Abstract
Osteoporosis, the progressive loss of bone mass resulting in fragility fractures, affects ∼75 million people in the United States, Europe and Japan. Bone mineral density (BMD) correlates with fracture risk and is widely used in clinical settings to predict fracture. Numerous studies have demonstrated that peak bone mass is highly heritable and consequently a number of genome-wide association studies (GWASs) have been conducted to identify the genes that regulate BMD. Traditional intercross mapping in the mouse has met with limited successes in the field of skeletal biology. With the advent of human GWAS, questions have arisen about the continued need for mouse models in genetics research. However, significant advances have been made in the field of mouse genetics, including new genetics resource populations and loci mapping techniques, which enable gene-level mapping resolution. In this review, we discuss the need for mouse models to help understand the skeletal biology underlying novel human GWAS findings, how loci discovered in the mouse can be used to complement GWAS analysis and highlight the recent advances made in the field of skeletal biology from the use of these new and developing resources. We conclude this paper with a discussion of the need for systems-level approaches in the skeletal biology field, with an emphasis on the need for pathway and network analyses.
Introduction
Osteoporosis, the progressive loss of bone mass leading to fractures, is a significant cause of morbidity and mortality worldwide. Fracture risk increases with age, and as the proportion of aged persons worldwide is increasing, this disease is likely to become an even greater public health burden.1 Bone mineral density (BMD) can be used to predict future fracture risk, and studies have demonstrated that over 80% of the variance in peak bone mass is due to heritable factors.2 For this reason, there has been significant interest in identifying the genes that regulate bone mass. The genome-wide association study (GWAS) approach has led to the identification of a number of validated loci for BMD.3
As the first wave of GWAS is completed, questions have arisen about what the next steps should be. In this review, we focus on the use of the mouse both as a discovery tool for finding smaller variance loci missed by GWAS and as a tool that can be used to complement GWAS. Although GWAS and related gene mapping studies can identify loci implicated in bone mass, additional information is required to understand the function of these loci in skeletal biology. In the first part of this review, we discuss the need for mouse models to validate and interpret novel GWAS findings. This is followed by a discussion of the current efforts to map candidate genes for bone phenotypes using mouse genetic resource populations. We conclude with a discussion regarding the need for systems genetics, pathway analysis and alternate methods to find genes to move the field of skeletal genetics forward.
The need for mouse models
A GWAS is a hypothesis-free method of identifying genetic loci associated with a heritable phenotype.4 Although genome wide association analyses can be done using data from mice,5,6,7,8,9 most frequently GWAS is employed as a loci discovery tool on the basis of the data from human subjects. The rationale behind GWAS is that common genetic variants cause common diseases.10 In short, a large cohort is genotyped using single-nucleotide polymorphisms (SNPs) and associations between genotype and phenotype are identified. Between several thousand and a few million SNPs are genotyped per individual and SNPs are chosen that represent common alleles.10 Although the benefits and limitations of GWAS are reviewed elsewhere (see Hardy et al.4), the GWAS represents a giant step forward in both genetic mapping power and resolution.
One misconception about GWAS is that this method identifies which genes are associated with a phenotype; however, GWAS identifies which loci are associated with a phenotype. As such, SNPs associated with a given GWAS loci may not be within a gene at all, but rather may be intergenic.4 In these instances, it may not be clear which gene is causal for the change in phenotype. Moreover, it is not clear that protein-coding genes alone need be the causative gene for any locus11 as non-coding RNAs also have cellular function. The rs7524102 locus on 1p36 is just such an example. The significant SNPs at this locus fall within an intergenic region between WNT4 and ZBTB40.12 Although preliminary data suggest that WNT4 is expressed in mouse osteoblasts,13 little is known about this gene with regard to basic skeletal biology and nothing is known about ZBTB40 in bone. It is here that the mouse, or another appropriate model system, is needed. These models are needed not only to determine which gene is causal, but also to identify the pathways that each locus interacts with and to interpret the cellular function of each locus.
Furthermore, it must be appreciated that SNPs genotyped in a GWAS may not be causal themselves, but may be in linkage with a causal polymorphism that was not assayed during genotyping.14 An appreciation for the location of the causative polymorphism(s) is important for understanding the underlying biology. It is well understood that the gene expression is controlled by local elements, as well as by more distant regulatory sequences15 and the causative SNPs may be located in such regulatory elements. For example, in the GEFOS GWAS, TNFSF11 (RANKL) and CTNNB1 (β-Catenin) were identified as genes associated with BMD12 and a plethora of data support the role of these two genes in basic skeletal biology.16,17 However, the significant SNPs for these loci were not within the coding region of either gene, and, consequently, the causative polymorphisms likely influence gene expression.
