Abstract
Multiple cellular, molecular, and biochemical changes contribute to the etiology and treatment outcome of contusion spinal cord injury (SCI). Dysregulation of microRNAs (miRNAs) has been found following SCI in recent studies. However, little is known about the functional significance of the unique role of miRNAs in SCI. We analyzed the miRNA expression patterns 1 and 3 days following rat SCI using miRNA microarray. Microarray data revealed that nine miRNAs were upregulated and five miRNAs were downregulated 1 day post-injury, and that three miRNAs were upregulated and five miRNAs were downregulated 3 days post-injury, in the sites of contused when compared with sham rat spinal cords. Because miR-21 was one of the miRNAs being most significantly upregulated, we investigated its function. Knockdown of miR-21 by antagomir-21 led to attenuated recovery in hindlimb motor function, increased lesion size, and decreased tissue sparing in rats. Compared with the negative control group, treatment with antagomir-21 significantly increased apoptosis following SCI. Pro-apoptosis genes Fas ligand (FasL), phosphatase and tensin homolog (PTEN), and programmed cell death protein 4 (PDCD4) were proved to be direct targets of miR-21 in many diseases and cell types. In vivo treatment with antagomir-21 increased the expression of FasL and PTEN, but did not affect PDCD4. These results suggested that miR-21 played an important role in limiting secondary cell death following SCI, and that the protective effects of miR-21 might have been the result of its regulation on pro-apoptotic genes. Thus, miR-21 may play an important role in the pathophysiology of SCI.
Key words: antagomir-21, apoptosis, miRNA, SCI
Introduction
Spinal cord injury (SCI) is one the most common and devastating injuries observed in spine and neurosurgery departments. It is usually caused by motor vehicle accidents, sports injuries, diving accidents, and violence. It can cause permanent disabilities such as paralysis and loss of movement or sensation. The treatment of SCI remains one of the greatest challenges for the basic science and clinical investigators. Although many therapies have been explored, all demonstrated limited efficacy thus far.1
The celluar mechanisms associated with SCI are complex and involve a multitude of signaling pathways and molecular dysfunctions.2,3 MicroRNAs (miRNAs or miRs) are endogenous, non-coding, single-stranded RNAs consisting of ∼22 nucleotides. They can regulate hundreds of gene expression via RNA-induced silencing complexes (RISC), targeting them to mRNAs where they inhibit translation or direct destructive cleavage.4 It was recently reported that miRNAs are estimated to regulate 60% of all genes in the human genome.5 As a consequence, they may widely influence the signaling networks leading to pathological responses after SCI. Therefore, miRs could become attractive novel therapeutic targets for the treatment of SCI.
Increasing evidence demonstrates that a large number of miRNAs are expressed in the central nervous system (CNS).6,7 Some miRNAs are involved in several neurological disorders including traumatic CNS injuries and neurodegenerative diseases.8–14 Interestingly, some studies have demonstrated that miR-21 was upregulated following several types of CNS injuries, such as traumatic brain injury and brain ischemia.12–15 Additionally, some studies have shown the involvement of miRs in the pathogenesis of SCI in a rat contusion SCI model,9–11 with miR-21 emerging as one of the most dysregulated miRs. Buller et al. observed that miR-21 overexpression protects neurons from ischemic neuronal death by targeting the tumor necrosis factor-α (TNF-α) family member Fas ligand gene (FasL), an important cell death-inducing ligand.15 In addition, some researchers found that miR-21 protects other organs following injury.16,17 However, the effects of miR-21 on SCI are still unknown.
In this study, we performed miRNA arrays to detect the expression patterns of miRs in a rat moderate contusion SCI model. We observed that miR-21 was one of the most dysregulated miRs after SCI. First of all, we demonstrated that knockdown of miR-21 by antagomir were associated with exacerbated functional deficit, less preservation of spinal cord tissue, and increased apoptotic cell. As direct targets of miR-21, proapoptotic genes, FasL, PTEN, and PDCD4 have been widely investigated in many diseases and cell types. Knockdown of miR-21 in vivo following SCI in rats, increased the expression of FasL and PTEN in the spinal cord but did not alter the expression of PDCD4. Therefore, our findings have implicated that miR-21 play an important role in the pathogenesis of SCI.
Methods
Animals
Adult male Sprague–Dawley (SD) rats, weighing 180–220 g, were provided by the Center of Experimental Animals, Central South University. All animal care, breeding, and testing procedures were approved by the Laboratory Animal Users Committee at Xiangya Hospital, Central South University, Changsha, China. All animals were housed in individual cages in a temperature- and light cycle-controlled environment with free access to food and water.
Establishment of contusion SCI model
Rats were anesthetized with an intraperitoneal injection of 10% chloral hydrate (3 mg/kg). A laminectomy was performed at thoracic vertebra level 10 (T10). Moderate contusion injury was induced using a modified Allen's weight drop apparatus (8 g weight at a vertical height of 40 mm, 8 g×40 mm) on the exposed dura of the spinal cord. Sham injured animals were only subjected to laminectomy.
Experimental design
Experiment 1: MiRNA microarray analyses and confirming the microarray data
The miRNA expression patterns 1 and 3 days following rat SCI were analyzed using miRNA microarray. Rats were randomly divided into four groups: sham-operated 1 day and 3 days, SCI 1 day and 3 days, n=3 per group. Then, quantitative real-time polymerase chain reaction (qRT-PCR) was performed to confirm the microarray data. Rats were randomly divided into four groups: sham-operated 1 day and 3 days, SCI 1 day and 3 days, n=5 per group. For miRNA microarray analysis, the sample size was 3; while for qRT-PCR, the sample size was 5.
Experiment 2: antagomir-21 experiments
The SCI rats were randomly divided into two groups, a negative control group and an anatgomir-21 group. In the negative control group (n=28), rats were subjected to SCI and treated intrathecally with negative control antagomir (1μL/h, 20 nmol/mL). In the antagomir-21 group (n=28), rats were subjected to SCI and treated intrathecally with antagomir-21 (1 μl/h, 20 nmol/mL) for 3 days. At the scheduled time points, rats in both groups were euthanized with an overdose of 10% chloral hydrate (10 mg/kg). Subsequently, the spinal cord was exposed, and a 10 mm long segment of the spinal cord centered at the injury epicenter was harvested. The time of euthanasia was determined according to the different parameters measured: motor function score (Basso, Beattie, and Bresnahan [BBB]) and lesion identification by cresyl violet staining at 28 days after SCI; Western blot analysis for FasL, PDCD4, and PTEN at 1 and 3 days after SCI; qRT-PCR analysis of miR-21 at 1 and 3 days after SCI; and immunohistochemistry staining, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, and in situ hybridization at 1 and 3 days after SCI (Table 1). The sample size selected for each group was estimated based on our pilot data in order to meet the power analysis requirement.
Table 1.
Antagomir-21 Experiments Design (Different Parameters Measured)
|
1 day |
3 days |
7 days |
14 days |
21 days |
28 days |
|---|---|---|---|---|---|
|
BBB (n=4) | |||||
| LI | |||||
| WB (FasL, PTEN and PDCD4) (n=4) | WB (FasL, PDCD4 and PTEN) (n=4) | ||||
| qRT-PCR (miR-21) (n=4) | qRT-PCR (miR-21) (n=4) | ||||
| ICH (PTEN & PDCD4), TUNEL & ISH (miR-21) (n=4) | ICH (PTEN & PDCD4), TUNEL & ISH (miR-21) (n=4) | ||||
Basso, Beattie, and Bresnahan score (BBB) for measuring hindlimb locomotor ability was repeat measured at 1, 3, 7, 14, 21, and 28 days post-injury. After the final behavioral tests, which occurred 28 days after surgery, the animals were terminally euthanized for the measure of the lesion identification.
LI: Lesion identification by Cresyl violet staining for assessing lesion size and spared tissue following behavioral analyses.
WB: Western blot analysis for quantifying the protein level of FasL, PTEN, and PDCD4.
qRT-PCR: Quantitative real-time polymerase chain reaction (miR-21) for determining the inhibitory effect of antagomir-21 on miR-21 in the spinal cord.
ICH: Immunohistochemistry staining for analyzing the expression of PTEN and PDCD4.
TUNEL: terminal deoxynucleotidyl transferase dUTP nick end labeling staining for detecting apoptosis cells
ISH: In situ hybridization for determining the inhibitory effect of antagomir-21 on miR-21 in the spinal cord.
