Abstract
The clinical application of human-induced pluripotent stem cells (hiPSCs) requires not only the production of Good Manufacturing Practice-grade (GMP-grade) hiPSCs but also the derivation of specified cell types for transplantation under GMP conditions. Previous reports have suggested that hiPSCs can be produced in the absence of animal-derived reagents (xenobiotics) to ease the transition to production under GMP standards. However, to facilitate the use of hiPSCs in cell-based therapeutics, their progeny should be produced not only in the absence of xenobiotics but also under GMP conditions requiring extensive standardization of protocols, documentation, and reproducibility of methods and product. Here, we present a successful framework to produce GMP-grade derivatives of hiPSCs that are free of xenobiotic exposure from the collection of patient fibroblasts, through reprogramming, maintenance of hiPSCs, identification of reprogramming vector integration sites (nrLAM-PCR), and finally specification and terminal differentiation of clinically relevant cells. Furthermore, we developed a primary set of Standard Operating Procedures for the GMP-grade derivation and differentiation of these cells as a resource to facilitate widespread adoption of these practices.
Keywords: Cellular therapy, Neural differentiation, Pluripotent stem cells, Reprogramming
Introduction
The clinical promise of stem cell biology is predicated on the ability to derive cells for use in cell-based therapeutics. To realize this potential, several key hurdles must be overcome [1]. First, functional cell types must be generated to homogeneity with the capability to replace lost or damaged tissue. Second, the transplanted cells should be as free from microbial contaminants or animal-derived materials (xenobiotics) as possible. Third, the donor cells may need to be either isogenic or at least immunologically compatible with the recipient. Two recent clinical trials using human embryonic stem cell-derived cellular products have successfully cleared the first two barriers according to preclinical testing and are now in stage I safety trials [2]. In addition, at least one group has reported the derivation of human embryonic stem cells (hESCs) under Good Manufacturing Practice (GMP) conditions [3]. However, because human embryonic stem cells are derived from embryos with no relation to the intended patient, it may be challenging to treat patients with derivatives of these cells that are not immunologically matched. Therefore, any cellular therapeutic treatment with ESC derivatives would potentially necessitate administration of immunosuppressive drugs with their associated toxicity-induced risk of renal dysfunction, hypertension, and cardiovascular disease, which contributes to graft loss and shortened life expectancy [4].
Recently, it has become widely accepted that many types of human somatic cells can be reprogrammed to a pluripotent state upon introduction of three to four transcription factors that are highly expressed in pluripotent stem cells [5–8]. The resulting human-induced pluripotent stem cells (hiPSCs) appear to share most of the molecular and functional characteristics of human embryonic stem cells [5, 9–11] but have the distinct advantage that they can be derived from many human tissues. Therefore, it is possible with hiPSCs to create patient-specific stem cells, from which patient-specific, immunologically compatible cell therapeutics can be derived. Standard protocols to obtain somatic cells from patients, reprogram the cells to pluripotency, and then differentiate the hiPSCs to a specific mature cell population typically require use of xenobiotic reagents or feeder cells from lower organisms, rendering these approaches incompatible with clinical application. Some progress has been made to derive hiPSCs cells under xeno-free (“xeno”, xenobiotics) conditions [12–15]. However, some used human feeders and/or human serum in their derivation of xeno-free iPSCs or derived iPSCs in the presence of xenobiotic-containing medium. Therefore, it is difficult to project how these methods could be applied in a clinical setting.
Futhermore, derivation of target cells from patient biopsy and differentiation techniques following reprogramming all typically are performed with xenobiotics, and no single study has shown that patient cells can be collected, reprogrammed, and differentiated all under defined, xeno-free conditions with commercially available reagents. In addition, most of these previous studies used retroviral integration of the reprogramming factors but did not provide evidence that these integrations were benign.
Here, we report that patient-specific neural progenitor cells (NPCs) and neurons can be derived from hiPSCs using methods that are completely devoid of xenobiotics and are GMP-compatible. These approaches apply novel techniques to derive fibroblasts from patients, reprogram them to a pluripotent state, and then differentiate them to NPCs and neurons, all in completely defined media and without animal products or feeder cells. All materials used in this study are also commercially available, facilitating GMP-grade quality control (QC). In addition, we used a polymerase chain reaction-sequencing (PCR-sequencing) technique to map the integration site for the reprogramming factors and demonstrate that these integrations did not affect gene expression. We further show that both the xeno-free iPSCs and their differentiated progeny cells are equivalent to counterparts derived in the presence of xenobiotics. To our knowledge, this is the first demonstration of xeno-free derivation and differentiation of iPS cells with commercially available reagents. Although previous efforts have described the feasibility of deriving xeno-free hESCs and hiPSCs [3, 12, 13, 15], SOPs were not provided to facilitate the broad application of these methods in a clinical setting. Here, we present GMP-compatible Standard Operating Procedures (SOPs) to facilitate clinical translation of hiPSCs technology.
