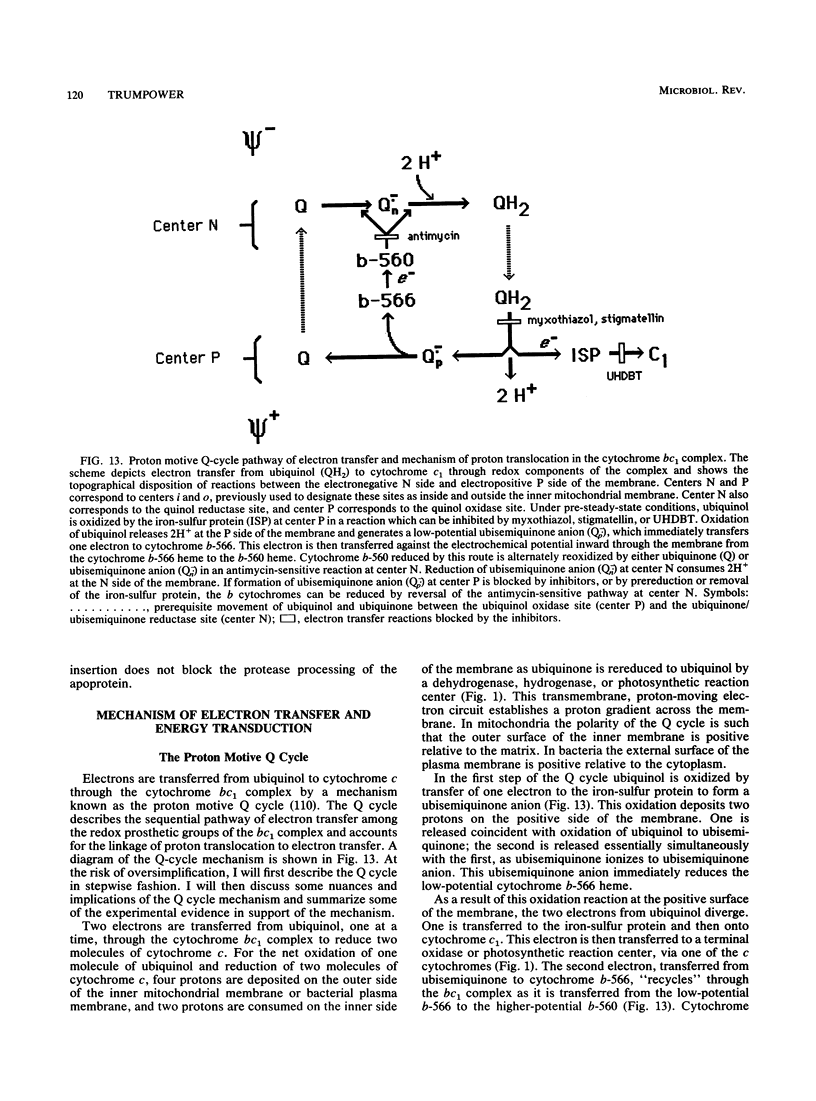
Abstract
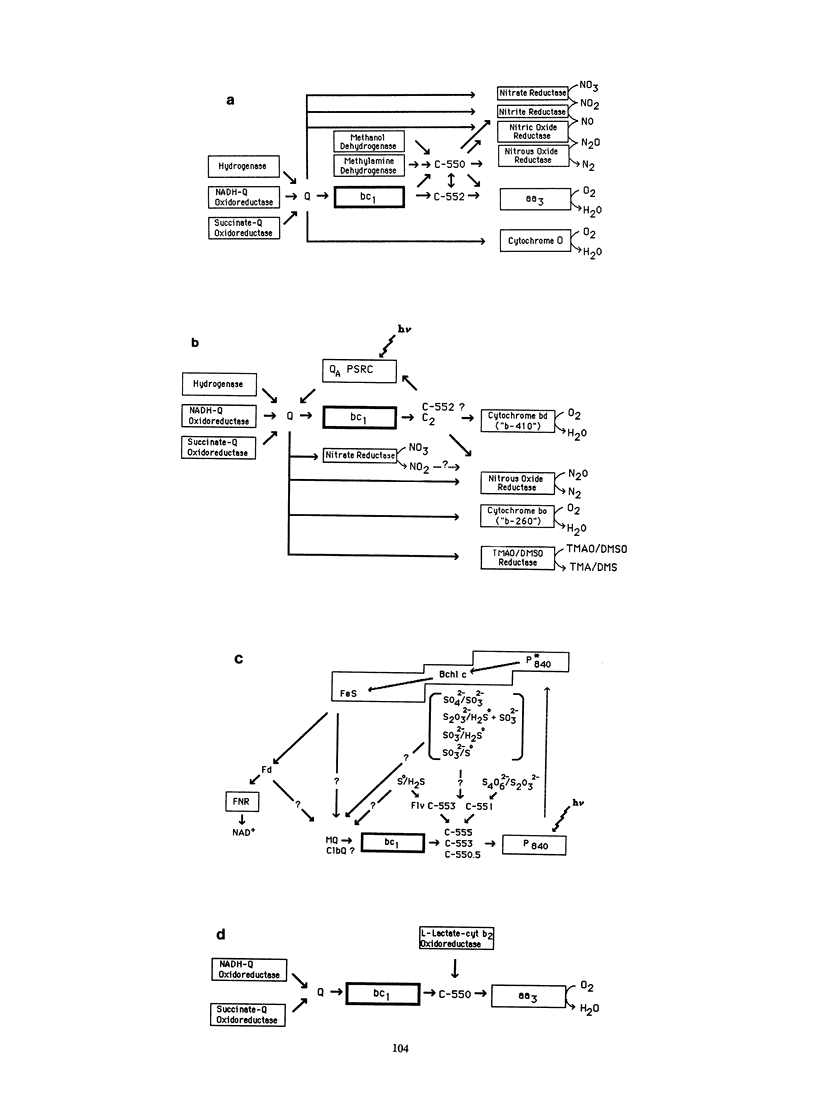
The cytochrome bc1 complex is the most widely occurring electron transfer complex capable of energy transduction. Cytochrome bc1 complexes are found in the plasma membranes of phylogenetically diverse photosynthetic and respiring bacteria, and in the inner mitochondrial membrane of all eucaryotic cells. In all of these species the bc1 complex transfers electrons from a low-potential quinol to a higher-potential c-type cytochrome and links this electron transfer to proton translocation. Most bacteria also possess alternative pathways of quinol oxidation capable of circumventing the bc1 complex, but these pathways generally lack the energy-transducing, protontranslocating activity of the bc1 complex. All cytochrome bc1 complexes contain three electron transfer proteins which contain four redox prosthetic groups. These are cytochrome b, which contains two b heme groups that differ in their optical and thermodynamic properties; cytochrome c1, which contains a covalently bound c-type heme; and a 2Fe-2S iron-sulfur protein. The mechanism which links proton translocation to electron transfer through these proteins is the proton motive Q cycle, and this mechanism appears to be universal to all bc1 complexes. Experimentation is currently focused on understanding selected structure-function relationships prerequisite for these redox proteins to participate in the Q-cycle mechanism. The cytochrome bc1 complexes of mitochondria differ from those of bacteria, in that the former contain six to eight supernumerary polypeptides, in addition to the three redox proteins common to bacteria and mitochondria. These extra polypeptides are encoded in the nucleus and do not contain redox prosthetic groups. The functions of the supernumerary polypeptides of the mitochondrial bc1 complexes are generally not known and are being actively explored by genetically manipulating these proteins in Saccharomyces cerevisiae.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alefounder P. R., Ferguson S. J. A periplasmic location for methanol dehydrogenase from Paracoccus denitrificans: implications for proton pumping by cytochrome aa3. Biochem Biophys Res Commun. 1981 Feb 12;98(3):778–784. doi: 10.1016/0006-291x(81)91179-7. [DOI] [PubMed] [Google Scholar]
- Anke T., Besl H., Mocek U., Steglich W. Antibiotics from basidiomycetes. XVIII. Strobilurin C and oudemansin B, two new antifungal metabolites from Xerula species (Agaricales). J Antibiot (Tokyo) 1983 Jun;36(6):661–666. doi: 10.7164/antibiotics.36.661. [DOI] [PubMed] [Google Scholar]
- Anke T., Hecht H. J., Schramm G., Steglich W. Antibiotics from basidiomycetes. IX. Oudemansin, an antifungal antibiotic from Oudemansiella mucida (Schrader ex Fr.) Hoehnel (Agaricales). J Antibiot (Tokyo) 1979 Nov;32(11):1112–1117. doi: 10.7164/antibiotics.32.1112. [DOI] [PubMed] [Google Scholar]
- Anraku Y. Bacterial electron transport chains. Annu Rev Biochem. 1988;57:101–132. doi: 10.1146/annurev.bi.57.070188.000533. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Beattie D. S., Clejan L., Chen Y. S., Lin C. I., Sidhu A. Orientation of complex III in the yeast mitochondrial membrane: labeling with [125I] diazobenzenesulfonate and functional studies with the decyl analogue of coenzyme Q as substrate. J Bioenerg Biomembr. 1981 Dec;13(5-6):357–373. doi: 10.1007/BF00743210. [DOI] [PubMed] [Google Scholar]
- Becker W. F., von Jagow G., Anke T., Steglich W. Oudemansin, strobilurin A, strobilurin B and myxothiazol: new inhibitors of the bc1 segment of the respiratory chain with an E-beta-methoxyacrylate system as common structural element. FEBS Lett. 1981 Sep 28;132(2):329–333. doi: 10.1016/0014-5793(81)81190-8. [DOI] [PubMed] [Google Scholar]
- Beckmann J. D., Ljungdahl P. O., Lopez J. L., Trumpower B. L. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J Biol Chem. 1987 Jun 25;262(18):8901–8909. [PubMed] [Google Scholar]
- Beckmann J. D., Ljungdahl P. O., Trumpower B. L. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. I. Construction of a RIP1 deletion strain and isolation of temperature-sensitive mutants. J Biol Chem. 1989 Mar 5;264(7):3713–3722. [PubMed] [Google Scholar]
- Berden J. A., Slater E. C. The allosteric binding of antimycin to cytochrome b in the mitochondrial membrane. Biochim Biophys Acta. 1972 Feb 28;256(2):199–215. doi: 10.1016/0005-2728(72)90053-9. [DOI] [PubMed] [Google Scholar]
- Berry E. A., Trumpower B. L. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985 Feb 25;260(4):2458–2467. [PubMed] [Google Scholar]
- Blumberg W. E., Peisach J. On the interpretation of electron paramagnetic resonance spectra of binuclear iron-sulfur proteins. Arch Biochem Biophys. 1974 Jun;162(2):502–512. doi: 10.1016/0003-9861(74)90210-0. [DOI] [PubMed] [Google Scholar]
- Boogerd F. C., van Verseveld H. W., Stouthamer A. H. Electron transport to nitrous oxide in Paracoccus denitrificans. FEBS Lett. 1980 May 5;113(2):279–284. doi: 10.1016/0014-5793(80)80609-0. [DOI] [PubMed] [Google Scholar]