MiRNA microarray analysis
Rats were anesthetized 1 or 3 days after surgery, and a 10 mm long segment of spinal cord, including the injury epicenter, was harvested and fresh-frozen in liquid nitrogen. Total RNA was isolated using TRIzol (Invitrogen, CA) and miRNeasy mini kit (Qiagen, West Sussex, UK) according to manufacturer's instructions. This efficiently recovered all RNA species, including miRNAs. RNA quality and quantity was measured using a nanodrop spectrophotometer (ND-1000, Nanodrop Technologies) and RNA integrity was determined by gel electrophoresis.
After RNA isolation from the samples, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) was used according to the manufacturer's guideline for miRNA labeling. One microgram of each sample was 3'-end-labeled with Hy3™ fluorescent label, using T4 RNA ligase according to the following procedure: RNA in 2.0 μL of water was combined with 1.0 μL of CIP buffer and CIP (Exiqon, Vedbaek, Denmark). The mixture was incubated for 30 min at 37°C, and was terminated by incubation for 5 min at 95°C. Then, 3.0 μL of labeling buffer, 1.5 μL of fluorescent label (Hy3™), 2.0 μL of dimethyl sulfoxide (DMSO), and 2.0 μL of labeling enzyme were added into the mixture. The labeling reaction was incubated for 1 h at 16°C, and terminated by incubation for 15 min at 65°C.
After stopping the labeling procedure, the Hy3™-labeled samples were hybridized on the miRCURY™ LNA Array (v.16.0) (Exiqon) according to the manufacturer's instruction. The total 25 μL mixture from Hy3™-labeled samples with 25 μL hybridization buffer was first denatured for 2 min at 95°C, incubated on ice for 2 min, and then hybridized to the microarray for 16–20 h at 56°C in a 12-Bay Hybridization System (Nimblegen Systems, Inc., Madison, WI), which provided an active mixing action and constant incubation temperature to improve hybridization uniformity and enhance signal. Following hybridization, the slides were retrieved, washed several times using Wash buffer kit (Exiqon), and finally dried by centrifugation for 5 min at 400 rpm. Then, the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA).
Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs with intensities ≥50 in all samples were chosen for calculating a normalization factor. Expressed data were normalized using median normalization. After normalization, miRNAs that had significant differential expression were identified through Volcano Plot filtering. Hierarchical clustering was performed using MEV software (v4.6, TIGR).
Intrathecal injection of antagomir-21 and negative control antagomir
The catheter and minipump implantation surgery were performed immediately after the SCI was performed. In brief, antagomir-21 (Ribobio, Guangzhou, China) or negative control antagomir (Ribobio, Guangzhou, China) was dissolved in saline (0.9%) at a concentration of 20 nmol/mL. They were filled into osmotic minipumps (model 1030D, Alzet, CA) and continuously infused into the spinal cords of SCI rats at a rate of 1 μL/h, as previously described.18 A partial laminectomy at T12/T13 was made for the placement of an intrathecal catheter. A small incision in the dura at the T12/T13 laminectomy was made with the point of a hypodermic needle. A 32-gauge catheter (cat. no. CS132G, lot no. 20422; ReCathCo, LLC, Allison Park, PA) was inserted through the dural incision and fed rostrally to lie immediately caudal to the T10 laminectomy. This catheter was connected to a 2 cm piece of PE10 tubing (ReCathCo©, LLC, Allison Park, PA), which was, in turn, connected to the osmotic minipump using a 4 cm piece of PE50 tubing. The catheter was secured with 4/0 silk threads to the bone and muscle. Osmotic minipumps were implanted subcutaneously. Muscles and the incision were sutured using 4/0 silk threads. In each rat, penicillin G (40,000 U, i.m.) was injected into the quadriceps femoris muscle during the surgery, and then once a day in both hindlimbs for 5 days. The entire surgery was performed in a warm environment. Finally, the rats were housed in cages and given free access to food and water. Distended bladders were emptied by manual massage on the lower abdomen twice a day until voluntary emptying returned.
BBB score
Locomotor activity was evaluated at 1, 3, 7, 14, 21, and 28 days post-injury using the BBB score, which measured locomotor ability for 4 min. Two independent and well-trained investigators observed the movement and scored the locomotor function according to the BBB scales as described previously.19 Investigators were blind to the treatment. The final score of each animal was obtained by averaging the values from both investigators.
Lesion identification by cresyl violet staining
After the final behavioral tests, which occurred 28 days after surgery, the animals were terminally anesthetized using an intraperitoneal injection of 10% chloral hydrate (10 mg/kg). Subsequently they were transcardially perfused with 250 mL of 0.9% NaCl (4°C) followed by 500 mL of 4% paraformaldehyde (PFA) (4°C) in 0.1M phosphate buffer saline (PBS, pH 7.4). A 1 cm segment of spinal cord including the injury epicenter was dissected and post-fixed in the same fixative for 24 h at 4°C. After fixation, the tissue blocks were embedded in paraffin. Transverse sections (10 μm thickness) were taken through the width of the spinal lesion site, and put onto Superfrost Plus Slides. Every 40th section of the lesion site sample was stained with 0.5% cresyl-violet acetate and imaged using a microscope (BH-2; Olympus, NY). Using Image-Pro Plus 6.0 (Media Cybernetics, USA) software, the lesion area and spared tissue area were outlined and quantified. Spared tissue was defined as the remaining areas where the normal anatomical structure of the spinal cord was preserved. The section with the lowest percentage of spared tissue was assigned as the injury epicenter. Transverse sections, with an interval of 400 μm rostral and caudal to this lesion epicenter, were analyzed up to a distance of 1600 μm away from the lesion epicenter for percentage tissue sparing.
Quantitative real-time RT-PCR
Total RNA from 10 mm long spinal cord segments containing the injury epicenter was extracted with TRIzol (Invitrogen, CA) according to manufacturer's instructions. Total RNA from each sample was reverse-transcribed to cDNA using the PrimeScript RT reagent Kit (TaKaRa, Tokyo, Japan) and qRT-PCR was performed using the SYBR Premix Ex Taq (TaKaRa, Tokyo, Japan) and miRNA-specific primers for miR-21 (Ribobio, Guangzhou, China). The relative microRNA levels were normalized to U6 expression for each sample. Analyses of gene expression was performed by the 2-▵▵Ct method.
Western blot
Rats (n=4) were euthanized with an overdose of 10% chloral hydrate (10 mg/kg) for the 1 and 3 days post-injury groups. A 1 cm long segment of spinal cord encompassing the injury site was then harvested for Western blot analyses. The harvested tissues were lysed, and their protein concentrations were determined using a BCA protein assay kit (Beyotime, Shanghai, China). Samples (30 μg of protein) were electrophoresed onto a 12% sodium dodecyl sulfate/polyacrylamide gel (SDS/PAGE), and transferred to PVDF membranes (Millipore, Mississauga, Canada). The membranes were blocked in 5% nonfat milk for 1 h at room temperature (RT). The membranes were then incubated overnight at 4°C with rabbit anti-FasL Polyclonal antibody (1:2000; Santa Cruz, CA), rabbit anti-PTEN Polyclonal antibody (1:500, Bioworld Technology, Louis Park, MN) and rabbit anti-PDCD4 Polyclonal antibody (1:2000, Proteintech, Chicago, IL), followed by horseradish peroxidase (HRP)-conjugated secondary antibody (1:10000) for 1 h at RT. Proteins were visualized by enhanced chemiluminescence. Relative intensities were determined with Quantity One 4.6.2 software (Bio-Rad, USA) and β-actin was used as the internal control. Data were given as mean±SD of the percentage ratio of the control.
Immunohistochemistry staining
Animals were euthanized 1 and 3 days after injury (n=4 for each group). Briefly, the rats were intracardially perfused with saline followed by ice-cold 4% PFA. A 10 mm segment of spinal cord encompassing the injury site was then harvested. After fixation, the tissue blocks were embedded in paraffin, and sectioned at 5 μm thickness. Paraffin-embedded sections were deparaffinized with xylene and hydrated through graded alcohols. Epitope unmasking was performed by microwave irradiation in 10 mM citrate buffer (pH 6.0) twice for 5 min at 800 W prior to cooling for 30 min. Subsequently, endogenous peroxidase was inactivated by incubation in 3% H2O2 for 15 min at RT. After rinsing with 0.01 M PBS, the sections were blocked with bovine serum (10%) in PBS for 30 min and then incubated overnight at 4°C with primary antibodies: rabbit anti-FasL Polyclonal antibody (1:200), rabbit anti-PTEN Polyclonal antibody (1:100) and rabbit anti-PDCD4 Polyclonal antibody (1:100). Subsequently, the sections were incubated with secondary antibodies (anti-rabbit IgG antibodies). Finally, the immunoreactivity was visualized by staining with diaminobenzidine (DAB) for 3 min, covered with a cover-slip, and analyzed under a light microscope.
