Abstract
Age-related macular degeneration (AMD) is a leading cause of vision loss and blindness among the elderly population in the industrialized world. One of the typical features of this pathology is the gradual death of retinal pigment epithelial (RPE) cells, which are essential for maintaining photoreceptor functions and survival. The etiology is multifactorial, and oxidative stress is clearly one of the key factors involved in disease pathogenesis.
Recent work has revealed the presence of phosphorylated signaling proteins in the vitreous humour of patients affected by AMD or other retinal diseases. While the location of these signaling proteins is typically the cell membrane or intracellular compartments, vitreous samples were proven to be cell-free.
To gain a better understanding of how these proteins can be shed into the vitreous, we used Reverse Phase Protein Arrays (RPMA) to analyze the protein and phosphoprotein content of exosomes shed by cultured ARPE-19 cells under oxidative stress conditions.
72 proteins were shown to be released by ARPE-19 cells and compartmentalized within exosomes. 41 of them were selectively detected in their post-translationally modified form (i.e., phosphorylated or cleaved) for the first time in exosomes. Sets of these proteins were linked together reflecting activation of pathway units within exosomes. A subset of (phospho)proteins were altered in exosomes secreted by ARPE-19 cells subjected to oxidative stress, compared to that secreted by control/non stressed cells. Stress-altered exosome proteins were found to be involved in pathways regulating apoptosis/survival (i.e, Bak, Smac/Diablo, PDK1 (S241), Akt (T308), Src (Y416), Elk1 (S383), ERK 1/2 (T202/Y204)) and cell metabolism (i.e., AMPK α1 (S485), Acetyl-CoA carboxylase (S79), LDHA).
Exosomes may thus represent the conduit through which membrane and intracellular signaling proteins are released into the vitreous. Changes in their (phospho)protein content upon stress conditions suggest their possible role in mediating cell-cell signaling during physio-pathological events; furthermore, exosomes may represent a potential source of biomarkers.
Keywords: ARPE-19 cells, exosomes, Reverse Phase Protein Arrays, Age Related Macular Degeneration, Oxidative stress
Introduction
Retinal age-related macular degeneration (AMD) is the leading cause of vision loss and blindness in individuals over age 60 in the industrialized world (Gehrs et al, 2006). The typical features of AMD include accumulation of extracellular deposits (drusen) in the macula, thickening of Bruch’s membrane, and gradual death of retinal pigment epithelial (RPE) cells and photoreceptors. As RPE cells and photoreceptors degenerate, there is a tendency for blood vessels to grow from their normal location in the choroid into an abnormal location beneath the retina. The abnormal blood vessels are dysfunctional and leaky, resulting eventually in progressive and severe loss of central vision.
Initial pathogenesis of AMD involves the degeneration of RPE cells, which progressively causes the death of rods and cones. RPE cells are essential for maintaining photoreceptor functions and survival: they are involved in the absorption of light energy, transport of metabolites and nutrients between photoreceptors and choriocapillaries, expression of growth factors for photoreceptors, and are responsible for phagocytizing shed photoreceptor outer segments, regulating the visual cycle and creating the blood-retinal barrier (Boulton and Dayhaw-Barker 2001; Kevany and Palczewski 2010).
The aetiology of AMD is multifactorial, including genetic, environmental, nutritional and behavioural factors (Ding et al, 2009; Zarbin, 2004; Kaarniranta and Salminen 2009; Jager et al, 2008); oxidative stress is clearly involved in the pathogenesis (Plafker, 2010; Qin, 2007). The retina is the tissue with the highest metabolic rate in the body (Yu and Cringle, 2001), and high oxygen consumption usually is accompanied by Reactive Oxygen Species (ROS) generation. RPE cells generate ROS due to phagocytosis of photoreceptor outer segments (Miceli et al, 1994), which also determines progressive accumulation of lipofuscin (Dorey et al, 1989), a mixture of non-degradable protein-lipid aggregates that leads to ROS generation upon irradiation with light (Wu et al, 2010; Wolf, 2003; Ng et al, 2008). Treatment with antioxidants reduces the progression of the pathology (Johnson, 2010). Inflammation is also linked to oxidative stress (Tschopp, 2011), and has an important role in AMD pathogenesis (Kaarniranta and Salminen, 2009).
Proteomic analysis of vitreous samples coming from patients affected by AMD or other retinal diseases and undergoing anti-VEGF therapy reported the presence of phosphorylated receptors and signaling proteins in the vitreous (Davuluri et al, 2009; Tamburro et al, 2010). Activated growth factor receptors had not been previously described in the vitreous; the relative abundance of certain proteins was dependent on the underlying retinal disease, and on whether patients were responders or non-responders to anti-VEGF therapy. These findings may thus constitute the basis of a new class of biomarkers for diagnosis and individualized therapy. These receptors normally reside in the cellular membrane, but the vitreous fluids in which they were identified were proven to be clear and cell-free. These findings raised the question of whether or not vitreous phosphoproteins or even portions of entire signal pathways were encapsulated within exosomes or microvesicles.
Increasing evidence points to exosomes and microvesicles as a novel alternative mechanism for protein export and intercellular communication, permitting exchange of proteins, lipids and nucleic acids between cells (Février and Raposo, 2004; Belting and Wittrup, 2008; Ahmed and Xiang, 2011). Exosomes are 40-100 nm membrane vesicles of endocytic origins. They form intracellularly by inward budding of the membrane of endosomes, leading to multivesicular bodies (MVBs); MVBs eventually fuse with the plasma membrane, releasing their internal vesicles (i.e., exosomes) in the extracellular medium. Exosomes from different cellular origins contain a common set of molecules (which are essential for their biogenesis, structure and trafficking), as well as cell type-specific components. They are secreted by most cell types in vitro, and found also in vivo in different body fluids (blood, urine, saliva, etc.) (Simpson et al, 2008; van Niel et al, 2006).
To gain a better understanding of how phosphoproteins can be shed into the vitreous, we used Reverse Phase Protein Arrays (RPMA) (Charboneau et al, 2002; Wilson et al, 2010) to analyze the protein and phosphoprotein content of exosomes shed by cultured ARPE-19 cells (a non transformed human RPE cell line) exposed to oxidative stress conditions.
Materials and methods
ARPE-19 cells culture
ARPE-19 cells (a non-transformed human RPE cell line) were obtained from American Type Culture Collection (ATCC, Manassas, VA). ARPE-19 cells were cultured in DMEM: F12 medium (ATCC) supplemented with 10% fetal bovine serum (FBS; from ATCC), at 37°C in a humidified 5.0 % CO2 atmosphere.
