Abstract
Exome sequencing is emerging as a popular approach to study the effect of rare coding variants on complex phenotypes. The promise of exome sequencing is grounded in theoretical population genetics and in empirical successes of candidate gene sequencing studies. Many projects aimed at common diseases are underway, and their results are eagerly anticipated. In this Perspective, using exome sequencing data from 438 individuals, we discuss several aspects of exome sequencing studies that we view as particularly important. We review processing and quality control of raw sequence data, evaluate the statistical properties of exome sequencing studies, discuss rare variant burden tests to detect association to phenotypes, and demonstrate the importance of accounting for population stratification in the analysis of rare variants. We conclude that enthusiasm for exome sequencing studies of complex traits should be combined with the caution that thousands of samples may be required to reach sufficient statistical power.
The promise of exome sequencing
Next-generation sequencing1–5 coupled with efficient DNA capture6–8 enable exome sequencing as a new approach to study the genetic basis of human phenotypes. A number of genes underlying Mendelian diseases have been mapped using this approach6, 9–15. Exome sequencing has also been applied to tumors16–20, where sample purity, read-mapping, and chromosomal rearrangements are critical and form a very distinctive set of issues. In this Perspective, we restrict our attention to complex traits. In complex trait genetics, exome sequencing studies bring to light rare coding variants that are undetected by microarray-based genome-wide association studies (GWAS). The promise of exome sequencing studies of complex traits is based on the success of candidate gene studies21–26 and has firm roots in population genetic theory27–35.
Large-scale GWAS of complex traits have consistently demonstrated that, with few exceptions, common variants have modest effects, often requiring tens of thousands of samples for their detection. Exome sequencing provides a complementary approach by comprehensively assessing the role of all coding variation, both common and rare. With incessant mutations occurring in each protein-coding gene (at a rate of ∼10−5 per gene for non-synonymous variants36–39) and fitness loss of less than 1% 29–31, 34 for most novel non-synonymous mutations, almost every gene is expected to harbor functionally important variants that can be tested through sequencing, even if these variants are rare. Therefore, the strong interest in exome sequencing stems from three factors: the potential to identify many genes underlying complex traits, straightforward functional annotation of coding variation, and cost being substantially lower (around 5 times) than whole-genome sequencing.
In this Perspective, we evaluate the extent of rare coding variation in empirical data, discuss data processing and quality control of raw sequence data, review analytical methods for detecting genotype-phenotype associations, their expected statistical power, and the potential for confounding due to population stratification. To illustrate our arguments, we used empirical whole-exome sequence data from 184 individuals from the International HIV Controllers Study40 and 254 control individuals from Schizophrenia (SCZ) exome sequencing study.
Assessment of rare coding variation in empirical data
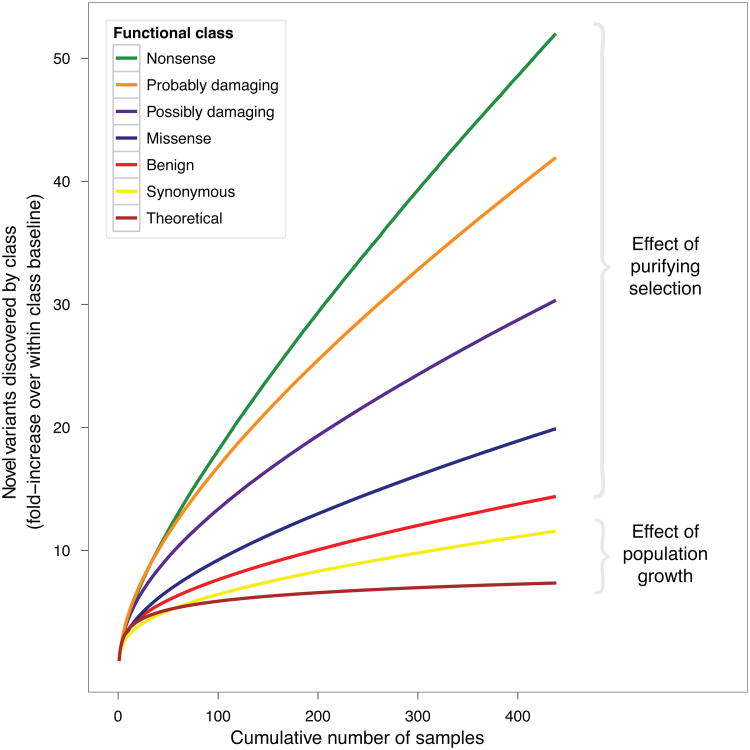
Exome sequencing data contain an abundance of rare coding variation and indicate that a large fraction of this variation is functional. Not only are there many more rare variants than common ones, but sequencing additional samples continues to uncover additional rare variants. In fact, as sample size increases, the number of observed variants grows much faster than predicted by the neutral model of constant population size41, 42 (Figure 1). This relative excess of rare variants can be, in part, attributed to recent population expansion43–45, but is also likely due to purifying selection. As a consequence, rare variation is enriched for evolutionarily deleterious, and thus functional, variants. Additionally, the proportion of non-synonymous variants is higher among rare than among common variants45. Finally, among rare variants, missense variants predicted46 to be damaging are more prevalent than variants predicted to be benign (Figure 1). These findings are consistent with studies that demonstrated that rare variants in protein-coding regions are under purifying selection35, 47–51. Because sequencing larger samples continuously uncovers functionally relevant variants, exome sequencing studies enable direct identification of causal variants (in contrast to GWAS that use linkage-disequilibrium patterns between common markers).
Figure 1.
Discovery of novel variants for increasing numbers of samples. For each functional class, the fold-increase over the number of variants in one sample for that class is plotted as a function of the number of samples in a sequencing experiment. For example, the number of nonsense variants discovered in 300 samples is 40 times greater than the average number discovered in a single sample while the number of synonymous variants is only 10 times greater (although the absolute number of nonsense variants is a relatively minor proportion of the total variation discovered); this effect is due to purifying selection. All classes of variants are discovered at rates exceeding what would be predicted under a neutral model of evolution in a population of constant size, an effect of population growth. The crossing between curves for synonymous variants and the theoretical prediction most likely is a signature of the out-of-Africa bottleneck. See Methods for additional details.
