Summary
Contamination from subcellular organelles and myelin has hindered attempts to characterize the lipidome of brain mitochondria. A high degree of mitochondrial purity is required for accurate measurements of the content and molecular species composition of mitochondrial lipids. We devised a discontinuous Ficoll and sucrose gradient procedure for the isolation and purification of brain mitochondria free from any detectable contamination. Shotgun lipidomics was used to analyze the lipid composition of the brain mitochondria. These procedures can be used to determine whether intrinsic lipid abnormalities underlie mitochondrial dysfunction associated with neurological and neurodegenerative diseases.
Keywords: Non-synaptic, synaptic, mitochondria, brain, lipidome
1. Introduction
The brain contains two major populations of mitochondria that can be isolated by discontinuous gradients. These populations include the non-synaptic (NS) mitochondria, which are largely derived from neuronal and glial cell bodies, and the synaptic (Syn) mitochondria, which originate from the synaptic nerve terminal of neurons (1, 2). These mitochondria populations differ in calcium homeostasis, enzymes involved in the TCA cycle, electron transport chain activities, and glutamate metabolism (2-5). Differences between NS and Syn mitochondria may underlie the complexity of neural metabolism (1). A better understanding of brain bioenergetics can be obtained with accurate information on the mitochondrial lipidome.
In contrast to extensive research on mitochondria DNA and proteins in disease pathology, little attention has focused on the role of mitochondrial membrane lipids (6). Mitochondrial lipids can influence electron transport chain activities and create a membrane environment conducive for an efficient proton gradient (7). In addition, mitochondrial lipids regulate membrane fluidity and can affect the morphological structure of mitochondria (8, 9). Also, lipid composition can influence numerous mitochondrial enzyme activities to include creatine kinase, adenine nucleotide transporter, voltage dependent ion channel, and carnitine acyltransferase (7, 10, 11). Thus, the maintenance of the mitochondrial lipidome is critical for mitochondrial functionality.
Shotgun lipidomics uses a high-throughput platform which allows for the rapid quantitation of lipid molecular species using tandem mass spectroscopy (12). This lipidomic approach has the capabilities of detecting glycerophospholipid subclasses which contain alkyl enyl, vinyl ether, or ester linkages (13-15). In addition, shotgun lipidomics has demonstrated increased detection sensitivity to quantitating cardiolipin, a mitochondrial specific phospholipid (16, 17). Cardiolipin is a complex phospholipid, containing 4 acyl chains, three glycerol moieties, and two phosphates groups (8). We also used shotgun lipidomics and tandem MS to measure all detectable molecular species of cardiolipin in the C57BL/6J mouse brain (1).
Difficulty in removing subcellular contamination from brain mitochondrial populations has hindered the accurate analysis of mitochondrial lipid composition. This is especially the case for lipid rich myelin membranes, which can remain bound to brain mitochondria thus distorting analysis of lipid composition and content. Mitochondria can be isolated by Percoll, Nycodenz, metrizamide, Ficoll, and sucrose discontinuous gradients, however, the purity of these mitochondrial preparations differ depending on the intended use of these mitochondrial fractions (18-24). We used discontinuous Ficoll and sucrose gradients to isolate NS and Syn brain mitochondria free from detectable contamination. This procedure will be useful for analyzing differences in brain mitochondria in a variety of neurodegenerative and neurological disorders that involve alterations in energy metabolism.
2. Materials
2.1. Equipment and Supplies
4° Celsius temperature controlled room
Potter Elvehjem homogenizer with a Teflon coated pestle (10 mL and 25 mL)
Hand-held drill (max speed 500 rpm)
Table top centrifuge
1.5mL Eppendorf tubes
15 mL and 50 mL conical tubes
Optima L-90K Ultracentrifuge
Sorvall SW 28 rotor
Ultra-Clear™ ultracentrifuge tubes (Part 344058 Beckman Coulter)
A triple-quadrupole mass spectrometer (Thermo Scientific TSQ Quantum Ultra, Plus, San Jose, CA, USA), equipped with an electrospray ion source and Xcalibur system software.
Culture borosilicate glass tube (16 × 100 mm)
2.2 Reagents
Mitochondrial isolation buffer: 0.32 M sucrose, 10 mM Tris-HCl, and 1 mM EDTA-K (pH 7.4)
Nanopure deionized water
TE buffer (1 mM EDTA-K and 10 mM Tris-HCl, pH 7.4)
20% Ficoll stock solution made with MIB, 12% and 7.5% Ficoll working solutions.