Materials and Methods
GMP Facility
We established a GMP-compliant facility with ISO 8 clean room standards equipped with class II and class III biosafety cabinets and all other standard tissue culture equipment certified as described below. Testing for adventitious agents was done by the UCLA Department of Pathology and Laboratory Medicine in clinically certified laboratories. TSS calibrated the biosafety cabinet and monitors and certified nonviable particles in the air as per the ISO 5 and 7 standards in the biosafety cabinet and the clean room twice a year. Rainin performed calibration and qualification for micropipettes. All incubators, refrigerators, freezers, and cryopreservation containers were equipped with an ISENSIX monitoring system that was calibrated twice a year and was monitored continuously for any fluctuations from the standard set points. Monotronics tested for viable particle count in the clean rooms and all the equipment used in the clean room area quarterly. We have an in-house qualification method which also involves cleaning and decontamination for water baths, incubators, refrigerators, freezers, and centrifuges. Biotech Services tested and certified water baths, centrifuges, incubators, refrigerators, freezers, and cryopreservation containers. Karyotyping was performed by Cell Line Genetics and HLA typing by the UCLA Immunogenetics Center.
Cell Culture Reagents
We procured xenobiotic-free cell culture reagents for the cell culture needed for the stages from derivation of fibroblasts from skin biopsies to their reprogramming into hiPSCs and the differentiation to NPCs, neurons, and glia. The following are the list of media and reagents used in our xeno-free process as described in the Results section: MSCGM-CD and ProFreeze CDM (Lonza, Basel, Switzerland, http://www.lonza.com), animal free origin collagenase (Worthington Biochemical Corporation, Lakewood, NJ, http://www.worthington-biochem.com), CELLstart, EZPassage Tool, HBSS-Ca/Mg free, D/F12, TrypLE, xeno-free B27, and N2 supplement (Invitrogen/Gibco, Carlsbad, CA, http://www.invitrogen.com), human serum (Innovative Research, Inc., Novi, Michigan, http://www.innov-research.com), basic fibroblast growth factor-2 (bFGF2; Peprotech, Rocky Hill, NJ, http://www.peprotech.com), TeSR2 (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com), and Nutristem Stemedia (Stemgent, San Diego, http://www.stemgent.com). Stemedia Nutristem consists of human recombinant insulin, human serum albumin, transferrin, human fibroblast growth factor, and transforming growth factor-β. TeSR2 includes high levels of bFGF together with transforming growth factor-β.
Collection of Patient Skin Biopsy and Derivation of Xeno-Free Fibroblast
Patient consent was obtained as per the UCLA IRB protocol (#08-09-038-01), and skin biopsies were obtained at the UCLA Medical Center using standard clinical methods and labeled with anonymous coding labels. The UCLA IRB and ESCRO approved consent form complied with U.S. Food and Drug Administration (FDA) and U.S. Department of Health and Human Services regulations, California law for research with pluripotent stem cells, and the National Academy of Sciences pluripotent stem cell research guidelines. The informed consent form was used to obtain permission from donors for the collection and reprogramming of the primary cells as well as possible future transplantation. There have been no problems obtaining informed consent from donors.
The punch biopsy samples were immersed in Dulbecco's modified Eagle's medium (DMEM)/F12 and transferred to the GMP-compatible BSCRC Cell Processing Laboratory under aseptic conditions. With use of a BSL-2 certified Biological Safety Cabinet, the biopsies were rinsed twice in HBSS and chopped into 1-mm pieces using a scalpel in 2% animal origin free collagenase solution. The fragments were incubated in a total volume of 4 ml for 90 minutes at 37°C in a 5% CO2 incubator. The tissue was collected and centrifuged at 300g for 5 minutes, the supernatant was aspirated, and the pellet was washed once with 10 ml of MSCGM-CD and centrifuged as described above. The pellets that contained the dissociated cells and tissue clumps were collected in 2 ml of MSCGM-CD medium and plated on a CELLstart-coated dish. The medium was changed once every 72 hours until the cell monolayer became 70% confluent, and then the cells were passaged using TrypLE. Single-cell suspensions of cells were then cryopreserved in ProFreeze CDM as per the manufacturer's protocol.