Variant calling and Quality Control filtering of exome sequencing datasets
An exome sequencing study starts with exome capture and sequencing of DNA samples followed by the identification of sequence variants. Exome capture may be realized on many platforms (e.g., Illumina HiSeq, Roche 454, ABI SOLiD) and through a variety of probe definitions (e.g., Agilent SureSelect, Nimblegen SeqCap EZ). Recent advances have enabled sequencing an entire exome or even several exomes at deep coverage in a single run of the sequencing instrument. However, exome capture technologies differ in what they target, how much they capture, and how consistently they do so8. Moreover, only 80-90% of the targeted regions are covered above 10×, which may leave 4–8Mb (or 1000–2000 genes) without sufficient coverage for variant detection.
Exome sequencing coverage has tremendous regional variation8. Some regions may be over-covered, representing true structural variation (e.g., segmental duplications for which only one copy of the region exists in the reference genome), or technical artifacts (e.g., greater abundance of capture probes, or overlapping probe definitions resulting in “double-capturing”). Similarly, some areas may be under-covered for biological reasons (e.g., segmental duplications where more than one copy exists in the reference sequence, preventing the aligner from placing the read uniquely) or for technical reasons (e.g., high GC content or density of variation, which impairs hybridization of probes). Furthermore, some “near-target” regions within 50 bp of the target boundary can have sufficient coverage to warrant inclusion in variant calling. Critically, whichever capture technology is used, either all samples should be processed using the same technology or the variability should be accounted for, e.g., by stratifying the study by technology (see section on population stratification).
For this Perspective, we generated whole-exome data (targeting 28 Mbp) from 438 samples (see Methods section) using a two-stage approach52, 53. First, we applied the data processing and variant calling protocol described previously54. Second, we applied post-SNP-calling Quality Control (QC) filters.
For quality control of the resulting SNPs, we used population-genetic statistics and properties of human genetic variation. Using those statistics helps to identify true variants because the properties of the mutational process37, 55 are different from errors of the sequencing technology.
We compared statistics computed in the 438-sample data set and in 37 whole-genome data sets released by Complete Genomics Inc. (CGI, see URLs), focusing only on the same genomic regions as the exome data. The CGI whole-genome dataset serves as a good comparison because whole-genome sequencing is not dependent on exome-capture technology. We further stratified these per-sample statistics into classes that are biologically interesting (functional class and CpG status) but may also exhibit different rates of technical artifacts. Table 1 shows that filtering is critical for achieving high-quality calls. Before filtering, the metrics show significant deviations from their expected values, which may indicate a high false-positive rate. After filtering, the statistics converge to those in the CGI dataset. The effectiveness of the filters is evident also from the comparison with human-chimpanzee divergence55.
Table 1.
SNP counts per Illumina-sequenced sample (computed as the median of the metrics value in each sample over 438 samples), localized to the exome, stratified by functional and biological criteria, and compared to SNP counts per Complete Genomics- sequenced sample (computed as the median of the metrics value in each sample over 37 samples). Before the application of filters, counts significantly differ from the expectations derived from the independently obtained comparison set (the data from Complete Genomics), indicating the presence of many false-positives. For example, in the SNPs identified as nonsense mutations, which initially appear 1.5-fold enriched for false-positives. As nonsense variants are rare, an unfiltered callset may contain many artifactual events that masquerade as nonsense variants. The QC filters help to align the metrics with the comparison set and the data from human-chimp divergence (see text).
| Filter | Counts (% filtered) | #Het (% filtered) | # Horn-derived (%filtered) | Ti/Tv | |
|---|---|---|---|---|---|
| Total | |||||
| unfiltered | 18,626 | 11,761 | 3,007 | 2.92 | |
|
| |||||
| filtered | 16,776 (10%) | 10,242 (13%) | 2,785 (7%) | 3.21 | |
|
| |||||
| comparison | 16,914 | 10,464 | 2,492 | 3.31 | |
|
| |||||
| By functional class | |||||
| silent | unfiltered | 9,536 | 5,933 | 1,601 | 4.80 |
|
| |||||
| filtered | 8,845 (7%) | 5,372 (9%) | 1,514 (5%) | 5.10 | |
|
| |||||
| comparison | 8,987 | 5,514 | 1,352 | 5.22 | |
|
| |||||
| missense | unfiltered | 8,698 | 5,557 | 1,350 | 1.92 |
|
| |||||
| filtered | 7,644 (12%) | 4,685 (15%) | 1,220 (9%) | 2.11 | |
|
| |||||
| comparison | 7,723 | 4,772 | 1,095 | 2.17 | |
|
| |||||
| nonsense | unfiltered | 70 | 60 | 9 | 1.31 |
|
| |||||
| filtered | 48 (31%) | 39 (35%) | 8 (11%) | 1.65 | |
|
| |||||
| comparison | 46 | 38 | 6 | 2.00 | |
|
| |||||
| By CpG status | |||||
| CpG | unfiltered | 2,213 | 1,539 | 422 | 4.82 |
|
| |||||
| filtered | 2,030 (8%) | 1,390 (9%) | 397 (6%) | 5.12 | |
|
| |||||
| comparison | 2,098 | 1,448 | 350 | 5.44 | |
|
| |||||
| non-CpG | unfiltered | 16,415 | 10,218 | 2,585 | 2.75 |
|
| |||||
| filtered | 14,752 (10%) | 8,852 (13%) | 2,338 (9%) | 3.03 | |
|
| |||||
| comparison | 14,822 | 9,901 | 2,145 | 3.11 | |
|
| |||||
The number of novel variant sites (defined here as not present in dbSNP 129) is another metric of SNP-call quality (Supplementary Table 1). Most novel variants have low frequency, and are especially enriched in the singletons and doubletons. Singletons and doubletons are particularly important to distinguish from false positives because technical artifacts or errors in data processing can easily manifest themselves as novel variation.
Statistics such as transition/transversion ratio (Ti/Tv) and the number of novel variants are useful as gross guides to the quality of the dataset and enable comparison of two sets of calls from the same dataset. However, precise expectations of these statistics are unknown because they depend on many factors, including uneven coverage, variability in DNA quality, or other sources of technical bias such as machine error. Therefore, interpreting small differences from expectation in these statistics is nontrivial. Genotyping validation provides an additional measure of callset quality, independent of the population-genetics statistics. Comparing genotyping data to sequencing data enables directly measuring callset quality by calculating the non-reference sensitivity (“NRS” — the rate at which non-reference sites in the genotyping data are recovered in the sequencing data) and non-reference discrepancy rate (“NRD” — the rate at which genotypes from sequencing and genotyping data differ). A genotyping assay should include sites at various allele frequencies, especially low frequencies (∼1%). When available, family data, particularly trios, can also be useful to assess callset quality.