1.6 M sucrose stock containing 1 mM EDTA-K and 10 mM Tris-HCl (pH 7.4); 0.8 M, 1.0M, 1.3M and 1.6M sucrose gradient working solutions
Bovine serum albumin (BSA)
Mitochondrial isolation buffer containing 0.5 mg/mL BSA
6 mM Tris-HCl (pH 8.1)
Lipid internal standards: 16:1-16:1 phosphatidylethanolamine, 14:1-14:1 phosphatidylcholine, T14:0 cardiolipin, 15:0-15:0 phosphatidylglycerol, 14:0-14:0 phosphatidylserine, 17:0 lysophosphatidylcholine, N12:0 sphingomyelin, and N17:0 ceramide
Chloroform
Methanol
Lithium chloride
3. Methods
3.1. Brain Mitochondrial Isolation
Mice of the C57BL/6 strain (4 months of age) were sacrificed by cervical dislocation and the cerebral cortex was dissected. Mitochondria were isolated in a cold room (4 °C) and all reagents were kept on ice. The isolation procedure employed a combination of gradients and strategies as previously described (2-4, 25-28) (Fig. 1). The cerebral cortexes (a pool of 6/sample) were diced on an ice cold metal plate and then placed in 12 mL of mitochondria isolation buffer (MIB; 0.32 M sucrose, 10 mM Tris-HCl, and 1 mM EDTA-K (pH 7.4)). The pooled cerebral cortexes were homogenized using a Potter Elvehjem homogenizer with a Teflon coated pestle attached to a hand-held drill. Samples were homogenized using 15 up and down strokes at 500 rpm. The homogenate was centrifuged at 1,000 × g for 5 min. The supernatant was collected and the pellet was washed twice by centrifugation, collecting the supernatants each time. The supernatants were pooled and centrifuged at 1,000 × g for 5 min. The collected supernatant was then spun at 14,000 × g for 15 min. The supernatant was discarded and the pellet, which contained primarily NS mitochondria, synaptosomes and myelin, was resuspended in 12 mL MIB and was layered on a 7.5%/12% discontinuous Ficoll gradient. Each Ficoll gradient layer contained 12 mL for a total volume of 36 mL. The Ficoll gradients were made from a 20% Ficoll stock with MIB. The gradient was centrifuged at 73,000 × g for 36 min (4 °C) in a Sorvall SW 28 rotor with slow acceleration and deceleration (Optima L-90K Ultracentrifuge). The centrifugation time used permitted sufficient acceleration and deceleration to achieve maximum g force (28), and to prevent synaptosomal contamination of the mitochondrial fraction below the 12% Ficoll layer. Crude myelin collected at the MIB/7.5% Ficoll interface was discarded. Synaptosomes were collected at the 7.5%/12% interface and were resuspended in MIB and centrifuged at 16,000 × g for 15 min. The Ficoll gradient purified NS mitochondria (FM) were collected as a pellet below 12% Ficoll.
Figure 1.
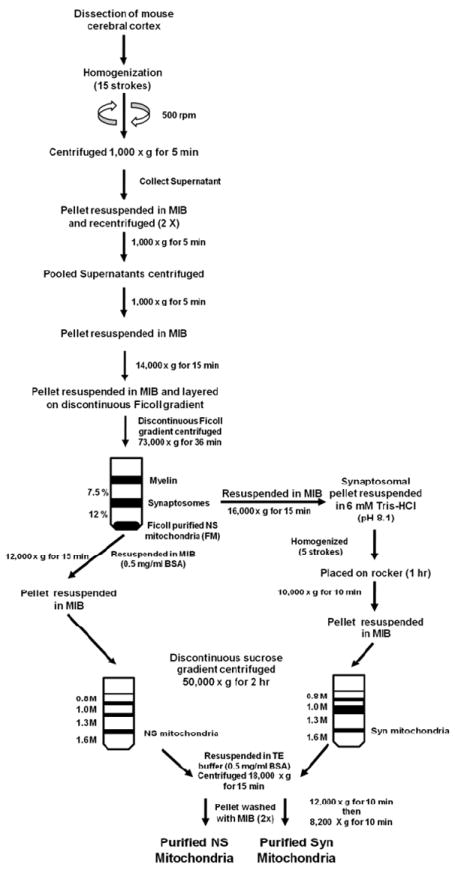
Procedure used for the isolation and purification of NS and Syn mitochondria from mouse cerebral cortex
3.2.1 Purification of NS Mitochondria
The FM pellet, containing NS mitochondria, was resuspended in MIB containing 0.5 mg/mL bovine serum albumin (BSA) and was centrifuged at 12,000 × g for 15 min. The resulting pellet was collected and resuspended in 6 mL of MIB. The resuspended FM pellet was layered on a discontinuous sucrose gradient containing 0.8 M/ 1.0 M/ 1.3 M/ 1.6 M sucrose. The volumes for the sucrose gradient were 6 mL/ 6 mL/ 10 mL/ 8 mL, respectively. The gradients were made from a 1.6 M sucrose stock containing 1 mM EDTA-K and 10 mM Tris-HCl (pH 7.4). The discontinuous sucrose gradient was centrifuged at 50,000 × g for 2 hr (4 °C) in a Sorvall SW 28 rotor using slow acceleration and deceleration to prevent disruption of the gradient. Purified NS mitochondria were collected at the interface of 1.3 M and 1.6 M sucrose. NS mitochondria were collected and resuspended in (1:3, v/v) TE buffer (1 mM EDTA-K and 10 mM Tris-HCl, pH 7.4) containing 0.5 mg/mL BSA and centrifuged at 18,000 × g for 15 min. The pellet was then resuspended in MIB and centrifuged at 12,000 × g for 10 min. The pellet was again resuspended in MIB and centrifuged at 8,200 × g for 10 min.
3.2.2 Purification of Syn mitochondria
Synaptosomes were burst by homogenization in 6 mM Tris-HCl (pH 8.1) using 5 up and down strokes. The homogenized synaptosomes were transferred to a 15 mL conical tube and then placed on a rocker for 1 hr (4 °C). The burst synaptosomes were centrifuged at 10,000 × g for 10 min. The pellet was resuspended in 6 mL of MIB. The resuspended pellet was layered on a discontinuous sucrose gradient and centrifuged following the same procedure as described above for NS mitochondria.