The problem of 'missing variance' has been an issue for most GWAS.18 In the GEFOS study mentioned above, 20 loci were identified that were significantly associated with BMD of either the lumbar spine or of proximal femur, yet these loci account for <5% of heritable variance in the phenotype.12 Owing to the large number of genetic tests being performed in GWAS, the potential for false discoveries is high. To compensate, a high multiple testing correction penalty is applied. The concern has been raised that this stringent cutoff is preventing the identification of true associations and that this may be a source of missing variance. By design, GWAS is hypothesis free and accordingly, all genomic regions are treated as equal. Given the rich amount of prior genetic mapping data available for many complex traits, methods using prior data to weight associations and minimize the multiple testing penalty have been proposed.19 The mouse BMD quantitative trait loci (QTL) display excellent concordance with human GWAS loci for these same phenotypes and to date, many more reproducible mouse BMD QTL have been identified than have human GWAS loci.20 A recent study from our group has shown that genetic mapping data from mice can be effectively used to find sub-significant loci associated with BMD by allowing for the identification of a 'prior' or narrow region of interest to be targeted for analysis in human data sets.21 This is yet another way in which data obtained from mouse models could be used to guide and enhance GWASs.
Genetic loci mapping in mice
The traditional approach for genetic loci mapping in mouse has been interval mapping. In short, either an intercross or a backcross between two strains of mice is constructed, the mice are genotyped at low marker density, they are phenotyped and QTL are identified.22 This method has been prolifically applied to the phenotype of BMD and ∼85 unique QTL have been reported.20 These types of mapping studies have suffered from poor mapping resolution23 and other issues, and only five candidate genes for BMD have been identified: Alox15, Darc, Trps1, Pparg and Sfrp4.24,25,26,27,28 Although two strain intercross and interval mapping is no longer the workhorse of mouse genetics that they once were, gene-mapping efforts have not ceased. Rather these efforts have evolved with the development of better methods and new genetic resource populations.
The Collaborative Cross.