Derivation of Xeno-Free iPS Cells
One × 105 fibroblast cells were plated in a CELLstart-coated well of a six-well plate in MSCGM-CD medium. After 8 hours, the cells were transduced with vector concentrate (7 × 106 TU/ml) in 1 ml of MSCGM-CD medium containing 10 μg/ml polybrene and incubated overnight at 37°C in 5% CO2 incubator. The vector-containing medium was aspirated, and the cells were rinsed three times with MSCGM-CD and further cultured for 3 days in the same medium. Cells were replated on the fifth day in 50:50 TeSR2/Nutristem containing 10 ng/ml bFGF in two 6-cm dishes coated with CELLstart and cultured until hiPSC-like colonies were formed, similarly to established protocols. The colonies were picked mechanically and cultured in CELLstart-coated dishes, passaged mechanically using the EZPassage tool as per the manufacturer's protocol. Briefly, cell medium was replaced, and the EZPassage tool was rolled in one direction throughout the plate with sufficient hand pressure to cut the colonies. The plate was then rotated 90° and the tool rolled once again throughout the plate. The colonies were collected by gentle pipetting using a serological pipette, transferred to a 15-ml tube, and passaged at the ratio of 1:6 into a new CELLstart-coated plate.
Teratoma Analysis
Teratomas were generated by injecting 5 × 106 to 10 × 106 undifferentiated hESCs resuspended in Matrigel into the testicles of adult male SCID-beige mice. Tumors were harvested 6–8 weeks after transplantation, processed to paraffin, and examined by histology using H&E staining. Surgery to generate teratomas was performed only after institutional approval from the UCLA Animal Research Committee (Assurance number A3196-01).
Differentiation of Xeno-Free NPCs and Neurons from Xeno-Free iPS Cells
Cells were mechanically passaged using the EZPassage tool and allowed to float as EBs in DMEM/F12 and N2 medium for 5 days. The cells were then collected and plated in DMEM/F12 and N2 in CELLstart-coated dishes and cultured for a further 7 days. By this time, rosettes formed in the plate. To the above medium, retinoic acid (RA) and xeno-free B27 were added, and the cells were cultured for 2 more weeks to obtain mature neurons. Alternatively, the rosettes were picked mechanically and replated onto CELLstart-coated dishes in B27- and RA-containing medium to obtain population of neurons and glia.
Immunocytochemistry
All immunocytochemistry analyses were performed as previously reported in [8] and [16], with antibodies against the following epitopes: OCT4, NANOG, SOX2, Tra1–81, SSEA3, MSI2, NESTIN, TUJ1, MAP2, S100, and GFAP.
Gene Expression Profiling
Microarray profiling was performed with Affymetrix Human HG-U133 2.0 Plus arrays as described in [8, 17]. Data represented as CEL files were imported into Genespring and normalized to the mean of all samples by Robust Multichip Algorithm (RMA). Probe sets that were not expressed at a raw value of >50 in at least 10% of samples were eliminated from further analysis. Uncentered, unsupervised Euclidian hierarchical clustering was performed to assess similarity of gene expression between the indicated samples.
Vector Integration Site Analysis
Vector integration site analysis was performed on 100 ng of purified genomic DNA from XiPSC lines using nonrestrictive PCR as described in [18]. The procedure was adapted to use Illumina/Solexa high-throughput sequencing by adding appropriate adaptor sequences on the 5′ ends of the PCR primers. Sequence read length was set at 100 nucleotides, which provided 47 nucleotides of vector sequence to confirm amplification of vector-genome junctions and 53 nucleotides of vector-adjacent genomic sequence. To avoid missing integrations into nonmappable regions of the genome, standard UNIX commands were used first to stack up and count identical 53 nt sequences detected in the several million sequence reads that were obtained. Abundant sequences were then aligned to the hg19 build of the human genome using the BLAT service of the UCSC genome browser. All of the abundant sequence reads that were detected could be mapped uniquely.
Results
Derivation of Patient Cells and Reprogramming Under GMP Conditions
To isolate fibroblasts from patient skin biopsies in the absence of animal products, punch biopsies were isolated, chemically dissociated with xeno-free collagenase, and allowed to grow in defined fibroblast medium (MSCGM-CD) for up to 3 weeks. Fibroblasts were expanded and banked in a defined freezing medium (ProFreeze CDM) prior to passage 3 to preserve reprogramming capability and to keep culture-induced genomic aberrations to a minimum (Fig. 1).