Comparing our callset with GWAS data in the same samples at overlapping sites suggested high sensitivity for common variants (98.6% NRS). To assess quality of low frequency variant calls in a comparable sequencing dataset we compared CGI data to Omni chip from the 1000 Genomes project (Supplementary Table 2). This comparison resulted in 95.65% NRS, 1.79% NRD and 1.12% NRD for novel variants.
Despite stringent QC, genotyping and sequencing errors are still present. Unfortunately, when stratifying variants based on putative functional consequences, the class of variation that is annotated to be most deleterious is also more heavily enriched for errors56. This underscores the importance of rigorous quality control.
It is critical that great care is taken to prevent technical biases and confounding in sequencing to avoid distorting association results. For instance, differences in how (rare and precious) case samples are handled compared to control samples may lead to systematic false-positives that masquerade as interesting associations. Likewise, simultaneous multi-sample variant calling on only cases or only controls may lead to differential detection of variants across batches, negatively impacting the accuracy of the allele frequency estimates and association analyses. Many other, often poorly understood or hidden, technical confounders (e.g., DNA preparation, exome-capture technology, machine type, read length, depth of coverage, SNP calling algorithm, QC filters) may influence the properties of exome-sequencing data. Therefore, although the use of shared controls (e.g., from the 1000 Genomes project) has been helpful in “filtering” approaches applied to Mendelian disorders6, 9, it is not likely to be applicable to association analysis of complex diseases.
Statistical methods for the analysis of rare variants
Analysis of rare variants requires statistical methods that are fundamentally different from association statistics used for testing common variants. There are two reasons for this. First, rare variants have to be combined in a gene (or pathway) for an association test to reach sufficient power57. For example, a causal SNP at a frequency of 1 in 500 and genotype relative risk of 10 in a sample of 200 cases and 200 controls, has 0.2% power to be detected at a conventional significance threshold for GWAS (P < 5 × 10−8). Second, functional and population genetics information can be added to the testing approach because exome sequencing comprehensively captures variation that can be annotated with such information.
Early candidate-gene sequencing studies for complex traits were based on the comparison of numbers of non-synonymous alleles exclusive to cases or controls (or samples at the extremes of the trait distribution)21, 26. This approach has limited power because it ignores common and low-frequency polymorphisms, as most such variants would be present in cases and controls. Recently, a number of statistical tests have been designed for rare-variant analysis. The Combined Multivariate and Collapsing (CMC) test58 jointly assesses the role of rare and common variation. For the common variants, traditional regression-based association is applied. For rare variation, an individual's predictor in a regression model is defined as 1 if the individual possesses at least one rare variant in the region (e.g., gene) and 0 otherwise. The Weighted-Sum Statistic (WSS) test59, creates a composite genotype score for all individuals. This score is the sum of alternate alleles weighted by the inverse of the binomial variance. A rank sum test is then performed on the genotype scores between phenotypic groups. The Kernel-Based Adaptive Cluster (KBAC) test60 also uses a weighting scheme that reflects apparent effect sizes of individual variants. An alternative approach to combine rare variants into a single test is to select an allele frequency threshold based on the observed data. This variable threshold (VT) approach61 was motivated by population genetic simulations that showed that there is no single optimal weighting scheme or allele frequency threshold. There are numerous other statistical tests for rare variants in complex traits (reviewed in refs.62–65).
In simulation studies64, most tests behave similarly in many situations. However, the results may depend on assumptions used in simulated data. The relative power to detect association depends on factors such as the number and proportion of causal variants, their population frequency, and their effect sizes, as well as directionality of effects, the number of genes contributing to the trait, and fraction of causal genetic variation located in the exome. Statistical tests were developed with various combinations of these factors in mind and therefore are likely to be sensitive to different disease architectures. For example, the simulation framework used in development of the WSS test assumes effect size proportional to 1/x(1 − x) (where x is the population frequency of the causal allele), while Sequence Kernel Association Test (SKAT)66 simulation framework uses effect size proportional to −log(x), and the VT test simulations uses a demographic history model with a range of possible values of strength of selection leading to different relationships between effect size and x. These simulations were designed to demonstrate the strengths of each methods under different effect-size distributions: the WSS is designed for effect sizes proportional to 1/x(1 − x), SKAT is designed for effect sizes proportional to beta density β(x; a1, a2) for prespecified a1 and a2, the C-alpha67 test is designed for effects going in opposite directions in the same region, while the VT test makes no assumptions about effect-size distributions.
When combining rare variants, all functional variants may either be assumed to influence the trait in the same direction, or some may be allowed to have opposite directions of effects. A biochemical argument can be made that most of non-synonymous variants are loss-of-function hypomorphs, while gain-of-function variants are infrequent. However, some genes (e.g., PCSK968) have variants of both kinds. Several tests allow for rare variants to have opposite effects on the trait (e.g., Step-up69, C-alpha, replication-based test70, SKAT). These tests are based either on the analysis of over-dispersion or on explicit linear models that determine the contribution of a variant to a score based on the direction of effect observed in the data.
Rare-variant tests can benefit from stratifying or weighting rare alleles by functional significance, as evidenced by simulations and sequencing studies of candidate genes61,64, 71–73. The power of rare-variant tests is strongly influenced by the fraction of causal variants among all variants analyzed and using functional information is an effective way to give greater weight to likely causal variants. For example, nonsense variants should be prioritized higher than nonconserved missense variants. Similarly, missense variants should be prioritized higher than synonymous variants. Functional consequences of variants can be predicted by examining effects of amino-acid changes using comparative sequence analysis and protein structure analysis. Many computational prediction and conservation74, 75 methods are available (reviewed in refs.76–79). The accuracy of those methods is around 80%80 and it is likely highest for rare variants (truly functional variants are most likely deleterious and kept at low frequencies by purifying selection, and so common variants are most likely neutral and nonfunctional). Therefore, using prediction methods enriches for functional variants and thus boosts the power of association tests. Because such predictions are not perfect, however, they should be used quantitatively by weighing variants, rather than qualitatively by filtering out variants. A number of tests allow including prediction scores into test statistics, e.g., VT test, KBAC, SKAT, Rare variant Weighted Aggregate Statistic (RWAS)72, Likelihood Ratio Test (LRT)73. The PLINK/SEQ suite includes precomputed PolyPhen-246 prediction scores for all possible missense changes in humans, which makes these scores readily applicable.