TUNEL Staining
Slides, prepared as described, were used for neuronal cell apoptosis analysis. Briefly, TUNEL staining was performed using the DeadEnd™ Colorimetric TUNEL System (Promega, USA). According to standard protocols, sections were de-waxed in xylene, rehydrated in graded alcohols, and placed in dH2O. Then, these sections were incubated in a 20 μg/ml proteinase K working solution for 15 min at RT. The slides were rinsed three times with PBS before being incubated in TUNEL reaction mixture for 1 h at 37°C. After rinsing with PBS three times for 5 min, sections were incubated with HRP-streptavidin reagent (1:200) in PBS for 30 min at RT. Sections were rinsed in PBS three times for 5 min each, and then incubated with 0.04% DAB and 0.03% H2O2 solution for 10 min. After rinsing with PBS three times for 5 min, sections were counterstained with hematoxylin. Then, sections were rinsed in distilled water two times for 5 min each, and cover-slipped with mounting medium. The number of TUNEL positive cells was counted. Each section was counted independently by two observers using a light microscope (×200 magnification). The quantitation method for estimating cells apoptosis after injury was as follows: 1) two spinal cord cross-sections for each animal were randomly selected for counting; 2) cell profiles were counted manually from the digital images in five randomly chosen fields for each section; and 3) the number of positive cells in each visual field and the ratio of positive cells were calculated.
In situ hybridization
Slides, prepared as described, were used for in situ hybridization. Briefly, sections were de-waxed in xylene, rehydrated in graded alcohols, and placed in diethyl pyrocarbonate (DEPC) H2O. Endogenous peroxidase was inactivated by incubation in 3% H2O2 for 15 min at RT. Sections were then digested in proteinase K (20 μg/ml) for 20 min, rinsed in NaCl/Tris, and then fixed in 4% PFA for 10 min. Following this, slides were rinsed with PBS twice for 5 min. Slices were blocked at RT for 2 h in hybridization buffer (50% formamide, 25% 5×saline sodium citrate (SSC), 10% 5×Denhardt's and 15% DEPC-H2O containing 200 ng/mL yeast RNA, 500 g/mL salmon sperm DNA and 20 mg/mL Roche blocking reagent), and then hybridized with 30 nmol of locked nucleic acid (LNA)-modified oligonucleotide probe (Exiqon, Woburn, MA) complementary to Rattus norvegicus (rno)-miR-21 and labeled with digoxigenin (DIG) at 52°C overnight. After hybridization, the slides were washed in 2×SSC twice at 37°C for 15 min followed by a wash in 0.5×SSC (15 min at 37°C), and 0.2×SSC (15 min at 37°C). The slides were then incubated with HRP-conjugated anti-DIG antibody. Sections were rinsed in PBS three times for 5 min, and peroxidase staining was visualized with DAB for 3 min. After rinsing with PBS three times for 5 min, sections were counterstained with hematoxylin. The quantitation method for estimating positive cells after injury was as follows: 1) two spinal cord cross-sections for each animal were randomly selected for counting; 2) cell profiles were counted manually from the digital images in five randomly selected fields for each section; and 3) the number of positive cells in each visual field were calculated.
Statistical analysis
All data were analyzed using SPSS 17.0 statistical software. For comparisons of BBB scores, repeated-measures analysis of variance (ANOVA) followed by Bonferroni post-hoc corrections were performed. Other data were tested using the independent Student's t test. All data were presented as mean±SEM, and the significance level was set at p≤0.05.
Results
Aberrant expression of miRNAs in injured spinal cord
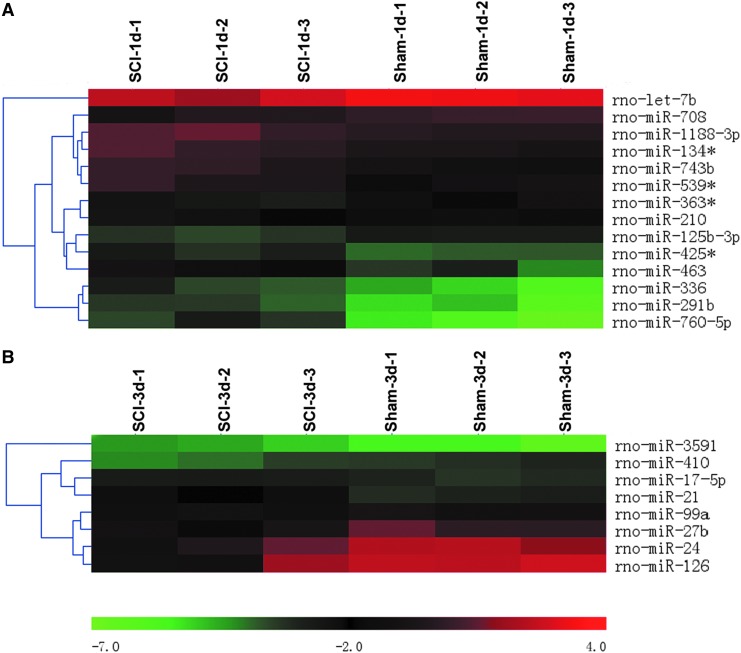
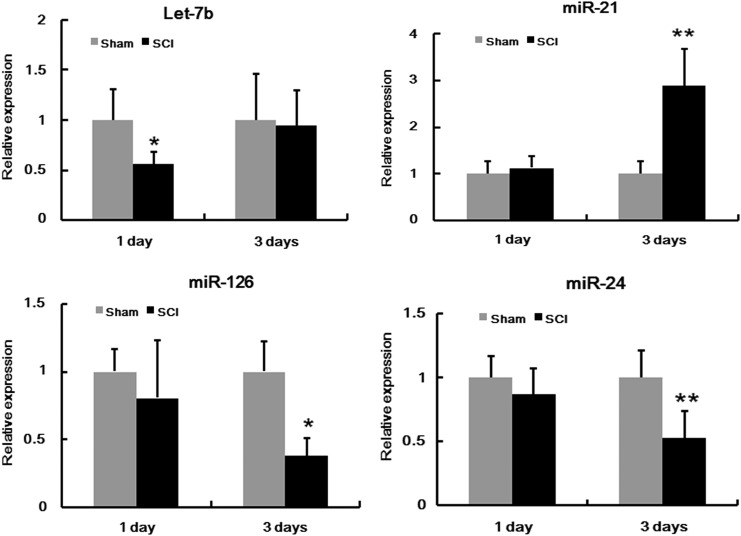
To determine the potential involvement of miRNAs in SCI, we used microarray analysis to determine miRNA levels in spinal cord after contusion SCI. Our data revealed that compared with the sham 1 day group, nine miRNAs were upregulated and five miRNAs were downregulated in the SCI 1 day group (Fig. 1A; Table 2); compared with sham 3 day group, three miRNAs were upregulated and five miRNAs were downregulated in the SCI 3 day group (Fig. 1B; Table 3). Four of the miRNAs that were abnormally expressed (Let-7b, miR-126, miR-24 and miR-21) were further confirmed by qRT-PCR (Fig. 2). Among them, miR-21 has been shown to have a protective effect in many injury models;14–16 therefore, we focused on miR-21 in SCI for further study.
FIG. 1.
Hierarchical cluster analyses of altered microRNAs (miRNAs) after spinal cord injury (SCI). Differentially expressed miRNAs in the sham and SCI group, 1 day post-injury (n=3/group) (A). Differentially expressed miRNAs in sham and SCI group 3 days post-injury (n=3/group) (B). The color code in the heat maps is linear, with green as the lowest and red as the highest. The miRNAs that were upregulated were shown in green to red, whereas the miRNAs that were downregulated were shown from red to green.