3.3 Mass Spectrometry
3.3.1 Materials
Synthetic phospholipids including 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (14:1-14:1 PtdCho), 1,2-dipalmitoleoyl-sn-glycero-3phosphoethanolamine (16:1-16:1 PtdEtn), 1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol (15:0-15:0 PtdGro), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (14:0-14:0 PtdSer), N-lauroryl sphingomyelin (N12:0 CerPCho), 1,1’,2,2’-tetramyristoyl cardiolipin (T14:0 Ptd2Gro), heptadecanoyl ceramide (N17:0 Cer), 1-heptadecanoyl-2-hydroxy-sn-Glycero-3-phosphocholine (17:0 LysoPtdCho) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). It should be noted that the prefix “N” denotes the amide-linked acyl chain. Deuterated cholesterol (d6-chol) was purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA, USA). All the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Burdick and Jackson, Muskegon, MI, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
3.3.2 Sample preparation for mass spectrometric analysis
An aliquot of the mitochondrial preparation was transferred to a disposable culture borosilicate glass tube (16 × 100 mm). Internal standards were added based on protein concentration and included 16:1-16:1 PtdEtn (100 nmol/mg protein), 14:1-14:1 PtdCho (45 nmol/mg protein), T14:0 Ptd2Gro (3 nmol/mg protein), 15:0-15:0 PtdGro (7.5 nmol/mg protein), 14:0-14:0 PtdSer (20 nmol/mg protein), 17:0 LysoPtdCho (1.5 nmol/mg protein), N12:0 CerPCho (20 nmol/mg protein), N17:0 Cer (5 nmol/mg protein). This allowed the final quantified lipid content to be normalized to the protein content and eliminated potential loss from the incomplete recovery. The molecular species of internal standards were selected because they represent < 0.1% of the endogenous cellular lipid mass as demonstrated by ESI/MS lipid analysis.
A modified Bligh and Dyer procedure was used to extract lipids from each mitochondrial preparation as previously described (29). Each lipid extract was reconstituted with a volume of 500 μL/mg protein (which was based on the original protein content of the samples as determined from protein measurement) in CHCl3/MeOH (1:1, v/v). The lipid extracts were flushed with nitrogen, capped, and stored at -20 °C for ESI/MS analysis. Each lipid solution was diluted approximately 50-fold immediately prior to infusion and lipid analysis.
3.3.3 Instrumentation and mass spectrometry
A triple-quadrupole mass spectrometer (Thermo Scientific TSQ Quantum Ultra, Plus, San Jose, CA, USA), equipped with an electrospray ion source and Xcalibur system software, was utilized as previously described (30). The first and third quadrupoles serve as independent mass analyzers using a mass resolution setting of peak width 0.7 Th while the second quadrupole serves as a collision cell for tandem MS. The diluted lipid extract was directly infused into the ESI source at a flow rate of 4 μL/min with a syringe pump. Lipid classes were analyzed in three different modes: negative-ion ESI, negative-ion ESI plus lithium hydroxide, and positive-ion ESI plus lithium hydroxide (Fig 2). Typically, a 2-min period of signal averaging in the profile mode was employed for each mass spectrum. For tandem MS, a collision gas pressure was set at 1.0 mTorr, but the collision energy varied with the classes of lipids as described previously (14, 30). Typically, a 2- to 5-min period of signal averaging in the profile mode was employed for each tandem MS spectrum. All the mass spectra and tandem MS spectra were automatically acquired by a customized sequence subroutine operated under Xcalibur software. Data processing of 2D MS analyses including ion peak selection, data transferring, peak intensity comparison, and quantitation was conducted using self-programmed MicroSoft Excel macros (30).
Figure 2.
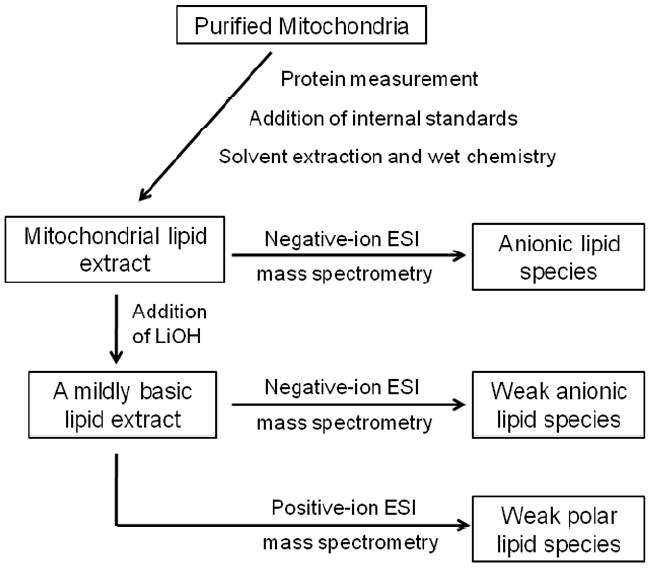
Schematic of the shotgun lipidomics procedure used for the analysis of the mitochondrial lipidome (modified from Han et al 2005 Mass Spectrom Reviews 24:367-412)
4. Results
4.1 Mitochondrial purity
Ficoll as well as sucrose discontinuous gradients were used to purify NS and Syn mitochondria (Fig. 1). As Ficoll gradient purified NS mitochondria (FM) contained markers for cytoskeletal (β-actin) and membrane (SNAP25, PCNA, tuberin, PLP, and calnexin) contamination, we further purified mitochondria using a discontinuous sucrose gradient. None of these markers were present in the Ficoll and sucrose discontinuous gradient purified mitochondria, which contained only mitochondrial-enriched markers representing the inner mitochondrial membrane (Complex IV, subunit IV) and the outer mitochondrial membrane (MAO-A) (Fig. 3A). Cholera toxin b immunostaining is a sensitive procedure for detecting gangliosides with the GM1a structure in cells and tissues (31). The toxin can have slight cross reactivity with GD1a. GM1a and a low amount of GD1a was found in the TH, My, and FM fractions, indicating the presence of myelin and microsomal membranes in these subcellular fractions (Fig. 3B). Only a trace amount of GM1a was detected in the NS mitochondria and no GM1a was detected in Syn mitochondria. These finding attest to the high degree of mitochondrial purity achieved with the isolation procedure.
