Abstract
Peroxisome proliferator-activated receptor-γ (PPARγ) is a member of the nuclear receptor family of transcription factors with important regulatory roles in cellular growth, differentiation and apoptosis. Using proteomic analysis, we demonstrated expression of PPARγ protein in a series of 260 newly diagnosed primary AML samples. Forced expression of PPARγ enhanced the sensitivity of myeloid leukemic cells to PPARγ agonists CDDO- or 15d15DPGJ2-induced apoptosis, through preferential cleavage of caspase-8. No effects on cell cycle distribution or differentiation were noted, despite prominent induction of p21 in PPARγ-transfected cells. In turn, antagonizing PPARγ function by siRNA or pharmacological PPARγ inhibitor significantly diminished apoptosis induction by CDDO. Overexpression of co-activator protein DRIP205 resulted in enhanced differentiation induction by CDDO in AML cells through PPARγ activation. Studies with DRIP205 deletion constructs demonstrated that the NR boxes of DRIP205 are not required for this co-activation. In a Phase I clinical trial of CDDO (RTA-401) in leukemia, CDDO induced an increase in PPARγ mRNA expression in 6 of 9 patient samples; of those, induction of differentiation was documented in 4, and of p21 in 3 patients, all expressing DRIP205 protein. In summary, these findings suggest that cellular levels of PPARγ regulate induction of apoptosis via caspase-8 activation, while the co-activator DRIP205 is a determinant of induction of differentiation, in response to PPARγ agonists in leukemic cells.
Keywords: PPARgamma, DRIP205, AML, CDDO, differentiation, apoptosis
Introduction
Peroxisome-proliferator-activated receptors (PPARs) are ligand-activated transcription factors that are members of the nuclear-hormone-receptor gene superfamily. To date, three PPAR subtypes have been identified, namely PPARα, PPARβ/δ and PPARγ (also termed as NR1C1, NR1C2 and NR1C3 (1). PPARγ is an important regulator of lipid and glucose homeostasis, of cellular differentiation and inflammation. There are three PPARγ isoforms, which differ at their 5-prime ends, each under the control of its own promoter (2). Most tissues express the PPARγ1 isoform, whereas the PPARγ2 isoform is specific to adipocytes. However, the receptor is expressed in many others tissues and cell types such as monocytes and macrophages, skeletal muscle, breast, prostate, colon, and type 2 alveolar pneumocytes.
PPARs form heterodimers with retinoid × receptors (RXRs). Transcriptional regulation by nuclear receptors (NRs), including PPARs and RXRs, involves the binding and recruitment of coactivators and/or mediators to target gene promoters. One component of the TRAP–Mediator complex, TRAP220 (the Thyroid Hormone Receptor Associated Protein, also known as ARC/DRIP205), is directly associated with the thyroid receptor (TR), Vitamin D receptor (VDR), and PPAR and facilitates NR-mediated transcription. Recent studies have further demonstrated a functional role of TRAP220 in the optimal VDR- and retinoic acid receptor (RAR)-mediated myelomonocytic differentiation processes in hematopoietic cells (3). It was also found to act as a pivotal co-activator for GATA-1 in erythroid development (4) and has been shown to play a role in differentiation and proliferation of keratinocytes in response to VDR stimulation (5, 6).
PPARγ ligands have been extensively investigated and are known to inhibit proliferation, induce differentiation or apoptosis in different cancer cell types including hematologic malignancies (7). Our studies have shown that PPARγ ligands alone or in combination with RXR-specific activators can inhibit clonal proliferation and induce differentiation of HL-60, U937, and THP1 human myeloid leukemic cell lines (8). 2-Cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), a synthetic triterpenoid, is a ligand for PPARγ (9) that induces growth arrest, apoptosis and/or differentiation of a variety of tumor cell types. We have shown that CDDO induces Bcl-2 downregulation, mitochondrial depolarization, and caspase activation in myeloid leukemic cells (10). It also potently enhances apoptosis induced by tumor necrosis factor in myeloid leukemia cells (11) and refractory CLL B cells (12). In breast cancer, CDDO-induced growth inhibition correlates with PPARγ transactivation and is mediated at least in part by up-regulation of p21cip1waf1 and down-regulation of cyclin D1 (13, 14). Notably, CDDO and its derivatives also induce differentiation of leukemic cells (15-17), the effects which in some cell types are synergistically enhanced by concomitant ligation of the RXR nuclear receptor. The multiple effects of CDDOs have been attributed to both, PPARγ-dependent and -independent mechanisms of action (18). One of the most powerful anti-inflammatory and anti-carcinogenic activities induced by CDDO and related compounds is activation of Nrf2/ARE signaling pathway, a major regulator of anti-oxidant cellular responses (for review, see Liby K, et al (19)).
In this study, we investigated the role of PPARγ and its co-activator DRIP205 on the apoptosis- and differentiation-inducing properties of CDDO in leukemic cells. The results demonstrate that the PPARγ ligands CDDO and 15d15DPGJ2 induce higher degrees of apoptosis in leukemic cells modified to overexpress PPARγ. Furthermore, we provide evidence that overexpression and recruitment of the co-activator DRIP205 contributes to the enhanced differentiation by this agent. Finally, we report induction of myeloid differentiation of PPARγ/DRIP205 co-expressing leukemic blasts from patients with AML treated with CDDO (RTA-401) in a first-in-man clinical trial.
Materials and Methods
Reagents and antibodies
CDDO was manufactured under the RAID Program and kindly provided by Drs. E. Sausville (NCI) and M. Sporn (Dartmouth Medical College). 15-deoxy-Δ12,14-15DPGJ2 (15d15DPGJ2) was purchased from Cayman Chemical Company (Ann Arbor, MI). N-(4′aminopyridyl)-2-chloro-5-nitrobenzamide (T007) (20) a selective PPARγ antagonist, was synthesized as described previously (21). Annexin V FITC was purchased from Roche Diagnostic Co. (Indianapolis, IN), and CD14 FITC, CD11b PE, and CD34 PE from BD Biosciences (San Jose, CA). Rabbit polyclonal Caspase-3 antibody (CPP32, 1:1000; PharMingen); mouse monoclonal anti-caspase-8 (1:1000) and rabbit polyclonal anti-caspase-9 (1:1000) were purchased from Cell Signaling Technology (Beverly, MA); rabbit polyclonal anti-TRAP220; mouse monoclonal anti-PPARγ (1:500) , mouse monoclonal poly( (adenosinediphosphate [ADP]-ribose) polymerase (anti-PARP) antibodies (1:200) and mouse monoclonal anti-PCNA (1:500) were from Santa Cruz Biotechnology (Santa Cruz, CA); mouse monoclonal anti-p21 antibody (1:1000) was from Calbiochem (La Jolla, CA); mouse monoclonal anti-p27 (1:1000), mouse monoclonal anti-HO-1(1:1000) and mouse monoclonal anti-Bip/GRP78(1:1000) antibodies were from BD Biosciences (San Jose, CA); mouse monoclonal anti-Flag and mouse monoclonal anti-β-actin (AC74) from Sigma Chemical Co (St. Louis, MO); and goat anti-mouse and goat anti-rabbit horseradish peroxidase-conjugated secondary antibodies from Bio-Rad (Hercules, CA).