Preparation of exosome-production medium
The culture medium used for preparing cell-conditioned media, from which exosomes were isolated, was depleted of contaminating FBS-derived exosomes according to the procedure described previously (Théry et al, 2006). Briefly, culture medium containing 20% FBS was centrifuged overnight (16 h) at 100,000g, 4°C. The supernatant was filter-sterilized and diluted with culture medium without FBS to 10% final FBS.
ARPE-19 viability assay
ARPE-19 cells were seeded at a density of 11,000 cells/cm2 onto 96-well plates. 3 days after seeding, cells were incubated for 24 h with exosome-production medium containing different concentrations of Rotenone (0 - 5 μM; Sigma) or Methyl Viologen (0 - 2000 μM; ACROS Organics). At the end of the treatments, cell viability was quantified by CellTiter-Blue™ assay (Promega), adding 20 μl/well of Resazurin dye solution and incubating in the dark at 37°C for 2 h. Fluorescence was recorded using a Microplate Reader (Fluoroskan, Ascent FL Labsystems; excitation wavelength 530 nm, emission 584 nm). All assay points were determined in triplicate, and experiments were repeated three times.
ARPE-19 cell treatments
ARPE-19 cells were seeded onto T75 culture flasks at a density of 11,000 cells/cm2, and grown three days until they reached confluency. After washing once with PBS and once with medium without FBS, cells were incubated 24 h in exosome-production medium with: - no further additions (control), or - 2.5 μM Rotenone, or - 62.5 μM Methyl Viologen. These concentrations were the maximum dose that did not affect cell viability, and thus were chosen to avoid the presence of contaminating apoptotic blebs in the final exosome preparation (Théry et al, 2001). To isolate sufficient amounts of exosomes, about 17.5 × 106 cells were used for each treatment; experiments were repeated three times for each condition.
Following cell treatments, conditioned medium was collected and stored at 4°C until exosome isolation. An aliquot of the cells (about 2.5 × 106) was washed twice with cold PBS, and lysed with 1 ml of extraction buffer (a 10% solution of TCEP (Pierce) in T-PER™ 2× (Pierce)/ SDS Tris-glycine 2× buffer (Invitrogen)). Cell lysates were immediately heated 7 min at 100°C, and stored at −80°C prior to microarray construction. The remaining cells (about 15× 106) were further incubated in exosome-production medium without any treatment/addition for another 96 h period; conditioned medium was then collected and stored at 4°C until exosome isolation, and cell lysates were prepared as described above.
Exosome isolation
Exosomes were isolated from conditioned media (60-70 ml from each treatment), according to the protocol described previously (Théry et al, 2006). Briefly, cell culture media were centrifuged at 4°C sequentially for 10 min at 300g, 20 min at 2,000g, and 30 min at 10,000g. After each centrifugation step, the pellet was removed and discarded. Exosomes were then precipitated by ultracentrifugation of the supernatant (70 min at 100,000g, Optima™ XL-80k Ultracentrifuge, Beckman, SW28 rotor), and washed by resuspending the pellet in PBS and centrifuging again for 70 min at 100,000g. Ultracentrifugation was performed using polyallomer tubes (Beckman).
Exosome lysates were prepared by suspending the final pellet in 20 μl of extraction buffer, and immediately heating the pellet for 7 minutes at 100°C. Exosome lysates were stored at −80°C prior to microarray construction.
AFM analysis
Exosomes were characterized by Atomic Force Microscopy (AFM) using an NSCRIPTOR™ DPN® System (NanoInk). After exosome isolation (see above), the pellet was resuspended in 50 μl PBS, deposited on freshly cleaved mica under humid atmosphere at room temperature for 15 minutes, and dried under nitrogen before measurement. Images were acquired in the AC mode using a silicon tip with a typical resonance frequency of 300 kHz and a radius smaller than 10 nm.
RPMA construction
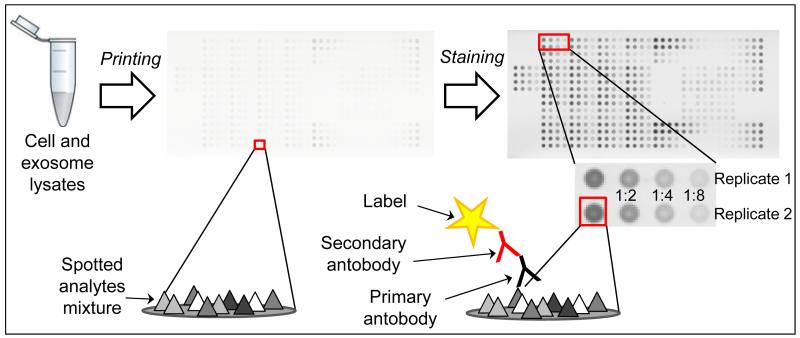
Cell and exosome lysates were printed undiluted on a glass-backed nitrocellulose array slide (20 × 50 mm; Schott) using an Aushon 2470 robotic arrayer (Aushon BioSystems) equipped with 350 μm pins, as previously described (VanMeter et al, 2007; Speer et al, 2007). Albumin was used for protein concentration calibration; it was printed in triplicate on the same slide, in eight-point dilution curves (0.08 - 1 mg/ml). Total protein concentration values per microarray spot was determined with Sypro Ruby protein stain (Invitrogen) per manufacturer’s directions, and imaged with a CCD camera (NovaRay; Alpha Innotech). The protein content of each sample was estimated by comparison to the albumin calibration curve. Lysates were then diluted appropriately to obtain a sample concentration of 0.5 mg/ml total protein in the neat spot, and printed on nitrocellulose array slides (Schott). Each sample was printed in duplicate; cell lysates were printed in a 4 point dilution curve (Fig. 1), while exosomes were printed only as neat sample. Cellular lysates prepared from Hela ± pervanadate, Hela + Heat Shock, A431 ± EGF, HepG2, T98G, Mouse Macrophage + LPS/IFNγ and MCF-7 were printed on each array for quality control assessments of (phospho)protein loading and detection. Slides were stored with desiccant (Drierite; WA Hammond) at −20°C prior to immunostaining.
Fig. 1. RPMA technology.
Sample lysates and controls are immobilized on a nitrocellulose-coated slide and probed with a primary and secondary antibody. The signal is amplified via horseradish peroxidase-mediated deposition of biotinyl tyramide. Cell lysates are prepared in a 2-fold dilution series, allowing analysis within the linear dynamic range for each sample-antibody pair.
RPMA detection
Immunostaining was performed on an automated slide stainer per manufacturer’s instructions (Autostainer CSA kit; Dako), as previously described (VanMeter et al, 2007; Speer et al, 2007). Each slide was incubated with a single primary antibody at room temperature for 30 min. The full list of primary antibodies used is reported in Supplementary Table 1. Antibodies were previously validated by Western Blot to ensure they give a single ( > 80% total signal) specific band (VanMeter et al, 2008). The negative control slide was incubated with antibody diluent instead of antibody. Secondary antibody was goat anti-rabbit IgG H+L (1:10,000) (Vector Labs), or goat anti-mouse IgG (1:25) (Dako). Subsequent signal detection was amplified via horseradish peroxidase mediated biotinyl tyramide deposition with chromogenic detection (Diaminobenzidine) per manufacturer’s instructions (Dako) (Fig. 1).