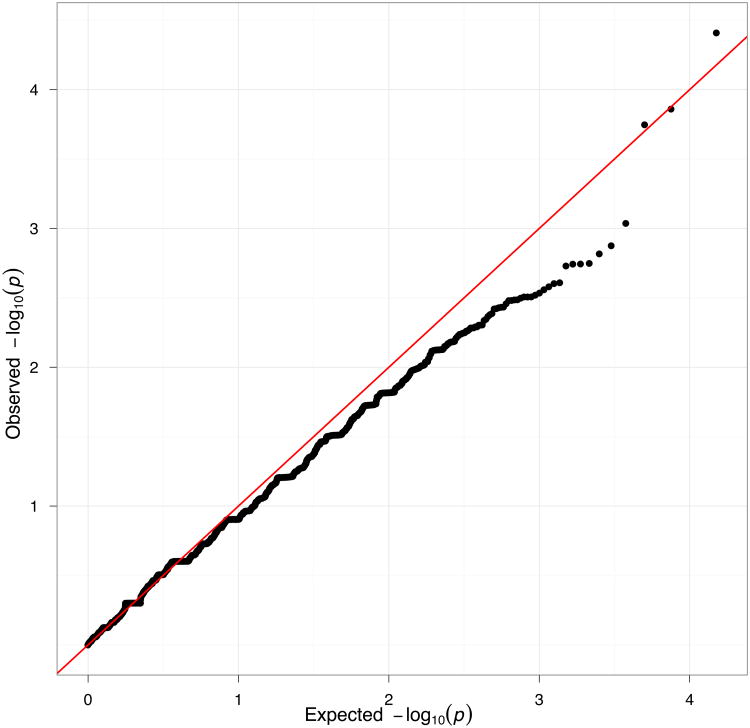
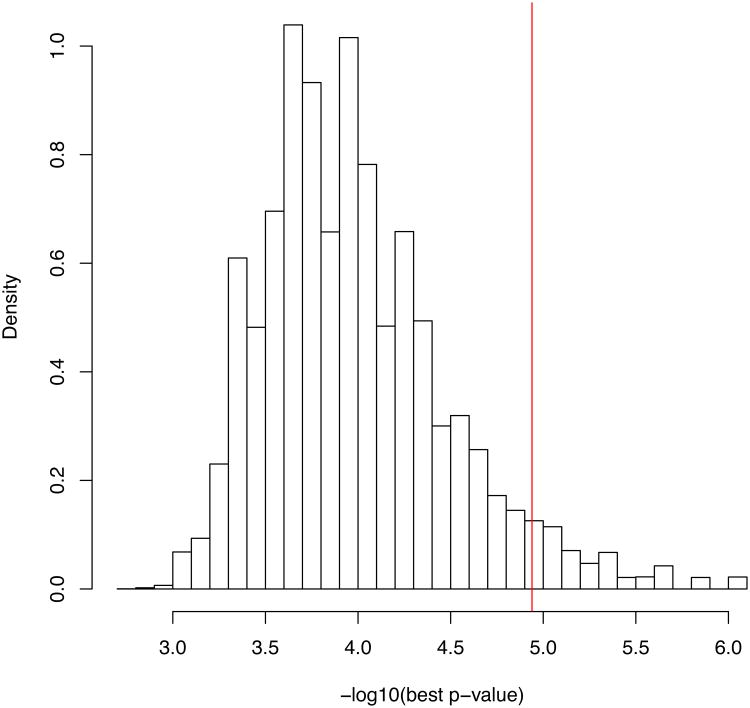
An important consideration for exome sequencing studies is selecting the significance threshold that accounts for multiple testing. A simple way is to adopt a Bonferroni correction for 20,000 independent tests (one test per each gene), which, for an experiment- wide significance of 0.05 gives a p-value threshold of 2.5 × 10−6 per gene. However, such a threshold may be overly conservative because it assumes that each tested gene has sufficient variation to achieve the asymptotic properties for the test statistic. For example, if only 2 individuals carry non-synonymous variants in a given gene, the difference between cases and controls never exceeds 2 total observations, and so the most significant p-value that can be achieved is around 0.25 assuming that these 2 variants are independent. Therefore, unless the study is large, association p-values will be generally less significant than expected under the null hypothesis. Figure 2a demonstrates this effect on the 438 whole exomes. The PLINK/SEQ suite computes from data the so-called i-stat, which is an estimate of the minimal achievable p-value for a gene. The i-stat can be used by setting a threshold (e.g., 10−3) and only correcting for the number of genes that have the i-stat below the threshold following the idea that for the genes with i-stat above the threshold there is no power to find an association. Another way to correct for multiple testing is to compute an experiment-wide significance threshold by permutations of phenotype labels, create the empirical distribution of minimal p-values for all genes across permutations, and compare the minimal p-value from the real data to that distribution (Figure 2b). This approach efficiently controls Type-I error and is less conservative than the Bonferroni correction. Importantly, the p-value threshold computed by permutations is dependent on both the study and on the statistical test. However, the experiment-wide correction via permutation is not robust to confounding and it is essential to assess the quality of the distribution of test statistics, for those genes that have i-stats less than the threshold, to ensure appropriate calibration of the distribution. Nevertheless, with increasing sample sizes, the dimensionality of the tests will also increase, and studies will be assessing close to 20,000 tests. Therefore, for large studies we consider the Bonferroni threshold to be preferable.
Figure 2.
Association analysis. (a) Q-Q plot of association p-values under the null hypothesis. (b) Distributions of lowest p-values under whole-exome permutations. The histograms show the distributions of the lowest p-values across permutations for the T5 test. The red vertical line indicates the 0.05 exome-wide significance level for the most significant gene (i.e., the most significant gene is exome-wide significant if its p-value is lower that the level indicated by the red line).
Statistical power of exome sequencing studies
Power of an exome-sequencing study is limited by the amount of variation in a gene. Therefore, power is higher for genes with more variants, for example longer genes or genes in regions of elevated mutation rate. Additionally, genes in which most variants are causal are easier to identify than those in which few variants are causal. In individual candidate gene sequencing studies estimates of this proportion ranged from 30% to 70%21, 22, 26. Consequently, the effect size is not only a property of an individual variant, but rather a reflection of the distribution of effects coupled with how those effects are interpreted via the test. Some statistical tests explicitly account for differences in power when evaluating evidence of association81.