The Collaborative Cross (CC) is an ambitious, multi-national, multi-institutional effort to create a large set of recombinant inbred (RI) strains, with each RI line derived from eight strains of mice. The generation of this panel has been undertaken to create a large enough population of mice with enough genetic diversity to allow for systems biology analyses of complex traits.29 The eight founder strains are five classical inbred strains: C57BL/6J, A/J, 129S1/SvImJ, NZO/H1LtJ and NOD/LtJ, and three wild-derived strains: WSB/EiJ, PWK/PhJ and CAST/EiJ.30 In short, offspring from a two strain mating (G1 generation) were mated with other G1 mice to produced G2 generation animals (descended from four strains). The G2 mice are interbred to generate G2:F1 mice (descended from all eight founder strains). These G2:F1 mice were then, and continue to be, interbred such that fully inbred RI lines are established.29
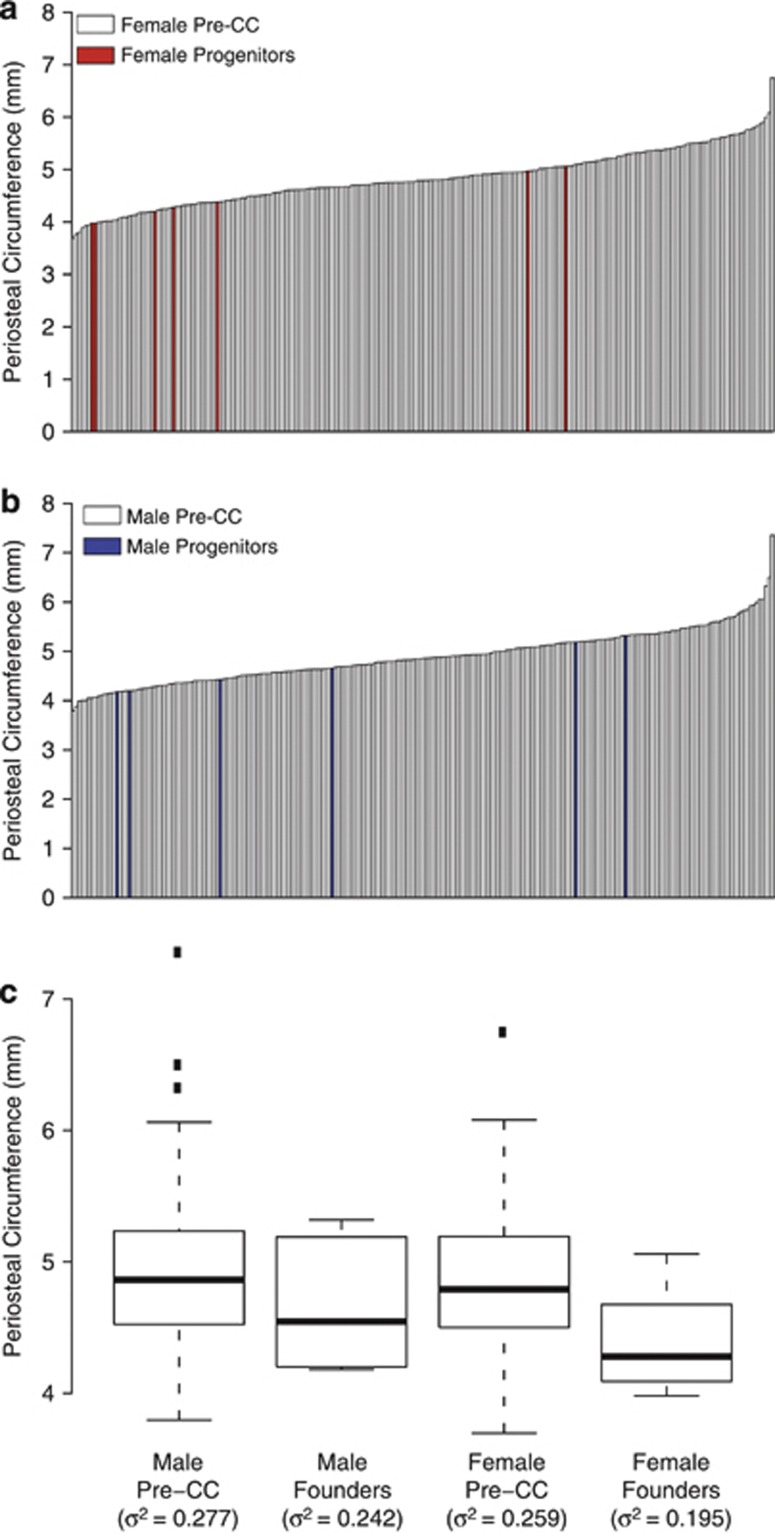
Some very interesting studies have now been completed using mice from the early stages of inbreeding the CC lines (so called 'pre-CC' mice). These studies demonstrate that the breadth of values for a given phenotype measured in the pre-CC is greater than observed in the founder strains alone31,32 (Figure 1). Moreover, these traits display greater breadth of phenotypic variation than is observed when measured in a genetic mapping panel derived from two progenitor strains.32 The first CC genetic loci have also now been mapped.31,32,33 As predicted from the computer simulations,34 QTL mapped in the pre-CC are an order of magnitude narrower than QTL mapped using an intercross mapping population.31,32 For example, a QTL for periosteal circumference mapped to chromosome 19 had a confidence interval width of only 0.96 Mb containing five candidate genes.32 The average confidence interval size of BMD QTL mapped using a traditional intercross population is ∼32 cM.20 Assuming that on average 1 cM equals 2 Mb and that there are on average 10 genes per Mb,35 this equals ∼640 candidate genes per QTL. Thus, five candidate genes for a QTL is nearly 2 orders of magnitude improvement in resolution. The power to map QTL utilizing the pre-CC is diminished relative to what is predicted for the completed lines33 and accordingly, there are high expectations for the final strains.
Figure 1. Periosteal circumference measured in the pre-CC.
Periosteal circumference of the mid-diaphysis of the femur, as measured by peripheral quantitative computed tomography, is presented for 224 female (a) and 225 male (b) pre-CC mice.32 As these data are from mice still carrying a large degree of heterozygosity, values for individual mice are shown. The data for the progenitor strains are also presented and these strains are represented by red (a) and blue (b) bars, respectively. Note, that data were not available from female NZO/H1LtJ (a) and male NZO/H1LtJ and A/J inbred mice (a). Data are also presented as box and whisker plot to emphasize the greater variance in BMD in the CC mice as compared with the founder strains (c).