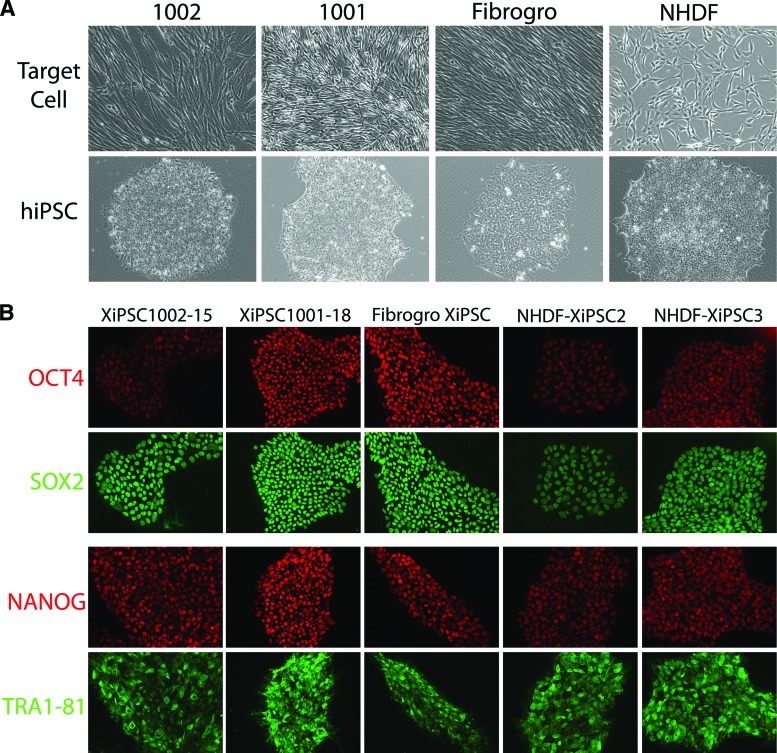
Figure 1.
Characterization of Good Manufacturing Practice-derived fibroblasts reprogrammed to hiPSC cells using pluripotency markers confirms their undifferentiated status. (A): Phase contrast images of fibroblasts and the lower panel show their respective immune-stained hiPSC lines. #1001 and #1002 represent the human skin biopsy-derived fibroblasts. Fibrogro is xenofree fibroblasts, and XiPSC2 and XiPSC3 are the xeno-free hiPSC clones derived from normal human dermal fibroblasts under Good Manufacturing Practices conditions. (B): Immunostaining for markers typical of human pluripotent stem cells indicated reprogramming of the target fibroblasts.
Fibroblasts were then reprogrammed with a modified version of the STEMCCA polycistronic lentivirus bearing the four reprogramming factors flanked by loxP sites [19]. The lentiviral vector was packaged by transfection into 293T cells (human origin) and collected in medium (DMEM) lacking xenobiotics. Although this method is generally compatible with GMP-grade vectors, we cannot rule out the possibility that residual xenobiotic contaminants were present in the vector from the prior culture of the 293T cells in fetal calf serum-containing medium. The presence of any harmful xenogenic contaminant in the final product can be tested for bovine and porcine pathogens before product release, a standard typical for regulatory approval. The medium containing the reprogramming vector was then concentrated approximately 1,000-fold and used to transduce the xeno-free fibroblast cultures as described elsewhere [20]. After transduction, the cells were transferred to plates coated with xeno-free substrate (CELLstart) and grown in a 1:1 ratio of two defined xeno-free media typically used to culture human embryonic stem cells without feeders (TeSR2 and Nutristem Stemedia). After sampling a variety of commercially available xeno-free hESC culture media, both alone or in various combinations, we found that the 1:1 ratio of these two media was the best formulation allowing reprogramming to occur in our system. After 2–3 weeks, clones were manually picked to fresh culture under the same conditions, and individual clones were expanded (Fig. 1). A representative SOP for reprogramming is shown in supplemental online Fig. 1.
Characterization of Reprogrammed Cells
The xeno-free hiPSC clones were characterized morphologically and molecularly. As shown in Figure 2, all the lines generated expressed OCT4, NANOG, and SOX2 and were positive for TRA1-81 and SSEA3 on their cell surface (Fig. 1). Additionally, injection of 5–10 × 106 cells into adult murine testicles led to the formation of teratomas containing representatives of all three germ layers (Fig. 2). These data clearly indicated that the xeno-free hiPSCs were pluripotent by the best assay available for human cells. Finally, each of the lines shown was subject to karyotyping to ensure its gross genomic stability. This analysis indicated that all lines described here had a normal karyotype (Fig. 2B).
Figure 2.
In vivo trilineage differentiation of Good Manufacturing Practice hiPSCs. (A): H&E staining and histologic analysis of 1001-18, 1002-15, Fibrogro, and XiPSC hiPSC lines show teratoma growth containing cells from the three distinct germ layers. Neural rosettes (ectoderm), glandular structures and gutlike endothelium (endoderm), and cartilage (mesoderm) were present. (B): Karyotyping analysis on the indicated lines demonstrated that all lines were grossly normal with respect to genomic stability after reprogramming.