Figure 3.
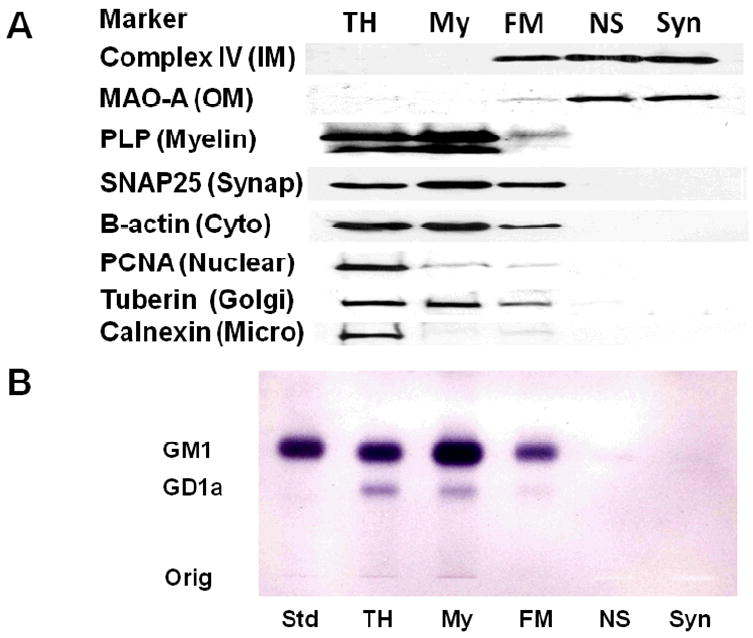
Distribution of protein markers on Western blots (A) and of gangliosides on thin-layer chromatography (B) in subcellular fractions from mouse cerebral cortex. Subcellular fractions included total homogenate (TH), crude myelin (My), Ficoll gradient purified NS mitochondria (FM), Ficoll and sucrose gradient purified non-synaptic mitochondria (NS), and Ficoll and sucrose gradient purified synaptic mitochondria (Syn). Western blots were performed to determine the distribution of specific protein markers for the inner mitochondria membrane (complex IV, subunit IV), outer mitochondrial membrane (monoamine oxidase A), myelin (proteolipid protein), synaptosomal membrane (SNAP25), cytoskeleton (β-actin), nuclear membrane (proliferating cell nuclear antigen), Golgi membrane (Tuberin), and microsomal membrane (calnexin). GM1a was visualized on TLC plates with cholera toxin b immunostaining as described in Methods. Std, is GM1a.
4.2 Mitochondrial lipid composition
We used shotgun lipidomics to evaluate lipid content and distribution of fatty acid molecular species in the NS and Syn mitochondria. The lipid classes were listed according to their relative abundance (Table 1). Although the content of most lipids was similar in the NS and Syn mitochondria, the content of Ptd2Gro was lower whereas the content of PtdSer and Cer were higher in the Syn mitochondria than in the NS mitochondria. The myelin-enriched lipids, sulfatides and cerebrosides, were not detected in either NS or Syn mitochondria.
Table 1.
Lipid composition of C57BL/6J mouse brain mitochondria
| Lipid | Non-synaptic | Synaptic |
|---|---|---|
| Ethanolamine glycerophospholipids | 187.4 ± 12.1 | 211.7 ± 21.3 |
| Phosphatidylethanolamine | 164.9 ± 10.0 | 184.6 ± 20.3 |
| Plasmenylethanolamine | 22.5 ± 2.2 | 27.0 ± 1.0 |
| Choline glycerophospholipids | 129.9 ± 7.7 | 156.3 ± 26.1 |
| Phosphatidylcholine | 119.6 ± 5.3 | 137.4 ± 17.2 |
| Plasmenylcholine | 1.2 ± 0.1 | 2.4 ± 1.1 |
| Plasmanylcholine | 9.1 ± 3.2 | 16.5 ± 8.5 |
| Cholesterol | 139.0 ± 46.7 | 126.7 ± 31.2 |
| Cardiolipin | 52.7 ± 4.5 | 39.9 ± 3.4 * |
| Phosphatidylinositol | 9.4 ± 0.8 | 10.2 ± 0.9 |
| Phosphatidylglycerol | 7.1 ± 0.5 | 6.4 ± 0.7 |
| Sphingomyelin | 5.3 ± 1.2 | 6.5 ± 0.6 |
| Phosphatidylserine | 4.6 ± 1.5 | 14.1 ± 3.0 * |
| Lysophosphatidylcholine | 2.7 ± 0.6 | 3.3 ± 0.4 |
| Ceramide | 0.7 ± 0.2 | 1.6 ± 0.2 ** |
Values are expressed as mean nmol/mg protein ± S.D. (N = 3)
Significantly different values from NS mitochondria at
: P < 0.02;
: P <0.005 as determined from the two-tailed t-test
No major changes in lipid molecular species were found between NS and Syn mitochondria. The major molecular species (>2%) of anionic, weak anionic, and weak polar lipids are found in Tables 2-4.
Table 2.