The advantages of the CC are many. One large burden for any genetics study is the cost involved in genotyping the mice. Once inbred, the genotypes of individual lines will be stable, precluding the need for future genotyping.30 However, genetic stability comes at a price. Every allele in the CC will be homozygous, which does not mirror the natural state36 and there may have been loss of allelic variation (due to genetic incompatibilities) as the inbreeding progressed. Another advantage is the anticipated size of this panel together with the genetic diversity captured within it by the inclusion of wild-derived strains. Despite our successes in mapping genetic loci using classical inbred strains of mice, these strains have limited genetic diversity.37 Although the wild-derived CAST/EiJ strain has been used in two separate studies for BMD QTL mapping,20 the WSB/EiJ strain, which is M. m. domesticus, and PWD/PhJ strain, which is essentially pure M. m. musculus,37 have not been used extensively in skeletal biology. The increase in genetic diversity seen in the CC may in part explain the number of novel QTL mapped for energy expenditure,33 and bodes well for BMD studies.
For GWASs that have been conducted using meta-analyzed data,12 the inherent differences in the comprising cohorts, which could be ethnically and geographically diverse in origin, could result in too much heterogeneity in the environmental data for these metadata be used for GxE studies. In contrast, as the CC population is genetically reproducible, this population has clear and distinct advantages over pre-existing mouse populations for gene by environment (GxE), aging and developmental genetics studies.30 For example, the CC could be used to set up studies where group A is exposed to an environmental variable (that is, a high-fat diet), whereas group B is not. Group A and B would be genetically identical to each other; however, each group could also have consistently high amounts of genetic variability. At the end of the experiment, all mice from both groups A and B would be examined for a phenotype of interest and genetic loci associated with that phenotype would be mapped. Loci mapped in both A and B would represent loci that are independent of the environmental condition, whereas loci mapped in only group A or B could be considered to interact with the environmental condition. This point is particularly relevant for studies of skeletal biology, where we know that environmental factors affect basic bone biology. Analyses of GxE are just now beginning for human GWAS, but these types of case versus control GxE mapping studies, with static genetics between the case and control groups, cannot be conducted with human subjects. Unlike for GxE studies done with humans, one (or a limited number of) environmental factor(s) at a time can be examined independently using mouse models, and these experiments do not rely on self-reporting, are not confounded by failure to comply with interventions such as diet, and so on, so in this way mouse models are simpler. Although mouse models may not be appropriate for the examination of all types of environmental variable, these 'simplified' GxE studies using the CC, or other pre-existing mouse populations, have the potential to identify key GxE interactions and may help direct simpler 'candidate gene by environment' studies in human subjects.
The Diversity Outcross.
The Diversity Outcross (DO) is an offshoot of the CC, but unlike the CC, the goal is to maintain alleles in a segregating state. The DO was started from 144 CC lines obtained at early stages of inbreeding and the DO is propagated using a random breeding strategy. Consequently, all DO mice carry alleles from the eight CC founder strains, but each mouse is a unique mixture of alleles and heterozygosity is maintained in the population, more closely mimicking the natural population. As with the pre-CC mice, phenotypic diversity is high in the DO, as is the genetic mapping resolution.36 However, this population has disadvantages. Specifically, every mouse must be genotyped at high resolution for mapping and the lack of genetic stability makes this panel less attractive for some experimental designs.
The Hybrid Mouse Diversity Panel.
The Hybrid Mouse Diversity Panel (HMDP) panel is composed of 29 inbred strains, as well as 71 RI lines of mice. These 71 lines come from three sets of two-founder strain RI panels, wherein one founder was C57BL/6J (B6). This is a 'use as is' panel of mice, in that all of the strains are 'completed' and can be readily purchased. Furthermore, high-density genotyping for all lines is available23 and gene expression data are available from femoral cortical bone from male mice for most HMDP strains.38 This panel is very different than the CC and the DO in a number of ways. First, as the panel is 'fixed,' in that the mice are not interbred in any way, no new allelic combinations are generated. Second, alleles from wild-derived strains of mice are under represented and as such the HMDP will not be able identify wild-allele-driven loci and may fail to map loci in regions where the classical strains are nearly all identical by decent.23,39 As with the CC and the DO, the mapping resolution of the HMDP is superior to that obtained with traditional two-strain-intercross mapping populations.23 Using a combined phenotype QTL and expression QTL approach, the Asxl2 gene was identified as underlying a BMD locus on mouse chromosome 12 in the HMDP. Furthermore, a role for this gene in osteoclast differentiation has been established, solidifying this gene as a true genetic regulator of bone mass,38 however, it has not yet been ascertained if genetic variation in this gene is associated with BMD in human subjects.