To confirm reprogramming to a pluripotent state under these xeno-free conditions at a molecular level, we performed gene expression profiling with these new lines and existing lines derived and grown under standard conditions. As shown in Figure 3, the xeno-free lines derived here were highly similar to both human embryonic stem cells and induced pluripotent stem cells derived under standard conditions by a variety of different standard analyses. Clustering by similarity of profiles suggested that the xeno-free lines were more similar to each other than any other pluripotent stem cells and suggested that perhaps different derivation or growth conditions between pluripotent stem cells imparts slightly different gene expression profiles (Fig. 3A). Pearson correlation analysis showed that the xeno-free hiPSCs were as similar to hESCs as hiPSCs derived and grown under standard conditions (xeno-free hiPSCs and hESCs, 0.980–0.992; standard hiPSCs with hESCs, 0.987–0.995) (Fig. 3B). In addition, we have not yet detected any functional difference between xeno-free and standard human pluripotent stem cells lines as all these lines perform similarly in the teratoma assay (Fig. 2) and directed differentiation (Fig. 4).
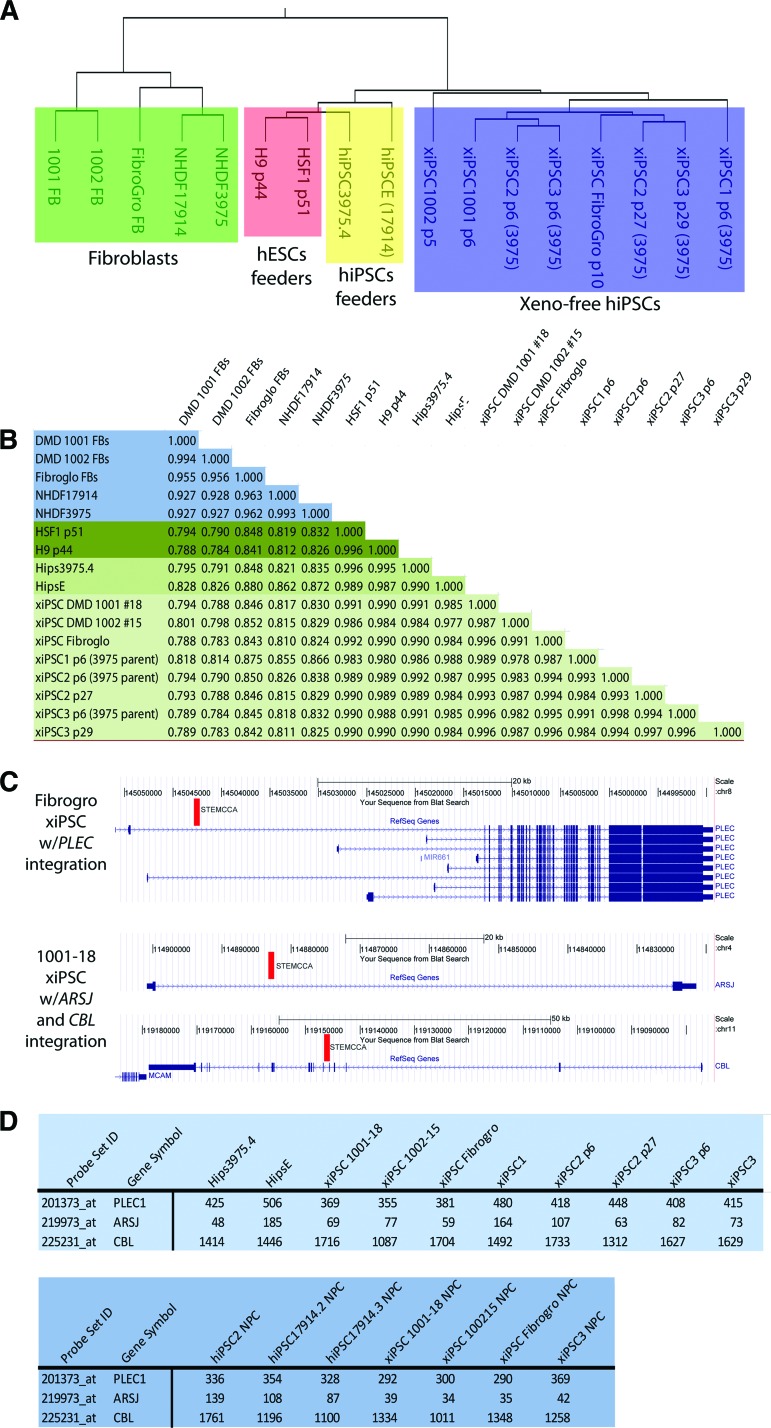
Figure 3.