Table 2.
microRNAs (miRNAs) Differentially Expressed in SCI Compared with Sham Group at 1 Day Post-Injury (n=3/Group)
| |
Intensities averaged over probes (normalized) |
Standard deviation |
|
|
||
|---|---|---|---|---|---|---|
| miRNA | SCI-1d | Sham-1d | SCI-1d | Sham-1d | Fold changes | p values |
| upregulated | ||||||
| miR-760-5p | 0.112 | 0.017 | 0.324 | 0.454 | 6.56 | 0.0114 |
| miR-463 | 0.429 | 0.097 | 0.329 | 0.423 | 4.43 | 0.0173 |
| miR-291b | 0.087 | 0.025 | 0.184 | 0.456 | 3.39 | 0.0058 |
| miR-336 | 0.101 | 0.030 | 0.407 | 0.452 | 3.37 | 0.0469 |
| miR-473b | 1.106 | 0.493 | 0.219 | 0.181 | 2.24 | 0.0146 |
| miR-539* | 0.962 | 0.482 | 0.269 | 0.308 | 2.00 | 0.0497 |
| miR-425* | 0.134 | 0.070 | 0.205 | 0.055 | 1.90 | 0.0165 |
| miR-134* | 1.372 | 0.720 | 0.283 | 0.103 | 1.90 | 0.0460 |
| miR-1188-3p | 1.719 | 0.959 | 0.257 | 0.079 | 1.79 | 0.0427 |
| miR-21 | 1.448 | 1.213 | 0.488 | 0.430 | 1.19 | 0.667 |
| downregulated | ||||||
| miR-363* | 0.168 | 0.418 | 0.153 | 0.301 | 0.40 | 0.0279 |
| Let-7b | 4.796 | 8.541 | 0.330 | 0.108 | 0.56 | 0.0240 |
| miR-708 | 0.809 | 1.315 | 0.224 | 0.087 | 0.62 | 0.0151 |
| miR-125b-3p | 0.095 | 0.153 | 0.106 | 0.029 | 0.62 | 0.0008 |
| miR-210 | 0.225 | 0.349 | 0.206 | 0.108 | 0.64 | 0.0229 |
| miR-126 | 5.374 | 8.491 | 0.783 | 0.292 | 0.63 | 0.331 |
| miR-24 | 3.864 | 5.056 | 0.746 | 0.085 | 0.76 | 0.518 |
Only fold changes that were significantly different (fold changes ≥1.5 and p<0.05) are provided.
SCI, spinal cord injury.
An miRNA expressed at low levels relative to the miRNA in the opposite arm of a hairpin.
Table 3.
microRNAs (miRNAs) Differentially Expressed in SCI Compared with Sham Group at 3 Days Post-Injury (n=3/Group)
| |
Intensities averaged over probes (normalized) |
Standard deviation |
|
|
||
|---|---|---|---|---|---|---|
| miRNA | SCI-3d | Sham-3d | SCI-3d | Sham-3d | Fold changes | p values |
| upregulated | ||||||
| miR-3591 | 0.116 | 0.031 | 0.347 | 0.469 | 3.71 | 0.0265 |
| miR-21 | 2.266 | 0.677 | 0.129 | 0.185 | 3.35 | 0.0009 |
| miR-17-5p | 0.876 | 0.568 | 0.037 | 0.181 | 1.54 | 0.0078 |
| downregulated | ||||||
| miR-126 | 3.777 | 9.160 | 0.791 | 0.061 | 0.41 | 0.0374 |
| miR-410 | 0.264 | 0.529 | 0.441 | 0.206 | 0.50 | 0.0454 |
| miR-24 | 4.064 | 7.740 | 0.363 | 0.144 | 0.53 | 0.0263 |
| miR-27b | 2.669 | 4.727 | 0.290 | 0.188 | 0.56 | 0.0390 |
| miR-99a | 1.838 | 2.974 | 0.353 | 0.070 | 0.62 | 0.0447 |
| let-7b | 5.117 | 6.582 | 0.131 | 0.214 | 0.78 | 0.179 |
Only fold changes that were significantly different (fold changes ≥1.5 and p<0.05) are provided.
SCI, spinal cord injury.
FIG. 2.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis confirming the microarray analyses of four microRNAs (miRNAs) 1 and 3 days after spinal cord injury (SCI). Relative expression indicates changes compared with the sham group at either 1 or 3 days after SCI. Bars represent means±SEM. *p<0.05; **p<0.01.
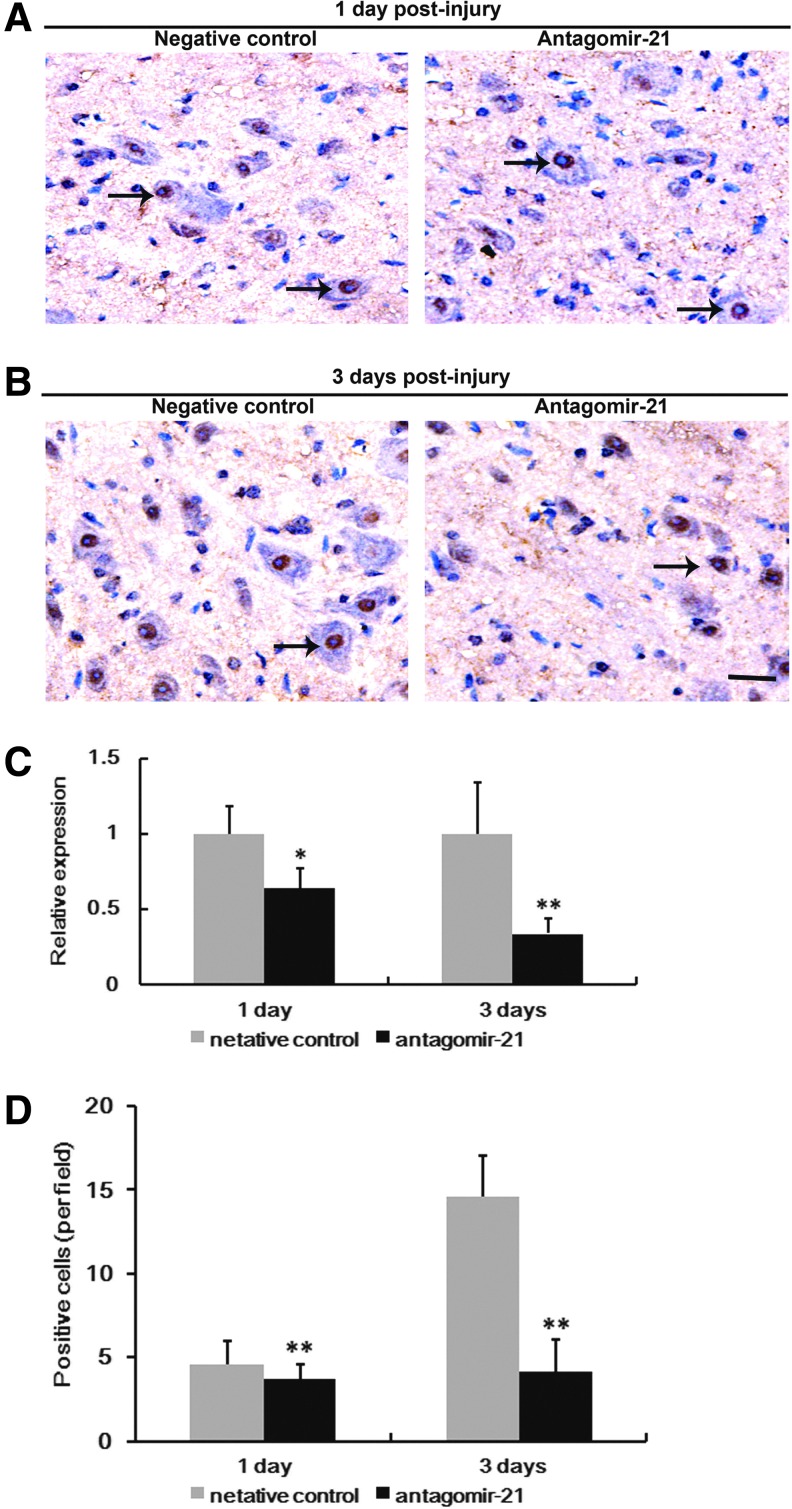
Angatomir-21 successfully inhibited miR-21 expression in vivo after SCI
To determine the inhibitory effect of antagomir-21 on miR-21 in the spinal cord in vivo, in situ hybridization using LNA-containing antisense oligonucleotide probes and qRT-PCR were performed. As shown in Figure 3, miR-21 was localized in the nuclei of several cells, in comparison with the negative control group. Antagomir-21 significantly reduced the number of miR-21 positive cells at day 1 and day 3 (Fig. 3A, B, and D). In agreement with in situ hybridization results, qRT-PCR also revealed that miR-21 expression was significantly lower at day 1 and day 3 (0.638±0.132-fold, p<0.05, n=4/group; 0.336±0.099 fold, p<0.01, n=4/group respectively) in RNA samples isolated from rats treated with antagomir-21 when compared with the negative control group (Fig. 3C).