- Borchart U., Machleidt W., Schägger H., Link T. A., von Jagow G. Isolation and amino acid sequence of the 9.5 kDa protein of beef heart ubiquinol:cytochrome c reductase. FEBS Lett. 1986 May 5;200(1):81–86. doi: 10.1016/0014-5793(86)80515-4. [DOI] [PubMed] [Google Scholar]
- Bowyer J. R., Dutton P. L., Prince R. C., Crofts A. R. The role of the Rieske iron-sulfur center as the electron donor to ferricytochrome c2 in Rhodopseudomonas sphaeroides. Biochim Biophys Acta. 1980 Oct 3;592(3):445–460. doi: 10.1016/0005-2728(80)90091-2. [DOI] [PubMed] [Google Scholar]
- Bowyer J. R., Edwards C. A., Ohnishi T., Trumpower B. L. An analogue of ubiquinone which inhibits respiration by binding to the iron-sulfur protein of the cytochrome b-c1 segment of the mitochondrial respiratory chain. J Biol Chem. 1982 Jul 25;257(14):8321–8330. [PubMed] [Google Scholar]
- Bowyer J. R., Edwards C. A., Trumpower B. L. Involvement of the iron-sulfur protein of the mitochondrial cytochrome b-c1 complex in the oxidant-induced reduction of cytochrome b. FEBS Lett. 1981 Apr 6;126(1):93–97. doi: 10.1016/0014-5793(81)81041-1. [DOI] [PubMed] [Google Scholar]
- Bowyer J. R., Trumpower B. L. Rapid reduction of cytochrome c1 in the presence of antimycin and its implication for the mechanism of electron transfer in the cytochrome b-c1 segment of the mitochondrial respiratory chain. J Biol Chem. 1981 Mar 10;256(5):2245–2251. [PubMed] [Google Scholar]
- Brandt U., Schägger H., von Jagow G. Characterisation of binding of the methoxyacrylate inhibitors to mitochondrial cytochrome c reductase. Eur J Biochem. 1988 May 2;173(3):499–506. doi: 10.1111/j.1432-1033.1988.tb14026.x. [DOI] [PubMed] [Google Scholar]
- Brasseur R. Calculation of the three-dimensional structure of Saccharomyces cerevisiae cytochrome b inserted in a lipid matrix. J Biol Chem. 1988 Sep 5;263(25):12571–12575. [PubMed] [Google Scholar]
- Brune D. C. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989 Jul 13;975(2):189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- Carr G. J., Page M. D., Ferguson S. J. The energy-conserving nitric-oxide-reductase system in Paracoccus denitrificans. Distinction from the nitrite reductase that catalyses synthesis of nitric oxide and evidence from trapping experiments for nitric oxide as a free intermediate during denitrification. Eur J Biochem. 1989 Feb 15;179(3):683–692. doi: 10.1111/j.1432-1033.1989.tb14601.x. [DOI] [PubMed] [Google Scholar]
- Carter K. R., Tsai A., Palmer G. The coordination environment of mitochondrial cytochromes b. FEBS Lett. 1981 Sep 28;132(2):243–246. doi: 10.1016/0014-5793(81)81170-2. [DOI] [PubMed] [Google Scholar]
- Coria R., García M., Brunner A. Mitochondrial cytochrome b genes with a six-nucleotide deletion or single-nucleotide substitutions confer resistance to antimycin A in the yeast Kluyveromyces lactis. Mol Microbiol. 1989 Nov;3(11):1599–1604. doi: 10.1111/j.1365-2958.1989.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Craske A., Ferguson S. J. The respiratory nitrate reductase from Paracoccus denitrificans. Molecular characterisation and kinetic properties. Eur J Biochem. 1986 Jul 15;158(2):429–436. doi: 10.1111/j.1432-1033.1986.tb09771.x. [DOI] [PubMed] [Google Scholar]
- Daldal F., Davidson E., Cheng S. Isolation of the structural genes for the Rieske Fe-S protein, cytochrome b and cytochrome c1 all components of the ubiquinol: cytochrome c2 oxidoreductase complex of Rhodopseudomonas capsulata. J Mol Biol. 1987 May 5;195(1):1–12. doi: 10.1016/0022-2836(87)90322-6. [DOI] [PubMed] [Google Scholar]
- Daldal F., Tokito M. K., Davidson E., Faham M. Mutations conferring resistance to quinol oxidation (Qz) inhibitors of the cyt bc1 complex of Rhodobacter capsulatus. EMBO J. 1989 Dec 20;8(13):3951–3961. doi: 10.1002/j.1460-2075.1989.tb08578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E., Daldal F. fbc operon, encoding the Rieske Fe-S protein cytochrome b, and cytochrome c1 apoproteins previously described from Rhodopseudomonas sphaeroides, is from Rhodopseudomonas capsulata. J Mol Biol. 1987 May 5;195(1):25–29. doi: 10.1016/0022-2836(87)90324-x. [DOI] [PubMed] [Google Scholar]
- De Haan M., van Loon A. P., Kreike J., Vaessen R. T., Grivell L. A. The biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. DNA sequence analysis of the nuclear gene coding for the 14-kDa subunit. Eur J Biochem. 1984 Jan 2;138(1):169–177. doi: 10.1111/j.1432-1033.1984.tb07896.x. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Verwiel P. E. Structure and activity of the prosthetic group of methanol dehydrogenase. Eur J Biochem. 1980;108(1):187–192. doi: 10.1111/j.1432-1033.1980.tb04711.x. [DOI] [PubMed] [Google Scholar]
- Dutton P. L., Leigh J. S. Electron spin resonance characterization of Chromatium D hemes, non-heme irons and the components involved in primary photochemistry. Biochim Biophys Acta. 1973 Aug 31;314(2):178–190. doi: 10.1016/0005-2728(73)90133-3. [DOI] [PubMed] [Google Scholar]
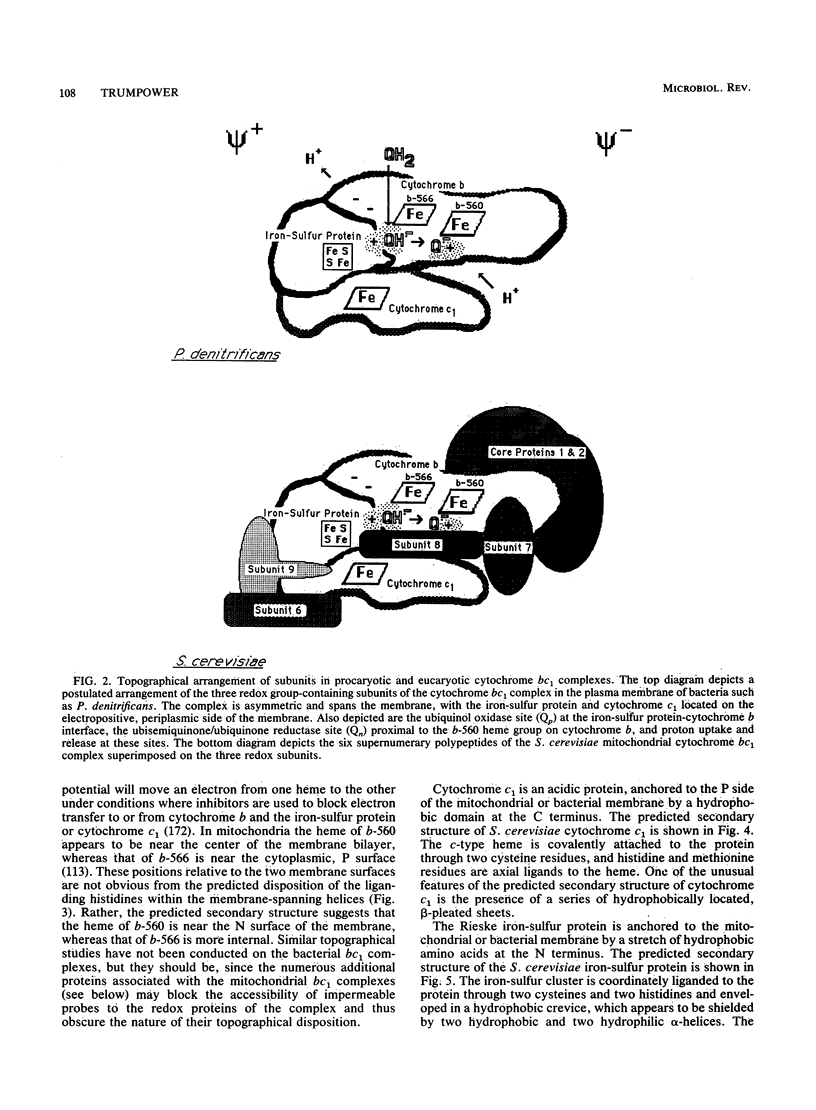