Cell lines and primary AML samples
HL-60, U937, MCF7 and SW480 were purchased from The American Type Culture Collection (ATCC, Rockville, MD). Bone marrow or peripheral blood samples from 260 AML or high-risk myelodysplastic syndrome (MDS) samples were assessed for PPARγ expression by Reverse Phase Protein Array (RPPA) (Supplemental methods and (22)). Samples were collected for the Leukemia Sample Bank at the University of Texas M.D. Anderson Cancer Center between 1/15/1998 and 3/9/2006, on Institutional Review Board (IRB) approved protocol Lab01-473 and consent was obtained according to the Declaration of Helsinki. Samples were analyzed under an IRB approved laboratory protocol (Lab05-0654).
Flow cytometric analysis of apoptosis and cell cycle
Early apoptotic events were detected by the flow cytometric measurement of externalized phosphatidylserine wit h t he Annexin-V-FLUOS Staining Kit from Roche Diagnostics (Indianapolis, IN). Cell cycle analysis was conducted using propidium iodide (PI) as described (23).
DNA fragmentation assay
Cells were washed twice with phosphate-buffered saline (PBS) and resuspended in lysis buffer (10mM Tris-HCl, 1 mM EDTA [ethylenediaminetetraacetic acid], 0.025% Triton X-100, pH 8.0) for 10 to 20 minutes on ice. Lysates were digested with RNaseA (50μ g/ml) and then proteinase K (2 mg/mL) for 1 hour each at 37°C. DNA samples were separated on 1.8% agarose gel containing ethidium bromide and visualized by UV light.
Differentiation assay
The differentiation of myeloid leukemic cells was determined by their ability to produce superoxide, as measured by the reduction of nitroblue tetrazolium (NBT) as described (24). The analysis of surface differentiation-specific cell surface antigens was measured by flow cytometry using the phycoerythrin-conjugated anti-CD11b and FITC-conjugated anti-CD14 monoclonal antibodies from Becton Dickinson (San Jose, CA).
Transient Transfection and Luciferase Activity Assay
Plasmids—SV40 β-galactosidase was obtained from Promega Corp., Madison, MI; pM vector was purchased from Clontech (Palo Alto, CA); and pMD, pMDm5, pMDm6, pMDm7, pMDm7Δ, and plasmids were generated by PCR (25). FLAG-tagged wt-PPARg construct was kindly provided by Dr. K. Chatterjee (Department of Medicine, University of Cambridge, Cambridge, United Kingdom). pcDNA3-DRIP205 expression plasmid was kindly provided by Dr. Leonard P. Freedman (Merck Research Laboratories, West Point, PA).
For transient transfection assays, SW480 cells were co-transfected with DNA using the FuGENE. In coactivation experiments, cells were co-transfected with 250 ng of SV40 β-galactosidase (used as an internal control), 1 μ g of the reporter construct containing three copies of the acyl-CoA oxidase PPRE cloned upstream of the TK-LUC reporter, PPREx3-LUC reporter (kindly provided by Dr. Ronald M. Evans ,The Salk Institute), and various amounts of DRIP205 or deletion mutant constructs. After 24h, transfected cells were treated with DMSO or CDDO 0.75μM for another 48-72h. Relative luciferase activity was calculated by dividing luciferase activity by β-galactosidase activity for each well.
Western blot analysis
Cells were lysed at a density of 1 × 106/50 μL in protein lysis buffer (0.25 M Tris-HCl, 2% sodium dodecylsulfate, 4% β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue) supplemented with a protease inhibitor cocktail (Roche Diagnostic Co.). For preparation of nuclear lysates, cells were washed in cold PBS and digitonin-permeabilized for 5 min on ice at a density of 20 million cells/ml in extraction buffer (250mM sucrose, 70mM KCL: 137mM NaCl: 4.3mM Na2HPO4: 1.4mM KH2PO4 (pH 7.2), 200 μ g/ml digitonin and protease inhibitors). Cells were centrifuged at 1000g for 5 min at 4°C, and pellet containing nuclear fraction was lysed as above. Cell lysates were then loaded onto a 10-12% SDS-PAGE gel (Bio-Rad). After electrophoresis, proteins were transferred to Hybond-P membranes (Amersham Pharmacia Biotech, Buckinghamshire, England), followed by immunoblotting. Signals were detected using a PhosphorImager (Storm 860, version 4.0; Molecular Dynamics, Sunnyvale, CA).
Clinical trial of CDDO in patients with AML
Patients with refractory/relapsed AML were treated with CDDO (RTA-401, Reata Pharmaceuticals , Irving, Tx) from 0.6 to 75 mg/m2/hr per hour × 5 days, in a phase 1 clinical trial, following informed consent according to the University of Texas M. D. Anderson Cancer Center and Princess Margaret Hospital - Ontario Cancer Institute guidelines (Suppl. Table 1). Bone marrow or peripheral blood samples mononuclear cells were separated by Ficoll-Hypaque (Sigma Chemicals) density-gradient centrifugation. Apoptosis was examined by simultaneous staining with CD34 APC (BD) and TMRM [tetramethylrhodamine methyl ester (Invitrogen) to measure mitochondrial inner transmembrane potential (ΔΨm). Cell differentiation was examined by simultaneous staining with CD34 APC (BD), CD33 PE-Cy7 (BD), CD14 FITC (BD), and CD11b PE (BD) at the indicated time points.
Quantitative real-time PCR
Bone marrow and peripheral blood samples obtained from AML patients were lysed with RNA Stat 60 (Tel-Test, Friendswood, TX). The RNA was subjected to purification by an RNeasy ion-exchange column (Qiagen, Valencia, CA) with oncolumn DNAse treatment. cDNA was prepared from 1.0 μg of total RNA per 20 μL mix containing 0.07μg/μL random-sequence hexamer primers, 0.5 mM dNTPs, 5 mM DTT, 0.2 u/μL SuperAsin RNAse inhibitor (Ambion, Austin, TX), and 10 u/μL SuperScript III (Invitrogen). Real time PCR was carried out using an ABI Prism 7700 instrument as described (26). For primer and probe sets to detect PPARγ, p21, NQO1 and housekeeping gene ABL1, we used TaqMan Gene Expression Assays Hs00234592_m1, Hs00355782_m1, Hs00168547_m1 and Hs00245445_m1, respectively. The abundance of each transcript of interest relative to that of ABL-1 was calculated as follows: relative expression (RE) =100 × 2 exp [−ΔCt], where ΔCt is the mean Ct of the transcript of interest less the mean Ct of the transcript for ABL.
Transfection of PPARγ small interfering RNA
Cells were transfected by the Amaxa electroporator Nucleofector I from Amaxa Biosystems (Gaithersburg, MD), using the Nucleofector Kit C (program W-001). Small interfering RNA (siRNA) PPARγ transfection was performed using validated Stealth RNAi VHS 40941 for duplex 1(siRNA1), VHS 40944 for duplex 2(siRNA2) (Invitrogen). Cells (106) were resuspended in 100μl of C cell nucleofector solution and transfected by electroporation with scramble LO GC Duplex Stealth RNAi Negative Control (12 935-200; Invitrogen) or with PPARγ siRNA. Forty eight hours after transfection, DMSO or CDDO was added to the cells for 24h, and protein expression was monitored by immunoblotting.