FIG. 3.
Angatomir-21 inhibits miR-21 expression in vivo following spinal cord injury (SCI). The expression of miR-21 was measured by in situ hybridization and quantitative real-time polymerase chain reaction (qRT-PCR) 1 and 3 days after SCI. miR-21 was localized in the nuclei of many cells following SCI (arrows). Compared with the negative control group, antagomir-21 significantly reduced the number of miR-21 positive cells 1 (A, D) and 3 days (B, D) following SCI. In agreement with in situ hybridization results, qRT-PCR also revealed that miR-21 expression was significantly lower at days 1 and 3 in RNA samples isolated from rats treated with antagomir-21 when compared with the negative control group (C). n=4/group/time point. Bars represent means±SEM. *p<0.05; **p<0.01. Scale bar=50 μm.
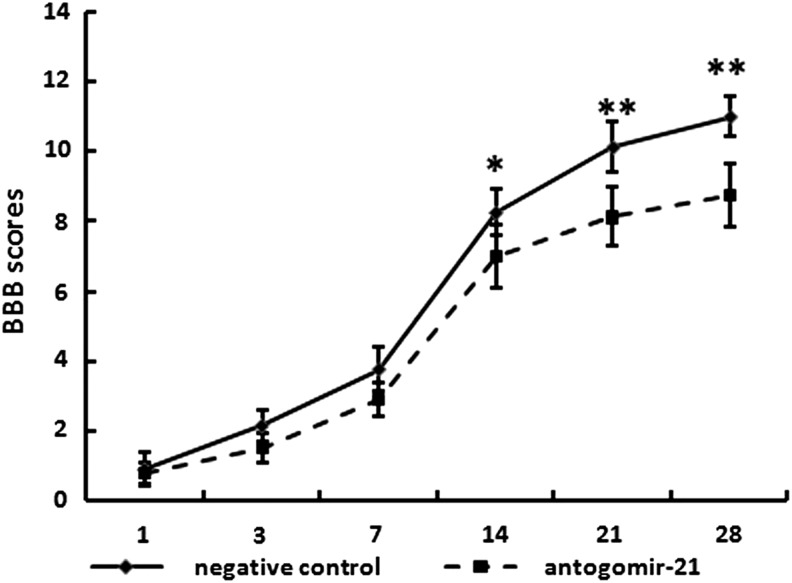
Inhibition of miR-21 exacerbated the functional deficit induced by SCI
After SCI, all rats were paralyzed in both hindlimbs. A spontaneous functional recovery after SCI in both groups was observed. As shown in Figure 4, hindlimb locomotor activity improved gradually during the evaluation period, as demonstrated by the increase in BBB scores in both groups. Compared with the negative control group, recovery was significantly attenuated in the antagomir-21 group from day 14 after the injury (day 14, p<0.05; days 21 and 28, p<0.01). Four weeks after SCI, the BBB average scores were 8.5±0.913 and 11±0.577 in the antagomir-21 and negative control groups, respectively (Fig. 4, p<0.01), indicating that antagomir-21 treatment exacerbated the movement of rats with SCI.
FIG. 4.
Inhibition of miR-21 exacerbated the functional deficit following spinal cord injury (SCI). Hindlimb recovery was assessed from day 1 to day 28 after SCI using Basso, Beattie, and Bresnahan (BBB) Score. Hindlimb dysfunction was exacerbated with treatment of antagomir-21 (n=4) when compared with negative control group (n=4). Bars represent means±SEM. *p<0.05; **p<0.01.
Inhibition of miR-21 aggravated tissue damage in the injured spinal cord
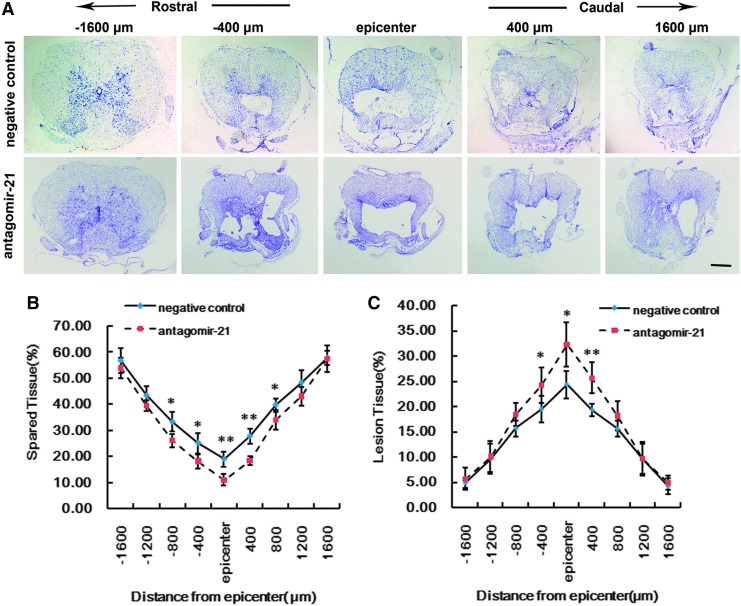
Cresyl violet staining was used to assess lesion size and spared tissue following behavioral analyses, at 28 days post-injury (n=4/group) (Fig. 5). Sections of thoracic spinal cord were analyzed and the ratio for ‘‘injured area/total area’’ from each section determined (Fig. 5A). The lesion was larger in antagomir-21-treated rats not only at the injury epicenter (Fig. 5C), but also in regions extending away from the epicenter, in both rostral and caudal directions. Compared with the negative control group, antagomir-21-treated rats had significantly smaller spared tissue areas at multiple distances from the lesion epicenter, both in rostral and caudal directions (Fig. 5B).
FIG. 5.
Inhibition of miR-21 aggravates tissue damage in injured spinal cord. Cresyl violet stained sections were analyzed for lesion size and spared tissue 28 days post-injury. Representative micrographs of spared tissue and lesion size in the injury site, and in spinal segments 1600 μm rostral and caudal to the injury, 28 days post-injury (A). Quantification of spared tissue within the injury site, and 1600 μm rostral and caudal to the epicenter, 28 days post-injury (B). Quantification of lesion size within the injury site, and 1600 μm rostral and caudal to the epicenter, 28 days post-injury (C). Antagomir-21 (n=4) treated rats were compared to negative control rats (n=4). Bars represent means±SEM. *p<0.05; **p<0.01. Scale bar=1 mm.
Reduced mir-21 expression increased apoptotic cell after SCI
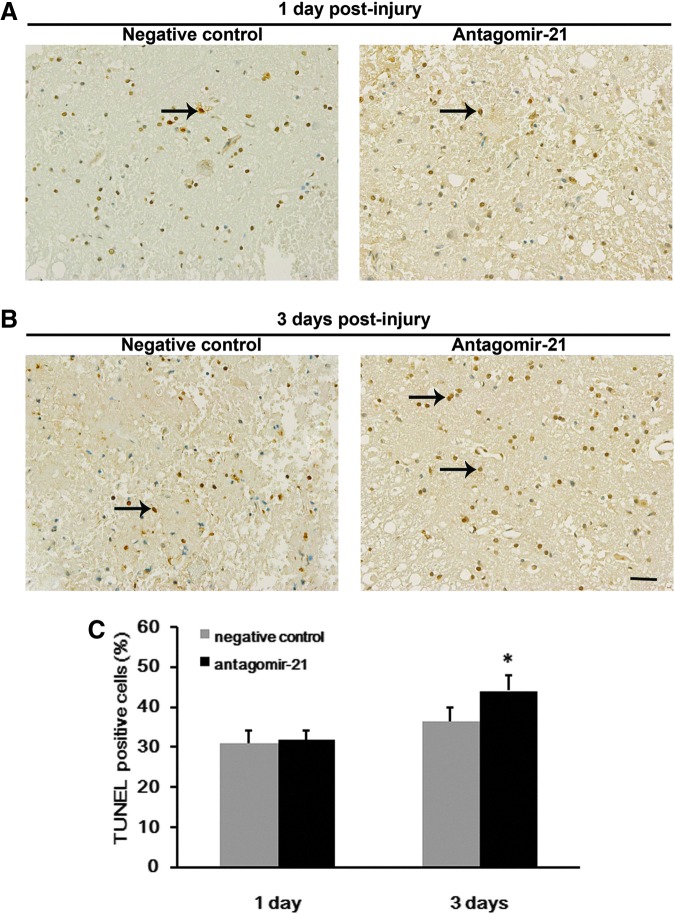
The effect of miR-21 on apoptotic cell after SCI was analyzed by TUNEL staining. Quantification of apoptosis was performed 1 and 3 days post-injury (n=4/group/time). Most of the TUNEL-positive stained cells were found within the gray matter, although some were present in the neighboring white matter. A higher number of positive stained cells were observed at day 3 after injury in both groups. Compared with the negative control group, there were significantly more TUNEL-positive cells in the antagomir-21 group at day 3 after SCI (Fig. 6).