- Edwards C. A., Bowyer J. R., Trumpower B. L. Function of the iron-sulfur protein of the cytochrome b-c1 segment in electron transfer reactions of the mitochondrial respiratory chain. J Biol Chem. 1982 Apr 10;257(7):3705–3713. [PubMed] [Google Scholar]
- Eilers M., Schatz G. Protein unfolding and the energetics of protein translocation across biological membranes. Cell. 1988 Feb 26;52(4):481–483. doi: 10.1016/0092-8674(88)90458-8. [DOI] [PubMed] [Google Scholar]
- Entian K. D. Glucose repression: a complex regulatory system in yeast. Microbiol Sci. 1986 Dec;3(12):366–371. [PubMed] [Google Scholar]
- Ferguson S. J. Aspects of the control and organization of bacterial electron transport. Biochem Soc Trans. 1982 Aug;10(4):198–200. doi: 10.1042/bst0100198. [DOI] [PubMed] [Google Scholar]
- Fink G. R. Pseudogenes in yeast? Cell. 1987 Apr 10;49(1):5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu Rev Cell Biol. 1989;5:153–180. doi: 10.1146/annurev.cb.05.110189.001101. [DOI] [PubMed] [Google Scholar]
- Gabellini N., Harnisch U., McCarthy J. E., Hauska G., Sebald W. Cloning and expression of the fbc operon encoding the FeS protein, cytochrome b and cytochrome c1 from the Rhodopseudomonas sphaeroides b/c1 complex. EMBO J. 1985 Feb;4(2):549–553. doi: 10.1002/j.1460-2075.1985.tb03663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini N. Organization and structure of the genes for the cytochrome b/c1 complex in purple photosynthetic bacteria. A phylogenetic study describing the homology of the b/c1 subunits between prokaryotes, mitochondria, and chloroplasts. J Bioenerg Biomembr. 1988 Feb;20(1):59–83. doi: 10.1007/BF00762138. [DOI] [PubMed] [Google Scholar]
- Gabellini N., Sebald W. Nucleotide sequence and transcription of the fbc operon from Rhodopseudomonas sphaeroides. Evaluation of the deduced amino acid sequences of the FeS protein, cytochrome b and cytochrome c1. Eur J Biochem. 1986 Feb 3;154(3):569–579. doi: 10.1111/j.1432-1033.1986.tb09437.x. [DOI] [PubMed] [Google Scholar]
- Gerth K., Irschik H., Reichenbach H., Trowitzsch W. Myxothiazol, an antibiotic from Myxococcus fulvus (myxobacterales). I. Cultivation, isolation, physico-chemical and biological properties. J Antibiot (Tokyo) 1980 Dec;33(12):1474–1479. doi: 10.7164/antibiotics.33.1474. [DOI] [PubMed] [Google Scholar]
- González-Halphen D., Lindorfer M. A., Capaldi R. A. Subunit arrangement in beef heart complex III. Biochemistry. 1988 Sep 6;27(18):7021–7031. doi: 10.1021/bi00418a053. [DOI] [PubMed] [Google Scholar]
- Grivell L. A. Protein import into mitochondria. Int Rev Cytol. 1988;111:107–141. doi: 10.1016/s0074-7696(08)61732-5. [DOI] [PubMed] [Google Scholar]
- Gupte S. S., Hackenbrock C. R. The role of cytochrome c diffusion in mitochondrial electron transport. J Biol Chem. 1988 Apr 15;263(11):5248–5253. [PubMed] [Google Scholar]
- Gurbiel R. J., Batie C. J., Sivaraja M., True A. E., Fee J. A., Hoffman B. M., Ballou D. P. Electron-nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989 May 30;28(11):4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- Gutweniger H., Bisson R., Montecucco C. Membrane topology of beef-heart ubiquinone-cytochrome c reductase (complex III). J Biol Chem. 1981 Nov 10;256(21):11132–11136. [PubMed] [Google Scholar]
- Hahn S., Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988 Apr 15;240(4850):317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- Haley J., Bogorad L. A 4-kDa maize chloroplast polypeptide associated with the cytochrome b6-f complex: subunit 5, encoded by the chloroplast petE gene. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1534–1538. doi: 10.1073/pnas.86.5.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Pfanner N., Nicholson D. W., Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989 Jan 18;988(1):1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Schmidt B., Wachter E., Weiss H., Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986 Dec 26;47(6):939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Hauska G., Hurt E., Gabellini N., Lockau W. Comparative aspects of quinol-cytochrome c/plastocyanin oxidoreductases. Biochim Biophys Acta. 1983 Jul 15;726(2):97–133. doi: 10.1016/0304-4173(83)90002-2. [DOI] [PubMed] [Google Scholar]
- Hauska G., Nitschke W., Herrmann R. G. Amino acid identities in the three redox center-carrying polypeptides of cytochrome bc1/b6f complexes. J Bioenerg Biomembr. 1988 Apr;20(2):211–228. doi: 10.1007/BF00768395. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988 Jun 3;53(5):795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Japa S., Zhu Q. S., Beattie D. S. Subunit VII, the ubiquinone-binding protein, of the cytochrome b-c1 complex of yeast mitochondria is involved in electron transport at center o and faces the matrix side of the membrane. J Biol Chem. 1987 Apr 25;262(12):5441–5444. [PubMed] [Google Scholar]
- Jin Y. Z., Tang H. L., Li S. L., Tsou C. L. The triphasic reduction of cytochrome b in the succinate-cytochrome c reductase. Biochim Biophys Acta. 1981 Oct 12;637(3):551–554. doi: 10.1016/0005-2728(81)90063-3. [DOI] [PubMed] [Google Scholar]