Each array was scanned, and spot-intensity analyzed. Spot intensity was integrated over a fixed area; local area background intensity was determined for each spot from the unprinted adjacent slide background (Image Quant v5.2; GE Healthcare). Non-specific background staining of the secondary antibody was subtracted, and data was normalized per microarray spot, according to Chiechi et al, 2012; a standardized single data value was generated for each sample on the array. NormFinder software was used to find the optimal normalization protein (Andersen et al, 2004).
Statistics
Significance in comparisons between groups was assessed using the non parametric Wilcoxon Rank Test (Wilcoxon-Mann-Whitney test; GraphPad Prism 6 software). We considered this test as the most appropriate to compare small size samples, since it does not require any assumption about the normality of the data distribution for a small N number.
Results
RPMA - Data generation and normalization
RPMA (Wilson et al, 2010; VanMeter et al, 2007; Speer et al, 2007) were used to identify the signaling pathways affected by oxidative stress in ARPE-19 cells, an immortalized human cell line that is widely used as an RPE model in vitro (Dunn et al, 1996), and to characterize for the first time the (phospho)protein content of exosomes shed by these cells (Fig. 1).
Antibodies used for RPMA staining were previously validated through Western Blot for their specificity. Only antibodies which gave a single band (> 80% total signal) specific for the target protein (or its specific post-translationally modified form) were used.
NormFinder software was used to find the protein with the greater expression stability among samples, which was then used to normalize data (Chiechi et al, 2012; Andersen et al, 2004). This was found to be β-actin (if only cells were considered) or cd63 (if only exosomes were considered). Therefore β-actin and cd63 normalization were preferred to compare protein levels between cells or between exosomes, respectively.
We avoided normalization to total protein, since it has been reported that culture media serum proteins associate with microvesicles (Palmisano et al, 2012), and thus may cause overestimation of sample total protein levels.
Exosome isolation and characterization
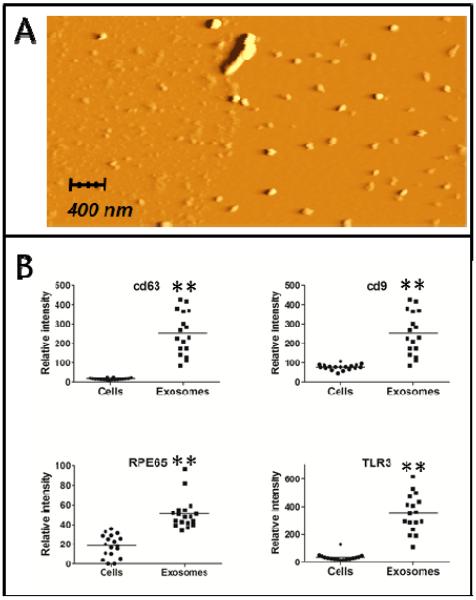
Exosomes shed by ARPE-19 cells were isolated with a series of centrifugation and ultracentrifugation steps, according to a commonly used protocol (Théry et al, 2006; see Materials & Methods). They were then characterized through Atomic Force Microscopy (AFM) (Palanisamy et al, 2010; Sharma et al, 2010); images revealed that the vast majority of the samples consisted of small vesicles with 80-110 nm diameter (Fig. 2A). A few larger particles were also present, which might be exosome aggregates (which are known to be difficult to avoid after ultracentrifugation; Théry et al, 2006), or larger microvesicles.
Fig. 2. Exosome characterization.
A: AFM image of exosomes isolated after a 24h treatment with Methyl Viologen. B: cd9, cd63, RPE-65 and TLR3 protein levels in ARPE-19 cells and exosomes (N = 18). **: significantly different from the cell lysates (Mann-Whitney test, p ≤ 0,001).
Exosomes were further characterized through RPMA analysis by the presence of the exosomal protein markers cd63 and cd9 (Théry et al, 2002). Other proteins were present in exosomes at higher levels, compared to the corresponding cell lysates (Fig. 2B and Tab. 1). Among these are RPE-65 (a typical marker of RPE cells) and TLR3. Since TLR3 is present mostly in endosomal membranes (Barton and Kagan, 2009; Kawai and Akira, 2010), its abundance supports the endocytic origin of these vesicles, and thus their identification as exosomes.
Tab. 1. Proteins detected in ARPE-19 exosomes and their relative abundance compared to cells.
Abundance of proteins found in exosomes is expressed as higher (+), lower (−) or non-significantly different (ns) compared to ARPE-19 cell lysates. Anti-phosphoprotein antibodies are expressed with the name of the protein followed by the position of phosphorylation; for caspases, the cleavage position is indicated.