FIG. 6.
Reduced mir-21 expression increases apoptotic cell death following spinal cord injury (SCI). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining was used to analyze neuronal apoptosis (arrow) after SCI. Representative microscopy images of spinal cord sections taken 1 (A) and 3 days (B) post-injury in negative control and antagomir-21 groups, respectively. The antagomir-21 group at 3 days after SCI (n=4) had a higher proportion of TUNEL-positive cells than did the negative control group (n=4) (C). Bars represent means±SEM. *p<0.05. Scale bar=50 μm.
Inhibition of miR-21 increased FasL and PTEN expression in vivo, but did not alter the expression of PDCD4
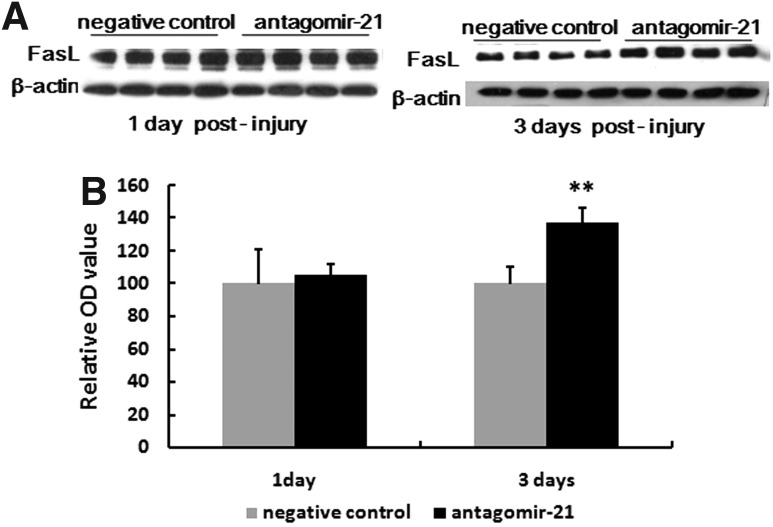
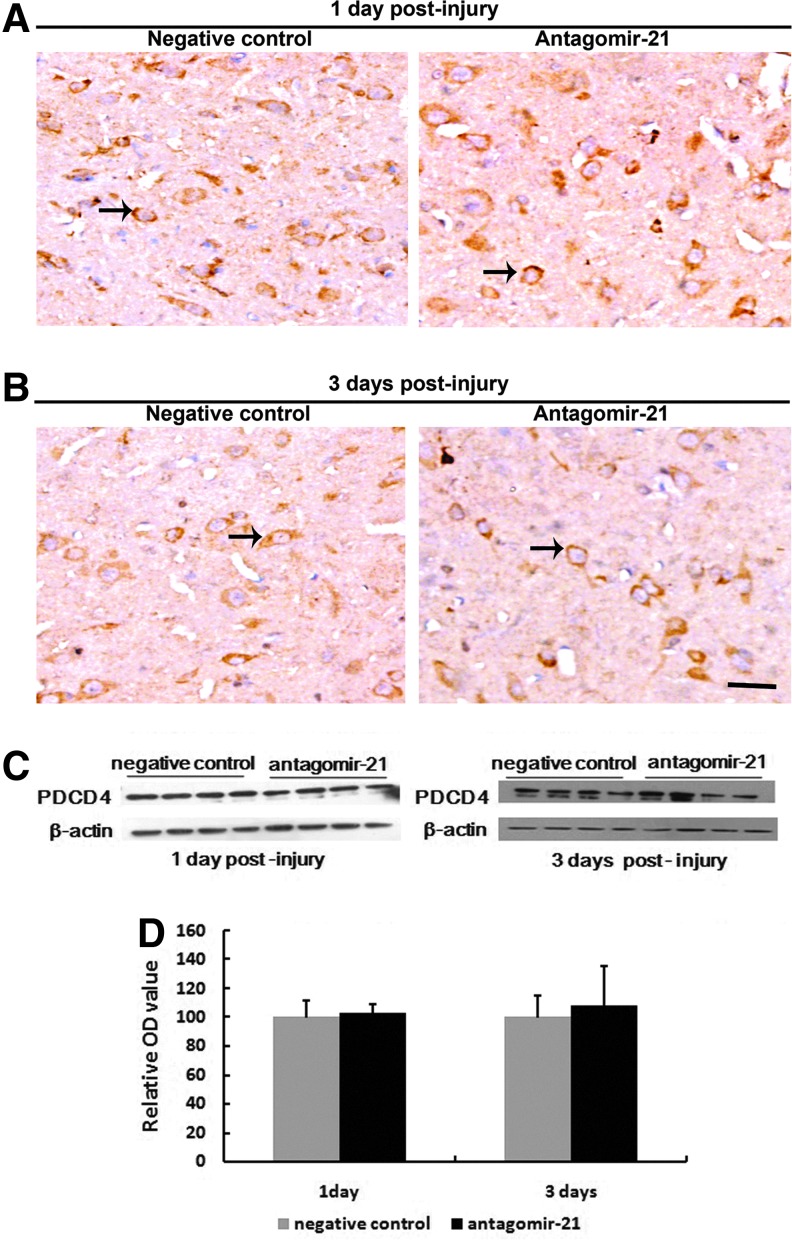
FasL, PTEN, and PDCD4 were validated as targets of miR-21. Therefore, we selected these as target genes that may be modulated by miR-21 in vivo. Protein level of FasL was quantified using Western blot. Compared with the negative control, treatment with antagomir-21 significantly increased FasL expression at day 3 (Fig. 7), but the expression of FasL was similar in both groups at day 1 (Fig. 7). Similarly, treatment with antagomir-21 increased PTEN expression compared with the negative control group at day 3 (Fig. 8C and D), but had no significant effect at day 1 in either group (Fig. 8C and D). In the immunohistochemistry staining results, PTEN was located in the cytoplasm, and PTEN was overexpressed after antagomir-21 treatment at day 3, but similar in both groups at day 1 (Fig. 8A and B), which was in agreement with Western blot data. PDCD4 is one of most analyzed miR-21 targets, but Western blot results showed that treatment with antagomir-21 had no effect on PDCD4 protein level following SCI (Fig. 9C and D). In the immunohistochemistry staining results, PDCD4 was located in the cytoplasm, and PDCD4 expression was similar in both groups at day 1 and day 3, which was in agreement with Western blot data.
FIG. 7.
Inhibition of miR-21 increased the expression of Fas ligand gene (FasL). The expression of FasL after SCI was detected by Western blot (A) in samples collected from antagomir-21 rats and negative control rats (n=4 per time point, respectively), and the intensity of each band was estimated by densitometric analysis (B). Compared with the negative control, treatment with antagomir-21 significantly increased FasL expression at day 3, but the expression of FasL was similar in both groups at day 1 (B). Bars represent means±SEM. **p<0.01.
FIG. 8.