Outbred mice.
Outbred stocks of mice are also starting to receive a lot of attention for GWASs.7,9,40 Although outbred stocks can be created,9 commercial outbreds are also useful.7,40 Most commercial outbred stocks are Swiss in origin and were derived from an extremely small founder population, suggesting that the number of private alleles in these colonies will be small, improving their usefulness for mapping.40 In theory, these mice are maintained as random bred colonies to maximize genetic variability. However, upon close examination, it has been determined that some stocks are actually inbred. A subset of the commercial outbred stocks have been identified that have low linkage disequilibrium, making them better suited for high-resolution mapping.40 One of these stocks is MF1. Farber et al.6 selectively genotyped the MF1 strain for the purposes of narrowing a traditionally mapped BMD QTL on mouse chromosome 11. Using this combined approach, they narrowed this QTL to two candidate genes: Wnt9a and Rasd1.
Additional resources.
In addition to the loci identification and characterization uses described in this review, projects are currently underway that leverage more 'traditional' transgenic mutant mouse models. The International Knockout Mouse Consortium (IKMC, http://www.knockoutmouse.org/) is tasked with developing knockout alleles for every gene in the mouse genome.41 In a complementary project, the International Mouse Phenotyping Consortium (IMPC, http://www.mousephenotype.org/) plans to phenotype the mice generated by the IKMC, in a high-throughput manner. The aim of the IMPC is to generate as much phenotyping data, on as many physiological systems as possible, in a high-throughput manner and as such will expand our genetic baseline knowledge of the laboratory mouse and provide resources for further mechanistic studies.42 This plethora of phenotyping data, including BMD data, will aid in identifying genes with direct actions on bone and those with indirect actions on bone (that is, a bone phenotype is observed that is caused by defects in another organ system). Combining this wealth of phenotyping data with new molecular and phenotypic measurement technologies, including high-throughput sequencing, chromatin immunoprecipitation, in vivo bone formation assays, and so on, are vital for downstream mechanistic studies of the proteins and pathways identified by genetics approaches. In this manner, the mouse will serve as an important mammalian genetic model for many more years.
The need to examine pathways, not just individual genes, to understand disease
A more general and fundamental problem with current genetics and biomedical research is that a reductive approach is applied to a limited set of 'candidate genes'. This may assume an inaccurate level of biological simplicity. Increasing evidence suggests that groups, suites, or pathways of proteins, RNAs, lipids and other small molecules work together to perform normal cellular functions, and that small perturbations to these components can cause systemic changes, potentially leading to impaired cellular function and a disease state. Genetic mapping research has identified many genes and pathways involving these genes that are responsible for phenotypic outcomes. However, in many cases, the specific genes responsible for the phenotypes may not be conserved between species,43,44 or even between strains of the same species; however, the pathways perturbed are often conserved across millions of evolutionary years. This suggests that rather than focusing on specific genetic alterations or expression changes in a single gene, research could benefit from considering data at a higher, so called 'systems' level.
This is especially true for many medical conditions, where subtle phenotypic variations can be just as important as dramatic phenotypic differences. In many situations, genetic or chemical manipulations that completely render a pathway or process inactive are likely to have detrimental outcomes, as extreme as embryonic lethality. For example, specific mutations that prevent the mineralization of the skeleton in model organisms are unlikely to contribute to osteoporosis, as carriers do not survive gestation. However, especially for highly polygenic diseases, such as osteoporosis, subtle defects in multiple genes and pathways appear to persist through development and potentially contribute to disease. But as discussed above, genetic mapping approaches such as GWAS often lack the statistical power to identify more subtle phenotypic effects, possibly overlooking clinically important modifiers. Further, some research supposes that identifying a 'causal' mutation directly implies a solution (that is, 'fixing' the mutation); however, given our limited capacity to manipulate cellular states, a more practical solution may involve indirectly 'fixing' the system through related genes. Thus, understanding entire pathways and networks of interacting molecules is required for improved diagnoses and treatments for complex genetic disorders, including osteoporosis. Fortunately, growing research in the areas of systems biology and functional genetics are developing approaches, tools and resources to address these outstanding problems.