- John P., Whatley F. R. Oxidative phosphorylation coupled to oxygen uptake and nitrate reduction in Micrococcus denitrificans. Biochim Biophys Acta. 1970 Sep 1;216(2):342–352. doi: 10.1016/0005-2728(70)90225-2. [DOI] [PubMed] [Google Scholar]
- Kallas T., Spiller S., Malkin R. Characterization of two operons encoding the cytochrome b6-f complex of the cyanobacterium Nostoc PCC 7906. Highly conserved sequences but different gene organization than in chloroplasts. J Biol Chem. 1988 Oct 5;263(28):14334–14342. [PubMed] [Google Scholar]
- Kallas T., Spiller S., Malkin R. Primary structure of cotranscribed genes encoding the Rieske Fe-S and cytochrome f proteins of the cyanobacterium Nostoc PCC 7906. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5794–5798. doi: 10.1073/pnas.85.16.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson B., Hovmöller S., Weiss H., Leonard K. Structural studies of cytochrome reductase. Subunit topography determined by electron microscopy of membrane crystals of a subcomplex. J Mol Biol. 1983 Apr 5;165(2):287–302. doi: 10.1016/s0022-2836(83)80258-7. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Balny C., King T. E. Role of the hinge protein in the electron transfer between cardiac cytochrome c1 and c. Equilibrium constants and kinetic probes. J Biol Chem. 1987 Jun 15;262(17):8103–8108. [PubMed] [Google Scholar]
- Knaff D. B., Buchanan B. B. Cytochrome b and photosynthetic sulfur bacteria. Biochim Biophys Acta. 1975 Mar 20;376(3):549–560. doi: 10.1016/0005-2728(75)90174-7. [DOI] [PubMed] [Google Scholar]
- Knaff D. B., Malkin R. Iron-sulfur proteins of the green photosynthetic bacterium Chlorobium. Biochim Biophys Acta. 1976 May 14;430(2):244–252. doi: 10.1016/0005-2728(76)90082-7. [DOI] [PubMed] [Google Scholar]
- Kriauciunas A., Yu L., Yu C. A., Wynn R. M., Knaff D. B. The Rhodospirillum rubrum cytochrome bc1 complex: peptide composition, prosthetic group content and quinone binding. Biochim Biophys Acta. 1989 Aug 17;976(1):70–76. doi: 10.1016/s0005-2728(89)80190-2. [DOI] [PubMed] [Google Scholar]
- Kucera I., Hedbávný R., Dadák V. Separate binding sites for antimycin and mucidin in the respiratory chain of the bacterium Paracoccus denitrificans and their occurrence in other denitrificans bacteria. Biochem J. 1988 Jun 15;252(3):905–908. doi: 10.1042/bj2520905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski B., Ludwig B. The genes of the Paracoccus denitrificans bc1 complex. Nucleotide sequence and homologies between bacterial and mitochondrial subunits. J Biol Chem. 1987 Oct 5;262(28):13805–13811. [PubMed] [Google Scholar]
- Kutoh E., Sone N. Quinol-cytochrome c oxidoreductase from the thermophilic bacterium PS3. Purification and properties of a cytochrome bc1(b6f) complex. J Biol Chem. 1988 Jun 25;263(18):9020–9026. [PubMed] [Google Scholar]
- Leonard K., Wingfield P., Arad T., Weiss H. Three-dimensional structure of ubiquinol:cytochrome c reductase from Neurospora mitochondria determined by electron microscopy of membrane crystals. J Mol Biol. 1981 Jun 25;149(2):259–274. doi: 10.1016/0022-2836(81)90301-6. [DOI] [PubMed] [Google Scholar]
- Lewis R. J., Prince R. C., Dutton P. L., Knaff D. B., Krulwich T. A. The respiratory chain of Bacillus alcalophilus and its nonalkalophilic mutant derivative. J Biol Chem. 1981 Oct 25;256(20):10543–10549. [PubMed] [Google Scholar]
- Li Y., De Vries S., Leonard K., Weiss H. Topography of the iron-sulphur subunit in mitochondrial ubiquinol:cytochrome c reductase. FEBS Lett. 1981 Dec 7;135(2):277–280. doi: 10.1016/0014-5793(81)80800-9. [DOI] [PubMed] [Google Scholar]
- Link T. A., Schägger H., von Jagow G. Analysis of the structures of the subunits of the cytochrome bc1 complex from beef heart mitochondria. FEBS Lett. 1986 Aug 11;204(1):9–15. doi: 10.1016/0014-5793(86)81378-3. [DOI] [PubMed] [Google Scholar]
- Ljungdahl P. O., Beckmann J. D., Trumpower B. L. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. II. Biochemical characterization of temperature-sensitive RIP1- mutations. J Biol Chem. 1989 Mar 5;264(7):3723–3731. [PubMed] [Google Scholar]
- Ljungdahl P. O., Pennoyer J. D., Robertson D. E., Trumpower B. L. Purification of highly active cytochrome bc1 complexes from phylogenetically diverse species by a single chromatographic procedure. Biochim Biophys Acta. 1987 May 6;891(3):227–241. doi: 10.1016/0005-2728(87)90218-0. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., De Haan M., Schoppink P. J., Berden J. A., Grivell L. A. Inactivation of the gene encoding the 11-kDa subunit VIII of the ubiquinol-cytochrome-c oxidoreductase in Saccharomyces cerevisiae. Eur J Biochem. 1988 Feb 15;172(1):179–184. doi: 10.1111/j.1432-1033.1988.tb13870.x. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., Grivell L. A. Nucleotide sequence of the gene encoding the 11-kDa subunit of the ubiquinol-cytochrome-c oxidoreductase in Saccharomyces cerevisiae. Eur J Biochem. 1987 Jun 1;165(2):419–425. doi: 10.1111/j.1432-1033.1987.tb11455.x. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., de Haan M., Bout A., Grivell L. A. Demarcation of a sequence involved in mediating catabolite repression of the gene for the 11 kDa subunit VIII of ubiquinol-cytochrome c oxidoreductase in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 11;16(13):5797–5811. doi: 10.1093/nar/16.13.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Bowyer J. R., Ohnishi T., Dutton P. L. Inhibition of electron transfer by 3-alkyl-2-hydroxy-1,4-naphthoquinones in the ubiquinol-cytochrome c oxidoreductases of Rhodopseudomonas sphaeroides and mammalian mitochondria. Interaction with a ubiquinone-binding site and the Rieske iron-sulfur cluster. J Biol Chem. 1983 Feb 10;258(3):1571–1579. [PubMed] [Google Scholar]
- McCarthy J. E., Ferguson S. J. Respiratory control and the basis of light-induced inhibition of respiration in chromatophores from Rhodopseudomonas capsulata. Biochem Biophys Res Commun. 1982 Aug 31;107(4):1406–1411. doi: 10.1016/s0006-291x(82)80155-1. [DOI] [PubMed] [Google Scholar]
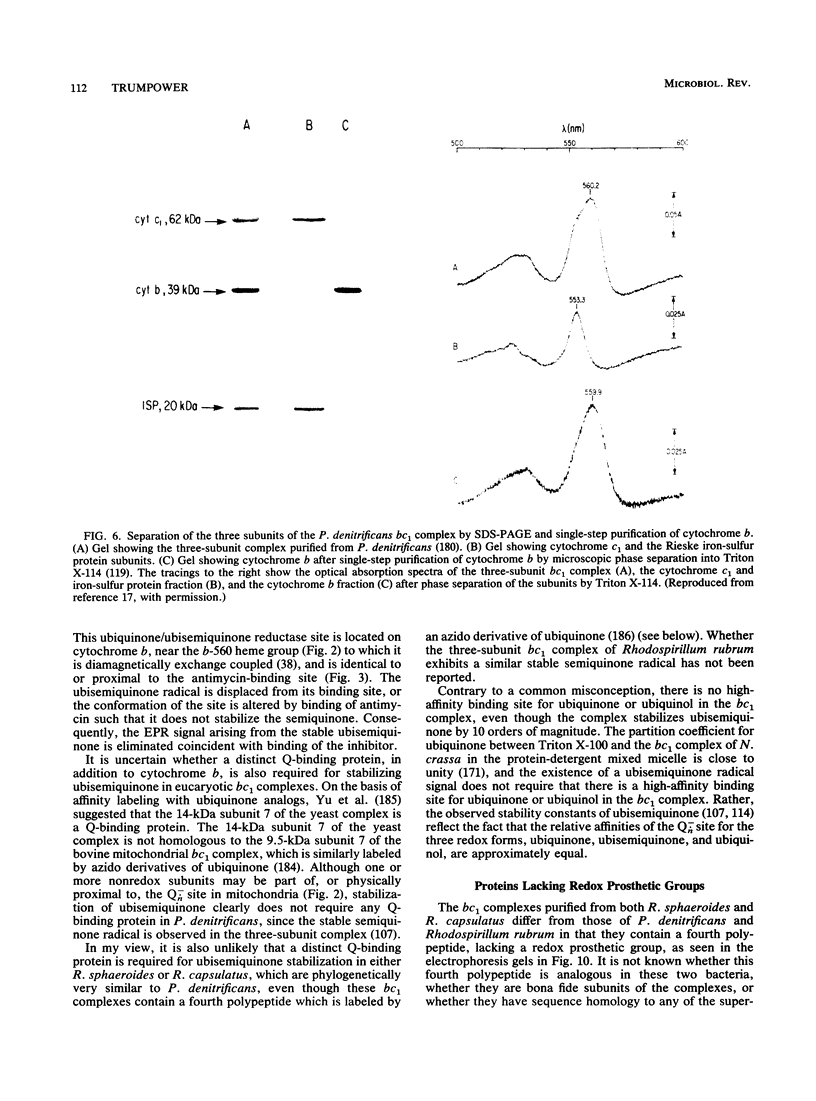
- Meinhardt S. W., Yang X. H., Trumpower B. L., Ohnishi T. Identification of a stable ubisemiquinone and characterization of the effects of ubiquinone oxidation-reduction status on the Rieske iron-sulfur protein in the three-subunit ubiquinol-cytochrome c oxidoreductase complex of Paracoccus denitrificans. J Biol Chem. 1987 Jun 25;262(18):8702–8706. [PubMed] [Google Scholar]
- Michelotti E. F., Hajduk S. L. Developmental regulation of trypanosome mitochondrial gene expression. J Biol Chem. 1987 Jan 15;262(2):927–932. [PubMed] [Google Scholar]
- Mitchell P. Possible molecular mechanisms of the protonmotive function of cytochrome systems. J Theor Biol. 1976 Oct 21;62(2):327–367. doi: 10.1016/0022-5193(76)90124-7. [DOI] [PubMed] [Google Scholar]
- Mitchell P. Protonmotive redox mechanism of the cytochrome b-c1 complex in the respiratory chain: protonmotive ubiquinone cycle. FEBS Lett. 1975 Aug 1;56(1):1–6. doi: 10.1016/0014-5793(75)80098-6. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Koerner T. J., Tzagoloff A. Characterization of the yeast HEM2 gene and transcriptional regulation of COX5 and COR1 by heme. J Biol Chem. 1987 Dec 15;262(35):16822–16829. [PubMed] [Google Scholar]
- Nałecz M. J., Bolli R., Azzi A. Molecular conversion between monomeric and dimeric states of the mitochondrial cytochrome b-c1 complex: isolation of active monomers. Arch Biochem Biophys. 1985 Feb 1;236(2):619–628. doi: 10.1016/0003-9861(85)90666-6. [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Schägger H., Meinhardt S. W., LoBrutto R., Link T. A., von Jagow G. Spatial organization of redox active centers in the bovine heart ubiquinol-cytochrome c oxidoreductase. J Biol Chem. 1989 Jan 15;264(2):735–744. [PubMed] [Google Scholar]
- Ohnishi T., Trumpower B. L. Differential effects of antimycin on ubisemiquinone bound in different environments in isolated succinate . cytochrome c reductase complex. J Biol Chem. 1980 Apr 25;255(8):3278–3284. [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Oudshoorn P., Van Steeg H., Swinkels B. W., Schoppink P., Grivell L. A. Subunit II of yeast QH2:cytochrome-c oxidoreductase. Nucleotide sequence of the gene and features of the protein. Eur J Biochem. 1987 Feb 16;163(1):97–103. doi: 10.1111/j.1432-1033.1987.tb10741.x. [DOI] [PubMed] [Google Scholar]
- Page M. D., Carr G., Bell L. C., Ferguson S. J. Structure, control and assembly of a bacterial electron transport system as exemplified by Paracoccus denitrificans. Biochem Soc Trans. 1989 Dec;17(6):991–993. doi: 10.1042/bst0170991. [DOI] [PubMed] [Google Scholar]
- Payne W. E., Trumpower B. L. A simple, one-step purification of cytochrome b from the bc1 complexes of bacteria. FEBS Lett. 1987 Mar 9;213(1):107–112. doi: 10.1016/0014-5793(87)81473-4. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Hartl F. U., Neupert W. Import of proteins into mitochondria: a multi-step process. Eur J Biochem. 1988 Aug 1;175(2):205–212. doi: 10.1111/j.1432-1033.1988.tb14185.x. [DOI] [PubMed] [Google Scholar]
- Prince R. C., Daldal F. Physiological electron donors to the photochemical reaction center of Rhodobacter capsulatus. Biochim Biophys Acta. 1987 Dec 17;894(3):370–378. doi: 10.1016/0005-2728(87)90115-0. [DOI] [PubMed] [Google Scholar]
- Rich P. R. Electron and proton transfers in chemical and biological quinone systems. Faraday Discuss Chem Soc. 1982;74:349–364. doi: 10.1039/dc9827400349. [DOI] [PubMed] [Google Scholar]
- Richardson D. J., McEwan A. G., Jackson J. B., Ferguson S. J. Electron transport pathways to nitrous oxide in Rhodobacter species. Eur J Biochem. 1989 Nov 20;185(3):659–669. doi: 10.1111/j.1432-1033.1989.tb15163.x. [DOI] [PubMed] [Google Scholar]
- Rigby S. E., Moore G. R., Gray J. C., Gadsby P. M., George S. J., Thomson A. J. N.m.r., e.p.r. and magnetic-c.d. studies of cytochrome f. Identity of the haem axial ligands. Biochem J. 1988 Dec 1;256(2):571–577. doi: 10.1042/bj2560571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D., Schatz G. Mitochondrial presequences. J Biol Chem. 1988 Apr 5;263(10):4509–4511. [PubMed] [Google Scholar]