| UniProt accession number |
Protein | Previous detection in exosomes (Exocarta database) |
Relative abundance |
|---|---|---|---|
| O00763 | Acetyl-CoA Carboxylase (S79) | only total protein | ns |
| Q9UE89 | β actin | yes | − * |
| P31749 | Akt (T308) | only total protein | + ** |
| Q13131 | AMPKα1 (S485) | only total protein | ns |
| Q9Y478 | AMPKβ1 (S108) | no | ns |
| P07355 | Annexin II | yes | ns |
| O14727 | Apaf-1 | yes | + ** |
| Q9H1Y0 | Atg5 | yes | − ** |
| O94817 | Atg12 | yes | + * |
| Q92934 | Bad (S112) | no | + * |
| Q92934 | Bad (S155) | no | ns |
| Q16611 | Bak | no | ns |
| Q07812 | Bax | yes | − ** |
| P10415 | Bcl-2 (T56) | no | + ** |
| Q07817 | Bcl-xL | no | |
| Q14457 | Beclin-1 | no | ns |
| P53004 | Biliverdin Reductase | yes | + ** |
| P42574 | Caspase-3, cleaved (D175) | no | ns |
| P55210 | Caspase-7, cleaved (D198) | no | + ** |
| P55211 | Caspase-9, cleaved (D330) | only total protein | + * |
| P21926 | cd9 | yes | + ** |
| P08962 | cd63 | yes | + ** |
| P10721 | c-Kit (Y703) | no | ns |
| P10721 | c-Kit (Y719) | no | ns |
| P23528 | Cofilin | yes | ns |
| P35354 | Cox-2 | no | + ** |
| P04049 | c-Raf (S338) | no | ns |
| P16220 | CREB (S133) | no | ns |
| P13073 | Cytochrome c oxidase- Complex IV, Sub. IV | yes | ns |
| P10606 | Cytochrome c oxidase- Complex IV, Sub.Vb | yes | ns |
| Q04637 | eIF4G (S1108) | only total protein | − ** |
| P19419 | Elk-1 (S383) | no | ns |
| P29474 | eNOS (S1177) | no | − * |
| P29474 | eNOS/NOS III (S116) | no | ns |
| P28482, P27361 | ERK 1/2 (T202/Y204) | only total protein | − ** |
| Q05397 | FAK (Y397) | no | ns |
| Q05397 | FAK (Y576/577) | no | ns |
| P49840 | GSK-3αβ (Y279/Y216) | no | ns |
| P09601 | Heme-Oxygenase-1 | no | − ** |
| P04792 | HSP27 Protein 1 | yes | − * |
| P07900 | HSP90 | yes | |
| P35228 | i-NOS | no | ns |
| P35568 | IRS-1 (S612) | only total protein | − * |
| P13473 | LAMP-2 | yes | ns |
| Q9GZQ8 | LC3B | yes | ns |
| P00338 | LDHA | yes | − ** |
| Q6LEN1 | Mn Superoxide Dismutase | yes | + * |
| P42345 | mTOR (S2481) | no | − ** |
| Q16539 | p38 MAP Kinase | no | − * |
| Q16539 | p38 MAP Kinase (T180/Y182) | no | ns |
| Q9GZT9 | PHD-2/Egln1 | no | − ** |
| P19838 | NF-kappaB | no | |
| P19838 | NF-kappaB p65 (S536) | no | + ** |
| Q15418 | p90RSK (S380) | only total protein | ns |
| P16234 | PDGFRβ (Y716) | only total protein | + ** |
| P16234 | PDGFRβ (Y751) | only total protein | ns |
| O15530 | PDK1 (S241) | no | − * |
| P60484 | PTEN (S380) | no | − ** |
| Q16518 | RPE65 | no | + ** |
| P62753 | S6 Ribosomal Protein (S240/244) | only total protein | − * |
| P45983 | SAPK/JNK (T183/Y185) | only total protein | − * |
| P29353 | Shc (Y317) | no | + * |
| Q92835 | SHIP1 (Y1020) | no | ns |
| Q9NR28 | Smac/Diablo | no | − ** |
| P12931 | Src Family (Y416) | only total protein | − * |
| P40763 | Stat3 (S727) | no | ns |
| P42229, P51692 | Stat5 (Y694) | no | − ** |
| O15455 | TLR3 | no | + ** |
| Q9NR96 | TLR9 | no | + ** |
| P29597 | Tyk2 (Y1054/1055) | only total protein | ns |
| P35968 | VEGFR 2 (Y951) | no | + ** |
| P35968 | VEGFR 2 (Y996) | no | − * |
significantly different from cell lysates (Mann-Whitney test, N=18; p ≤ 0,05).
significantly different from cell lysates (Mann-Whitney test, N=18; p ≤ 0,001).
To compare protein levels in cells vs exosomes, we normalized data to a third protein, LC3B. This was found to be the most stably expressed protein (Chiechi et al, 2012; Andersen et al, 2004) if cells and exosomes were considered together. LC3B is intended as the total LC3B pool, and not as the lipidated form LC3B II, which is a marker for autophagy. Furthermore, conclusions drawn normalizing data to LC3B were consistent with data obtained with β-actin or cd63 normalization.
ARPE-19 exosomes contain a novel set of signaling phosphoproteins
RPMA analysis of exosomes and corresponding cell lysates was performed over 75 endpoints representing key disease related pathways associated with autophagy, oxidative stress, cell adhesion, angiogenesis, and pro-survival/apoptotic pathways (Supplementary Table 1).
RPMA analysis of exosomes revealed 72 proteins. We consulted Exocarta, a compendium for proteins, RNA molecules and lipids identified in exosomes (Mathivanan ans Simpson, 2009; Mathivanan et al, 2012), and compared the proteins already present in the database (4563 proteins in February, 2013) with those found in exosomes during this study. 13 of the proteins analyzed here were detected in exosomes for the first time, and 41 were specifically detected in their phosphorylated or cleaved form for the first time. Table 1 reports the complete list of the proteins detected in exosomes during this study, their previous detection, and their relative abundance in exosomes compared to cells.
An intriguing aspect of these results was the detection of proteins in their phosphorylated state; since phosphorylation reflects the activation state of signaling pathways, phosphoprotein content may be critical for defining a role of exosomes in intercellular signaling. Compared to control, the levels of certain proteins were altered in exosomes shed by stressed ARPE-19 cells. Among these were phosphorylated members of pathways regulating survival, proliferation, apoptosis and cell metabolism (Tab. 2).
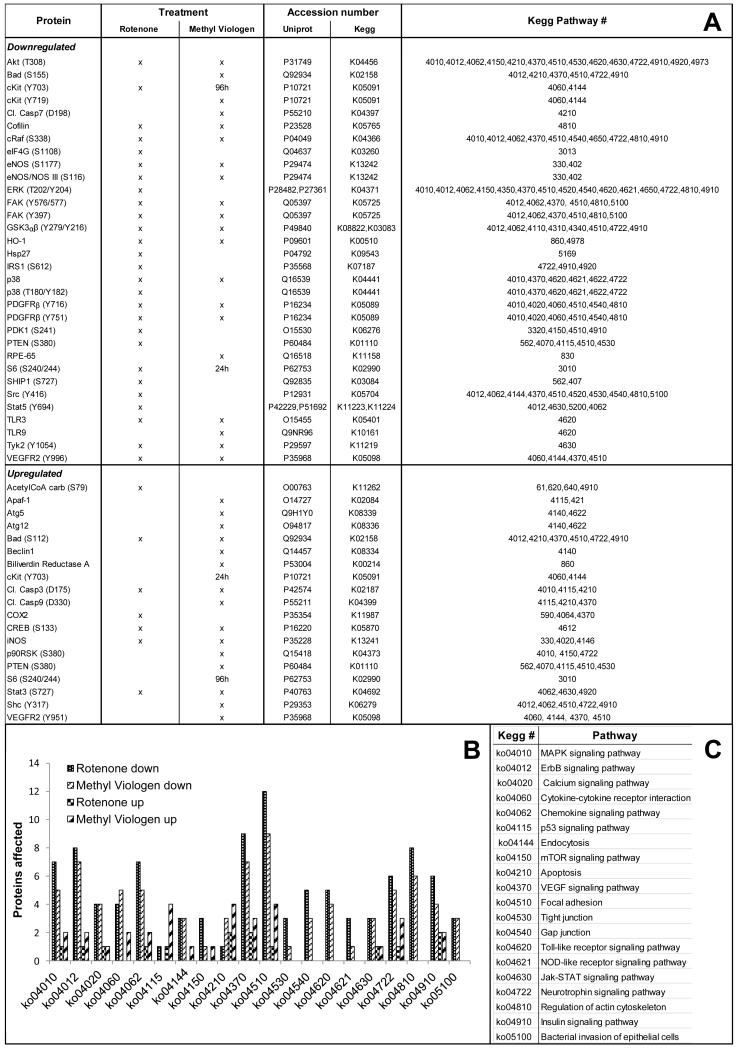
Tab. 2. Down- and upregulated proteins in ARPE-19 exosomes after Rotenone or Methyl Viologen treatment.