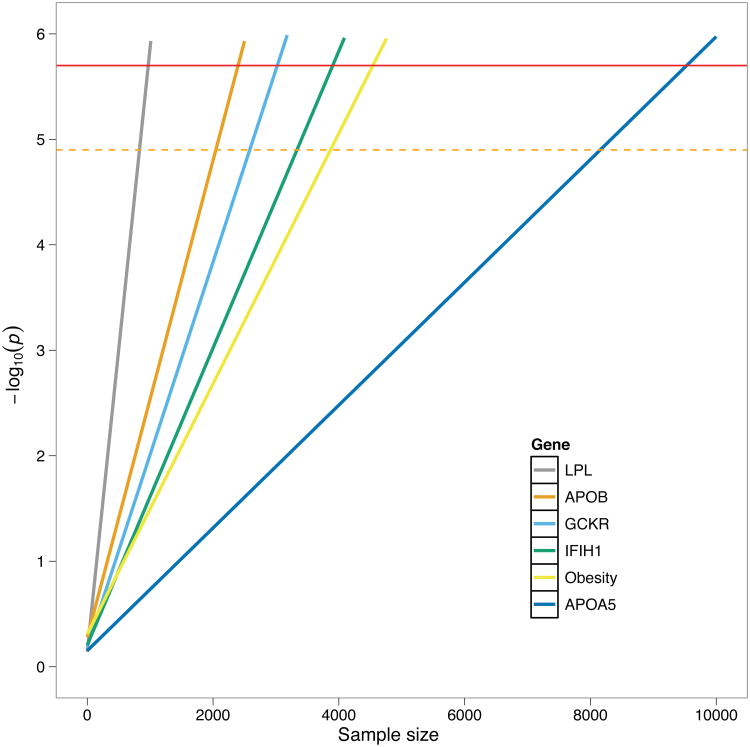
Given the sample sizes, the likely effect sizes and frequencies of causal variants, and the proportion of causal variants in a gene, do current exome sequencing studies have sufficient power to detect genes underlying complex phenotypes? The enthusiasm about exome sequencing studies stems, in part, from successful candidate gene sequencing studies, and so we sought to test whether exome sequencing would be expected to have sufficient power to detect genes discovered by the candidate gene approach. So far, no published candidate gene study reported p-values that would be significant on the background of the complete exome (Table 2). This is particularly striking because some candidate gene studies used much larger sample sizes (thousands of individuals) than ongoing exome sequencing studies (hundreds of individuals). This demonstrates that current exome sequencing studies are underpowered to detect genes with the allelic distribution and effect sizes similar to the published examples. Indeed, extrapolation of effect sizes and frequencies from published studies shows (Figure 3) that thousands of individuals are required to reach acceptable statistical power. This analysis is consistent with an earlier study based on population genetic simulations that concluded that as many as 10,000 individuals at phenotypic extremes would be needed to achieve satisfactory power30. The very first GWAS82–84 were also highly underpowered but the combination of falling costs and combining studies in meta-analyses enabled rapid creation of well-powered studies and many discoveries. Similarly, with the falling cost of sequencing and targeted enrichment85, exome sequencing will soon be affordable to many research groups, and we expect that consortia will form to facilitate pooling of exome sequencing data, thus enabling better powered studies and a new wave of discoveries.
Table 2.
Summary of gene burden test results for rare variant studies. This table summarizes gene burden test results from published candidate gene resequencing studies. The table shows that only one signal for the LPL gene is strongly associated (P = 2.47 × 10−5) but does not attain a genome-wide significance of P < 2.5 × 10−6 (P < 0.05 after applying a Bonferroni correction for 20,000 genes tested). This highlights the importance of sequencing large numbers of samples. Abbreviations: a: allele count for non-reference allele, b: as reported in published study, RVE rare variant exclusive test. Counts for ref.26 and ref.21 are as reported in published study. Counts for ref.22 are based on “functional mutation carriers” as described in the published study. Counts for ref.25 and ref.24 are based on SNPs with minor allele frequency (MAF) < 0.01. Counts for ref.23 are based on SNPs with MAF < 0.01 in controls only. All p-values are 2-sided tests unless reported otherwise in published study.
| Trait | Gene | Test | aAC low | AC high | n | P | Ref |
|---|---|---|---|---|---|---|---|
| TG | ANGPTL4 | Fisher's exact | 13 | 2 | 1775 | b0.016 | 26 |
| TG | ANGPTL5 | Fisher's exact | 9 | 1 | 1775 | b0.022 | |
| HDL | ABCA1 | RVE | 28 | 4 | 519 | b<0.0001 | 21 |
| APOA1 | 1 | 0 | 519 | ||||
| LCAT | 6 | 1 | 519 | ||||
| BP | SLC12A1/3, KCNJ1 | Fisher's exact | 9 | 1 | 626 | 0.02 | 22 |
| Obesity | Obesity | Fisher's exact | 73 | 97 | 757 | 0.061 | 25 |
| T1D | IFIH1 | Fisher's exact | 21 | 39 | 960 | 0.025 | 24 |
| HyperTG | APOA5 | Fisher's exact | 1 | 5 | 765 | 0.25 | 23 |
| GCKR | Fisher's exact | 5 | 20 | 765 | 0.024 | ||
| LPL | Fisher's exact | 8 | 44 | 765 | 2.47×10−5 | ||
| APOB | Fisher's exact | 39 | 85 | 765 | 0.008 |
Figure 3.
Extrapolation of gene burden results. Horizontal solid red line shows Bonferroni genome-wide significance threshold of P = 2.5 × 10−6. Horizontal dashed line shows the threshold derived from whole-exome permutations (Figure 2b). For larger sample sizes, the permutation threshold would be closer to the Bonferroni threshold, asymptotically approaching it as the sample sizes increase.
Replication to confirm association
To discover robust associations, replication in exome sequencing studies will be critical. Because small early studies will be inevitably underpowered, no gene may achieve exome-wide statistical significance. In such cases, unless strict correction for multiple tests is performed, researchers should resist the temptation to apply a battery of statistical tests, each with various weighing schemes and variant selection. We strongly argue that an association can only be considered real if it has been replicated. A reasonable replication strategy is to select a few genes (e.g., 10), based on the strength of association86 from the discovery stage and prior biological plausibility. Sequencing and rare-variant associations must then be performed on new samples, using multiple-test correction threshold applied only to the (smaller) set of candidate genes.