Comparative gene expression analysis of Good Manufacturing Practice hiPSCs with hiPSCs and hESCs grown on feeders. (A): Block in green indicates fibroblasts, orange for hESCs on feeders, yellow for hiPSCs on feeders, and blue for the different xeno-free iPSCs analyzed. (B): Pearson analysis shows very minor differences in the global expression of genes between the xeno-free xiPSCs and iPSCs derived on feeders at different passage numbers. (C): Mapping of integration sites for the floxable polycistron used to reprogram shows that the STEMCCA integration is found either once or twice in two lines, and in each case is found in the intron of a gene (indicated by red bar). (D): Expression analysis indicates that these integrations did not influence expression of these genes.
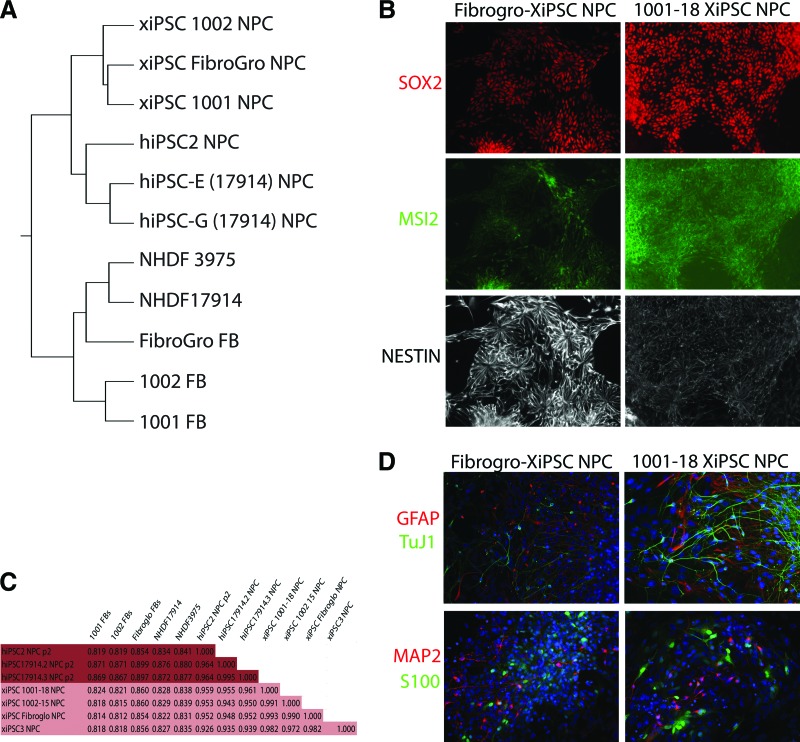
Figure 4.
Xeno-free differentiation of reprogrammed cells: (A): Gene expression clustering of xenofree-derived NPCs compared with NPCs derived under murine feeder cell-supported conditions from standard iPSCs and hESCs. (B): Immunocytochemical analysis of Fibrogro-XiPSC and 1001-18-XiPSC derived NPCs. (C): Pearson correlation analysis of microarray data from xeno-free and xenobiotic produced counterparts of iPSCs and hESCs derived NPCs. (D): Fibrogro-XiPSC and 1001-18-XiPSC derived NPCs were differentiated by growth factor withdrawal creating neurons and glia. Abbreviations: FB, fibroblast; NPCs, neural precursor cells; HDNF, normal human dermal fibroblast.
For clinical application of hiPSCs to become a reality, the reprogrammed cells must either be generated free of genomic integration of the reprogramming genes or have any genomic integration sites of the reprogramming vector(s) carefully documented to evaluate the potential for trans-activation of adjacent genes and/or insertional mutagenesis. Recent evidence suggests that the integrated reprogramming factors are not typically re-expressed upon differentiation of hiPSCs [21]. However, it is possible that altered expression at the loci of integration could have deleterious effects, and thus should be monitored before clinical application of hiPSCs. Traditional integration mapping techniques use restriction enzymes to digest genomic DNA to install linkers carrying reverse PCR priming sites into the genomic DNA. Because these restriction-based techniques suffer from blind spots in the genome due to the uneven distribution of restriction sites, we used a modified nonrestrictive linear amplification-mediated PCR (nrLAM-PCR) protocol coupled with Illumina high-throughput sequencing to map integrations of the reprogramming vector [18].
One hiPSC line was produced from the Fibrogro human fibroblast line (Millipore, Billerica, MA, http://www.millipore.com). We detected one integration of the polycistron in the Fibrogro hiPSC line, which occurred in an intron of the PLEC gene. Interestingly, PLEC has at least seven isoforms that differ only in their transcription start sites, and this integration is within the transcription unit of only two of these seven isoforms. Microarray expression data for PLEC show no difference in expression across all of the lines at all time points tested, suggesting that the vector integration does not interfere with PLEC expression, at least in undifferentiated iPSCs and neuronal progenitor cells (Fig. 3C).