Results
PPARγ protein expression in AML
We have previously demonstrated that PPARγ protein is expressed in both myeloid and lymphoid leukemic cell lines (8). Expression of PPARγ protein was analyzed in 260 newly diagnosed leukemia-enriched AML/MDS samples. PPARγ protein was variably expressed in different AML subtypes (Suppl. Fig. 1). There was no difference in overall survival among patients expressing high or low levels PPARγ (not shown).
CDDO induces PPARγ activation in AML cells
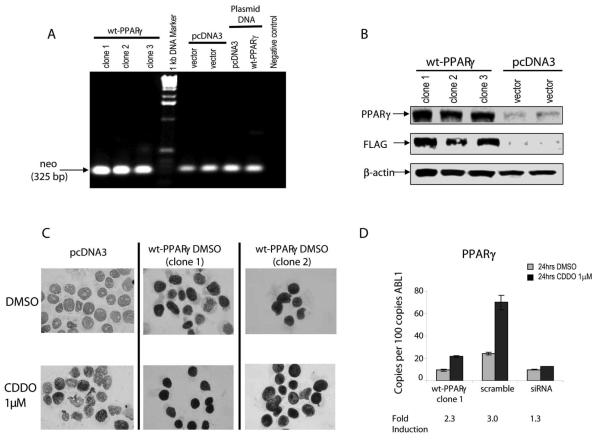
We next characterized the relationship between the PPARγ expression and the ability of pharmacological PPARγ ligands to affect growth and survival of leukemic cells. To this end, we established stable transfectants of U937 cells expressing empty vector (pcDNA3) or Flag-tagged wt-PPARγ (Fig. 1A-C) Overexpression of PPARγ facilitated growth of cells but the differences did not reach statistical significance (p=0.1, Suppl. Fig. 2A). Exposure of cells to the PPARγ ligand CDDO further enhanced PPARγ protein expression in both, vector control and transfected cells (Fig. 1C). As shown in Fig. 1D, CDDO induced PPARγ mRNA expression in wt-PPARγ-overexpressing cells or in the same cells transfected with scrambled siRNA (2.3- and 3-fold, respectively), and this induction was largely abrogated in PPARγ-siRNA-transfected cells. This data suggest that PPARγ ligation with CDDO induces expression of its cognate receptor in PPARγ-dependent fashion.
Fig. 1.
A. RT-PCR of neomycin gene expression in pcDNA3 or wt-PPARγ transfected clones. B. PPARγ and FLAG protein expression in transfected cells was detected by immunoblotting. C. Leukemic cells transfected with pcDNA3 or wt-PPARγ were treated with 1 μM CDDO for 5 hours, and PPARγ protein levels were examined by immunocytochemistry. D. PPARγ transcript was measured in wt-PPARγ transfected cells (clone 1) or in the same cells transfected with scramble or PPARγ siRNA, by real-time PCR. Results are reported as the mean number of transcripts per hundred transcripts of ABL1. Error bars denote the standard error of the mean.
Relationship between PPARγ expression and growth-inhibitory responses to PPARγ agonists
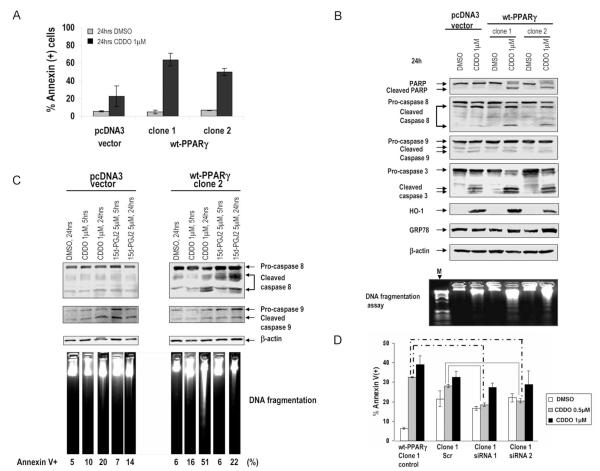
Next, we examined the functional consequences of PPARγ induction and overexpression in response to the receptor activation by CDDO or by the structurally different PPARγ agonist 15dPGJ2. Vector control (pcDNA3) or wt-PPARγ transfected U937 cells were treated with 1μ M CDDO for 24 hrs. Overexpression of PPARγ significantly enhanced the sensitivity of leukemic cells to apoptosis induced by CDDO or 15dPGJ2 (Fig. 2A and Suppl. Fig. 2B). We next analyzed modulation of apoptosis and cell cycle-regulating proteins in PPARγ-transfected clones. Following treatment with CDDO for 24 hours, cells overexpressing wt-PPARγ exhibited significantly decreased pro-caspase 3 and increased cleavage of caspase 3 and its substrate PARP (Fig. 2B). In accordance with these results, CDDO induced a higher degree of endonucleolytic DNA cleavage in PPARγ-overexpressing cells. Analysis of the upstream (initiator) caspases demonstrated appearance of the active (p18) fragment of caspase-8 in these cells, while caspase-9 was similarly cleaved in both, vector control and PPARγ-overexpressing cells (Fig. 2B). In a time-course experiment no caspase-8 cleavage and DNA fragmentation was observed in vector-treated cells, while both ligands induced caspase-8 cleavage and DNA fragmentation at 5 and 24hrs in PPARγ-overexpressing cells (Fig. 2C). In contrast, caspase-9 cleavage did not substantially differ between control and PPARγ-transfected cells. Activation of caspase-8 may proceed through CHOP-dependent transcriptional upregulation of DR5/TRAIL-R2 triggered by ER stress and unfolded protein response (UPR) (27), and CDDO slightly increased the level of ER stress marker protein Bip/GRP78 in PPARγ-overexpressing cells (Fig. 2B). Knocking down exogenously transfected PPARγ by two different siRNA constructs abolished apoptosis induced by the lower CDDO (0.5μM) but did not significantly affect it at higher (1μM) concentrations (Fig. 2D), indicating the contribution of PPARγ-dependent and - independent mechanisms of cell death. Likewise, the pharmacological PPARγ inhibitor T007 partially protected PPARγ-overexpressing, but not control cells, from CDDO-induced apoptosis (Suppl. Fig. 2C). CDDO potently induced expression of stress responsive inducible enzyme hemeoxygenase-1 (HO-1) (Fig. 2B) and of one of the critical Nrf2 target genes NADPH quinine oxidoreductase-1 (NQO-1) (Suppl. Fig. 3A) in both, control and PPARγ-overexpressing cells.
Fig. 2.