List of the down- and up-regulated signaling proteins in exosomes shed by treated ARPE-19 cells, compared to control (Mann-Whitney test, p ≤ 0.1).
| Protein | UniProt accession number |
Kegg accession number |
Kegg Pathway # | same change in corresponding cells |
|---|---|---|---|---|
| Downregulated | ||||
| Akt (T308) | P31749 | K04456 | 4010,4012,4062,4150,4210,4370,4510,4530,4620,4630,4722,4910,4920,4973 | x |
| Apaf-1 | O14727 | K02084 | 4115,421 | |
| Atg5 | Q9H1Y0 | K08339 | 4140,4622 | |
| Elk-1 (S383) | P19419 | K04375 | 4010,4012,4510,4910 | |
| ERK 1/2 (T202/Y204) | P28482, P27361 | K04371 | 4010,4012,4062,4150,4350,4370,4510,4520,4540,4620,4621,4650,4722,4810,4910 | x |
| p38 MAP Kinase (T180/Y182) | Q16539 | K04441 | 4010,4370,4620,4621, 4622,4722 | x |
| PDK1 (S241) | O15530 | K06276 | 3320,4150,4510,4910 | x |
| Src Family (Y416) | P12931 | K05704 | 4012,4062,4144,4370,4510,4520,4530,4540,4810,5100 | x |
| TLR9 | Q9NR96 | K10161 | 4620 | x |
| Upregulated | ||||
| Acetyl-CoA Carboxylase (S79) | O00763 | K11262 | 61,620,640,4910 | x |
| AMPKα1 (S485) | Q13131 | K07198 | 4140, 4150, 4910, 4920 | |
| Bak | Q16611 | K14021 | 4141 | |
| LDHA | P00338 | K03778 | 620 | |
| Smac/Diablo | Q9NR28 | K10522 | 4210 |
Oxidative stress modifies signaling protein content of ARPE-19 cells
Since oxidative stress plays a key role in AMD pathogenesis, and to take into account possible differences in cellular response to different agents (Weigel et al, 2002; Lu et al, 2006), ARPE-19 cells were treated with Methyl Viologen, a pro-oxidant that leads to superoxide radical generation (Suntres, 2002), or alternatively with Rotenone, a known inhibitor of mitochondrial Complex I. Inhibition of the respiratory chain leads to an increased production of mitochondrial ROS, with consequent mitochondrial DNA damage and further ROS generation due to defective respiratory chain (Koopman et al, 2010); moreover, an increase in defective mitochondria can cause overloading of lysosomes in RPE cells, stimulation of autophagy, and further ROS production (Wang et al, 2009).
RPMA analysis of cell lysates exposed to Rotenone or Methyl Viologen showed altered levels of many (phospho)proteins involved in signaling pathways regulating apoptosis (i.e., cleaved caspases 3 and 9; Bad (S112)), proliferation and survival (i.e., cRaf (S338), Akt (T308), CREB (S133)), angiogenesis (i.e., PDGFRβ (Y716) and (Y751), VEGFR (Y951)), adhesion (i.e., FAK (Y576/577) and (Y397)), metabolism (i.e., GSK3 αβ (Y279/Y216), AcetylCoA carboxylase (S79)), inflammation (i.e., COX2, Stat3 (S727)) (Fig. 3). RPMA analysis did not detect cleaved PARP (an hallmark of apoptosis; Oliver FJ et al, 1998) in all ARPE-19 cell samples, while it was present in quality control cell lysates printed at similar concentrations in the same slide.
Fig. 3. Down- and upregulated proteins in ARPE-19 cells after Rotenone or Methyl Viologen treatment.
Down- or up-regulated signaling proteins in treated ARPE-19 cells, compared to control (Mann-Whitney test, p ≤ 0.1). A. List of affected proteins; B. Proteins grouped in signaling pathways; C. Pathway description.
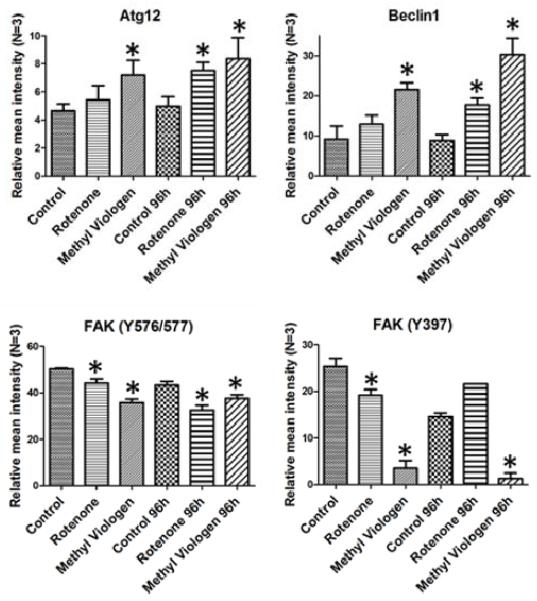
Some of the alterations observed in cell lysates exposed to oxidative stress reflect signaling perturbations observed during AMD (Fig. 4). For example, levels of autophagy associated Beclin-1 and Atg12 were increased; autophagy markers have been found in drusen of AMD patients, and stressed RPE cells have been reported to increase their autophagic activity (Wang et al, 2009).
Fig. 4. Proteins altered in stressed ARPE-19 cells.
Protein levels (Relative mean intensity ± SEM) in ARPE-19 cells treated with Rotenone or Methyl Viologen. *: significantly different from the corresponding control lysates (Mann-Whitney test, N=3, p ≤ 0,1).
Phosphorylated forms of FAK (FAK (Y576/577): Maa and Leu, 1998; FAK (Y397): Schlaepfer et al, 1999) were decreased. FAK is an important cell signaling mediator involved in cell migration, survival and cell cycle regulation; its phosphorylation was reported to be essential for RPE phagocytic activity, and it is decreased/compromised in age-related retinal diseases (Nandrot et al, 2004; Finnemann, 2003).