Population stratification
Population stratification—systematic ancestry differences between cases and controls—is a well-studied confounder in genetic association studies87. In GWAS, commonly used approaches to correct for stratification include stratifying by population cluster (Structured Association), principal components analysis (PCA) and mixed models87–90. Genomic Control may also be applied, but it is generally more useful for assessing stratification than correcting for stratification87, 91.
An important question is whether population stratification can confound exome sequencing studies, and if so, how to correct for stratification in this context. Although excess-of-rare-variant tests are fundamentally different than single-variant tests, the possibility of stratification still exists because different ancestries within a structured population sample (e.g., African and European ancestry in African Americans, or northern European and southern European ancestry in European Americans) may have different allele frequency spectra due to their different demographic histories. For example, in an exome sequencing study in African Americans in which disease cases have more African ancestry than controls, one expects to see an excess of rare variants in cases, because African chromosomes carry more rare variants92.
We created a hypothetical case-control exome sequencing study involving real sequencing data and simulated phenotype data using 438 individuals, split in two populations (see Methods). To induce population stratification, we assigned case-control status to each sample randomly with a bias to take more cases from one population, and more controls from the other population. Association tests indicated inflated rates of spurious statistically significant p-values. We corrected for population stratification by modifying the permutation scheme to account for subpopulations. This correction was effective at controlling Type-I errors in all association tests.
Our simulations demonstrate that exome-sequencing studies can be affected by population stratification, which may produce spurious associations. We have shown that a simple permutation scheme is sufficient to correct for population stratification when discrete clusters corresponding to genome-wide ancestry are known or can be inferred by applying standard methods to GWAS chip data88, 89, 93. The permutation scheme is appealing in that it generalizes most burden of multiple rare variants tests, however, some tests may also be amenable to the use of PCA covariates in instances in which population structure is best described by continuous clines rather than discrete clusters89.
Conclusion
Exome sequencing studies bring the promise of comprehensive testing of coding variation in an unbiased manner. However, we expect that initial studies will be underpowered, and we have highlighted a number of technical issues that could bias the interpretation and analysis of rare variant data, especially novel variants. We expect that thousands of exomes are going to be required to achieve sufficient statistical power to robustly detect associations of rare variation with complex traits. Issues we discussed in this Perspective are also relevant to future whole-genome sequencing studies, in which the analysis of protein-coding variation will remain the same as in the case of exomes.
Focusing exclusively on exome is an especially serious limitation in complex trait genetics, where noncoding genetic variation is believed to play a larger role than in Mendelian genetics or in somatic cancer genetics. However, there are clear reasons to start with exomes. First, statistical approaches combining multiple rare variants are problematic in non-coding regions because there is no easily identifiable set of sites harboring variants with unidirectional phenotypic effects. Second, variants in regulatory regions are likely to have smaller effect sizes. In contrast, protein coding genes provide a well-defined and interpretable target for mutations in the locus. These mutations create variants that, in a well-powered study, highlight association of the locus with the trait. Thus, although focusing on the exome is unlikely to explain all of heritability, it has the potential to highlight genes involved in complex traits.
Despite challenges discussed in this Perspective, the observation that a large trove of functionally significant coding variants exists in the human population brings hope that the exome sequencing approach will ultimately help identify many loci important for complex traits and diseases.
Methods
Simulating discovery of novel variants
To calculate the discovery rate of novel variants for increasing numbers of samples, first all exome samples are arranged in a random order. Then, samples are analyzed sequentially, starting with the first sample, and the cumulative set of identified variants is computed. For every subsequent sample, a variant site is considered novel if that site has not been identified as variant in the cumulative set of preceding samples. The fold-increase over baseline (where the baseline for each class is the number of variants discovered in the first sample) is plotted in Figure 1. To avoid sampling bias, random resampling is performed and the overall mean is calculated. Nonsense, Missense, and Synonymous classes are based on RefSeq annotations. The Missense class is further divided into “Probably damaging”, “Possibly damaging”, and “Benign” subclasses according to PolyPhen-2 predictions46. The “Theoretical” line plots the expected number of segregating sites under a neutral model of evolution in a population of constant size41.
Data generation
Reads were aligned to the reference genome using Burrows-Wheeler Aligner (BWA)94, PCR duplicate reads were removed using Picard (see Web Resources), base quality scores were recalibrated using the Genome Analysis Toolkit (GATK), and alignments near putative indels were refined using GATK. The resulting data was run through the GATK to discover and genotype SNP candidates.
QC filters
We used the following QC filters: a (1) quality-score-vs.-depth filter, which excludes variants whose depth-normalized discovery confidence does not exceed 2.0; (2) a homopolymer-run filter, which excludes variants that have an alternate allele that matches the allele in an immediately adjacent homopolymer-run of length greater than 5; (3) a strand-bias filter, which excludes variants whose alternate allele is preferentially found on one of the two available read orientations at the site, and (4) an indel-mask filter, which excludes variants discovered at sites that overlap with indels.
Association analysis
Case/control status was assigned randomly and a T5 test for burden of rare variants was executed on all genes (T5 is a variant of the CMC test58 that considers only non-synonymous variants with minor allele frequencies below 5%, uses the total count of alternative minor alleles in cases as the test statistic, and assigns significance by permuting phenotype labels). The overall deflation in significant p-values (i.e., there are fewer genes associated at any significance level than expected by chance, is due to low counts of variants in genes. Results were similar for T1 version of CMC, as well as for WSS59, and VT tests61. This pattern is expected in studies with small sample sizes (below around 1000 individuals). Whole-exome permutations can be used to establish exome-wide significance in such cases.
Whole-exome permutations
Phenotype labels of full exomes were permuted 1,000,000 times, i.e., permuted phenotype affected all genes in an individual. In each permutation, the lowest exome-wide p-value was computed. It took fewer than 1000 computing hours to run 8 statistical tests on the 1 million whole-exome permutations of 15,122 genes in 438 individuals. The computation is very easy to parallelize and thus quite affordable using cluster or cloud computing.
Power calculations
Data was extrapolated from results from five candidate-genes and one obesity gene set from published studies (Table 2). Fisher's exact test was used to calculate p-values after sample size extrapolations.
Population stratification
We induced population stratification in a hypothetical exome sequencing study involving real sequencing data and simulated phenotype data using 184 individuals from the HIV and 254 individuals from Schizophrenia (SCZ) exome sequencing studies. We observed that there were exome-wide differences in allele frequencies between the populations, which we quantified by estimating the FST between HIV and SCZ samples using exome sequencing data95. FST was estimated using the EIGENSOFT software. Using variants with minor allele frequencies at least 5%, we observed an FST value of 0.003, which is consistent with the different European ancestries of the HIV (European-American) and SCZ (Swedish) samples and with previous estimates of genetics distances between European populations96. We considered the possibility that the observed differences between HIV and SCZ samples could be due to differential bias resulting from differences in sample collection, sequencing, or data processing97, but view this as unlikely because we applied identical data processing and QC procedures to both sample sets and because QC metrics revealed no systematic differences between the sample sets.