Mass content of major molecular species of anionic lipids as detemined by shotgun lipidomics
| Cardiolipin
| |||
|---|---|---|---|
| [M-2H]- | Major Species | Non-synaptic | Synaptic |
| 713.0 | 18:2-18:1-18:1-16:1 | 0.71 ± 0.02 | 0.53 ± 0.04 |
| 18:1-18:1-18:1-16:2 | |||
| 18:2-18:1-18:0-16:2 | |||
| 18:2-18:2-18:0-16:1 | |||
| 714.0 | 18:1-18:1-18:1-16:1 | 1.47 ± 0.23 | 1.03 ± 0.12 |
| 18:2-18:1-18:1-16:0 | |||
| 715.0 | 18:1-18:1-18:1-16:0 | 0.68 ± 0.04 | 0.61 ± 0.08 |
| 18:0-18:1-18:1-16:1 | |||
| 724.0 | 20:4-18:2-18:1-16:1 | 0.95 ± 0.08 | 0.66 ± 0.08 |
| 725.0 | 20:4-18:2-18:1-16:0 | 2.22 ± 0.17 | 1.56 ± 0.22 |
| 20:4-18:1-18:1-16:1 | |||
| 726.0 | 20:4-18:1-18:1-16:0 | 1.74 ± 0.13 | 1.31 ± 0.09 |
| 20:3-18:1-18:1-16:1 | |||
| 20:3-18:1-18:1-16:1 | |||
| 727.0 | 18:2-18:1-18:1-18:1 | 2.48 ± 0.33 | 1.59 ± 0.18 |
| 728.0 | 18:1-18:1-18:1-18:1 | 3.72 ± 0.72 | 2.217 ± 0.15 |
| 735.0 | 20:4-20:4-18:2-16:1 | 0.56 ± 0.06 | 0.39 ± 0.02 |
| 736.0 | 20:4-20:4-18:1-16:1 | 1.41 ± 0.16 | 1.10 ± 0.12 |
| 737.0 | 20:4-20:4-18:1-16:0 | 2.11 ± 0.07 | 1.54 ± 0.17 |
| 22:6-18:1-18:1-16:1 | |||
| 22:6-18:2-18:1-16:0 | |||
| 738.0 | 20:4-18:2-18:1-18:1 | 2.68 ± 0.37 | 2.02 ± 0.12 |
| 22:6-18:1-18:1-16:0 | |||
| 739.0 | 20:4-18:1-18:1-18:1 | 3.30 ± 0.45 | 2.48 ± 0.23 |
| 740.0 | 20:4-18:1-18:1-18:0 | 1.07 ± 0.14 | 0.75 ± 0.03 |
| 20:3-18:1-18:1-18:1 | |||
| 748.0 | 20:4-20:4-18:2-18:2 | 1.38 ± 0.09 | 1.11 ± 0.17 |
| 20:4-20:4-20:4-16:0 | |||
| 22:6-20:4-18:1-16:1 | |||
| 22:6-22:6-16:0-16:0 | |||
| 22:6-18:2-18:2-18:2 | |||
| 749.0 | 20:4-20:4-18:2-18:1 | 1.99 ± 0.19 | 1.46 ± 0.20 |
| 750.0 | 20:4-20:4-18:1-18:1 | 3.24 ± 0.24 | 2.31 ± 0.28 |
| 751.0 | 22:6-18:1-18:1-18:1 | 2.74 ± 0.10 | 2.36 ± 0.26 |
| 20:4-20:3-18:1-18:1 | |||
| 752.0 | 22:6-18:1-18:1-18:0 | 0.54 ± 0.17 | 0.46 ± 0.07 |
| 760.0 | 22:6-20:4-18:2-18:2 | 0.76 ± 0.05 | 0.58 ± 0.09 |
| 22:6-20:4-20:4-16:0 | |||
| 22:6-22:6-18:1-16:1 | |||
| 761.0 | 22:6-20:4-18:2-18:1 | 1.72 ± 0.08 | 1.27 ± 0.05 |
| 762.0 | 22:6-20:4-18:1-18:1 | 2.94 ± 0.23 | 2.36 ± 0.13 |
| 763.0 | 22:6-20:4-18:1-18:0 | 1.03 ± 0.05 | 0.78 ± 0.07 |
| 22:6-20:3-18:1-18:1 | |||
| 773.0 | 22:6-20:4-20:4-18:1 | 1.42 ± 0.12 | 1.10 ± 0.10 |
| 774.0 | 22:6-22:6-18:1-18:1 | 1.81 ± 0.09 | 1.56 ± 0.17 |
| 22:6-20:4-20:3-18:1 | |||
| 786.0 | 22:6-22:6-20:4-18:1 | 1.33 ± 0.08 | 1.11 ± 0.16 |
| 22:6-20:4-20:4-20:3 | |||
| 797.0 | 22:6-22:6-22:6-18:1 | 0.50 ± 0.04 | 0.50 ± 0.05 |
| 22:6-22:6-20:4-20:3 | |||
|
Phosphatidylinositol
| |||
| [M-H]- | Major Species | Non-synaptic | Synaptic |
|
| |||
| 857.5 | 16:0-20:4 | 1.12 ± 0.09 | 1.17 ± 0.11 |
| 881.5 | 18:2-20:4 | 0.23 ± 0.01 | 0.26 ± 0.06 |
| 16:0-22:6 | |||
| 885.5 | 18:0-20:4 | 6.68 ± 0.41 | 7.04 ± 0.69 |
| 909.5 | 18:0-22:6 | 0.37 ± 0.02 | 0.48 ± 0.03 |
|
Phosphatidylglycerol
| |||
| [M-H]- | Major Species | Non-synaptic | Synaptic |
|
| |||
| 719.5 | 16:0-16:1 | 0.29 ± 0.02 | 0.31 ± 0.02 |
| 721.5 | 16:0-16:0 | 0.49 ± 0.04 | 0.49 ± 0.03 |
| 743.5 | 16:1-18:2 | 0.22 ± 0.04 | 0.19 ± 0.04 |
| 745.5 | 16:0-18:2 | 0.54 ± 0.10 | 0.48 ± 0.05 |
| 747.5 | 16:0-18:1 | 3.41 ± 0.13 | 3.07 ± 0.43 |
| 769.5 | 18:2-18:2 | 0.82 ± 0.04 | 0.64 ± 0.10 |
| 771.5 | 18:1-18:2 | 0.29 ± 0.03 | 0.26 ± 0.02 |
| 773.5 | 18:1-18:1 | 0.47 ± 0.12 | 0.38 ± 0.02 |
| 775.5 | 18:0-18:1 | 0.55 ± 0.10 | 0.60 ± 0.14 |
|
Phosphatidylserine
| |||
| [M-H]- | Major Species | Non-synaptic | Synaptic |
|
| |||
| 786.5 | 18:0-18:2 | 0.29 ± 0.07 | 0.47 ± 0.11 |
| 788.5 | 18:0-18:1 | 0.24 ± 0.21 | 1.61 ± 0.77 |
| 810.5 | 18:0-20:4 | 0.10 ± 0.08 | 0.35 ± 0.15 |
| 834.5 | 18:0-22:6 | 3.13 ± 1.22 | 9.59 ± 1.60 |
| 836.5 | 18:0-22:5 | 0.22 ± 0.27 | 0.31 ± 0.20 |
| 838.6 | 20:0-20:4 | 0.28 ± 0.07 | 0.79 ± 0.04 |
| 18:0-22:4 | |||
Table 4.