One new area of growing research incorporates the systems biology 'pathway-level' view directly into data analysis for genetic mapping studies. For example, so-called Pathway-Wide or Network-Based Association Studies aim to analyze molecular phenotypes and genetic alterations at the level of groups or pathways of related genes.45,46 This reduces the size of the hypothesis space, increasing statistical power, and it also improves the interpretability of results. Further developments of these techniques include identifying pathways de novo from high-throughput molecular assays,47 which could enable the entire molecular network of a disease state to be rapidly, and automatically, identified. These methods have been applied to the studies of cancer and mammalian cell biology, and are now ripe for future applications in skeletal biology.
In addition to building better methods to conduct new studies of disease, a growing number of approaches and resources are available that take advantage of the existing wealth of genetic and genomic information in the literature and in public data repositories to predict associations between genes, pathways and phenotypes. Many of these projects have applied techniques from machine learning and data mining fields to search and analyze data collections to identify reliable data sources, and then to infer statistically likely relationships between genes and proteins given all available data. The result of this data integration is often presented as networks or graphs, which 'summarize' the input data used to generate them.
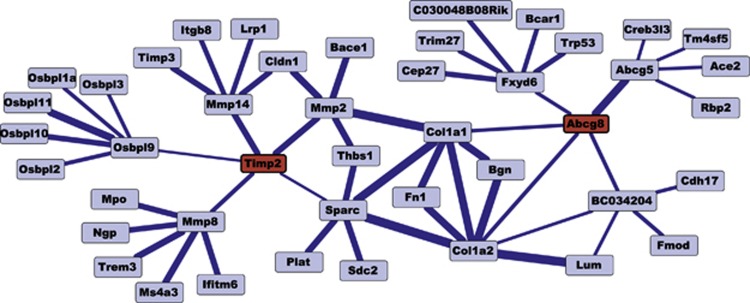
Functional integration of biological data were originally developed primarily for single-celled organisms,48,49 but more recent efforts have focused on skeletally relevant models, including mouse50,51 and human.52 These graphs can be further automatically analyzed, or manually explored by researchers, to quickly identify relevant experimental information from the literature, place new results into a broader context, and to form new hypotheses regarding protein function. This initial generation of systems biology tools and resources shows great promise, but they are still developing in maturity, scope and biological accuracy. A key limitation is that methods often treat mammalian genetic systems as homogenous, by overlooking the vital role of cellular context (that is, cell type, tissue niche, developmental stage, and so on) on molecular functions. For example, several recent efforts to integrate high-throughput mouse data into functional networks,51,53 a recent competition to predict mouse protein function from high-throughput data,54 and many others, all make the simplifying, and false, assumption that all genes perform the same function, regardless of cell type, conditions or other factors. As such, recent mammalian networks often show bias by identifying genes repeatedly investigated (often oncogenes or tumor suppressors, such as P53 or BRCA1) as the key, most connected features of mammalian networks. Consequently, for studies of specific pathway dynamics in the bone, these efforts should be regarded with some caution. This is not to say that these approaches are not useful, but that the assumptions in the data and models underlying them should be considered before their application. In fact, based on a generalized mouse integrated network, one recent study from our research group53 predicted several novel BMD genes, two of which (Timp2 and Abcg8) we validated in vivo despite the fact that neither was a candidate from any previous genetic mapping study50 (Figure 2).
Figure 2. A network view of functional relationships centered around Timp2 and Abcg8.
Nodes indicate genes, and the thickness of lines indicates relationship confidence. Both Timp2 and Abcg8 are connected to several genes that demonstrate a bone mineralization phenotype or are otherwise implicated in osteoporosis.
The systems biology community is beginning to recognize the potential benefits of incorporating additional contextual information into computational models, and ongoing efforts are focusing on the construction of functional networks for specific cellular contexts. For example, from our own work, the StemSight project (http://stemsight.org) is creating functional networks for self-renewal processes in various mammalian stem cell types, beginning with mouse embryonic stem cells, and the mouse MAP project (http://mousemap.princeton.edu) is developing tissue-specific functional networks for over 100 tissues (including bone and other skeletally-relevant tissues) in the laboratory mouse. As relatively few efforts are currently focused specifically on bone biology, as these methods mature, available resources will expand.