- Roise D., Theiler F., Horvath S. J., Tomich J. M., Richards J. H., Allison D. S., Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988 Mar;7(3):649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno J. C., Xu Y., Osgood M. P., Kim C. H., King T. E. Thermodynamic and spectroscopic characteristics of the cytochrome bc1 complex. Role of quinone in the behavior of cytochrome b562. J Biol Chem. 1989 Sep 15;264(26):15398–15403. [PubMed] [Google Scholar]
- Schneider J. C., Guarente L. The untranslated leader of nuclear COX4 gene of Saccharomyces cerevisiae contains an intron. Nucleic Acids Res. 1987 Apr 24;15(8):3515–3529. doi: 10.1093/nar/15.8.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes P. B., Smith L. Composition and properties of the membrane-bound respiratory chain system of Micrococcus denitrificans. Biochim Biophys Acta. 1968 Feb 12;153(2):363–375. doi: 10.1016/0005-2728(68)90081-9. [DOI] [PubMed] [Google Scholar]
- Schoppink P. J., Hemrika W., Berden J. A. The effect of deletion of the genes encoding the 40 kDa subunit II or the 17 kDa subunit VI on the steady-state kinetics of yeast ubiquinol-cytochrome-c oxidoreductase. Biochim Biophys Acta. 1989 May 8;974(2):192–201. doi: 10.1016/s0005-2728(89)80372-x. [DOI] [PubMed] [Google Scholar]
- Schulte U., Arretz M., Schneider H., Tropschug M., Wachter E., Neupert W., Weiss H. A family of mitochondrial proteins involved in bioenergetICS and biogenesis. Nature. 1989 May 11;339(6220):147–149. doi: 10.1038/339147a0. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G., Borchart U., Machleidt W. Amino-acid sequence of the smallest protein of the cytochrome c1 subcomplex from beef heart mitochondria. Hoppe Seylers Z Physiol Chem. 1983 Mar;364(3):307–311. [PubMed] [Google Scholar]
- Scolnik P. A., Marrs B. L. Genetic research with photosynthetic bacteria. Annu Rev Microbiol. 1987;41:703–726. doi: 10.1146/annurev.mi.41.100187.003415. [DOI] [PubMed] [Google Scholar]
- Sedmera P., Musílek V., Nerud F., Vondrácek M. Mucidin: its identity with strobilurin A. J Antibiot (Tokyo) 1981 Aug;34(8):1069–1069. [PubMed] [Google Scholar]
- Sidhu A., Beattie D. S. Purification and polypeptide characterization of complex III from yeast mitochondria. J Biol Chem. 1982 Jul 10;257(13):7879–7886. [PubMed] [Google Scholar]
- Siedow J. N., Power S., de la Rosa F. F., Palmer G. The preparation and characterization of highly purified, enzymically active complex III from baker's yeast. J Biol Chem. 1978 Apr 10;253(7):2392–2399. [PubMed] [Google Scholar]
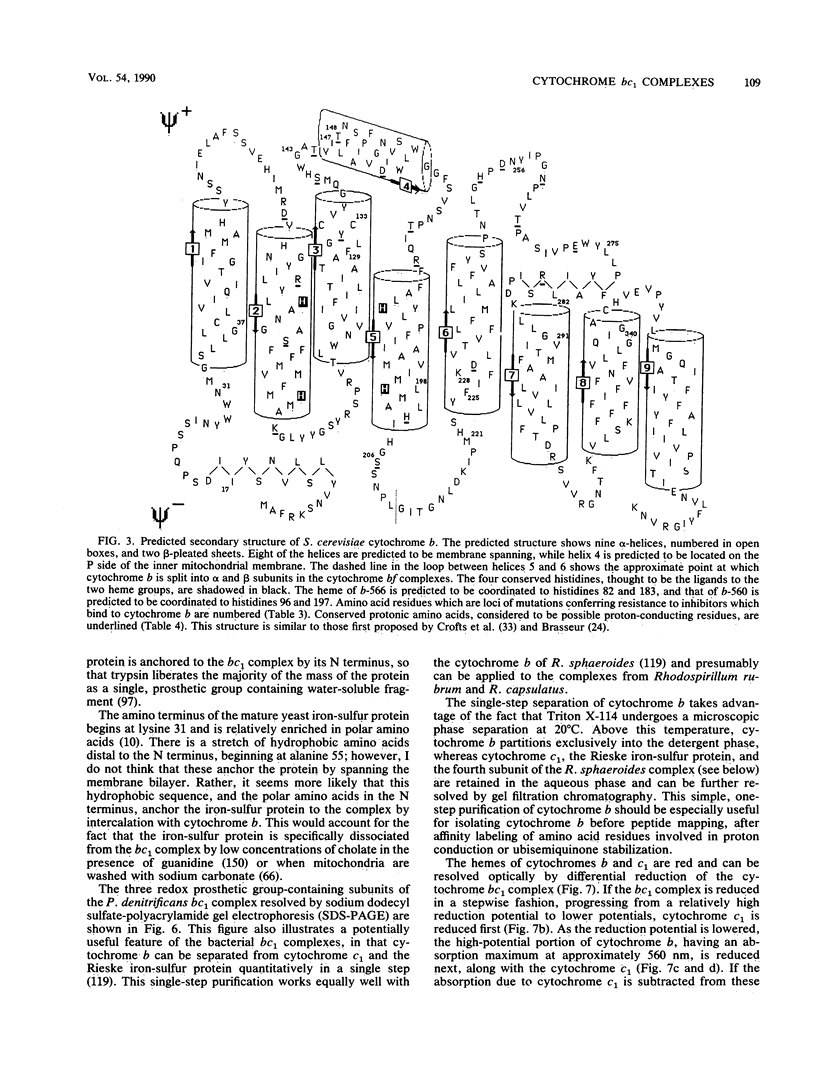
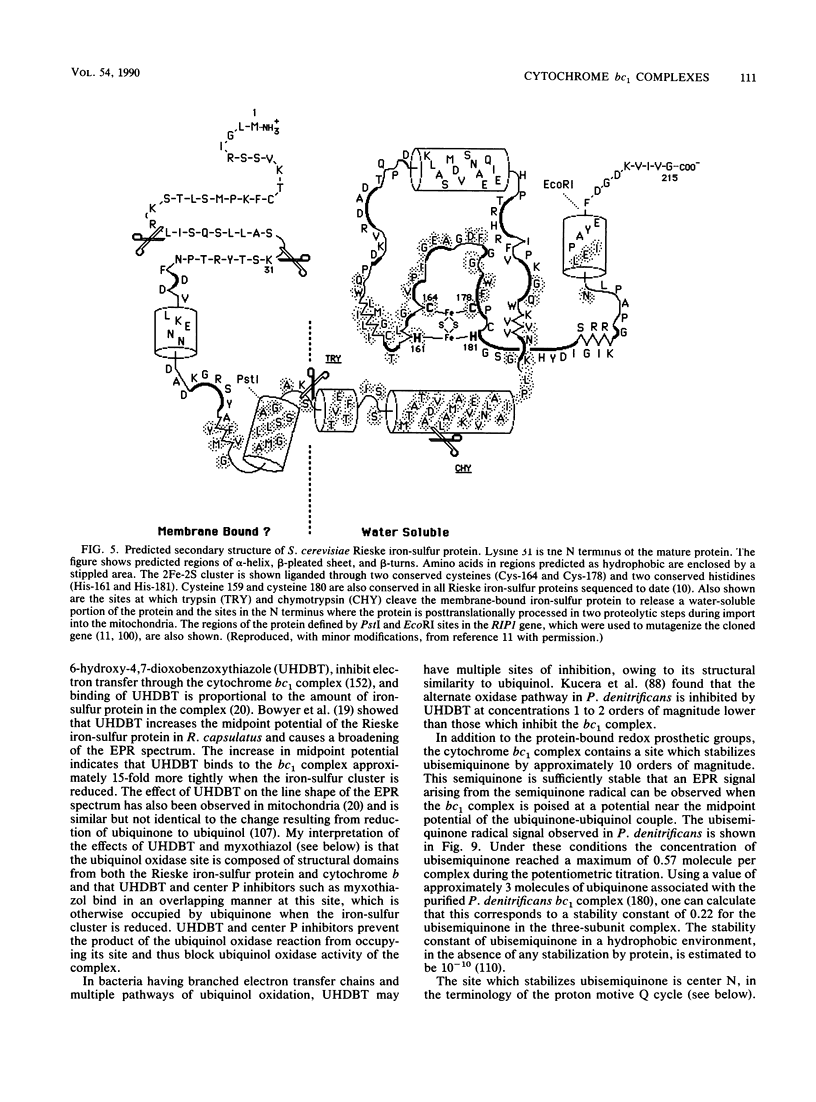
- Simpkin D., Palmer G., Devlin F. J., McKenna M. C., Jensen G. M., Stephens P. J. The axial ligands of heme in cytochromes: a near-infrared magnetic circular dichroism study of yeast cytochromes c, c1, and b and spinach cytochrome f. Biochemistry. 1989 Oct 3;28(20):8033–8039. doi: 10.1021/bi00446a010. [DOI] [PubMed] [Google Scholar]
- Soberón M., Williams H. D., Poole R. K., Escamilla E. Isolation of a Rhizobium phaseoli cytochrome mutant with enhanced respiration and symbiotic nitrogen fixation. J Bacteriol. 1989 Jan;171(1):465–472. doi: 10.1128/jb.171.1.465-472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone N., Sekimachi M., Kutoh E. Identification and properties of a quinol oxidase super-complex composed of a bc1 complex and cytochrome oxidase in the thermophilic bacterium PS3. J Biol Chem. 1987 Nov 15;262(32):15386–15391. [PubMed] [Google Scholar]
- Tang H. L., Trumpower B. L. Triphasic reduction of cytochrome b and the protonmotive Q cycle pathway of electron transfer in the cytochrome bc1 complex of the mitochondrial respiratory chain. J Biol Chem. 1986 May 15;261(14):6209–6215. [PubMed] [Google Scholar]
- Teintze M., Slaughter M., Weiss H., Neupert W. Biogenesis of mitochondrial ubiquinol:cytochrome c reductase (cytochrome bc1 complex). Precursor proteins and their transfer into mitochondria. J Biol Chem. 1982 Sep 10;257(17):10364–10371. [PubMed] [Google Scholar]
- Telser J., Hoffman B. M., LoBrutto R., Ohnishi T., Tsai A. L., Simpkin D., Palmer G. Evidence for N coordination to Fe in the [2Fe-2S] center in yeast mitochondrial complex III. Comparison with similar findings for analogous bacterial [2Fe-2S] proteins. FEBS Lett. 1987 Apr 6;214(1):117–121. doi: 10.1016/0014-5793(87)80024-8. [DOI] [PubMed] [Google Scholar]
- Thierbach G., Reichenbach H. Myxothiazol, a new antibiotic interfering with respiration. Antimicrob Agents Chemother. 1981 Apr;19(4):504–507. doi: 10.1128/aac.19.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny-Meyer L., Stax D., Hennecke H. An unusual gene cluster for the cytochrome bc1 complex in Bradyrhizobium japonicum and its requirement for effective root nodule symbiosis. Cell. 1989 May 19;57(4):683–697. doi: 10.1016/0092-8674(89)90137-2. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L., Edwards C. A., Ohnishi T. Reconstitution of the iron-sulfur protein responsible for the g = 1.90 electron paramagnetic resonance signal and associated cytochrome c reductase activities to depleted succinate-cytochrome c reductase complex. J Biol Chem. 1980 Aug 10;255(15):7487–7493. [PubMed] [Google Scholar]