Mass content of major molecular species of weak polar lipids as detemined by shotgun lipidomics
| Choline Glycerophospholipids
| |||
|---|---|---|---|
| [M+Li]+ | Major Species | Non-synaptic | Synaptic |
| 740.6 | D16:0-16:0 | 10.00 ± 1.27 | 13.58 ± 1.32 |
| 766.6 | D16:0-18:1 | 33.31 ± 1.31 | 37.34 ± 3.07 |
| 768.6 | D16:0-18:0 | 2.47 ± 0.26 | 3.36 ± 0.93 |
| 782.7 | A16:0-20:0 | 7.42 ± 2.81 | 13.18 ± 7.40 |
| 788.6 | D18:2-18:2 | 23.37 ± 1.28 | 23.13 ± 2.55 |
| D16:0-20:4 | |||
| 792.6 | D18:0-18:2 | 4.15 ± 0.42 | 3.89 ± 0.28 |
| D18:1-18:1 | |||
| 794.6 | D18:0-18:1 | 4.39 ± 0.46 | 6.43 ± 0.99 |
| 812.6 | D16:0-22:6 | 13.94 ± 0.79 | 14.08 ± 1.45 |
| D18:2-20:4 | |||
| 814.6 | D18:1-20:4 | 5.66 ± 0.72 | 5.51 ± 1.07 |
| D16:0-22:5 | |||
| 816.6 | D18:2-20:2 | 11.36 ± 0.90 | 12.21 ± 2.03 |
| D18:0-20:4 | |||
| 840.6 | D18:0-22:6 | 2.07 ± 1.96 | 4.14 ± 1.02 |
| D20:2-20:4 | |||
|
Sphingomyelin
| |||
| [M+Li]+ | Major Species | Non-synaptic | Synaptic |
|
| |||
| 709.6 | N16:0 | 0.27 ± 0.32 | 0.27 ± 0.13 |
| 735.6 | N18:1 | 0.26 ± 0.02 | 0.29 ± 0.08 |
| 737.6 | N18:0 | 1.99 ± 0.38 | 2.76 ± 0.17 |
| 765.6 | N20:0 | 2.33 ± 0.41 | 2.61 ± 0.32 |
| 793.7 | N22:0 | 0.16 ± 0.27 | 0.32 ± 0.13 |
| 821.7 | N24:0 | 0.20 ± 0.14 | 0.08 ± 0.02 |
|
Lysophosphatidylcholine
| |||
| [M+Na]+ | Major Species | Non-synaptic | Synaptic |
|
| |||
| 490.3 | 14:0 | 0.06 ± 0.01 | 0.04 ± 0.02 |
| 504.3 | A16:0 | 0.06 ± 0.03 | 0.04 ± 0.01 |
| 516.3 | 16:1 | 0.09 ± 0.04 | 0.13 ± 0.07 |
| 518.3 | 16:0 | 0.89 ± 0.21 | 1.22 ± 0.14 |
| 542.3 | 18:2 | 0.06 ± 0.02 | 0.03 ± 0.02 |
| 544.3 | 18:1 | 0.41 ± 0.13 | 0.54 ± 0.12 |
| 546.4 | 18:0 | 0.32 ± 0.08 | 0.44 ± 0.08 |
| 566.3 | 20:4 | 0.16 ± 0.09 | 0.16 ± 0.03 |
| 574.4 | 20:0 | 0.04 ± 0.02 | 0.07 ± 0.09 |
| 590.3 | 22:6 | 0.24 ± 0.12 | 0.27 ± 0.03 |
| 592.3 | 22:5 | 0.08 ± 0.03 | 0.09 ± 0.03 |
Discussion
The analysis of highly purified brain mitochondria by shotgun lipidomics presents an innovated high-throughput approach to analyzing the mitochondrial lipidome. The major obstacle that arises from analyzing brain mitochondria is obtaining a highly purified fraction free from contamination. Although numerous types of discontinuous gradients can be utilized, we found that a Ficoll gradient in addition to a sucrose discontinuous gradient could obtain a highly purified brain mitochondria preparation suitable for lipidomic analysis (1).
