregation of TG synthesis to LDs may also serve to protect the ER from stress due to the accumulation of lipid synthesis intermediates, such as diacylglycerol or phosphatidic acid. In addition, segregation of GPAT and AGPAT activities to LDs could separate TG synthesis from ER-based phospholipid synthesis, catalyzed for example by diacylglycerol-cholinephosphotransferase (CPT), which generates PC from DG (and CDP-choline) and remains in the ER under oleate-loading conditions (Krahmer et al., 2011).
Besides large LDs, a class of smaller LDs that does not contain GPAT4 and the other TG synthesis enzymes form during incubation with oleate-containing medium. Possibly, these LDs segregate from the ER once they reach a particular size, which prevents their further growth and explains their relatively uniform size. A subset of these LDs later acquires GPAT4 and other TG synthesis enzymes to subsequently expand.
The formation of these smaller LDs may reflect an overflow mechanism that operates when the capacity of LD-localized TG synthesis is overwhelmed, and this pathway’s function may be to protect the ER from accumulating lipotoxic intermediates. DGAT1 depletion in Drosophila cells leads to reduced TG levels, and the absence, specifically, of the smaller LDs, with only one or very few large LDs remaining. This shows that DGAT1 is required for the formation of the small class of LDs. Since some LDs initially formed by DGAT1 expand at a later time point, DGAT1 depletion also affects overall LD number.
Mammalian genomes encode two enzymes with high similarity to Drosophila GPAT4. In most tissues, GPAT3 and GPAT4 constitute about 90% of total GPAT activity (Coleman and Lee, 2004). So far, only GPAT4 function has been analyzed in knockout mice (Beigneux et al., 2006; Chen et al., 2008; Nagle et al., 2008; Vergnes et al., 2006). Absence of GPAT4 leads to subdermal lipodystrophy, as well as a strong reduction of TG content in adipose tissue and in liver, where GPAT4 is highly expressed. Interestingly, the absence of GPAT4 also leads to a resistance to diet-induced obesity, which suggests that it could be a drug target (Cao et al., 2012). It will be interesting to test whether this effect is due to the inability to expand LDs (e.g., in liver), thus limiting the storage of fat and the accumulation of lipotoxic intermediates produced by the TG synthesis pathway downstream of GPAT4 activity.
EXPERIMENTAL PROCEDURES
Cellular Fractionation
Cells were harvested, washed with ice-cold PBS, resuspended in 2 ml of buffer (250 mM sucrose, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, Roche complete protease inhibitor tablet), lysed by a tissue homogenizer, and lysates were cleared by 5 min centrifugation at 1,000 × g and 15 min at 12,000 × g. The supernatant was fractionated into floating LDs, soluble, and membrane fractions by using a two-layer gradient (bottom, 5 ml: 250 mM sucrose, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, Roche complete protease inhibitor tablet; upper layer, 5 ml: 50 mM sucrose, 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, Roche complete protease inhibitor tablet) centrifuged at 100,000 × g for 12 hr. Protein concentration was determined using BCA (Thermo Scientific).
TLC and Lipid Measurements
Lipids were extracted (Folch et al., 1957), separated on silica TLC plates (Merck) with chloroform/methanol/water (65:25:4) for phospholipids or n-heptane/isopropyl ether/acetic acid (60:40:4) for neutral lipids, and detected by cerium molybdate staining. Bands were identified by comparing with standards. GPAT4 activity assays were performed as described by Chen et al. (2008).
LD Size Measurements
Cells were treated with 1 mM oleate, stained with BODIPY, and subsequently imaged. The diameter of the 20 largest LDs within one cell was measured using ImageJ (http://rsbweb.nih.gov/ij/). A minimum of 25 cells was analyzed for each individual condition. Density plots were computed using R (http://www.r-project.org/).