- Trumpower B. L., Edwards C. A. Purification of a reconstitutively active iron-sulfur protein (oxidation factor) from succinate . cytochrome c reductase complex of bovine heart mitochondria. J Biol Chem. 1979 Sep 10;254(17):8697–8706. [PubMed] [Google Scholar]
- Trumpower B. L. Evidence for a protonmotive Q cycle mechanism of electron transfer through the cytochrome b-c1 complex. Biochem Biophys Res Commun. 1976 May 3;70(1):73–80. doi: 10.1016/0006-291x(76)91110-4. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L. Function of the iron-sulfur protein of the cytochrome b-c1 segment in electron-transfer and energy-conserving reactions of the mitochondrial respiratory chain. Biochim Biophys Acta. 1981 Dec 4;639(2):129–155. doi: 10.1016/0304-4173(81)90008-2. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L., Haggerty J. G. Inhibition of electron transfer in the cytochrome b-c, segment of the mitochondrial respiratory chain by a synthetic analogue of ubiquinone. J Bioenerg Biomembr. 1980 Aug;12(3-4):151–164. doi: 10.1007/BF00744680. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L., Katki A. Controlled reduction of cytochrome b in succinate-cytochrome c reductase complex by succinate in the presence of ascorbate and antimycin. Biochem Biophys Res Commun. 1975 Jul 8;65(1):16–23. doi: 10.1016/s0006-291x(75)80055-6. [DOI] [PubMed] [Google Scholar]
- Trumpower B. L. New concepts on the role of ubiquinone in the mitochondrial respiratory chain. J Bioenerg Biomembr. 1981 Apr;13(1-2):1–24. doi: 10.1007/BF00744743. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Macino G., Sebald W. Mitochondrial genes and translation products. Annu Rev Biochem. 1979;48:419–441. doi: 10.1146/annurev.bi.48.070179.002223. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Wu M. A., Crivellone M. Assembly of the mitochondrial membrane system. Characterization of COR1, the structural gene for the 44-kilodalton core protein of yeast coenzyme QH2-cytochrome c reductase. J Biol Chem. 1986 Dec 25;261(36):17163–17169. [PubMed] [Google Scholar]
- Van Loon A. P., De Groot R. J., De Haan M., Dekker A., Grivell L. A. The DNA sequence of the nuclear gene coding for the 17-kd subunit VI of the yeast ubiquinol-cytochrome c reductase: a protein with an extremely high content of acidic amino acids. EMBO J. 1984 May;3(5):1039–1043. doi: 10.1002/j.1460-2075.1984.tb01924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon A. P., Kreike J., De Ronde A., Van der Horst G. T., Gasser S. M., Grivell L. A. Biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. Characterization of precursor forms of the 44-kDa, 40-kDa and 17-kDa subunits and identification of individual messenger RNAs for these and other imported subunits of the complex. Eur J Biochem. 1983 Oct 3;135(3):457–463. doi: 10.1111/j.1432-1033.1983.tb07673.x. [DOI] [PubMed] [Google Scholar]
- Van Spanning R. J., Wansell C., Harms N., Oltmann L. F., Stouthamer A. H. Mutagenesis of the gene encoding cytochrome c550 of Paracoccus denitrificans and analysis of the resultant physiological effects. J Bacteriol. 1990 Feb;172(2):986–996. doi: 10.1128/jb.172.2.986-996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verner K., Schatz G. Protein translocation across membranes. Science. 1988 Sep 9;241(4871):1307–1313. doi: 10.1126/science.2842866. [DOI] [PubMed] [Google Scholar]
- Von Jagow G., Gribble G. W., Trumpower B. L. Mucidin and strobilurin A are identical and inhibit electron transfer in the cytochrome bc1 complex of the mitochondrial respiratory chain at the same site as myxothiazol. Biochemistry. 1986 Feb 25;25(4):775–780. doi: 10.1021/bi00352a006. [DOI] [PubMed] [Google Scholar]
- Weber S., Wolf K. Two changes of the same nucleotide confer resistance to diuron and antimycin in the mitochondrial cytochrome b gene of Schizosaccharomyces pombe. FEBS Lett. 1988 Sep 12;237(1-2):31–34. doi: 10.1016/0014-5793(88)80165-0. [DOI] [PubMed] [Google Scholar]
- Weiss H., Kolb H. J. Isolation of mitochondrial succinate: ubiquinone reductase, cytochrome c reductase and cytochrome c oxidase from Neurospora crassa using nonionic detergent. Eur J Biochem. 1979 Aug 15;99(1):139–149. doi: 10.1111/j.1432-1033.1979.tb13240.x. [DOI] [PubMed] [Google Scholar]
- Weiss H., Wingfield P. Enzymology of ubiquinone-utilizing electron transfer complexes in nonionic detergent. Eur J Biochem. 1979 Aug 15;99(1):151–160. doi: 10.1111/j.1432-1033.1979.tb13241.x. [DOI] [PubMed] [Google Scholar]
- West I. C., Mitchell P., Rich P. R. Electron conduction between b cytochromes of the mitochondrial respiratory chain in the presence of antimycin plus myxothiazol. Biochim Biophys Acta. 1988 Mar 30;933(1):35–41. doi: 10.1016/0005-2728(88)90053-9. [DOI] [PubMed] [Google Scholar]