In a hiPSC line derived from a normal donor, we detected two proviral integrant sites. The first occurred in the sole intron of the ARSJ gene, whereas the second was found in intron 5 of the CBL gene. Similar to PLEC in the Fibrogro line, neither of these genes in the donor-derived hiPSC line differed noticeably in expression in the cellular contexts and time points that were examined by microarray analysis (Fig. 3C). It is also important to note that the polycistron used is flanked by Loxp sites, allowing for excision of the factors with introduction of Cre recombinase activity. So although we show here that integration in these lines appears benign, these integrations could also be removed post facto.
Specification and Differentiation of Reprogrammed Cells Under GMP Conditions
To determine the potential of these xeno-free clones to generate clinically useful cell derivatives that are amenable to purification, the lines were subjected to neural specification. To extend the GMP-compatible nature of these potential products, fully defined, xeno-free components were used to drive neural specification, including culture media, growth, and differentiation factors. As shown in Figure 4, neural progenitor cells were derived that expressed the standard markers of this lineage (SOX2, MSI2, and NESTIN). To completely determine the specific nature of these NPCs, they were profiled by microarray for their gene expression. Their expression profiles were compared with NPCs derived under standard conditions from hESCs and hiPSCs grown on murine feeder cells. Clustering (Fig. 4A) and Pearson analysis (Fig. 4C) demonstrated that the xeno-free methods used here generated NPCs that are highly similar to their counterparts derived under standard conditions (Pearson, xeno-NPCs to standard NPCs: 0.926–0.959).
NPCs are capable of generating both neurons and glial cells. To ensure that the NPCs generated here under GMP-compatible conditions also were capable of generating differentiated cells types of the nervous system, we further developed xeno-free differentiation strategies. With use of defined media and growth factors produced under xeno-free conditions, the NPCs made from xeno-free hiPSCs were differentiated toward neurons and glia. Immunostaining for markers of both neurons (TUJ1 and MAP2) and astrocytes (S100 and GFAP) indicated that these NPCs were in fact capable of generating such cells (Fig. 4D).
Generation of Standard Operating Procedures
Finally, to facilitate the general application of these procedures and their eventual clinical application, we have generated SOPs for each of the methods outlined here (supplemental online Fig. S1). SOPs for the GMP laboratory have been broadly classified into five categories: Administrative, Material Management, Equipment, Manufacturing, and Quality Control and Assurance. We have generated a general platform for SOPs that addresses the administration, material management, equipment qualification, maintenance and quality control, and quality assurance procedures. The SOPs for manufacturing procedures have been customized specifically for the hiPSCs that are derived through our current development procedures. We have generated a full draft set of manufacturing SOPs that includes derivation and characterization of fibroblasts from patient skin biopsies that will be used for generation of hiPSCs, derivation and characterization of hiPSCs, and their cryopreservation as “master” and “working” cell banks.
Conclusion
To our knowledge, this is the first description of procedures necessary to take patient biopsies and generate NPCs, neurons, and glia through a pluripotent intermediate in the complete absence of either xenobiotics or feeder cells, with commercially available reagents. As new xeno-free media formulations, and even clinical grade feeders, are now regularly brought to market, it is possible that the conditions used here are not the only feasible methods to achieve this end. This is the first report to document the site of integration for the reprogramming factors after xeno-free reprogramming and demonstrate that such integrations can be benign. As a result, this work can serve as a starting point for clinical application of hiPSCs. The presentation here of clinical-grade SOPs could help to ensure that these methods are applied consistently and appropriately.
The procedures and SOPs outlined here are modular; therefore, as reliable methods are introduced to improve this protocol, including the use of nonintegrative methods to reprogram cells to the pluripotent state, new specific SOPs detailing these approaches can easily be integrated into the existing framework. Recent evidence demonstrated that human somatic cells can be reprogrammed to a pluripotent state by introduction of the reprogramming factors by protein, mRNA, miRNA, plasmid DNA transfection, adenovirus, and Sendai virus. Any of these nonintegrative methods could easily be incorporated into the existing protocols and SOPs described here.
Of course, future clinical application of hiPSCs may not require derivation under completely xeno-free conditions, as a clinical trial was recently initiated using derivatives of an hESC line that was initially derived with animal products and grown on murine feeders [2]. However, the process by which hiPSCs would need to be “cleaned up” to make them GMP compliant takes significant resources and, more importantly, considerable time. Instead, with the framework provided here, the testing and quality control mandated by regulatory agencies could be streamlined.
In the future, it will be crucial to establish standard QC tests for pluripotent stem cells in clinical translation. However, the establishment of standard QC procedures is challenging because criteria need to be established for their identity, purity, safety, and genetic stability. We have used established standard immunostaining and FACS methods to verify their identity and purity. We have also established tests for adventitious agents to ensure that these cells are free from any contaminating microorganisms or their by-products. In supplemental online Figure 2, we outline our work flow to demonstrate how each step in the process relies on successful translation of the previous step, and where QC becomes critical in the decision to proceed.