Discussion
It has been previously reported that RPE cells are able to secrete exosomes, and exosome markers have been found in drusen in eyes with AMD (Wang et al, 2009). However, to our knowledge this is the first extensive (phospho)proteomic characterization of RPE-secreted exosomes. A previous proteomic study was performed on RPE membrane blebs (Alcazar et al, 2009), but these are a different class of vesicles. Blebs are bigger than exosomes, originate directly from the plasma membrane and can be precipitated from conditioned medium through 15 min of a 100g centrifugation. Exosomes, on the other hand, have an endocytic origin, and can be precipitated only after 1 hour of a 100,000g ultracentrifugation of the medium (previously deprived of larger vesicles through sequential centrifugation steps at lower, increasing speeds).
RPMA technology is an optimal proteomic method for exosome analysis because it is quantitative, high throughput, multiplexed, high in sensitivity and can be applied to proteins from extremely small amounts of input material (Wilson et al, 2010; Mueller et al, 2010). RPMA is most often used for measuring post-translational modifications; the sensitivity of detection for the RPMAs is such that low-abundance phosphorylated protein isoforms can be measured from a spotted lysate representing fewer than 10 cell equivalents (VanMeter et al, 2008). Compared to Western Blot, ELISA, or traditional proteomic technologies such as two-dimensional gel electrophoresis, RPMA are thus particularly suitable for analysis of small sample amounts (i.e., biopsies or vitreous samples) and detection of low abundance post-translationally modified proteins in exosomes.
Exosomes have been extensively characterized for their protein, mRNA, miRNA and lipid content (see the ExoCarta Database) (Mathivanan and Simpson, 2009; Mathivanan et al, 2012); however, to our knowledge only 2 previous papers deal with post-translationally modified proteins included in these vesicles (Gonzales et al, 2009: urinary exosomes; Palmisano et al, 2012: beta cell-derived microparticles and exosomes). Moreover, the present study quantitatively evaluated a comprehensive list of exosome phosphoproteins that are members of disease relevant signal pathways.
Several proteins and phosphoproteins have been detected in exosomes for the first time in this study. Among the 72 proteins detected in exosomes, 13 were not previously reported to be present in exosomes, and 41 were specifically detected for the first time in their post-translationally modified form (phosphorylated or cleaved). Some of them were the same phosphorylated receptors that were found in the vitreous of patients: PDGFRβ (Y751) and (Y716), VEGFR2 (Y996), c-kit (Y703) (Davuluri et al, 2009).
The relative abundance of the proteins detected in exosomes was often different from that in the corresponding cells. Importantly, in addition to the exosome markers cd9 and cd63, ARPE-19 secreted exosomes contain high levels of activated signaling proteins, such as phosphorylated Akt (T308), VEGFR2 (Y951) or PDGFRβ (Y751). Another protein that is highly enriched in exosomes is TLR3; Toll Like Receptors (TLRs) are important for innate immunity, recognizing highly conserved molecular patterns that are common to broad pathogen classes (Kawai and Akira, 2010). Cellular localization of TLRs has important consequences for ligand accessibility and downstream signaling events (Barton and Kagan, 2009); TLR3, as well as other TLRs involved in recognition of nucleic acids, is tipically located in endolysosomes. The finding here that exosomes contain high levels of TLR3 suggests a new possible way of action for TLR3, and might also have physiological and therapeutical implications for AMD. TLR3 has in fact been reported to be involved in sequence- and target-independent angiogenesis suppression (Kleinman et al, 2008; Cho et al, 2009) and retinal degeneration (Kleinman et al, 2012) induced by siRNAs in a mouse model of choroidal neovascularization. Furthermore, human choroidal neovascular membranes from AMD patients were shown to express TLR3 exclusively in RPE cells (Maloney et al, 2009); in view of the differences in RPE staining for TLR3 among samples, a possible role for this receptor in neovascular AMD was hypothesized.
Exposure of cells to oxidative stress induced in exosomes a change in the levels of various phosphorylated signaling proteins. Sets of these proteins are linked together in pathways regulating cell proliferation and survival (i.e., PDK1 (S241), Akt (T308), Elk1 (S383), ERK1/2 (T202/Y204), Src family (Y416)), cell death (i.e., Smac/Diablo, Bak), and cell metabolism (i.e., AMPK α1 (S485), Acetyl-CoA Carboxylase (S79), LDHA).
Our data suggests that exosomes do not simply reflect the average protein composition of the cell. The specific recruitment of proteins into exosomes is a matter of big interest, but it is still only partially understood. Among the factors involved in the sorting of cargo are specific interactions with lipids and proteins on membrane sub-domains, and interactions with tetraspanins (i.e., cd9, cd63); also the ESCRT complex, implicated in MVBs formation, is involved in selective cargo sorting (Fevrier and Raposo, 2004; Pant et al, 2012).
ROS, in particular H2O2, act as intracellular messengers; they can have a proliferative effect (Groeger et al, 2009) or, on the other hand, they can induce apoptosis (Circu and Aw, 2010) and, at higher levels, necrosis (Vanlangenakker et al, 2008). The final effect depends on the cellular context and on the quantitative and temporal aspects of ROS production (e.g. Gough and Cotter, 2011). In our case, a decreased activation of prosurvival pathways and an increase of some pro-apoptotic proteins were detected both in stressed cells and exosomes; however, the prooxidant treatment was not strong enough to induce a decrease in cell viability.
Exosomes from treated cells were found to contain a set of proteins that reflect a metabolic cell response to redox stress; ROS cause AMPK activation, which in turn phosphorylates and inactivates AcetylCoA Carboxylase (Ha et al, 1994) and up-regulates glycolytic enzymes such as lactate dehydrogenase (LDHA) (Wu and Wei, 2012).
It is known that exosomes can transfer functional proteins, miRNAs and mRNAs to recipient cells (Pant et al, 2012). Our finding that exosomes contain a large set of proteins in their phosphorylated form suggests that these vesicles have the potential to alter and/or regulate signaling cascades in recipient cells. Furthermore, exosomes may represent the way through which non-secreted (phospho)proteins are shed by retinal cells into the vitreous; they may thus constitute the route through which potential biomarkers reach easy-to-access body fluids, constituting candidate analytes for early diagnosis and/or personalized therapy. Despite the presence of the overlying neural retina, we hypothesize that exosomes secreted by RPE cells have the potential to reach the vitreous in vivo, thanks to their small size. This hypothesis is supported by in vivo data with nanoparticles, developed for slow-release drug delivery and/or gene delivery, and similar in size to exosomes (about 100-150 nm in diameter). It was reported that, after intra-vitreous injection, nanoparticles are able to migrate through the retinal layers and reach the RPE cells (Bejjani et al 2005; Birch and Qi Liang 2007; Bourges et al 2003). A retrograde flow (from RPE cells to the vitreous) is thus plausible.