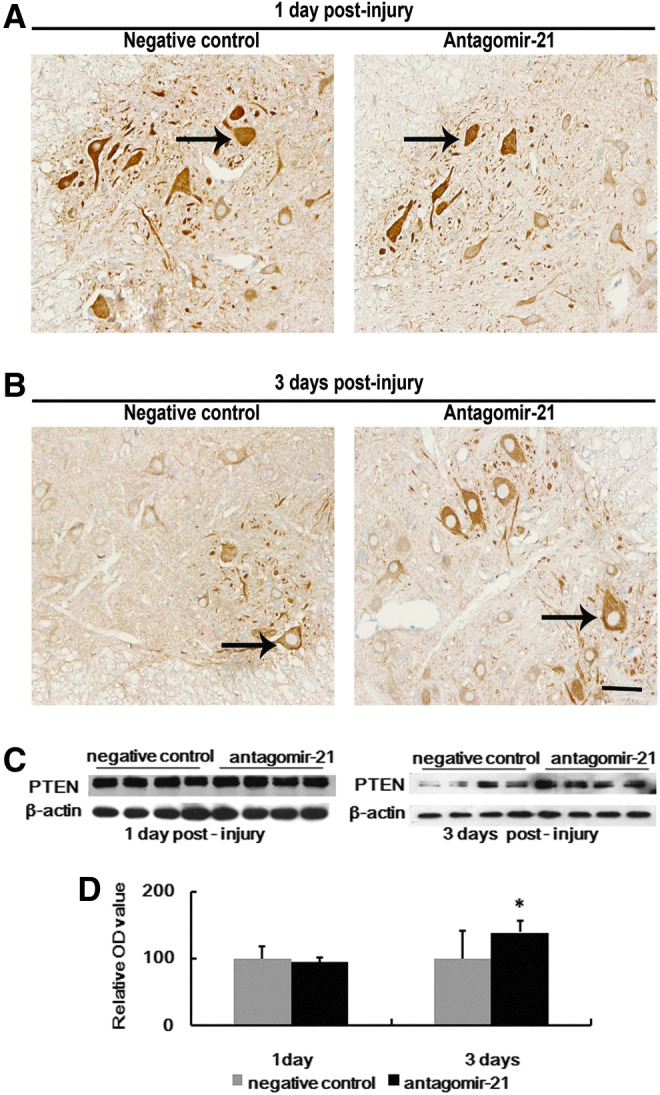
Inhibition of miR-21 increases the expression of phosphatase and tensin homolog (PTEN). The expression of PTEN after SCI was detected by immunohistochemistry staining (A, B) in spinal cord collected from each group (antagomir-21, n=4; negative control, n=4). PTEN was localized in the cytoplasm of neurons (A, B; arrows). The staining was similar in both groups at day 1 (A). Antagomir-21 treatment resulted in deeper staining compared with the negative control group at day 3 (B). The expression of PTEN protein after SCI was detected by Western blot in the sample from each group (n=4 per group per time point) (C, 1), and the intensity of each band was estimated by densitometric analysis (C, D). Treated with antagomir-21 increased PTEN expression compared with the negative control group at day 3 (C, D), but had no significant effect at day 1 in either group (C, D). Bars represent means±SEM. *p<0.05. Scale bar=50 μm.
FIG. 9.
Inhibition of miR-21 has no affect on the expression of PDCD4. The expression of PDCD4 after spinal cord injury (SCI) was detected by immunohistochemistry staining (A, B) in spinal cord collected from each group (antagomir-21, n=4 per time point; negative control, n=4 per time point). PDCD4 was localized in the cytoplasm of various cells (A, B; arrows). The staining was similar in both groups at day 1 (A) and day 3 (B). The expression of PDCD4 protein after SCI was detected by Western blot in the sample from each group (n=4 per group per time point) (C, D), and the intensity of each band was estimated by densitometric analysis (C, D). Treatment with antagomir-21 had no effect on PDCD4 protein level following SCI (C, D). Bars represent means±SEM. Scale bar=100 μm.
Discussion
SCI induces widespread molecular and biochemical changes, including altered mRNA and protein expression, inflammatory activation, production of free radicals, axonal plasticity, and neuronal cell death.20,21 Many studies have been performed to advance our understanding of the changes in mRNA and protein expression following SCI, as well as to explain how these changes relate to SCI pathophysiology. Recently, studies showed that individual miRNAs can regulate hundreds of genes simultaneously by targeting RISCs to mRNAs where they acted to inhibit translation or direct destructive cleavage.22 Therefore, altering the expression of miRNAs may greatly affect SCI pathophysiology and functional outcome. By using miRNA microarray analyses, we found some miRNAs that had altered expression after SCI, and the expression of four of them (miR-21, −126, −24 and let-7b) were further confirmed by qRT-PCR analysis.
It should be noted that several other studies have demonstrated aberrant expression of miR-21 in many injury models. For example, in the rat cerebral cortex, microarray analyses demonstrated that the levels of many miRNAs were altered at several time points (6–72 h) after traumatic brain injury, and that miR-21 was upregulated at all times.12 Similarly, Redell et al. reported that the expressions of many miRNAs were altered in the hippocampus after traumatic brain injury, and miR-21 expression was significantly upregulated in the hippocampus with expression levels peaking 3 days post-injury, and returned to near normal levels 15 days post-injury.23 After middle cerebral artery occlusion of rats, Buller et al. observed that miR-21 levels increased approximately threefold in neurons isolated from the ischemic boundary zone, when compared with homologous contralateral neurons, at both 2 and 7 days after injury.15 miR-21 was significantly downregulated in infarcted areas, but was upregulated in border areas following acute myocardial infarction (AMI).17 Recently, several studies demonstrated that miR-21 was upregulated following contusion SCI,9–11 similar to our results demonstrating that miR-21 was one of the aberrantly expressed miRs after SCI.
MiR-21 has been shown to be a strong anti-apoptotic factor.24,25 Knockdown of miR-21 in cultured glioblastoma cells triggered activation of caspases, and led to increased apoptotic cell death.24 In a cardiac cell injury model, Cheng et al. found that H2O2-induced cardiac cell death and apoptosis were increased by an miR-21 inhibitor, and was decreased by pre-miR-21, indicating that miR-21 functions to attenuate apoptosis during cardiac cell injury.16 In vivo, overexpression of miR-21 decreased myocardial infarct size and decreased the dimension of left ventricles after AMI.17 In this study, we found that miR-21 was significantly upregulated 3 days post-injury, and that inhibition of miR-21 expression exacerbated the functional deficit, aggravated tissue damage, and increased apoptotic cell death induced by SCI. These results indicate that miR-21 has a protection effect in SCI by inhibiting neuronal cell death.
MiR-21 exerted its anti-apoptotic effect by targeting pro-apoptotic genes. FasL, a member of the TNF-α family of ligand, was one of validated targets of miR-21. miR-21 was shown to reduce the death of ischemic cortical neurons by downregulating FasL.15 Overexpression of miR-21 in transgenic mouse heart inhibited ischemia-induced upregulation of FasL expression, limited infarct size, attenuated apoptosis, and ameliorated heart failure.26 In a variety of neurological disorders (including SCI), FasL was elevated.27–29 FasL had been shown to play an important role in apoptotic cell death following SCI.27,29–30 Our data showed that inhibition of miR-21 expression in vivo upregulated the FasL expression. It has been demonstrated that miR-21 played an anti-apoptotic role by targeting FasL following SCI.
PTEN is another direct target of miR-21. MiR-21-mediated regulation of PTEN expression had been shown in several cell types.31–33 Moreover, PTEN deletion promoted compensatory sprouting of uninjured corticospinal tract (CST) axons, enabled successful regeneration of a cohort of injured CST axons past the spinal cord lesion after SCI,34 and enhanced neural survival.35,36 Our results showed that inhibiting the expression of miR-21 increased the PTEN expression, and induced a larger lesion size with less spared tissue. These findings indicated that miR-21 might promote neurogenesis and enhance neural survival by targeting PTEN following SCI.
PDCD4 is currently one of the most widely studied miR-21 targets. PDCD4 is a tumor suppressor that is downregulated in many forms of cancer, and upregulated during apoptosis.37–39 PDCD4 was identified as a direct miR-21 target in multiple cell types and diseases. Studies showed increased PDCD4 expression following miR-21 inhibition in vitro, as well as direct miR-21 binding to the 3' region of PDCD4 mRNA.40 Negative regulation of PDCD4 by miR-21 contributed to its protective effect against ischemia/reperfusion injury in the heart.16 Delayed ischemic preconditioning-regulated miR-21 protected kidney against ischemia/reperfusion injury via anti-apoptosis through its target PDCD4.41 However, in our study we found that expression of PDCD4 was not altered by inhibiting the miR-21 expression. This suggests that although a miRNA may have hundreds of putative mRNA targets, the regulation may depend upon tissue- or cell-specificity.