To induce population stratification, we randomly assigned 80% of samples from HIV samples and 20% of SCZ samples as cases, and remaining samples as controls. We then used case-control labels to run four association tests: fixed-threshold approach (T1 and T5 versions of the CMC test58), WSS59, and VT test61. We quantified the evidence of population stratification by considering the most significant p-value (of 15,122 genes) and the proportion of p-values < 0.05, and < 0.01. As seen in the null distribution (Figure 2), it is expected that, due to low counts, p-values will have a deficiency of statistically significant signals. Before correction for population stratification, however, our metrics indicate an excess of statistically significant signals. For example, for T5, the most significant p-value was <0.000001, and the proportions of p-values were 0.0595 at level 0.05, and 0.0136 at level 0.01. Results were similar for the other statistical tests, and for other proportions of HIV samples assigned as cases (we experimented with 90%, 80%, and 70%, as well as 30%, 20%, and 10%). We note that when the proportion of HIV individuals assigned as cases was above 50% the induced inflation was higher than when the proportion was below 50%, which could be due to a population genetic excess of rare variants in Swiss and European-American samples as compared to Swedish samples.
To correct for stratification, we modified the script that implements association tests (see Web resources) to employ a permutation scheme in which case/control status was permuted within each population (HIV and SCZ), assuming known population labels. This permutation scheme does not change the computational cost of the study. The results show that the permutation procedure adequately controlled for population stratification, removing the excess of significant signals. For example, for T5, the most significant p-value after correction was 0.0001 and the proportions of p-values were 0.0340 at level 0.05, and 0.0060 at level 0.01. As mentioned previously, the deficiency of statistically significant signals is due to low counts and is consistent with the null distribution (Figure 2). Results were similar for the other statistical tests, and for other proportions of HIV samples assigned as cases. These results show that the permutation-based correction was effective at controlling Type-I errors.
Supplementary Material
Acknowledgments
This work was made possible, in part, by National Institutes of Health Grant 5R01MH084676, and in part, by the International HIV Controllers Study, supported by the Collaboration for AIDS Vaccine Discovery of the Bill and Melinda Gates Foundation (to P.I.W.d.B.), and the AIDS Clinical Trials Group, supported by NIH grants AI069513, AI34835, AI069432, AI069423, AI069477, AI069501, AI069474, AI069428, AI69467, AI069415, Al32782, AI27661, AI25859, AI28568, AI30914, AI069495, AI069471, AI069532, AI069452, AI069450, AI069556, AI069484, AI069472, AI34853, AI069465, AI069511, AI38844, AI069424, AI069434, AI46370, AI68634, AI069502, AI069419, AI068636, RR024975, and AI077505. The sequencing of the SCZ control individuals was funded by 3 sources: NIH grant RC2MH089905, the Herman Foundation, and the Stanley Medical Research Institute. N.O.S. was supported in part by National Institutes of Health Training Grant T32-HL07604-25, Brigham and Women's Hospital, Division of Cardiovascular Medicine. B.M.N. was supported by 1R01MH089208-01. The authors are grateful to Samuela Pollack for assistance with EIGENSOFT. The views expressed in this presentation do not necessarily represent the views of the NIMH, NIH, HHS, or the United States Government.
Footnotes
- http://www.completegenomics.com/sequence-data/download-data (Complete Genomics dataset)
- http://genetics.bwh.harvard.edu/rare_variants (R script used for all association analyses in this Perspective. Contains T1, T5, WSS and VT tests, optionally weighted PolyPhen-2 predictions)
- http://www.hsph.harvard.edu/faculty/alkes-price/software (EIGEN-SOFT software)
- http://picard.sourceforge.net/index.shtml (Picard utilities for manipulation of Sequence Alignment/Map, or SAM, files)
- http://bio-bwa.sourceforge.net (Burrows-Wheeler Aligner, or BWA)
- http://atgu.mgh.harvard.edu/plinkseq (PLINK/SEQ library helps with management, QC, and analysis of exome sequencing data, including several statistical tests mentioned in the text)
Competing Interests: The authors declare that they have no competing financial interests.
Author contributions: A.K. developed the computational analysis pipeline and analyzed data, K.G. performed upstream quality control and analysis of sequencing data, R.D. performed the power analysis, N.O.S. performed the assessment of rare variants in empirical data, B.M.N. contributed to statistical analyses, P.J.M. assisted with data analysis. T.L. participated in designing the study. P.S., P.F.S., J.L.M., C.M.H., P.L., P.M., P.I.W.D.B., N.G. and S.M.P. contributed data. A.L.P., P.I.W.D.B., and S.R.S. conceived and designed the study. A.L.P., P.I.W.D.B., S.M.P., and S.R.S. supervised the work. A.K., K.G., R.D., N.O.S., B.N.M., Y.Y.S., A.L.P., P.I.W.D.B., and S.R.S. wrote the manuscript. All authors approved the manuscript.
Data access. The SCZ control data can be accessed via the Database of Genotypes and Phenotypes (dbGAP), accession code phs000473.v1.p1. To access the HIV data, investigators can submit a brief concept sheet detailing their study design, research questions and other needs. The concept sheet with detailed instructions can be downloaded from: http://cfar.globalhealth.harvard.edu/fs/docs/icb.topic938249.files/Harvard%20CFAR%20Concept%20Sheet%20Template%20.docx
Please e-mail completed forms to Pamela Richtmyer (prichtmyer@partners.org). Requests will be reviewed on the basis of scientific merit, feasibility and overlap with ongoing concept sheets/investigations.