Conclusions
ARPE-19 cells secrete through exosomes a great variety of signaling proteins, and the exosome content is perturbed by exposing ARPE-19 cells to oxidative stress under conditions that do not cause cell death.
RPMA permits the highly specific quantitation of post-translationally modified proteins, even when present at low abundance. Since phosphorylation reflects the activation state of signaling pathways, characterization of the phosphoprotein content may be critical for defining the role of exosomes in intercellular signaling.
13 proteins, 38 phosphoproteins and 3 cleaved proteins were detected in exosomes for the first time during this study; interestingly, many of these were also detected in the vitreous of patients with AMD (Davuluri et al, 2009; Espina et al, unpublished results). This strengthens the idea that these small vesicles may represent the way by which cells in the retina secrete intracellular/non soluble/receptor proteins into the vitreous. Detection of exosomes markers (cd-9, cd-63) in the vitreous would confirm this hypothesis.
Supplementary Material
Highlights.
ARPE-19 cells secrete through exosomes a great variety of signaling proteins
Reverse Phase Protein Arrays technology was used to characterize exosome content
72 proteins were detected in exosomes, 41 in post-translationally modified form
Exosome content is perturbed by exposing ARPE-19 cells to oxidative stress
Phosphoprotein content may be critical for the signaling role of exosomes
Acknowledgements
This work was funded by the University of Padova, George Mason University and the grant 1R21EY018942-01 to LAL from NIH/NEI. We thank Prof. S. Garbisa and Dr. M. Zoratti for their support and interest and for useful discussions.
Abbreviations
- AFM
Atomic Force Microscopy
- AMD
Age-related Macular Degeneration
- ESCRT
Endosomal Sorting Complex Required for Transport
- FBS
Fetal Bovine Serum
- MVB
Multi Vesicular Bodies
- ROS
Reactive Oxygen Species
- RPE
Retinal Pigment Epithelium
- RPMA
Reverse Phase Protein Arrays
- TLR
Toll Like Receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed K, Xiang J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mol Med. 2011;15:1458–73. doi: 10.1111/j.1582-4934.2010.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar O, Hawkridge AM, Collier TS, Cousins SW, Bhattacharya SK, Muddiman DC, Marin-Castano ME. Proteomics characterization of cell membrane blebs in human retinal pigment epithelium cells. Mol Cell Proteomics. 2009;8:2201–11. doi: 10.1074/mcp.M900203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejjani RA, BenEzra D, Cohen H, Rieger J, Andrieu C, Jeanny JC, Gollomb G, Behar-Cohen FF. Nanoparticles for gene delivery to retinal pigment epithelial cells. Mol Vis. 2005;11:124–32. [PubMed] [Google Scholar]
- Belting M, Wittrup A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J Cell Biol. 2008;183:1187–91. doi: 10.1083/jcb.200810038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch DG, Liang FQ. Age-related macular degeneration: a target for nanotechnology derived medicines. Int J Nanomedicine. 2007;2:65–77. doi: 10.2147/nano.2007.2.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton M, Dayhaw-Barker P. The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye (Lond) 2001;15:384–9. doi: 10.1038/eye.2001.141. [DOI] [PubMed] [Google Scholar]
- Bourges JL, Gautier SE, Delie F, Bejjani RA, Jeanny JC, Gurny R, BenEzra D, Behar-Cohen FF. Ocular drug delivery targeting the retina and retinal pigment epithelium using polylactide nanoparticles. Invest Ophthalmol Vis Sci. 2003;44:3562–9. doi: 10.1167/iovs.02-1068. [DOI] [PubMed] [Google Scholar]
- Charboneau L, Tory H, Chen T, Winters M, Petricoin EF, Liotta L, Paweletz C. Utility of reverse phase protein arrays: applications to signalling pathways and human body arrays. Brief Funct Genomic Proteomic. 2002;1:305–15. doi: 10.1093/bfgp/1.3.305. [DOI] [PubMed] [Google Scholar]
- Chiechi A, Mueller C, Boehm KM, Romano A, Benassi MS, Picci P, Liotta LA, Espina V. Improved data normalization methods for reverse phase protein microarray analysis of complex biological samples. Biotechniques. 2012;0:1–7. doi: 10.2144/000113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WG, Albuquerque RJ, Kleinman ME, Tarallo V, Greco A, Nozaki M, Green MG, Baffi JZ, Ambati BK, De Falco M, Alexander JS, Brunetti A, De Falco S, Ambati J. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proc Natl Acad Sci U S A. 2009;106:7137–42. doi: 10.1073/pnas.0812317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–62. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G, Espina V, Petricoin Er, Ross M, Deng J, Liotta L, Glaser B. Activated VEGF receptor shed into the vitreous in eyes with wet AMD: a new class of biomarkers in the vitreous with potential for predicting the treatment timing and monitoring response. Arch Ophthalmol. 2009;127:613–21. doi: 10.1001/archophthalmol.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Patel M, Chan C. Molecular pathology of age-related macular degeneration. Prog Retin Eye Res. 2009;28:1–18. doi: 10.1016/j.preteyeres.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey C, Wu G, Ebenstein D, Garsd A, Weiter J. Cell loss in the aging retina. Relationship to lipofuscin accumulation and macular degeneration. Invest Ophthalmol Vis Sci. 1989;30:1691–9. [PubMed] [Google Scholar]
- Dunn K, Aotaki-Keen A, Putkey F, Hjelmeland L. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–69. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- Février B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–21. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Finnemann S. Focal adhesion kinase signaling promotes phagocytosis of integrin-bound photoreceptors. EMBO J. 2003;22:4143–54. doi: 10.1093/emboj/cdg416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrs K, Anderson D, Johnson L, Hageman G. Age-related macular degeneration-emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–71. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–79. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough DR, Cotter TG. Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2011;2:e213. doi: 10.1038/cddis.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger G, Quiney C, Cotter TG. Hydrogen peroxide as a cell-survival signaling molecule. Antioxid Redox Signal. 2009;11:2655–71. doi: 10.1089/ars.2009.2728. [DOI] [PubMed] [Google Scholar]
- Ha J, Daniel S, Broyles SS, Kim KH. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J Biol Chem. 1994;269:22162–8. [PubMed] [Google Scholar]
- Jager R, Mieler W, Miller J. Age-related macular degeneration. N Engl J Med. 2008;358:2606–17. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- Johnson E. Age-related macular degeneration and antioxidant vitamins: recent findings. Curr Opin Clin Nutr Metab Care. 2010;13:28–33. doi: 10.1097/MCO.0b013e32833308ff. [DOI] [PubMed] [Google Scholar]