Differences can exist in content, molecular species, glycerophospholipid subclass, or total fatty acid distribution in mitochondrial lipids. These differences can be readily detected using a shotgun lipidomics platform with minimal sample processing (14). Changes in any or all aspects of the mitochondrial lipidome will likely change membrane fluidity, efficiency of the proton gradient, regulation of specific enzyme activities, as well as the overall bioenergetic efficiency of mitochondria (7). Alterations in brain energy metabolism in neurological and neurodegenerative diseases is well established (32-34). Analysis of the brain mitochondrial lipidome in diseased tissues or mouse models can provide new insight into the role of mitochondria lipid alterations during disease pathogenesis.
The development of a high-throughput approach to analyze the brain mitochondrial lipidome is a new tool to study the cause(s) for altered energy metabolism in diseased brain (35). We have also used this approach to provide lipidomic evidence supporting the Warburg theory of cancer in a series of mouse brain tumors (36). It is our opinion that new insight on brain function and pathogenesis will be realized with further investigations of the brain mitochondrial lipidome.
Table 3.
Mass content of major molecular species of weak anionic lipids as detemined by shotgun lipidomics
| Ethanolamine Glycerophospholipids
| |||
|---|---|---|---|
| [M-H]- | Major Species | Non-synaptic | Synaptic |
| 716.5 | D16:0-18:1 | 5.11 ± 1.14 | 4.82 ± 0.68 |
| 742.5 | D18:0-18:2 | 5.07 ± 0.62 | 5.09 ± 0.88 |
| D18:1-18:1 | |||
| D16:0-20:2 | |||
| 746.5 | P16:0-22:6 | 9.03 ± 1.39 | 9.89 ± 1.72 |
| D18:0-18:0 | |||
| P18:2-20:4 | |||
| 762.5 | D16:0-22:6 | 9.39 ± 0.81 | 11.08 ± 1.00 |
| 764.5 | D16:0-22:5 | 8.78 ± 0.52 | 8.35 ± 0.86 |
| D18:1-20:4 | |||
| 766.5 | D18:0-20:4 | 37.49 ± 2.42 | 36.39 ± 4.86 |
| D16:0-22:4 | |||
| 768.6 | D18:1-20:2 | 11.49 ± 2.19 | 16.42 ± 1.11 |
| D16:0-22:3 | |||
| D18:0-20:3 | |||
| 774.5 | P18:0-22:6 | 10.37 ± 1.44 | 13.25 ± 1.63 |
| P18:1-22:5 | |||
| D18:0-20:0 | |||
| 776.6 | P18:0-22:5 | 7.29 ± 2.19 | 8.24 ± 2.24 |
| P18:1-22:4 | |||
| 788.5 | D18:1-22:6 | 5.95 ± 0.44 | 6.47 ± 0.55 |
| 790.5 | D18:0-22:6 | 58.18 ± 1.87 | 58.66 ± 8.62 |
| D18:1-22:5 | |||
| 792.6 | D18:1-22:4 | 3.62 ± 0.29 | 4.90 ± 0.38 |
| D18:0-22:5 | |||
| 794.6 | D20:0-20:4 | 6.53 ± 0.46 | 15.63 ± 2.58 |
| D18:1-22:3 | |||
| D18:0-22:4 | |||
| 796.6 | D20:0-20:3 | 6.85 ± 1.34 | 9.14 ± 0.48 |
| D18:0-22:3 | |||
|
Ceramide
| |||
| [M-H]- | Major Species | Non-synaptic | Synaptic |
|
| |||
| 564.5 | N18:0 | 0.66 ± 0.15 | 1.54 ± 0.19 |
| 592.6 | N20:0 | 0.04 ± 0.02 | 0.03 ± 0.00 |
| 646.6 | N24:1 | 0.02 ± 0.02 | 0.04 ± 0.02 |
| 648.6 | N24:0 | 0.02 ± 0.00 | 0.01 ± 0.01 |
References
- 1.Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. J Neurochem. 2008;106:299–312. doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai JC, Walsh JM, Dennis SC, Clark JB. Synaptic and non-synaptic mitochondria from rat brain: isolation and characterization. J Neurochem. 1977;28:625–31. doi: 10.1111/j.1471-4159.1977.tb10434.x. [DOI] [PubMed] [Google Scholar]
- 3.Brown MR, Sullivan PG, Geddes JW. Synaptic mitochondria are more susceptible to Ca2+overload than nonsynaptic mitochondria. J Biol Chem. 2006;281:11658–11668. doi: 10.1074/jbc.M510303200. [DOI] [PubMed] [Google Scholar]
- 4.Dagani F, Gorini A, Polgatti M, Villa RF, Benzi G. Synaptic and non-synaptic mitochondria from rat cerebral cortex. Characterization and effect of pharmacological treatment on some enzyme activities related to energy transduction. Farmaco [Sci] 1983;38:584–94. [PubMed] [Google Scholar]
- 5.Villa RF, Gorini A, Geroldi D, Lo Faro A, Dell’Orbo C. Enzyme activities in perikaryal and synaptic mitochondrial fractions from rat hippocampus during development. Mech Ageing Dev. 1989;49:211–25. doi: 10.1016/0047-6374(89)90072-9. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found Symp. 2001;235:247–63. doi: 10.1002/0470868694.ch20. discussion 263-6. [DOI] [PubMed] [Google Scholar]
- 7.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 8.Hoch FL. Cardiolipins and biomembrane function. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 9.Stuart JA, Gillis TE, Ballantyne JS. Remodeling of phospholipid fatty acids in mitochondrial membranes of estivating snails. Lipids. 1998;33:787–793. doi: 10.1007/s11745-998-0271-1. [DOI] [PubMed] [Google Scholar]