Conclusions
Mouse models continue to be valuable for research into the genetic underpinnings of osteoporosis. Early successes have been had in the field of osteoporosis using GWAS approaches, however, the future of genetic research in bone biology will likely still involve genetic mapping in the mouse. The newly available mouse genetic resource populations are far superior to traditional mapping panels and the preliminary use of these panels has already been successfully demonstrated. Animal model mapping studies should be thought of as complementary to, not in competition with, GWAS. Our forays into systems genetics have taught us the folly of the assumption that 'complex' genetic diseases can be easily dissected by genetic screening and statistical analysis. GWAS, despite all of its successes, should be thought of as more hypothesis generating than as a final solution. The reality of interdependent biological complexity requires that we measure and analyze data on multiple scales, with multiple levels of abstraction. Our challenge lies in integrating these measurements and observations into a complete picture of biology, at a level of granularity so that we can understand and correct deviations from normal function observed in a specific patient. Of course, this is no easy feat, but the combination of genetic, genomic, proteomic, functional, biomedical, and computational approaches will begin to achieve this goal. Skeletal biology is well positioned for this data analysis revolution, as clinically relevant bone measurements are easy to obtain, and as multiple vertebrate model organisms are gaining traction with the computational community.
Acknowledgments
We would like to thank Mr J Hammer for his assistance in figure preparation. This publication was made possible by Grant Numbers AR060234 and AR060981 from NIAMS/NIH.
Footnotes
The authors declare no conflict of interest.
References
- Cummings SR, Melton LJ. Epidemiology and outcomes of osteoporotic fractures. Lancet 2002;359:1761–1767. [DOI] [PubMed] [Google Scholar]
- Ralston SH, Uitterlinden AG. Genetics of osteoporosis. Endocr Rev 2010;31:629–662. [DOI] [PubMed] [Google Scholar]
- International Bone and Mineral Society (IBMS) BoneKEy. BoneKEy Genetics, (acessed on 1 November 2011). (Available at: http://www.bonekey-ibms.org/misc/Genetics/GeneticsPortal.dtl). [Google Scholar]
- Hardy J, Singleton A. Genome-wide association studies and human disease. N Engl J Med 2009;360:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD et al. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 2010;185:1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber C, van Nas A, Ghazalpour A, Aten J, Doss S, Sos B et al. An integrative genetics approach to identify candidate genes regulating BMD: combining linkage, gene expression, and association. J Bone Miner Res 2009;24:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Kang H, Farber C, Wen P-Z, Brozell A et al. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet 2008;4:e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y et al. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet 2006;38:888–895. [DOI] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 2006;38:879–887. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. Faseb J 2011;25:444–448. [DOI] [PubMed] [Google Scholar]
- Rivadeneira F, Styrkársdottir U, Estrada K, Halldórsson BV, Hsu YH, Richards JB et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet 2009;151:528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak W, Shao X, Dunstan CR, Seibel MJ, Zhou H. Biphasic glucocorticoid-dependent regulation of Wnt expression and its inhibitors in mature osteoblastic cells. Calcif Tissue Int 2009;85:538–545. [DOI] [PubMed] [Google Scholar]
- Marian AJ. Molecular genetic studies of complex phenotypes. Transl Res 2012;159:64–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW. Genome-scale techniques highlight the epigenome and redefine fundamental principles of gene regulation. J Bone Miner Res 2011;26:1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone 2007;40:251–264. [DOI] [PubMed] [Google Scholar]
- Piters E, Boudin E, Van Hul W. Wnt signaling: a win for bone. Arch Biochem Biophys 2008;473:112–116. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet 2009;10:241–251. [DOI] [PubMed] [Google Scholar]
- Roeder K, Bacanu SA, Wasserman L, Devlin B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet 2006;78:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackert-Bicknell CL, Karasik D, Li Q, Smith R, Hsu Y, Churchill GA et al. Mouse BMD quantitative trait loci show improved concordance with human genome wide association loci when recalculated on a new, common mouse genetic map. J Bone Min Res 2010;25:1808–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackert-Bicknell CL, Demissie S, Tsaih SW, Beamer WG, Cupples LA, Paigen BJ et al. Genetic variation in TRPS1 may regulate hip geometry as well as bone mineral density. Bone 2012;50:1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW. Review of statistical methods for QTL mapping in experimental crosses. Lab Anim 2001;30:44–52. [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res 2010;20:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackert-Bicknell CL, Demissie