Light Microscopy
Cells were prepared and imaged, including staining with BODIPY, as described (Krahmer et al., 2011). For immunostaining, cells were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at room temperature. Fixed cells were permeabilized using a buffer containing 0.05% NP40, 0.05% saponin, and 0.1% BSA in PBS for 3 min. Fixed cell-coated coverslips were rinsed in PBS, blocked in blocking buffer (0.05% saponin, and 5% normal goat serum in PBS) for 1 hr, incubated with primary antibodies diluted in blocking buffer for 1 hr, washed three times with washing buffer (0.05% saponin, and 0.2% BSA in PBS) for 5 min, incubated with fluorescently labeled secondary antibody (Sigma-Aldrich) for 1 hr, washed three times and rinsed in PBS, followed by BODIPY staining, as indicated. McArdle cells were imaged live on a Nikon Eclipse TE2000 inverted microscope with a Yokogawa CSU-X1 spinning disk and 1003 ApoTIRF 1.4 NA objective.
STED imaging was performed with a TCS STED (Leica Microsystems) equipped with a pulsed 640 nm diode laser (PDL 800-B; PicoQuant) for excitation and an 80 MHz Ti:Sapphire laser (Mai Tai; Spectra Physics) tuned to 770 nm for depletion (~180 mW). Fluorescence from the sample was band-pass filtered (FF01-685/40; Semrock) and detected by an avalanche photodiode through a 0.5 Airy unit pinhole. Images of 1,024 3 1,024 pixels (with a 30 nm pixel size) were acquired at a scan speed of 1,000 lines/s with 16 line averages.
EM
Cells were fixed in 2.5% glutaraldehyde/2% PFA in 0.1 M Na cacodylate buffer (pH 7.4) with 2% sucrose for 1 hr. Following rinsing in Na cacodylate buffer, cells were pelleted in 2% agar and postfixed in 1% osmium tetroxide in 0.1 M Na cacodylate for 1 hr and en bloc stained in 2% uranyl acetate in maleate buffer (pH 5.2) for 1 hr.
For pre-embedding immunolabeling and silver enhancement, cells were fixed on coverslips in 2% PFA rinsed in PBS and incubated with rabbit anti-GPAT4 antibody 1:50, rinsed, and incubated with anti-rabbit NANOGOLD (Nanoprobes) 1:100 for 30 min at room temperature. After rinsing, coverslips were fixed in 1% glutaraldehyde, rinsed, and silver enhanced using HQ Silver (Nanoprobes) for 5 min. All samples were rinsed in PBS, dehydrated through an ethanol series, embedded in Epon812 (Electron Microscopy Sciences), and hardened in capsules at 60°C overnight.
For high-pressure freezing, unfixed samples were frozen using a Leica EM HPM100 at 2,000 psi. Samples were freeze substituted in a Leica AFS at −90°C using 0.1% uranyl acetate/acetone for 50 hr, rinsed in acetone, and infiltrated for 10 hr at −45°C with Lowicryl HM20 resin (EMS). Samples were placed in gelatin capsules and UV hardened at −45°C. Hardened blocks were sectioned using a Leica UltraCut UC7, and 60 nm ultrathin sections were collected on formvar/carbon-coated nickel grids and counterstained using uranyl acetate and lead citrate. Grids were imaged in a Tencai Biotwin (FEI) transmission electron microscope at 80 kV, and digital images were taken using Morada CCD and iTEM (Olympus) software.
Animal Experiments
All procedures were approved by the Purdue Animal Care and Use Committee. For imaging of intestinal tissues, all mice had food removed for 4 hr, were gavaged with 200 ml olive oil, and were euthanized at 2 hr postoil bolus administration. Fresh intestinal tissues (5 mm, S2, representing upper jejunum) were cut longitudinally and laid flat for luminal imaging. All tissues were imaged within 3 hr after euthanasia. CARS imaging was performed as previously described by Zhu et al. (2009).