A. U937( pcDNA3) and U937 (pcDNA3-wt-PPARγ) cells were cultured in the presence of CDDO or vehicle for 24h, and apoptosis induction was analyzed by Annexin V flow cytometry at 24 hours. B. U937( pcDNA3) and U937 (pcDNA3-wt-PPARγ) cells were grown in the presence of 1μM CDDO or vehicle for 24h. Expression of PARP, caspase-3,-8, -9, HO-1 and GRP78 was analyzed by immunoblotting, and effects of CDDO on DNA fragmentation were determined. C. Caspase-8, and -9 processing and DNA fragmentation at 5 hours and 24 hours was measured. D. U937-wt-PPARγ cells were transiently transfected with scramble or PPARγ siRNA (siRNA1 and siRNA2), at a final concentration of 67nM. Seventy-two hours after transfection, cells were exposed to indicated concentrations of CDDO for 24 hrs. Induction of apoptosis was measured by annexinV flow cytometry. The data represent average results from three independent experiments. Solid lines, paired t-test comparing apoptosis in scrambled siRNA-transfected cells with apoptosis in PPARγ siRNA transfectants (p<0.01); dashed lines, paired t-test comparing apoptosis in untransfected cells with PPARγ siRNA transfectants (p<0.001).
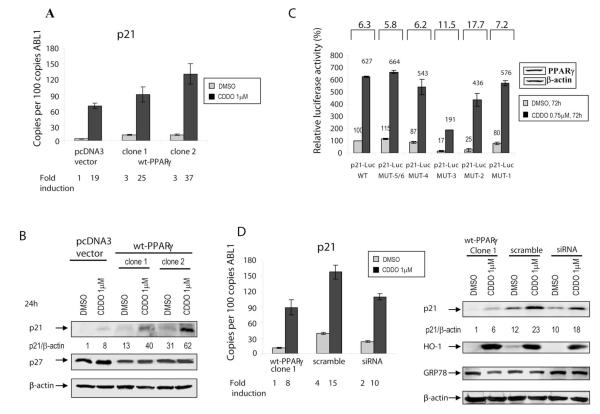
PPARγ agonists are known to induce expression of cell cycle inhibitory protein p21Waf1/CIP1, and we have previously shown that CDDO induced p21 mRNA and protein in breast cancer cells (14). Consistent with these findings, CDDO induced p21 mRNA in leukemic cells (Fig. 3A). This increase in p21 transcription was evident in both, control cells and in cells overexpressing PPARγ (Fig. 3A, B). The expression of p27KIP1 was unchanged, and no significant differences in cell cycle distribution were noted between PPARγ-overexpressing compared to control cells (Suppl. Fig. 2D). Although p21 promoter contains a potential conserved consensus PPARγ response element (PPRE), CDDO may also increase transcription of p21 indirectly, via increased binding of Sp1, Sp3, and Sp4 transcription factors to the GC-rich regions of the p21 promoter. To determine the mechanism of transcriptional activation of p21 by CDDO, we conducted promoter assays by transient transfection of SW480 cells expressing endogenous PPARγ (insert Fig. 3C) with full-length p21 luciferase promoter plasmid and plasmids containing point mutations in GC-rich elements 1-6 (Mut1-Mut6) of the proximal p21 promoter (28). CDDO induced an approximately 6-fold increase in p21-wt promoter activity (Fig. 3C, p21-Luc WT). Interestingly, CDDO was capable of transactivating all constructs containing point mutations. To determine if CDDO induces p21 expression via PPARγ, we measured p21 levels following depletion of PPARγ by siRNA. As demonstrated in Fig. 3D, inhibition of PPARγ using siRNA failed to block CDDO-induced levels of p21 mRNA or protein expression. This data indicate that CDDO causes induction of p21 levels in PPARγ-independent fashion. Consistent with documented properties of triterpenoids and other electrophilic compounds to activate the Keap1/Nrf2 antioxidant pathway, CDDO robustly induced HO-1 (Fig. 3D) and NQO1 (Suppl. Fig. 3B) in cells with functional or silenced PPARγ, indicating that these responses are likewise PPARγ-independent. No appreciable change in Bip/GRP78 expression was noted in control or transfected cells (Fig. 3D), indicating no significant contribution of ER stress to pro-apoptotic effects of CDDO in this cell system.
Fig. 3.
A. U937( pcDNA3) and U937 (pcDNA3-wt-PPARγ) cells were cultured in the presence of 1μM CDDO or vehicle for 24h, and p21 transcript was measured by real-time PCR. B. Cells were harvested at 24h, and p21/p27 levels were determined by immunoblotting. C. The fold transactivation of the p21 promoter by CDDO (0.75μM, 72 hrs). SW480 colon cancer cells were co-transfected with the wt- or mutant −2325/+8 p21 promoter-luciferase reporter vectors. Cells were co-transfected with β-galactosidase for normalization of the transfection variability. The fold transactivation of each p21 reporter construct is shown on top of the graph. D. Analysis of p21 levels in wt-PPARγ clone 1 electroporated with scrambled or PPARγ siRNA, 48h after transfection. Following 48 hrs, DMSO or CDDO were added for the next 24 hrs, and after 24hrs p21, HO-1, GRP78 and β-actin levels were determined by quantitative TaqMan PCR and Western blot analyses.
PPARγ ligands enhance DRIP205 co-activator binding to PPARγ
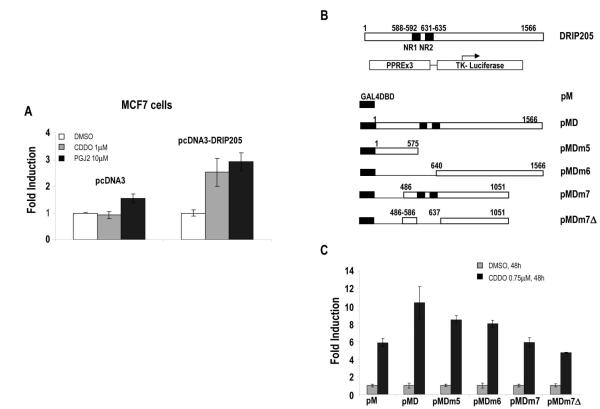
PPARγ ligands recruit the co-activator DRIP205 to PPARγ Furthermore (29) nuclear factor coactivators are known to mediate tissue-specific effects. To ascertain whether DRIP205 overexpression will affect transcriptional activity of PPARγ, the ability of CDDO and 15dPGJ2 to induce (PPRE)3-tk-luc reporter was examined in MCF-7 cells transfected with full-length DRIP205 plasmid. Transient co-transfection with (PPRE)3-tk-luc reporter and full-length pcDNA3-DRIP205 plasmid resulted in higher levels of PPARγ transactivation induced by 15dPGJ2 and CDDO (1.9 and 2.7-fold, respectively, Fig. 4A).
Fig. 4.
A. MCF-7 cells were transiently co-transfected with 1μg of pPPRE-TK-LUC and 4μg of pcDNA3 or pcDNA3-DRIP205, treated with indicated concentrations of CDDO or 15dPGJ2 for 92.5 hours, after which PPARγ transactivation was determined by relative luciferase activity calculated by dividing luciferase activity by protein concentration for each well. Results are shown as mean ± SEM for three separate experiments. B. Map of GAL4-DRIP fusion proteins. C. Coactivation of PPARγ by GAL4-DRIP fusion proteins. SW480 cells were co-transfected with 1000 ng of pPPRE3-LUC, 250 ng of β-galactosidase, 500 ng of pM empty or pM DRIP deletion mutants, treated with DMSO and 0.75μM CDDO, and luciferase activity was determined. Results are shown as mean ± SEM for three separate experiments.