- 10.Rostovtseva TK, Bezrukov SM. VDAC regulation: role of cytosolic proteins and mitochondrial lipids. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell AM, Chan SH. Mitochondrial membrane cholesterol, the voltage dependent anion channel (VDAC), and the Warburg effect. J Bioenerg Biomembr. 2008 doi: 10.1007/s10863-008-9138-x. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass spectrometry reviews. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 13.Yang K, Zhao Z, Gross RW, Han X. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS ONE. 2007;2:e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Gross RW. Shotgun lipidomics: multidimensional MS analysis of cellular lipidomes. Expert review of proteomics. 2005;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- 15.Han X, Holtzman DM, McKeel DW., Jr Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem. 2001;77:1168–80. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 16.Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Shotgun Lipidomics Reveals the Temporally Dependent, Highly Diversified Cardiolipin Profile in the Mammalian Brain: Temporally Coordinated Postnatal Diversification of Cardiolipin Molecular Species with Neuronal Remodeling. Biochemistry. 2008 doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47:864–79. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zischka H, Lichtmannegger J, Jagemann N, Jennen L, Hamoller D, Huber E, Walch A, Summer KH, Gottlicher M. Isolation of highly pure rat liver mitochondria with the aid of zone-electrophoresis in a free flow device (ZE-FFE) Methods Mol Biol. 2008;424:333–48. doi: 10.1007/978-1-60327-064-9_26. [DOI] [PubMed] [Google Scholar]
- 19.Sims NR. Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem. 1990;55:698–707. doi: 10.1111/j.1471-4159.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- 20.Sims NR, Anderson MF. Isolation of mitochondria from rat brain using Percoll density gradient centrifugation. Nat Protoc. 2008;3:1228–39. doi: 10.1038/nprot.2008.105. [DOI] [PubMed] [Google Scholar]
- 22.Taylor SW, Warnock DE, Glenn GM, Zhang B, Fahy E, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. An alternative strategy to determine the mitochondrial proteome using sucrose gradient fractionation and 1D PAGE on highly purified human heart mitochondria. J Proteome Res. 2002;1:451–8. doi: 10.1021/pr025533g. [DOI] [PubMed] [Google Scholar]
- 23.Stocco DM, Hutson JC. Characteristics of mitochondria isolated by rate zonal centrifugation from normal liver and Novikoff hepatomas. Cancer Res. 1980;40:1486–92. [PubMed] [Google Scholar]
- 24.Graham JM. Purification of a crude mitochondrial fraction by density-gradient centrifugation. Curr Protoc Cell Biol. 2001;Chapter 3(Unit 3 4) doi: 10.1002/0471143030.cb0304s04. [DOI] [PubMed] [Google Scholar]
- 25.Lai JC, Clark JB. Preparation and properties of mitochondria derived from synaptosomes. The Biochemical journal. 1976;154:423–432. doi: 10.1042/bj1540423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mena EE, Hoeser CA, Moore BW. An improved method of preparing rat brain synaptic membranes. Elimination of a contaminating membrane containing 2’,3’-cyclic nucleotide 3’-phosphohydrolase activity. Brain Res. 1980;188:207–31. doi: 10.1016/0006-8993(80)90569-7. [DOI] [PubMed] [Google Scholar]
- 27.Rendon A, Masmoudi A. Purification of non-synaptic and synaptic mitochondria and plasma membranes from rat brain by a rapid Percoll gradient procedure. J Neurosci Methods. 1985;14:41–51. doi: 10.1016/0165-0270(85)90113-x. [DOI] [PubMed] [Google Scholar]
- 28.Battino M, Bertoli E, Formiggini G, Sassi S, Gorini A, Villa RF, Lenaz G. Structural and functional aspects of the respiratory chain of synaptic and nonsynaptic mitochondria derived from selected brain regions. J Bioenerg Biomembr. 1991;23:345–363. doi: 10.1007/BF00762227. [DOI] [PubMed] [Google Scholar]
- 29.Cheng H, Guan S, Han X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: implications for the role of altered triacylglycerols in diabetic neuropathy. J Neurochem. 2006;97:1288–300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X, Yang J, Cheng H, Ye H, Gross RW. Toward fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal Biochem. 2004;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Brigande JV, Platt FM, Seyfried TN. Inhibition of glycosphingolipid biosynthesis does not impair growth or morphogenesis of the postimplantation mouse embryo. J Neurochem. 1998;70:871–882. doi: 10.1046/j.1471-4159.1998.70020871.x. [DOI] [PubMed] [Google Scholar]
- 32.Petrozzi L, Ricci G, Giglioli NJ, Siciliano G, Mancuso M. Mitochondria and neurodegeneration. Biosci Rep. 2007;27:87–104. doi: 10.1007/s10540-007-9038-z. [DOI] [PubMed] [Google Scholar]
- 33.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 34.Calabrese V, Scapagnini G, Giuffrida Stella AM, Bates TE, Clark JB. Mitochondrial involvement in brain function and dysfunction: relevance to aging, neurodegenerative disorders and longevity. Neurochem Res. 2001;26:739–64. doi: 10.1023/a:1010955807739. [DOI] [PubMed] [Google Scholar]
- 35.Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sciences. 1995;56:1151–1171. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 36.Kiebish MA, Han X, Cheng H, Chuang JH, Seyfried TN. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: Lipidomic evidence supporting the Warburg theory of cancer. J Lipid Res. 2008 doi: 10.1194/jlr.M800319-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]