Conclusion
In conclusion, miRNA signatures after SCI have revealed that multiple miRs are aberrantly expressed. Among them, miR-21 dysregulation contributes to SCI. Knockdown of miR-21 in vivo exacerbates the functional deficit, aggravates tissue damage, and increases apoptotic cell death in rats following SCI. Moreover, miR-21 most likely exerts its anti-apoptotic effect by targeting pro-apoptotic genes FasL and PTEN. These results suggest that miR-21 may play an important role in the pathophysiology of SCI. MiRNAs are highly promiscuous regulators with a cumulative impact on dozens to hundreds of gene targets; therefore, the exact role of miR-21 in SCI needs to be explored.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (No. 81171698) and the Hunan Provincial Natural Science Foundation of China (No. 09JJ3047).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Thuret S. Moon L.D. Gage F.H. Therapeutic interventions after spinal cord injury. Nat. Rev. Neurosci. 2006;7:628–643. doi: 10.1038/nrn1955. [DOI] [PubMed] [Google Scholar]
- 2.Carlson G.D. Gorden C. Current developments in spinal cord injury research. Spine J. 2002;2:116–128. doi: 10.1016/s1529-9430(01)00029-8. [DOI] [PubMed] [Google Scholar]
- 3.Dumont R.J. Okonkwo D.O. Verma S. Hurlbert R.J. Boulos P.T. Ellegala D.B. Dumont A.S. Acute spinal cord injury, part I: pathophysiologic mechanisms. Clin. Neuropharmacol. 2001;24:254–264. doi: 10.1097/00002826-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Friedman R.C. Farh K.K. Burge C.B. Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bak M. Silahtaroglu A. Møller M. Christensen M. Rath M.F. Skryabin B. Tommerup N. Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosik K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R. Zuccato C. Belyaev N.D. Guest D.J. Cattaneo E. Buckley N.J. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol. Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Liu N.K. Wang X.F. Lu Q.B. Xu X.M. Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strickland E.R. Hook M.A. Balaraman S. Huie J.R. Grau J.W. Miranda R.C. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yunta M. Nieto–Díaz M. Esteban F.J. Caballero–López M. Navarro–Ruíz R. Reigada D. Pita–Thomas D.W. del Águila A. Muñoz–Galdeano T. Maza R.M. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One. 2012;7:e34534. doi: 10.1371/journal.pone.0034534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lei P. Li Y. Chen X. Yang S. Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 13.Redell J.B. Liu Y. Dash P.K. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J. Neurosci. Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L. Dong LY. Li YJ. Hong Z. Wei WS. miR–21 represses FasL in microglia and protects against microglia-mediated neuronal cell death following hypoxia/ischemia. Gila. 2012;60:1888–1895. doi: 10.1002/glia.22404. [DOI] [PubMed] [Google Scholar]
- 15.Buller B. Liu X. Wang X. Zhang R.L. Zhang L. Hozeska–Solgot A. Chopp M. Zhang Z.G. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y. Zhu P. Yang J. Dong S. Wang X. Chun B. Zhuang J. Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc. Res. 2010;87:431–439. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong S. Cheng Y. Yang J. Li J. Liu X. Wang X. Wang D. Krall TJ. Delphin ES. Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009;284:29,514–29,525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caggiano A.O. Zimber M.P. Ganguly A. Blight A.R. Gruskin E.A. Chondroitinase ABCI improves locomotion and bladder function following contusion injury of the rat spinal cord. J. Neurotrauma. 2005;22:226–239. doi: 10.1089/neu.2005.22.226. [DOI] [PubMed] [Google Scholar]
- 19.Basso D.M. Beattie M.S. Bresnahan J.C. A sensitive and reliable locomotor rating scale for open field testing in rats. J. Neurotrauma. 1995;12:1–21. doi: 10.1089/neu.1995.12.1. [DOI] [PubMed] [Google Scholar]
- 20.Di Giovanni S. Knoblach S.M. Brandoli C. Aden S.A. Hoffman E.P. Faden A.I. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann. Neurol. 2003;53:454–468. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- 21.De Biase A. Knoblach S.M. Di Giovanni S. Fan C. Molon A. Hoffman E.P. Faden A.I. Gene expression profiling of experimental traumatic spinal cord injury as a function of distance from impact site and injury severity. Physiol. Genomics. 2005;22:368–381. doi: 10.1152/physiolgenomics.00081.2005. [DOI] [PubMed] [Google Scholar]
- 22.Baek D. Villen J. Shin C. Camargo F.D. Gygi S.P. Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redell J.B. Zhao J. Dash P.K. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J. Neurosci. Res. 2011;89:212–221. doi: 10.1002/jnr.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan J.A. Krichevsky A.M. Kosik K.S.l. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y. Liu W. Chao T. Zhang Y. Yan X. Gong Y. Qiang B. Yuan J. Sun M. Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 26.Yed D. He M. Hong C. Gao S. Rane S. Yang Z. Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J. Biol. Chem. 2010;285:20,281–20,290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casha S. Yu W.R. Fehlings M.G. Oligodendroglial apoptosis occurs along degenerating axons and is associated with FAS and p75 expression following spinal cord injury in the rat. Neuroscience. 2001;103:203–218. doi: 10.1016/s0306-4522(00)00538-8. [DOI] [PubMed] [Google Scholar]
- 28.Chli C. Benveniste E.N. Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses. Brain Res. Rev. 2004;44:65–81. doi: 10.1016/j.brainresrev.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Demjen D. Klussmann S. Kleber S. Zuliani C. Stieltjes B. Metzger C. Hirt U.A. Walczak H. Falk W. Essig M. Edler L. Krammer P.H. Martin–Villalba A. Neutralization of CD95 ligand promotes regeneration and functional recovery after spinal cord injury. Nat. Med. 2004;10:389–395. doi: 10.1038/nm1007. [DOI] [PubMed] [Google Scholar]
- 30.Yoshino O. Matsuno H. Nakamura H. Yudoh K. Abe Y. Sawaim T. Uzuki M. Yonehara S. Kimura T. The role of Fas-mediated apoptosis after traumatic spinal cord injury. Spine. 2004;29:1394–1404. doi: 10.1097/01.brs.0000129894.34550.48. [DOI] [PubMed] [Google Scholar]
- 31.Meng F. Henson R. Lang M. Wehbe H. Maheshwari S. Mendell J.T. Jiang J. Schmittgen T.D. Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 32.Ji R. Cheng Y. Yue J. Yang J. Liu X. Chen H. Dean D.B. Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 33.Haverty P.M. Fridlyand J. Li L. Getz G. Beroukhim R. Lohr S. Wu T.D. Cavet G. Zhang Z. Chant J. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 34.Liu C. Wu J. Xu K. Cai F. Gu J. Ma L. Chen J. Neuroprotection by baicalein in ischemic brain injury involves PTEN/AKT pathway. J. Neurochem. 2010;112:1500–1512. doi: 10.1111/j.1471-4159.2009.06561.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q.G. Wu D.N. Han D. Zhang G.Y. Critical role of PTEN in the coupling between PI3K/Akt and JNK1/2 signaling in ischemic brain injury. FEBS Lett. 2007;581:495–505. doi: 10.1016/j.febslet.2006.12.055. [DOI] [PubMed] [Google Scholar]
- 36.Shi G.D. Ouyang Y.P. Shi J.G. Liu Y. Yuan W. Jia L.S. PTEN deletion prevents ischemic brain injury by activating the mTOR signaling pathway. Biochem. Biophys. Res. Commun. 2011;404:941–945. doi: 10.1016/j.bbrc.2010.12.085. [DOI] [PubMed] [Google Scholar]
- 37.He Q. Cai L. Shuai L. Li D. Wang C. Liu Y. Li X. Li Z. Wang S. Ars2 is overexpressed in human cholangiocarcinomas and its depletion increases PTEN and PDCD4 by decreasing MicroRNA-21. Mol. Carcinog. 2013;52:286–296. doi: 10.1002/mc.21859. [DOI] [PubMed] [Google Scholar]
- 38.Asangani I.A. Rasheed S.A. Nikolova D.A. Leupold J.H. Colburn N.H. Post S. Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H. Ozaki I. Mizuta T. Hamajima H. Yasutake T. Eguchi Y. Ideguchi H. Yamamoto K. Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 40.Frankel L.B. Christoffersen N.R. Jacobsen A. Lindow M. Krogh A. Lund A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 41.Xu X. Kriegel A.J. Liu Y. Usa K. Mladinov D. Liu H. Fang Y. Ding X. Liang M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012;82:1167–1175. doi: 10.1038/ki.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]