Previous studies have shown that the NR boxes in DRIP205/TRAP220 contribute to the physical and functional interactions of these co-activators with NRs (30;31). Their role in co-activation of PPARγ was further investigated in SW-480 co-transfected with (PPRE)3-tk-luc reporter and N- or C-terminal GAL4-DRIP205 deletion constructs (Fig. 4B). The full-length DRIP205 expression plasmid encodes for 1566 amino acids, which is identical to amino acids 16–1581 of the TRAP220 coding sequence. CDDO significantly induced activity in cells transfected with (PPRE)3-tk-luc and GAL4-DRIP205 (wild-type). In addition, significant coactivation, albeit to a lesser degree, was observed for several GAL4-DRIP205 chimeras (pMDm5, pMDm7, pMDm7 , and pMDm6), two of which express N-terminal (pMDm5) or C-terminal (pMDm6) regions of DRIP205 but do not contain the central NR box sequences (Fig. 4C). These results confirm that the NR boxes of DRIP205 may contribute to but are not required for co-activation, and indicate that multiple domains of DRIP205 are involved in interactions with PPARγ.
DRIP205 contributes to myelomonocytic differentiation of leukemic cells in response to CDDO
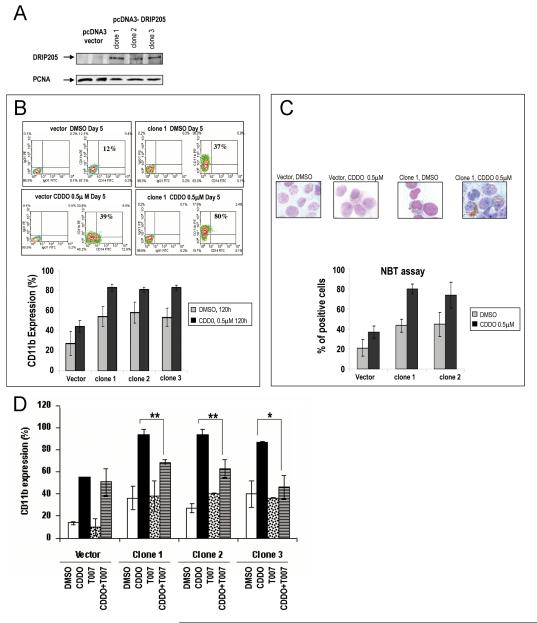
To determine the functional role of DRIP205 in the context of PPARγ ligation in leukemic cells, we investigated effects of CDDO in HL-60 cells stably transfected with DRIP205 plasmid (three separate clones, Fig. 5A and Suppl. Fig. 4A). No difference in cell growth (Suppl. Fig. 4B), cell cycle progression (Suppl. Fig. 4C) or apoptosis (Suppl. Fig. 4D) was found in cells overexpressing DRIP205, cultured alone or exposed to CDDO. In contrast, CDDO induced a higher degree of myelo-monocytic differentiation in DRIP205-overexpressing cells as shown by increased expression of CD11b (p<0.01, Fig. 5B) and by NBT assay (36.7±6.1 vs 79.7±4.7%, p< 0.001, Fig. 5C).
Fig. 5.
A. Nuclear fractions of HL-60 cells tranfected with vector (pcDNA3) or pcDNA3-DRIP205 constructs were lysed and analyzed by western blot analysis. PCNA was used as a loading control for nuclear lysates. B, C. HL-60 cells tranfected with vector (pcDNA3) or pcDNA3-DRIP205 constructs were treated with 0.5 μ M CDDO for 120 hours, and induction of myelomonocytic differentiation was determined by CD11b flow cytometry (%CD11b(+) cells, (B)) or NBT assay (C). Data represent average results from three independent experiments. D. HL-60 cells tranfected with vector (pcDNA3) or pcDNA3-DRIP205 constructs were pretreated with 4 μ M T007, a PPARγ antagonist, for 1 hour, followed by 0.75 μ M CDDO for 120 hours. Data represent average results (mean±SD) of three independent experiments. Induction of differentiation was determined by CD11b flow cytometry. ** denotes p=0.01, and *p=0.02.
To determine if the enhanced differentiation was mediated through PPARγ, we assessed CD11b expression in cells pre-treated with pharmacological antagonist PPARγ T007. As shown in Fig. 5D, blocking PPARγ transactivation significantly diminished CDDO-induced myelomonocytic differentiation in DRIP205-overexpressing but not in vector-transduced control cells.
CDDO induces expression of markers of differentiation and apoptosis in leukemic blasts of patients treated in Phase I clinical trial
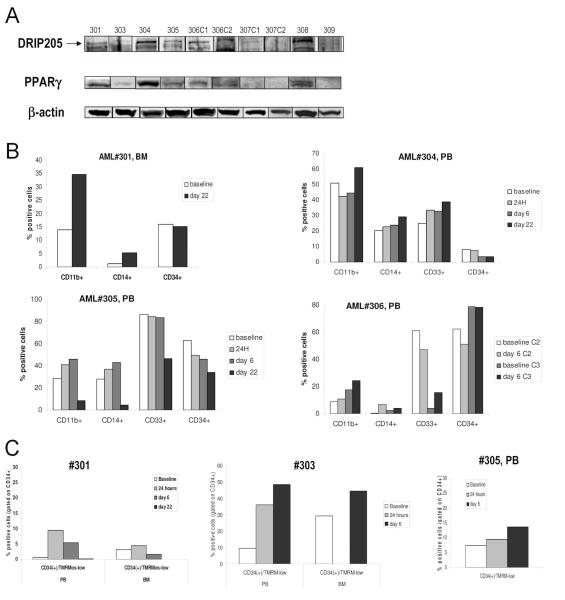
In a “first in man” clinical Phase I trial of with increasing doses of CDDO (RTA-401, escalated from 0.6 mg/m2/hr to 75 mg/m2/hr) in patients with relapsed/refractory AML, we investigated the in vivo differentiating and pro-apoptotic effects of CDDO and correlated these changes with PPARγ and DRIP205 expression, in cells from 9 patients by quantitative TaqMan PCR (PPARγ) and immunoblotting (PPARγ, DRIP205). Clinical characteristics of the patients are summarized in Suppl. Table 1. PPARγ mRNA was expressed in all samples at baseline albeit at different levels. PPARγ and DRIP205 proteins were expressed in samples from 7 out of 9 patients studied; no expression of either protein was detected in samples from patients #307 and #309 (Fig. 6A). Following 6 days of continuous CDDO administration, PPARγ mRNA was induced >2-fold in 4 patient samples (Suppl. Table 2). In 4 of the 9 patients, an increase in CD11b+ and CD14+ cells and a concomitant reduction of immature cells expressing CD34 or CD33 was observed (#301, 304, 305 and 306, Fig. 6B). Examples of flow cytometric profiles are shown in the Suppl. Fig. 5. Baseline expression of PPARγ protein was highest in samples from patients #301 and #304 (Fig. 6A), and in all four patients increase of PPARγ mRNA was demonstrated (1.5, 2.4, 1.8 and 2.2-fold, respectively, Suppl. Table 2). In these, p21 mRNA was induced >2-fold in samples #304, 305 and 306. No change in differentiation markers was observed in patients #307 and #309 with no detectable baseline PPARγ or DRIP205 proteins. Moderate induction of apoptosis documented as loss of mitochondrial membrane potential in circulating CD34+ cells was observed in samples from 3 patients (#301, #303 and #305); in sample#303 corresponding apoptosis induction was seen in Day 6 bone marrow CD34+ cells (Fig. 6C). Examples of flow cytometric profiles are shown in the Suppl. Fig. 6. Clinically, patients did not fulfill protocol response criteria, differential counts did not change significantly and MTD was not reached, at the low dose levels in this Phase I study.