Unfortunately, no universal standards have been established to date to compare human pluripotent stem cells or their derivatives. The currently available method to establish the pluripotent status of these stem cells is the teratoma assay. However, the immune-deficient status of the animals, the number of cells required to make teratoma, the number of repetitions, the site of injection, and the meaning for any negative results need to be predefined. It is possible that high-throughput surrogates for the teratoma assay and other QC criteria will be developed, perhaps even bioinformatic approaches using genomic data [22]. Finally, characterization of the final transplantable cell product is at least as important as characterization of the undifferentiated stem cell itself for clinical translational purposes. Hence, significant effort and coordination will be required to establish appropriate QC tests, which should be customized for each type of specific final cell product.
Acknowledgments
We acknowledge the hESC/iPS core facilities (EEBSCRC), the clinical array core (Pathology, UCLA); Jinghua Tang, Rachel Kim, Anne Lindgren, Edward Kuoy, and Dinithi Senadheera for their expert technical support and support of the GMP laboratory (EEBSCRC); the EEBSCRC administration for their assistance in regulatory issues. Isolation and culture of fibroblasts from patient samples was in part funded from grants from the NIH (1RC1AR058333), the Department of Defense (85280103), the Foundation to Eradicate Duchenne, and the UCLA Muscular Dystrophy Core Center (5P30AR057230) including a supplement and seed grant from the UCLA P30. This work was also supported by the following: CIRM: New Cell Lines awards to J.Z. (RL1-00681-1) and A.C. (RL1-00636), a CIRM Young Investigator Award to K.P. (RN1-00564); the NIH (P01-GM081621 to J.A.Z.), and by the EEBSCRC.
Author Contributions
S.K., P.L., S.F.A., and A.R.C.: performed research; D.K., A.P., A.C., J.B., M.P., J.A.Z., K.P., and W.E.L.: manuscript writing.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Lowry WE, Quan WL. Roadblocks en route to the clinical application of induced pluripotent stem cells. J Cell Sci. 2010;123:643–651. doi: 10.1242/jcs.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trounson A. New perspectives in human stem cell therapeutic research. Bmc Med. 2009;7:29. doi: 10.1186/1741-7015-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crook JM, Peura TT, Kravets L, et al. The generation of six clinical-grade human embryonic stem cell lines. Cell Stem Cell. 2007;1:490–494. doi: 10.1016/j.stem.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Filler G. Challenges in pediatric transplantation: the impact of chronic kidney disease and cardiovascular risk factors on long-term outcomes and recommended management strategies. Pediatr Transplant. 2011;15:25–31. doi: 10.1111/j.1399-3046.2010.01439.x. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 7.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 8.Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin MH, Pellegrini M, Plath K, et al. Molecular analyses of human induced pluripotent stem cells and embryonic stem cells. Cell Stem Cell. 2010;7:263–269. doi: 10.1016/j.stem.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock C, Kiskinis E, Verstappen G, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Piza I, Richaud-Patin Y, Vassena R, et al. Reprogramming of human fibroblasts to induced pluripotent stem cells under xeno-free conditions. Stem Cells. 2010;28:36–44. doi: 10.1002/stem.248. [DOI] [PubMed] [Google Scholar]
- 13.Ross PJ, Suhr ST, Rodriguez RM, et al. Human-induced pluripotent stem cells produced under xeno-free conditions. Stem Cells Dev. 2010;19:1221–1229. doi: 10.1089/scd.2009.0459. [DOI] [PubMed] [Google Scholar]
- 14.Swistowski A, Peng J, Liu Q, et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells. 2010;28:1893–1904. doi: 10.1002/stem.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajala K, Lindroos B, Hussein SM, et al. A defined and xeno-free culture method enabling the establishment of clinical-grade human embryonic, induced pluripotent and adipose stem cells. PLoS One. 2010;5:e10246. doi: 10.1371/journal.pone.0010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karumbayaram S, Novitch BG, Patterson M, et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tchieu J, Kuoy E, Chin MH, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paruzynski A, Arens A, Gabriel R, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 19.Sommer CA, Stadtfeld M, Murphy GJ, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper AR, Patel S, Senadheera S, et al. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J Virol Methods. 2011;177:1–9. doi: 10.1016/j.jviromet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson M, Chan DN, Ha I, et al. Defining the nature of human pluripotent stem cell progeny. Cell Res. 2011 doi: 10.1038/cr.2011.133. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller FJ, Schuldt BM, Williams R, et al. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]