S, Marin de Evsikova C, Hsu YH, DeMambro VE, Karasik D et al. PPARG by dietary fat interaction influences bone mass in mice and humans. J Bone Miner Res 2008;23:1398–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edderkaoui B, Baylink DJ, Beamer WG, Wergedal JE, Porte R, Chaudhuri A et al. Identification of mouse duffy antigen receptor for chemokines (Darc) as a BMD QTL gene. Genome Res 2007;17:577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimori N, Stylianou IM, Korstanje R, Marion MA, Li R, Donahue LR et al. Quantitative trait loci for bone mineral density in an SM/J by NZB/BlNJ intercross population and identification of Trps1 as a probable candidate gene. J Bone Miner Res 2008;23:1529–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RF, Allard J, Avnur Z, Nikilcheva T, Rotstein D, Carlos AS et al. Regulation of bone mass in mice with the lipoxygenase gene Alox15. Science 2004;303:229–232. [DOI] [PubMed] [Google Scholar]
- Nakanishi R, Shimizu M, Mori M, Akiyama H, Okudaira S, Otsuki B et al. Secreted frizzled-related protein 4 is a negative regulator of peak BMD in SAMP6 mice. J Bone Miner Res 2006;21:1713–1721. [DOI] [PubMed] [Google Scholar]
- Chesler E, Miller D, Branstetter L, Galloway L, Jackson B, Philip V et al. The collaborative cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome 2008;19:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey C, Allayee H, Angel J. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 2004;36:1133–1137. [DOI] [PubMed] [Google Scholar]
- Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res 2011;21:1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Sokoloff G, Ackert-Bicknell CL, Striz M, Branstetter L, Beckmann MA et al. Genetic analysis in the Collaborative Cross breeding population. Genome Res 2011;21:1223–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes WF, Aylor DL, Miller DR, Churchill GA, Chesler EJ, de Villena FP et al. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am J Physiol Endocrinol Metab 2011;300:E1124–E1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R. Simulating the Collaborative Cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 2006;172:1783–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway WM, Healy B, Smink LJ, Rainbow D, Wicker LS. New tools for defining the ′genetic background′ of inbred mouse strains. Nat Immunol 2007;8:669–673. [DOI] [PubMed] [Google Scholar]
- Svenson K, Gatti D, Valdar W, Welsh CE, Cheng R, Chesler EJ et al. The mouse diversity outbred population. Genetics 2012;190:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nature Gen 2011;43:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E et al. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genetics 2011;7:e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Gen 2007;39:1100–1107. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Nicod J, Bhomra A, Davidson S, Cleak J, Farinelli L et al. Commercially available outbred mice for genome-wide association studies. PLoS Genet 2010;6:e1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. Mouse library set to be knockout. Nature 2011;474:262–263. [DOI] [PubMed] [Google Scholar]
- Schofield PN, Hoehndorf R, Gkoutos GV. Mouse genetic and phenotypic resources for human genetics. Hum Mutat 2012;33:826–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M. When orthologs diverge between human and mouse. Brief Bioinform 2011;12:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci USA 2010;107:6544–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ala-Korpela M, Kangas AJ, Inouye M. Genome-wide association studies and systems biology: together at last. Trends Genet 2011;27:493–498. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am J Hum Genet 2010;86:6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano A, Butte A, Friend S, Ideker T, Schadt EE. Integrative network-based association studies: leveraging cell regulatory models in the post-GWAS era. Nature Proc 2011. (10.1038/npre.2011.5732.1). [Google Scholar]
- Greene CS, Troyanskaya OG. Integrative systems biology for data-driven knowledge discovery. Semin Nephrol 2010;30:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein DA. Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae). Proc Natl Acad Sci USA 2003;100:8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Ackert-Bicknell CL, Kell B, Troyanskaya OG, Hibbs MA. Functional genomics complements quantitative genetics in identifying disease-gene associations. PLoS Comput Biol 2010;6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Krumpelman C, Marcotte EM. Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol Vol 2008;9 (Suppl 1): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Haley EM, Hibbs MA, Dumeaux V, Barrett DR, Coller HA et al. Exploring the human genome with functional maps. Genome Res 2009;19:1093–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Myers CL, Lu R, Lemischka IR, Bult CJ, Troyanskaya OG. A genome wide functional network for the laboratory mouse. PLoS Comput Biol 2008;4:e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Castillo L, Tasan M, Myers CL, Lee H, Joshi T, Zhang C et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol 2008;9 (Suppl 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]