Fig. 6.
A. Peripheral blood (PB) or bone marrow (BM) samples from patients enrolled in the Phase 1 clinical trial were lysed and probed with DRIP205 and PPARγ by Western blot. β-actin was used as a loading control. In baseline sample from patient#2, not enough material was available for immunblotting. B. Patients were treated with CDDO (RTA-401) during a Phase I clinical trial, and cells were collected from the peripheral blood (PB) or bone marrow (BM) and assessed for expression of surface markers CD11b, CD14, CD33 and CD34 by flow cytometry at the indicated times (see also suppl. Table 2). Four of nine patients (patients #301, 304, 305, 306) showed alterations of these parameters during the observed period. PB baseline percentages are not available from patient #301, therefore, BM percentages are provided. C. Cells from the PB or BM were counterstained with CD34-APC and TMRM. Three of five patients (patients #301, 303, 305) showed alterations of these parameters during the observed period. Data are presented as percentage of CD34+ cells that have lost mitochondrial membrane potential (TMRM-low). BM baseline percentages are not available from patient #305.
Discussion
PPARγ ligands inhibit cancer cell proliferation, induce apoptosis and/or differentiation in multiple tumor types, and these effects have been attributed to both, PPARγ-dependent and - independent mechanisms. In this study, we evaluated the role of PPARγ and one of its cellular co-activators, DRIP205, in the pro-apoptotic and differentiating properties of PPARγ agonists CDDO and 15dPGJ2. A high-throughput RPPA technique demonstrated high levels of PPARγ expression in 260 primary AML samples. To functionally characterize the relationship between baseline PPARγ levels and cellular effects of PPARγ agonists in leukemic cells, we generated stably transfected myeloid leukemic cells overexpressing the receptor. U937 cells induced to overexpress wt-PPARγ were more sensitive to the pro-apoptotic effects of PPARγ ligands CDDO and 15dPGJ2 compared to vector-transduced cells. These pro-apoptotic effects were significantly inhibited by silencing PPARγ with siRNA or by blocking PPARγ activation with the pharmacological antagonist T007, consistent with previously published findings of PPARγ-dependent and -independent mechanisms of action of this class of agents. Time-course analysis demonstrated that high PPARγ levels facilitated cleavage of caspase-8 and -3 (but not of caspase-9) which resulted in accelerated PARP cleavage, DNA fragmentation and apoptosis. Of note, several reports indicated the ability of CDDOs to activate the extrinsic apoptotic pathway and sensitize to TRAIL, via diverse molecular mechanisms including FLIP downregulation (32), JNK-mediated induction of TRAIL receptor expression (33) and inhibition of NFκ-B-dependent anti-apoptotic proteins (11). Conversely, data reported by us and others demonstrate that CDDO and its more potent derivative CDDO-Me promoted the release of cytochrome c from isolated mitochondria, suggesting that CDDOs directly target the mitochondria to trigger the intrinsic pathway of cell death (34, 35). Data presented here suggest a proximal role for caspase-8 downstream of ligand-activated PPARγ, whereas direct mitochondrial effects of CDDO observed at higher concentrations are likely PPARγ-independent, possibly by modifying the mitochondrial proteins through nucleophilic attack and Michaels addition (36). Unlike in non-small lung cancer cells (27). CDDO did not induce significant ER stress response, hence making the upregulation of DR5 unlikely mechanism of caspase-8 activation. The exact mechanistic link between PPARγ transactivation and activation of the extrinsic apoptotic pathway in AML remains to be determined.
It has recently been demonstrated that synthetic triterpenoids are potent activators of Nrf2/ARE signaling in a variety of cell types, resulting in marked induction of a variety of anti-oxidative genes and detoxifying enzymes (37-39). In our studies, CDDO promptly upregulated expression of HO-1 and NQO1 in leukemia cells engineered to overexpress or silence PPARγ. These observations are consistent with the notion that these responses are likely mediated by chemical structure of CDDO and other electrophilic compounds capable to modify cysteine residues on KEAP1 protein (37), and represent important PPARγ-independent activities of this class of compounds.
PPARγ agonists including CDDO modulate cell cycle progression in multiple tumor types (13, 14). Our present data show that CDDO induced expression of p21waf1/CIP protein in leukemic cells. This induction was observed in parental cells, in cells overexpressing PPARγ, or in cells transfected with PPARγ siRNA. These findings indicate that the ability of CDDO to activate p21 promoter is likely mediated via PPARγ-independent mechanisms. Since induction of p21 expression is frequently mediated via increased binding of Sp1, Sp3, and Sp4 transcription factors to the GC-rich regions of the p21 promoter, we utilized the constructs containing point mutations in the GC elements. In contrast to our previous study in pancreatic cells (40) we were unable to identify the specific site required for PPARγ-dependent activation of p21. Of note, p21 is regulated by many different pathways and transcription factors and CDDO could conceivably mediate its effects on p21 expression through an alternate pathway. Surprisingly, induction of p21 protein expression did not translate into a discernible cell cycle arrest in leukemic cells. The observation that CDDO preferentially induces apoptosis rather than cell cycle arrest in AML cells attests to the cell-type dependent properties of these agents, likely related to the distinct mitochondrial architecture of leukemic cells compared to solid tumor cells. Whether functional consequences of p21 overexpression other than control of cell cycle, such as regulation of apoptosis, differentiation or transcriptional activation (41) are operational in leukemic cells, remains to be investigated. Notably, p21 mRNA induction was observed in samples from 3 of the 9 patients treated with very low doses of CDDO (RTA-401) in a Phase I clinical trial.
Emerging evidence suggests the critical importance of the cellular context, in particular the composition of tissue-specific co-activators and co-repressors, in the biological responses to nuclear receptor agonists. PPARγ is known to interact with both the p160/SRC-1 family of coactivators and the multisubunit DRIP/Mediator co-activator complex. Our results show that CDDO induced significant activity in cells transfected with (PPRE)3-tk-luc and full-length DRIP205. In addition, significant coactivation was observed utilizing several N- and C-terminal domain mutants of DRIP205, and this coactivation did not require the NR boxes (Fig. 4B). These results suggest that multiple domains of DRIP205 are involved in interactions with PPARγ, similar to findings reported for ERα coactivation by DRIP205 (25). Interestingly, recent structural and functional analyses indicate that a direct interaction of PPARγ with DRIP/Mediator complex through the NR motifs of DRIP205 is not required for PPARγ-stimulated adipogenesis (42).