Pull-Down Analysis
S2 cells were harvested, washed with ice-cold PBS, and resuspended in lysisbuffer (150 mM NaCl, 50 mM Tris [pH 7.4], 1 mM EDTA, 0.5% Triton X-100, and Roche complete protease inhibitor). Pull-downs were performed using antibodies against endogenous GPAT4 and Roche protein A-agarose beads according to the manual. In the last step, proteins were denatured using 8 M urea. Eluates were analyzed as described by Hubner et al. (2010). In short, peptides were separated using an Easy nLC system with a segmented gradient of 5%–75% solvent B over 111 min with a constant flow of 250 nl min−1, coupled to an LTQ-Orbitrap Velos mass spectrometer via a nanoscale LC interface (spray voltage, 2.2 kV; temperature, 200°C; all Thermo Fisher Scientific). Full-scan spectra (m/z, 300–1,750) were acquired in positive ion mode with a resolution of 30,000 at a m/z of 400 after accumulation of 1,000,000 ions. Up to ten of the most intense ions were analyzed by HCD. Precursor ion charge-state screening was enabled, and all unassigned charge states, as well as singly charged peptides, were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 60 s and a relative mass window of 10 ppm. Orbitrap measurements were performed enabling the lock mass option for real-time internal calibration using the 445.120025 ion for survey scans to improve mass accuracy. Data were acquired using the Xcalibur software (Thermo Fisher Scientific) and MaxQuant, version 1.2.0.19 (http://maxquant.org/downloads.htm; Cox and Mann, 2008). The data were searched against the Drosophila melanogaster database concatenated with reversed copies of all sequences. Carbamidomethylated cysteines were set as fixed, whereas oxidation of methionine and N-terminal acetylation and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Maximum allowed mass deviation for MS/MS peaks and missed cleavages were 20 ppm and three, respectively. Maximum false discovery rates (FDRs) were 0.01 both on peptide and protein levels. Minimum required peptide length was six residues. Proteins with at least two peptides were considered identified. The mass spectrometry data associated with this manuscript may be downloaded from http://ProteomeCommons.org Tranche using the following hash: HdYbRlfWvLAG8/PK7f3sgxBfvUxm+76wmFZfp9PrGjjDhFBCUYQY6J 8VhOeu5poU1uMeT/7EAT1BXMT2ABrgwskv4PsAAAAAAAAG8g==.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nora Kory for critical comments on the manuscript and Gary Howard for editorial assistance. We also thank Dr. Karen Reue for providing Gpat4−/− MEFs, Dr. Scot Stone and Dr. Ping Zhou for generating DGAT-overexpressing hepatoma cells, and Dr. A. Rachid Thiam for help with statistical analysis. F.W. is a fellow of the Boehringer Ingelheim Fonds. This work was supported by grants R01 GM097194 (to T.C.W.), R01 GM099844 (to R.V.F.), and R01 DK56598 (to R.A.C.), by the Gladstone Institutes (to R.V.F.), National Institute of General Medical Sciences F32GM096859 (to T.J.G.), AHA NCRP Scientist Development Grant 0835203 N (to K.K.B.), and AHA Predoctoral Fellowship 11PRE5140017 (to A.U.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures, three tables, and two movies, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2013.01.013.
J.B. declares significant financial interest in Vutara, Inc.