- Kaarniranta K, Salminen A. Age-related macular degeneration: activation of innate immunity system via pattern recognition receptors. J Mol Med. 2009;87:117–23. doi: 10.1007/s00109-008-0418-z. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kevany B, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman ME, Kaneko H, Cho WG, Dridi S, Fowler BJ, Blandford AD, Albuquerque RJ, Hirano Y, Terasaki H, Kondo M, Fujita T, Ambati BK, Tarallo V, Gelfand BD, Bogdanovich S, Baffi JZ, Ambati J. Short-interfering RNAs induce retinal degeneration via TLR3 and IRF3. Mol Ther. 2012;20:101–8. doi: 10.1038/mt.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Karikó K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–7. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman W, Nijtmans L, Dieteren C, Roestenberg P, Valsecchi F, Smeitink J, Willems P. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal. 2010;12:1431–70. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- Lu L, Hackett SF, Mincey A, Lai H, Campochiaro PA. Effects of different types of oxidative stress in RPE cells. J Cell Physiol. 2006;206:119–25. doi: 10.1002/jcp.20439. [DOI] [PubMed] [Google Scholar]
- Maa MC, Leu TH. Vanadate-dependent FAK activation is accomplished by the sustained FAK Tyr-576/577 phosphorylation. Biochem Biophys Res Commun. 1998;251:344–9. doi: 10.1006/bbrc.1998.9464. [DOI] [PubMed] [Google Scholar]
- Maloney SC, Antecka E, Orellana ME, Fernandes BF, Odashiro AN, Eghtedari M, Burnier MN., Jr. Choroidal neovascular membranes express toll-like receptor 3. Ophthalmic Res. 2010;44:237–41. doi: 10.1159/000313989. [DOI] [PubMed] [Google Scholar]
- Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue):D1241–4. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Simpson R. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- Miceli M, Liles M, Newsome D. Evaluation of oxidative processes in human pigment epithelial cells associated with retinal outer segment phagocytosis. Exp Cell Res. 1994;214:242–9. doi: 10.1006/excr.1994.1254. [DOI] [PubMed] [Google Scholar]
- Mueller C, Liotta LA, Espina V. Reverse phase protein microarrays advance to use in clinical trials. Mol Oncol. 2010;4:461–81. doi: 10.1016/j.molonc.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot E, Kim Y, Brodie S, Huang X, Sheppard D, Finnemann S. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J Exp Med. 2004;200:1539–45. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Gugiu B, Renganathan K, Davies M, Gu X, Crabb J, Kim S, Rózanowska M, Bonilha V, Rayborn M, Salomon R, Sparrow J, Boulton M, Hollyfield J, Crabb J. Retinal pigment epithelium lipofuscin proteomics. Mol Cell Proteomics. 2008;7:1397–405. doi: 10.1074/mcp.M700525-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–9. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong D. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano G, Jensen SS, Le Bihan MC, Laine J, McGuire JN, Pociot F, Larsen MR. Characterization of membrane-shed micro-vesicles from cytokine-stimulated beta-cells using proteomics strategies. Mol Cell Proteomics. 2012;11:230–43. doi: 10.1074/mcp.M111.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–94. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker S. Oxidative stress and the ubiquitin proteolytic system in age-related macular degeneration. Adv Exp Med Biol. 2010;664:447–56. doi: 10.1007/978-1-4419-1399-9_51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S. Oxidative damage of retinal pigment epithelial cells and age-related macular degeneration. Drug Develop Res. 2007;68:213–225. [Google Scholar]
- Schlaepfer DD, Hauck CR, Sieg DJ. Signaling through focal adhesion kinase. Prog Biophys Mol Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- Sharma S, Rasool H, Palanisamy V, Mathisen C, Schmidt M, Wong D, Gimzewski J. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–6. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R, Jensen S, Lim J. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–99. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- Speer R, Wulfkuhle J, Espina V, Aurajo R, Edmiston K, Liotta L, Petricoin EF. Development of reverse phase protein microarrays for clinical applications and patient-tailored therapy. Cancer Genomics Proteomics. 2007;4:157–64. [PubMed] [Google Scholar]
- Suntres Z. Role of antioxidants in paraquat toxicity. Toxicology. 2002;180:65–77. doi: 10.1016/s0300-483x(02)00382-7. [DOI] [PubMed] [Google Scholar]
- Tamburro D, Facchiano F, Petricoin EF, Liotta LA, Zhou W. Mass spectrometry-based characterization of the vitreous phosphoproteome. Proteomics Clin Appl. 2010;4:839–46. doi: 10.1002/prca.201000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–18. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Tschopp J. Mitochondria: Sovereign of inflammation? Eur J Immunol. 2011;41:1196–202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Vanlangenakker N, Vanden Berghe T, Krysko DV, Festjens N, Vandenabeele P. Molecular mechanisms and pathophysiology of necrotic cell death. Curr Mol Med. 2008;8:207–20. doi: 10.2174/156652408784221306. [DOI] [PubMed] [Google Scholar]
- VanMeter A, Rodriguez A, Bowman E, Jen J, Harris C, Deng J, Calvert V, Silvestri A, Fredolini C, Chandhoke V, Petricoin EF, Liotta L, Espina V. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol Cell Proteomics. 2008;7:1902–24. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanMeter A, Signore M, Pierobon M, Espina V, Liotta L, Petricoin EF. Reverse-phase protein microarrays: application to biomarker discovery and translational medicine. Expert Rev Mol Diagn. 2007;7:625–33. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- Wang A, Lukas T, Yuan M, Du N, Tso M, Neufeld A. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel A, Handa J, Hjelmeland L. Microarray analysis of H2O2-, HNE-, or tBH-treated ARPE-19 cells. Free Radic Biol Med. 2002;33:1419–32. doi: 10.1016/s0891-5849(02)01082-1. [DOI] [PubMed] [Google Scholar]
- Wilson B, Liotta L, Petricoin EF. Monitoring proteins and protein networks using reverse phase protein arrays. Dis Markers. 2010;28:225–32. doi: 10.3233/DMA-2010-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G. Lipofuscin and macular degeneration. Nutr Rev. 2003;61:342–6. doi: 10.1301/nr.2003.oct.342-346. [DOI] [PubMed] [Google Scholar]
- Wu SB, Wei YH. AMPK-mediated increase of glycolysis as an adaptive response to oxidative stress in human cells: implication of the cell survival in mitochondrial diseases. Biochim Biophys Acta. 2012;1822:233–47. doi: 10.1016/j.bbadis.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Wu Y, Yanase E, Feng X, Siegel M, Sparrow J. Structural characterization of bisretinoid A2E photocleavage products and implications for age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7275–80. doi: 10.1073/pnas.0913112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Cringle S. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog Retin Eye Res. 2001;20:175–208. doi: 10.1016/s1350-9462(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Zarbin M. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.