DRIP205 is involved in the Vitamin D-triggered regulation of gene transcription during keratinocyte differentiation (6), and overexpression of DRIP205 was observed in some cancer cell lines (43). CDDOs have shown to induce differentiation in myeloid leukemia cells (16, 17, 44) and in this study CDDO induced a higher degree of myelo-monocytic differentiation in DRIP205-overexpressing HL-60 cells, a process mediated through PPARγ. While we recently reported that one of the mechanisms of differentiation induction by CDDO involves modulation of CEBPα expression and function (45), our data shown here provide first evidence that high cellular levels of the co-activator DRIP205 can enhance the differentiation induced by PPARγ ligation and is therefore an important determinant of tissue-specific effects of PPARγ agonists. We here report that leukemic blasts from patients treated in a phase I clinical trial of CDDO (RTA-401) express DRIP205 in 7/9 samples, all of which expressed PPARγ mRNA and protein. Further, sequential studies demonstrated increased expression of the differentiation markers CD11b and/or CD14 in 4 patients. In these patients, CDDO induced PPARγ transcription. Even though the numbers are too small to draw definitive conclusions, the data suggest that CDDO activates PPARγ in a subset of patients with AML in vivo, whose cells express DRIP205. We did not observe correlation between PPARγ levels and apoptosis induction, possibly due to very low levels of CDDO in this Phase I study. Alternatively, this finding may indicate that PPARγ-dependent functions of CDDO may manifest primarily through differentiation induction rather than apoptotic responses in primary AML cells. Recently, the RXR agonist Bexarotene was shown to induce differentiation in non-APL patients with AML who were treated with this agent in a Phase I trial (46). Taking into consideration multiple studies that demonstrate that addition of RXR ligands synergistically enhance the differentiating and growth-suppressive effects of PPARγ ligands (8, 47), the combined use of these agents appears worth testing in the therapy of AML. The ongoing efforts by the Nuclear Receptor Signaling Atlas (NURSA) consortium to profile co-activator/co-repressors in primary AML may assist in identifying patients who are likely to benefit from PPARγ/RXR ligation strategies.
Supplementary Material
Acknowledgments
Grant support: Leukemia and Lymphoma Society grant R64149-07 (MK)
Leukemia Spore (NCI: (1 P50 CA100632-01) (MA)
Leukemia and Lymphoma Society grant 6089 and NIH PO1 grant CA-55164 (SMK)
Cancer Center Support Grant P30 CA016672 34 (KRC)
References
- 1.Nuclear Receptors Nomenclature Committee A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–3. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 2.Fajas L, Auboeuf D, Raspe E, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–89. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 3.Urahama N, Ito M, Sada A, et al. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells. 2005;10:1127–37. doi: 10.1111/j.1365-2443.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 4.Stumpf M, Waskow C, Krotschel M, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A. 2006;103:18504–9. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawker NP, Pennypacker SD, Chang SM, Bikle DD. Regulation of human epidermal keratinocyte differentiation by the vitamin D receptor and its coactivators DRIP205, SRC2, and SRC3. J Invest Dermatol. 2007;127:874–80. doi: 10.1038/sj.jid.5700624. [DOI] [PubMed] [Google Scholar]
- 6.Oda Y, Ishikawa MH, Hawker NP, Yun QC, Bikle DD. Differential role of two VDR coactivators, DRIP205 and SRC-3, in keratinocyte proliferation and differentiation. J Steroid Biochem Mol Biol. 2007;103:776–80. doi: 10.1016/j.jsbmb.2006.12.069. [DOI] [PubMed] [Google Scholar]
- 7.Konopleva M, Andreeff M. Role of peroxisome proliferator-activated receptor-gamma in hematologic malignancies. Curr Opin Hematol. 2002;9:294–302. doi: 10.1097/00062752-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Konopleva M, Elstner E, McQueen TJ, et al. Peroxisome proliferator-activated receptor gamma and retinoid × receptor ligands are potent inducers of differentiation and apoptosis in leukemias. Mol Cancer Ther. 2004;3:1249–62. [PubMed] [Google Scholar]
- 9.Wang Y, Porter WW, Suh N, et al. A synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), is a ligand for the peroxisome proliferator-activated receptor gamma. Mol Endocrinol. 2000;14:1550–6. doi: 10.1210/mend.14.10.0545. [DOI] [PubMed] [Google Scholar]
- 10.Konopleva M, Tsao T, Estrov Z, et al. The synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces caspase-dependent and -independent apoptosis in acute myelogenous leukemia. Cancer Res. 2004;64:7927–35. doi: 10.1158/0008-5472.CAN-03-2402. [DOI] [PubMed] [Google Scholar]
- 11.Stadheim TA, Suh N, Ganju N, Sporn MB, Eastman A. The novel triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) potently enhances apoptosis induced by tumor necrosis factor in human leukemia cells. J Biol Chem. 2002;277:16448–55. doi: 10.1074/jbc.M108974200. [DOI] [PubMed] [Google Scholar]
- 12.Pedersen IM, Kitada S, Schimmer A, et al. The triterpenoid CDDO induces apoptosis in refractory CLL B cells. Blood. 2002;100:2965–72. doi: 10.1182/blood-2002-04-1174. [DOI] [PubMed] [Google Scholar]
- 13.Konopleva M, Zhang W, Shi YX, et al. Synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces growth arrest in HER2-overexpressing breast cancer cells. Mol Cancer Ther. 2006;5:317–28. doi: 10.1158/1535-7163.MCT-05-0350. [DOI] [PubMed] [Google Scholar]
- 14.Lapillonne H, Konopleva M, Tsao T, et al. Activation of peroxisome proliferator-activated receptor gamma by a novel synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces growth arrest and apoptosis in breast cancer cells. Cancer Res. 2003;63:5926–39. [PubMed] [Google Scholar]
- 15.Ji Y, Lee HJ, Goodman C, et al. The synthetic triterpenoid CDDO-imidazolide induces monocytic differentiation by activating the Smad and ERK signaling pathways in HL60 leukemia cells. Mol Cancer Ther. 2006;5:1452–8. doi: 10.1158/1535-7163.MCT-06-0136. [DOI] [PubMed] [Google Scholar]
- 16.Konopleva M, Tsao T, Ruvolo P, et al. Novel triterpenoid CDDO-Me is a potent inducer of apoptosis and differentation in acute myelogenous leukemia. Blood. 2002;99:326–35. doi: 10.1182/blood.v99.1.326. [DOI] [PubMed] [Google Scholar]
- 17.Suh N, Wang Y, Honda T, et al. A novel synthetic oleanane triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, with potent differentiating, antiproliferative, and anti-inflammatory activity. Cancer Res. 1999;59:336–41. [PubMed] [Google Scholar]
- 18.Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I, Safe S. 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor gamma-dependent and -independent pathways. Mol Pharmacol. 2005;68:119–28. doi: 10.1124/mol.105.011437. [DOI] [PubMed] [Google Scholar]