REFERENCES
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguado B, Campbell RD. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J. Biol. Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and endoplasmic reticulum of Saccharomyces cerevisiae. J. Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Beigneux AP, Vergnes L, Qiao X, Quatela S, Davis R, Watkins SM, Coleman RA, Walzem RL, Philips M, Reue K, Young SG. Agpat6—a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J. Lipid Res. 2006;47:734–744. doi: 10.1194/jlr.M500556-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Barber T, Coxey RA, Takeda T, Rondinone CM, Theodorakis JL, Greenberg AS, Londos C. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 1995;36:1211–1226. [PubMed] [Google Scholar]
- Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 2012;287:2273–2279. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Cao G, Konrad RJ, Li SD, Hammond C. Glycerolipid acyltransferases in triglyceride metabolism and energy homeostasis-potential as drug targets. Endocr. Metab. Immune Disord. Drug Targets. 2012;12:197–206. doi: 10.2174/187153012800493459. [DOI] [PubMed] [Google Scholar]
- Chen YQ, Kuo MS, Li S, Bui HH, Peake DA, Sanders PE, Thibodeaux SJ, Chu S, Qian YW, Zhao Y, et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J. Biol. Chem. 2008;283:10048–10057. doi: 10.1074/jbc.M708151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004;43:134–176. doi: 10.1016/s0163-7827(03)00051-1. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 2011;3:a004838. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Kinoshita T, Homma KJ, Onoduka J, Mori M, Yamaguchi S, Makita M, Higashi Y, Yamashita A, Takano T. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- Gong J, Sun Z, Wu L, Xu W, Schieber N, Xu D, Shui G, Yang H, Parton RG, Li P. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 2011;195:953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Jambunathan S, Yin J, Khan W, Tamori Y, Puri V. FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One. 2011;6:e28614. doi: 10.1371/journal.pone.0028614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Hilger M, Kory N, Wilfling F, Stoehr G, Mann M, Farese RV, Walther TC. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics. 2013 doi: 10.1074/mcp.M112.020230. Published online January 14, 2013. http://dx.doi.org/10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed]
- Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Lehner R, Kuksis A. Triacylglycerol synthesis by purified triacylglycerol synthetase of rat intestinal mucosa. Role of acyl-CoA acyltransferase. J. Biol. Chem. 1995;270:13630–13636. doi: 10.1074/jbc.270.23.13630. [DOI] [PubMed] [Google Scholar]
- Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR. Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 1999;10:51–58. doi: 10.1006/scdb.1998.0275. [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim. Biophys. Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One. 2010;5:e15030. doi: 10.1371/journal.pone.0015030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle CA, Vergnes L, Dejong H, Wang S, Lewin TM, Reue K, Coleman RA. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 2008;49:823–831. doi: 10.1194/jlr.M700592-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG. Evidence for the existence of isoenzymes of glycerol phosphate acyltransferase. Biochem. J. 1979;177:283–288. doi: 10.1042/bj1770283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J. Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Poppelreuther M, Rudolph B, Du C, Großmann R, Becker M, Thiele C, Ehehalt R, Füllekrug J. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 2012;53:888–900. doi: 10.1194/jlr.M024562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao GA, Johnston JM. Purification and properties of triglyceride synthetase from the intestinal mucosa. Biochim. Biophys. Acta. 1966;125:465–473. doi: 10.1016/0005-2760(66)90035-x. [DOI] [PubMed] [Google Scholar]
- Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J. Cell Sci. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- Saggerson ED, Carpenter CA, Cheng CH, Sooranna SR. Subcellular distribution and some properties of N-ethylmaleimide-sensitive and-insensitive forms of glycerol phosphate acyltransferase in rat adipocytes. Biochem. J. 1980;190:183–189. doi: 10.1042/bj1900183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Farese RV., Jr. Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. J. Biol. Chem. 2006;281:40273–40282. doi: 10.1074/jbc.M607986200. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Levin MC, Zhou P, Han J, Walther TC, Farese RV., Jr. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 2009;284:5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele C, Spandl J. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Turró S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernández MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- Vergnes L, Beigneux AP, Davis R, Watkins SM, Young SG, Reue K. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. J. Lipid Res. 2006;47:745–754. doi: 10.1194/jlr.M500553-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr. The life of lipid droplets. Biochim. Biophys. Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner G, Formanek H, Theimer RR. The ontogeny of lipid bodies (spherosomes) in plant cells. Planta. 1981;151:109–123. doi: 10.1007/BF00387812. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, Jr., Mak HY. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehmer JK, Bartz R, Liu P, Anderson RG. Identification of a novel N-terminal hydrophobic sequence that targets proteins to lipid droplets. J. Cell Sci. 2008;121:1852–1860. doi: 10.1242/jcs.012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehmer JK, Bartz R, Bisel B, Liu P, Seemann J, Anderson RG. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J. Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lee B, Buhman KK, Cheng JX. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J. Lipid Res. 2009;50:1080–1089. doi: 10.1194/jlr.M800555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.