- Widger W. R., Cramer W. A., Herrmann R. G., Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci U S A. 1984 Feb;81(3):674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström M. K., Berden J. A. Oxidoreduction of cytochrome b in the presence of antimycin. Biochim Biophys Acta. 1972 Dec 14;283(3):403–420. doi: 10.1016/0005-2728(72)90258-7. [DOI] [PubMed] [Google Scholar]
- Witte C., Jensen R. E., Yaffe M. P., Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988 May;7(5):1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn R. M., Gaul D. F., Shaw R. W., Knaff D. B. Identification of the components of a putative cytochrome bc1 complex in Rhodopseudomonas viridis. Arch Biochem Biophys. 1985 May 1;238(2):373–377. doi: 10.1016/0003-9861(85)90177-8. [DOI] [PubMed] [Google Scholar]
- Yang X. H., Trumpower B. L. Protonmotive Q cycle pathway of electron transfer and energy transduction in the three-subunit ubiquinol-cytochrome c oxidoreductase complex of Paracoccus denitrificans. J Biol Chem. 1988 Aug 25;263(24):11962–11970. [PubMed] [Google Scholar]
- Yang X. H., Trumpower B. L. Purification of a three-subunit ubiquinol-cytochrome c oxidoreductase complex from Paracoccus denitrificans. J Biol Chem. 1986 Sep 15;261(26):12282–12289. [PubMed] [Google Scholar]
- Yu L., Chiang Y. L., Yu C. A., King T. E. A trypsin-resistant heme peptide from cardiac cytochrome c1. Biochim Biophys Acta. 1975 Jan 30;379(1):33–42. doi: 10.1016/0005-2795(75)90006-9. [DOI] [PubMed] [Google Scholar]
- Yu L., Mei Q. C., Yu C. A. Characterization of purified cytochrome b-c1 complex from Rhodopseudomonas sphaeroides R-26. J Biol Chem. 1984 May 10;259(9):5752–5760. [PubMed] [Google Scholar]
- Yu L., Yang F. D., Yu C. A. Interaction and identification of ubiquinone-binding proteins in ubiquinol-cytochrome c reductase by azido-ubiquinone derivatives. J Biol Chem. 1985 Jan 25;260(2):963–973. [PubMed] [Google Scholar]
- Yu L., Yang F. D., Yu C. A., Tsai A. L., Palmer G. Identification of ubiquinone-binding proteins in yeast mitochondrial ubiquinol-cytochrome c reductase using an azido-ubiquinone derivative. Biochim Biophys Acta. 1986 Mar 12;848(3):305–311. doi: 10.1016/0005-2728(86)90204-5. [DOI] [PubMed] [Google Scholar]
- Zannoni D. Effect of oxygen limitation on the formation of the electron transport system of the phytopathogenic fluorescent bacterium Pseudomonas cichorii. J Bioenerg Biomembr. 1986 Dec;18(6):461–470. doi: 10.1007/BF00743144. [DOI] [PubMed] [Google Scholar]
- de Vries S., Marres C. A. The mitochondrial respiratory chain of yeast. Structure and biosynthesis and the role in cellular metabolism. Biochim Biophys Acta. 1987;895(3):205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- de Vries S. The pathway of electron transfer in the dimeric QH2: cytochrome c oxidoreductase. J Bioenerg Biomembr. 1986 Jun;18(3):195–224. doi: 10.1007/BF00743464. [DOI] [PubMed] [Google Scholar]
- de la Rosa F. F., Palmer G. Reductive titration of CoQ-depleted Complex III from Baker's yeast. Evidence for an exchange-coupled complex between QH . and low-spin ferricytochrome b. FEBS Lett. 1983 Oct 31;163(1):140–143. doi: 10.1016/0014-5793(83)81181-8. [DOI] [PubMed] [Google Scholar]
- di Rago J. P., Colson A. M. Molecular basis for resistance to antimycin and diuron, Q-cycle inhibitors acting at the Qi site in the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J Biol Chem. 1988 Sep 5;263(25):12564–12570. [PubMed] [Google Scholar]
- di Rago J. P., Coppée J. Y., Colson A. M. Molecular basis for resistance to myxothiazol, mucidin (strobilurin A), and stigmatellin. Cytochrome b inhibitors acting at the center o of the mitochondrial ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J Biol Chem. 1989 Aug 25;264(24):14543–14548. [PubMed] [Google Scholar]
- di Rago J. P., Perea X., Colson A. M. DNA sequence analysis of diuron-resistant mutations in the mitochondrial cytochrome b gene of Saccharomyces cerevisiae. FEBS Lett. 1986 Nov 24;208(2):208–210. doi: 10.1016/0014-5793(86)81019-5. [DOI] [PubMed] [Google Scholar]
- van Loon A. P., de Groot R. J., van Eyk E., van der Horst G. T., Grivell L. A. Isolation and characterization of nuclear genes coding for subunits of the yeast ubiquinol-cytochrome c reductase complex. Gene. 1982 Dec;20(3):323–337. doi: 10.1016/0378-1119(82)90201-3. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jagow G., Link T. A. Use of specific inhibitors on the mitochondrial bc1 complex. Methods Enzymol. 1986;126:253–271. doi: 10.1016/s0076-6879(86)26026-7. [DOI] [PubMed] [Google Scholar]
- von Jagow G., Ljungdahl P. O., Graf P., Ohnishi T., Trumpower B. L. An inhibitor of mitochondrial respiration which binds to cytochrome b and displaces quinone from the iron-sulfur protein of the cytochrome bc1 complex. J Biol Chem. 1984 May 25;259(10):6318–6326. [PubMed] [Google Scholar]
- von Jagow G., Ohnishi T. The chromone inhibitor stigmatellin--binding to the ubiquinol oxidation center at the C-side of the mitochondrial membrane. FEBS Lett. 1985 Jun 17;185(2):311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]