- 19.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7:357–69. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 20.Lee G, Elwood F, McNally J, et al. T0070907, a selective ligand for peroxisome proliferator-activated receptor gamma, functions as an antagonist of biochemical and cellular activities. J Biol Chem. 2002;277:19649–57. doi: 10.1074/jbc.M200743200. [DOI] [PubMed] [Google Scholar]
- 21.Qin C, Morrow D, Stewart J, et al. A new class of peroxisome proliferator-activated receptor gamma (PPARgamma) agonists that inhibit growth of breast cancer cells: 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2004;3:247–60. [PubMed] [Google Scholar]
- 22.Tibes R, Qiu Y, Lu Y, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 23.Milella M, Kornblau SM, Estrov Z, et al. Therapeutic targeting of the MEK/MAPK signal transduction module in acute myeloid leukemia. J Clin Invest. 2001;108:851–9. doi: 10.1172/JCI12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabe Y, Konopleva M, Kondo Y, et al. PPAR gamma-active triterpenoid CDDO enhances ATRA-induced differentiation in APL. Cancer Biology & Therapy. 2007;6:1967–77. doi: 10.4161/cbt.6.12.4982. [DOI] [PubMed] [Google Scholar]
- 25.Wu Q, Burghardt R, Safe S. Vitamin D-interacting protein 205 (DRIP205) coactivation of estrogen receptor alpha (ERalpha) involves multiple domains of both proteins. J Biol Chem. 2004;279:53602–12. doi: 10.1074/jbc.M409778200. [DOI] [PubMed] [Google Scholar]
- 26.Mullican SE, Zhang S, Konopleva M, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nature Medicine. 2007;13:730–5. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 27.Zou W, Yue P, Khuri FR, Sun SY. Coupling of endoplasmic reticulum stress to CDDO-Me-induced up-regulation of death receptor 5 via a CHOP-dependent mechanism involving JNK activation. Cancer Res. 2008;68:7484–92. doi: 10.1158/0008-5472.CAN-08-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–84. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 29.Yang W, Rachez C, Freedman LP. Discrete roles for peroxisome proliferator-activated receptor gamma and retinoid × receptor in recruiting nuclear receptor coactivators. Mol Cell Biol. 2000;20:8008–17. doi: 10.1128/mcb.20.21.8008-8017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burakov D, Wong CW, Rachez C, Cheskis BJ, Freedman LP. Functional interactions between the estrogen receptor and DRIP205, a subunit of the heteromeric DRIP coactivator complex. J Biol Chem. 2000;275:20928–34. doi: 10.1074/jbc.M002013200. [DOI] [PubMed] [Google Scholar]
- 31.Rachez C, Gamble M, Chang CP, Atkins GB, Lazar MA, Freedman LP. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–26. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suh WS, Kim YS, Schimmer AD, et al. Synthetic triterpenoids activate a pathway for apoptosis in AML cells involving downregulation of FLIP and sensitization to TRAIL. Leukemia. 2003;17:2122–9. doi: 10.1038/sj.leu.2403112. [DOI] [PubMed] [Google Scholar]
- 33.Zou W, Liu X, Yue P, et al. c-Jun NH2-terminal kinase-mediated up-regulation of death receptor 5 contributes to induction of apoptosis by the novel synthetic triterpenoid methyl-2-cyano-3,12-dioxooleana-1, 9-dien-28-oate in human lung cancer cells. Cancer Res. 2004;64:7570–8. doi: 10.1158/0008-5472.CAN-04-1238. [DOI] [PubMed] [Google Scholar]
- 34.Samudio I, Konopleva M, Hail N, Jr., et al. 2-Cyano-3,12-dioxooleana-1,9-dien-28-imidazolide (CDDO-Im) directly targets mitochondrial glutathione to induce apoptosis in pancreatic cancer. J Biol Chem. 2005;280:36273–82. doi: 10.1074/jbc.M507518200. [DOI] [PubMed] [Google Scholar]
- 35.Samudio I, Konopleva M, Pelicano H, et al. A novel mechanism of action of methyl-2-cyano-3,12 dioxoolean-1,9 diene-28-oate: direct permeabilization of the inner mitochondrial membrane to inhibit electron transport and induce apoptosis. Mol Pharmacol. 2006;69:1182–93. doi: 10.1124/mol.105.018051. [DOI] [PubMed] [Google Scholar]
- 36.Brookes PS, Morse K, Ray D, et al. The triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid and its derivatives elicit human lymphoid cell apoptosis through a novel pathway involving the unregulated mitochondrial permeability transition pore. Cancer Res. 2007;67:1793–802. doi: 10.1158/0008-5472.CAN-06-2678. [DOI] [PubMed] [Google Scholar]
- 37.Dinkova-Kostova AT, Liby KT, Stephenson KK, et al. Extremely potent triterpenoid inducers of the phase 2 response:correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci U S A. 2005;102:4584–9. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yates MS, Kwak MK, Egner PA, et al. Potent protection against aflatoxin-induced tumorigenesis through induction ofNrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–94. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, Calingasan NY, Thomas B, et al. Neuroprotective effects of the triterpenoid, CDDO methyl amide, a potent inducer of Nrf2-mediated transcription. PLoS One. 2009;4:e5757. doi: 10.1371/journal.pone.0005757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong J, Samudio I, Liu S, Abdelrahim M, Safe S. Peroxisome proliferator-activated receptor gamma-dependent activation of p21 in Panc-28 pancreatic cancer cells involves Sp1 and Sp4 proteins. Endocrinology. 2004;145:5774–85. doi: 10.1210/en.2004-0686. [DOI] [PubMed] [Google Scholar]
- 41.Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 2003;13:65–70. doi: 10.1016/s0962-8924(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 42.Ge K, Cho YW, Guo H, et al. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–91. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Y, Qi C, Jain S, et al. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc Natl Acad Sci U S A. 1999;96:10848–53. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito Y, Pandey P, Place A, et al. The novel triterpenoid 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid induces apoptosis of human myeloid leukemia cells by a caspase-8-dependent mechanism. Cell Growth Differ. 2000;11:261–7. [PubMed] [Google Scholar]
- 45.Koschmieder S, D’Alo F, Radomska H, et al. CDDO induces granulocytic differentiation of myeloid leukemic blasts through translational up-regulation of p42 CCAAT enhancer binding protein alpha. Blood. 2007;110:3695–705. doi: 10.1182/blood-2006-11-058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai DE, Luger SM, Kemner A, et al. Evidence of myeloid differentiation in non-M3 acute myeloid leukemia treated with the retinoid × receptor agonist bexarotene. Cancer Biol Ther. 2007;6:18–21. doi: 10.4161/cbt.6.1.3619. [DOI] [PubMed] [Google Scholar]
- 47.Sporn MB, Suh N. Chemoprevention: an essential approach to controlling cancer. Nat Rev Cancer. 2002;2:537–43. doi: 10.1038/nrc844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.