References
- 1.Fuller CW, et al. The challenges of sequencing by synthesis. Nature Biotechnology. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 2.Rusk N, Kiermer V. Primer: Sequencing—the next generation. Nature Methods. 2008;5:15. doi: 10.1038/nmeth1155. [DOI] [PubMed] [Google Scholar]
- 3.Metzker ML. Sequencing technologies the next generation. Nature Reviews Genetics. 2009;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 4.Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 5.Clarke J, et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotechnology. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 6.Ng SB, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature Genetics. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Human Molecular Genetics. 2010;19:R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedges DJ, et al. Comparison of Three Targeted Enrichment Strategies on the SOLiD Sequencing Platform. PLoS ONE. 2011;6:e18595. doi: 10.1371/journal.pone.0018595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human ex- omes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierce SB, et al. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. The American Journal of Human Genetics. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krawitz PM, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nature Genetics. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 12.Wang JL, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain: A journal of neurology. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 13.Ng SB, Nickerson DA, Bamshad MJ, Shendure J. Massively parallel sequencing and rare disease. Human Molecular Genetics. 2010;19:R119–R124. doi: 10.1093/hmg/ddq390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musunuru K, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. The New England Journal of Medicine. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoischen A, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nature Genetics. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Q, et al. Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome Biology. 2010;11:R114. doi: 10.1186/gb-2010-11-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nature Genetics. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF com- plex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal N, et al. Exome Sequencing of Head and Neck Squamous Cell Carcinoma Reveals Inactivating Mutations in NOTCH1. Science. 2011;28 doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang H, et al. Exome Sequencing Reveals Comprehensive Genomic Alterations across Eight Cancer Cell Lines. PLoS ONE. 2011;6:e21097. doi: 10.1371/journal.pone.0021097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 22.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nature Genetics. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nature Genetics. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahituv N, et al. Medical sequencing at the extremes of human body mass. The American Journal of Human Genetics. 2007;80:779–791. doi: 10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo S, et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. Journal of Clinical Investigation. 2009;119:70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? The American Journal of Human Genetics. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Human Molecular Genetics. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 29.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. The American Journal of Human Genetics. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyko AR, et al. Assessing the Evolutionary Impact of Amino Acid Mutations in the Human Genome. PLoS Genetics. 2008;4:13. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williamson SH, et al. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7882–7887. doi: 10.1073/pnas.0502300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eyre-Walker A, Woolfit M, Phelps T. The Distribution of Fitness Effects of New Deleterious Amino Acid Mutations in Humans. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yampolsky LY, Kondrashov FA, Kondrashov AS. Distribution of the strength of selection against amino acid replacements in human proteins. Human Molecular Genetics. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- 35.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human Mutation. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 38.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y, et al. Human Y Chromosome Base-Substitution Mutation Rate Measured by Direct Sequencing in a Deep-Rooting Pedigree. Current Biology. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereyra F, et al. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science. 2010;1551:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ewens WJ. The sampling theory of selectively neutral alleles. Theoretical Population Biology. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- 42.Kimura M. Molecular evolutionary clock and the neutral theory. Journal of Molecular Evolution. 1987;26:24–33. doi: 10.1007/BF02111279. [DOI] [PubMed] [Google Scholar]
- 43.Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coventry A, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nature Communications. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nature Genetics. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 46.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7 doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halushka MK, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genetics. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 48.Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genetics. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 49.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 50.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends in Genetics. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 51.Sunyaev S, et al. Prediction of deleterious human alleles. Human Molecular Genetics. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 52.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hellmann I, et al. Selection on Human Genes as Revealed by Comparisons to Chimpanzee cDNA. Genome Research. 2003;13:831–837. doi: 10.1101/gr.944903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacArthur DG, Tyler-Smith C. Loss-of-function variants in the genomes of healthy humans. Human Molecular Genetics. 2010;19:R125–R130. doi: 10.1093/hmg/ddq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 58.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. The American Journal of Human Genetics. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Madsen BE, Browning SR. A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic. PLoS Genetics. 2009;5:11. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu DJ, Leal SM. A Novel Adaptive Method for the Analysis of Next-Generation Sequencing Data to Detect Complex Trait Associations with Rare Variants Due to Gene Main Effects and Interactions. PLoS Genetics. 2010;6:14. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price AL, et al. Pooled Association Tests for Rare Variants in Exon-Resequencing Studies. The American Journal of Human Genetics. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nature Reviews Genetics. 2010;11:773–785. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annual Review of Genetics. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 64.Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genetic Epidemiology. 2011 doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stitziel NO, Kiezun A, Sunyaev SR. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biology. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu MC, et al. Rare Variant Association Testing for Sequencing Data Using the Sequence Kernel Association Test (SKAT) The American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neale BM, et al. Testing for an Unusual Distribution of Rare Variants. PLoS Genetics. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kotowski IK, et al. A Spectrum of PCSK9 Alleles Contributes to Plasma Levels of Low-Density Lipoprotein Cholesterol. The American Journal of Human Genetics. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffmann TJ, Marini NJ, Witte JS. Comprehensive Approach to Analyzing Rare Genetic Variants. PLoS ONE. 2010;5:9. doi: 10.1371/journal.pone.0013584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ionita-Laza I, Buxbaum JD, Laird NM, Lange C. A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease. PLoS Genetics. 2011;7:e1001289. doi: 10.1371/journal.pgen.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tavtigian SV, et al. Rare, evolutionarily unlikely missense substitutions in ATM confer increased risk of breast cancer. The American Journal of Human Genetics. 2009;85:427–446. doi: 10.1016/j.ajhg.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sul JH, Han B, He D, Eskin E. An optimal weighted aggregated association test for identification of rare variants involved in common diseases. Genetics. 2011;188:181–188. doi: 10.1534/genetics.110.125070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sul JH, Han B, Eskin E. Increasing Power of Groupwise Association Test with Likelihood Ratio Test; Proceedings of the Fifteenth Annual Conference on Research in Computational Biology; Vancouver. 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Research. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cooper GM, et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nature Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annual Review of Genomics and Human Genetics. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 77.Jordan DM, Ramensky VE, Sunyaev SR. Human allelic variation: perspective from protein function, structure, and evolution. Current Opinion in Structural Biology. 2010;20:342–350. doi: 10.1016/j.sbi.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Human Mutation. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 79.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causing variants in a wealth of genomic data. Nature Reviews Genetics. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 80.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Human Mutation. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nature Reviews Genetics. 2009;10:681–690. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 82.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 83.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 84.Burton P, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 86.Lipman PJ, et al. On the follow-up of genome-wide association studies: an overall test for the most promising SNPs. Genetic Epidemiology. 2011;35:303–309. doi: 10.1002/gepi.20578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nature Reviews Genetics. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Price AL, et al. Principal components analysis corrects for stratification in genome- wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 90.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 92.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nature Genetics. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Durbin R. Fast and accurate short read alignment with Burrows Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nature Reviews Genetics. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature Genetics. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.