Abstract
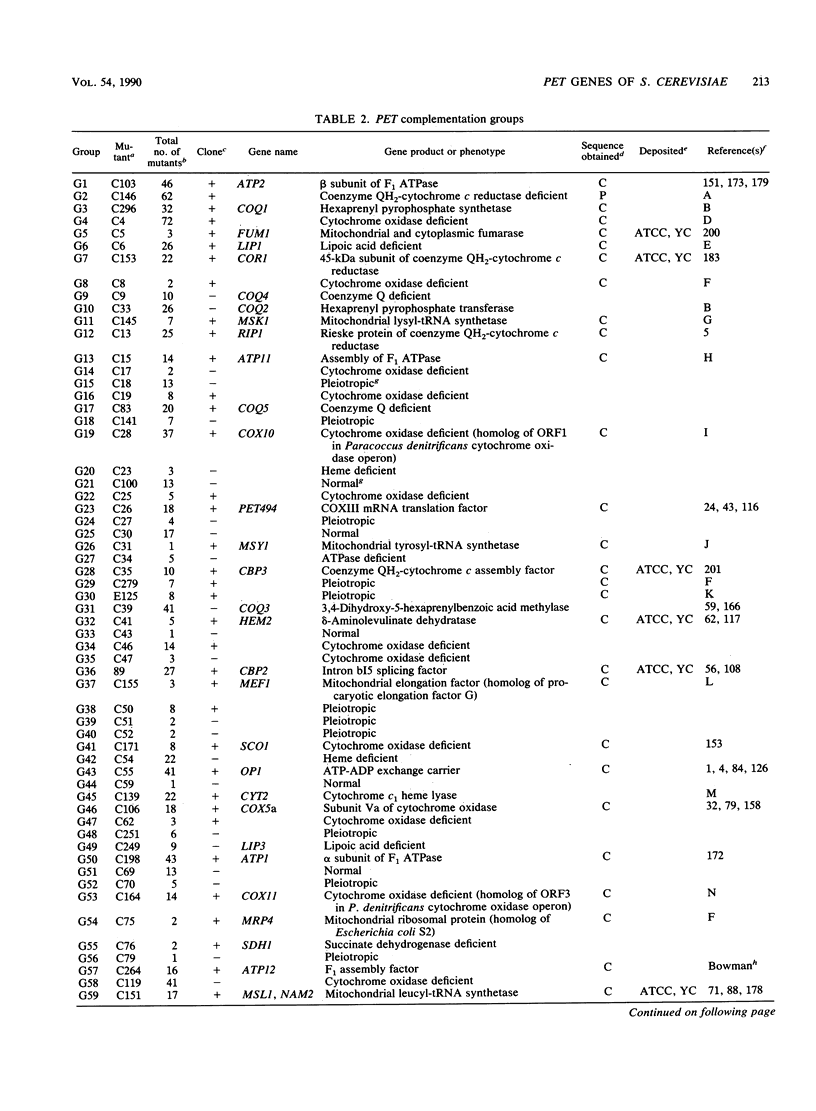
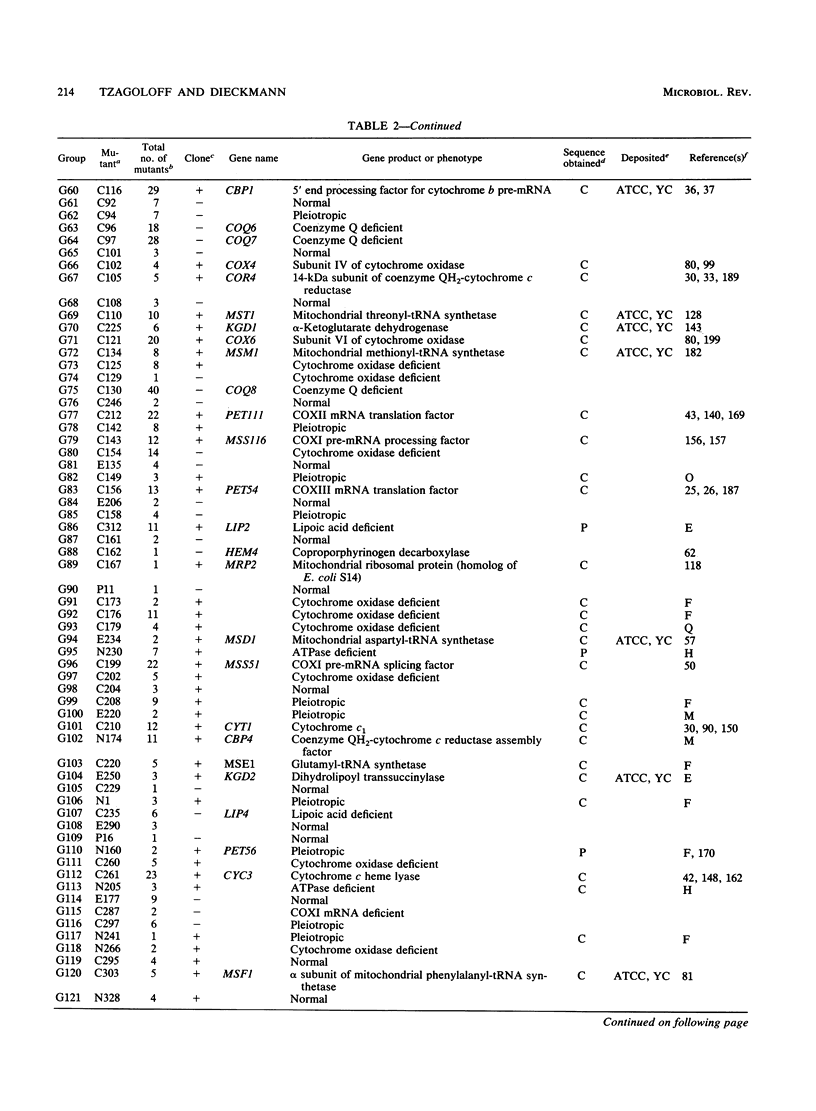
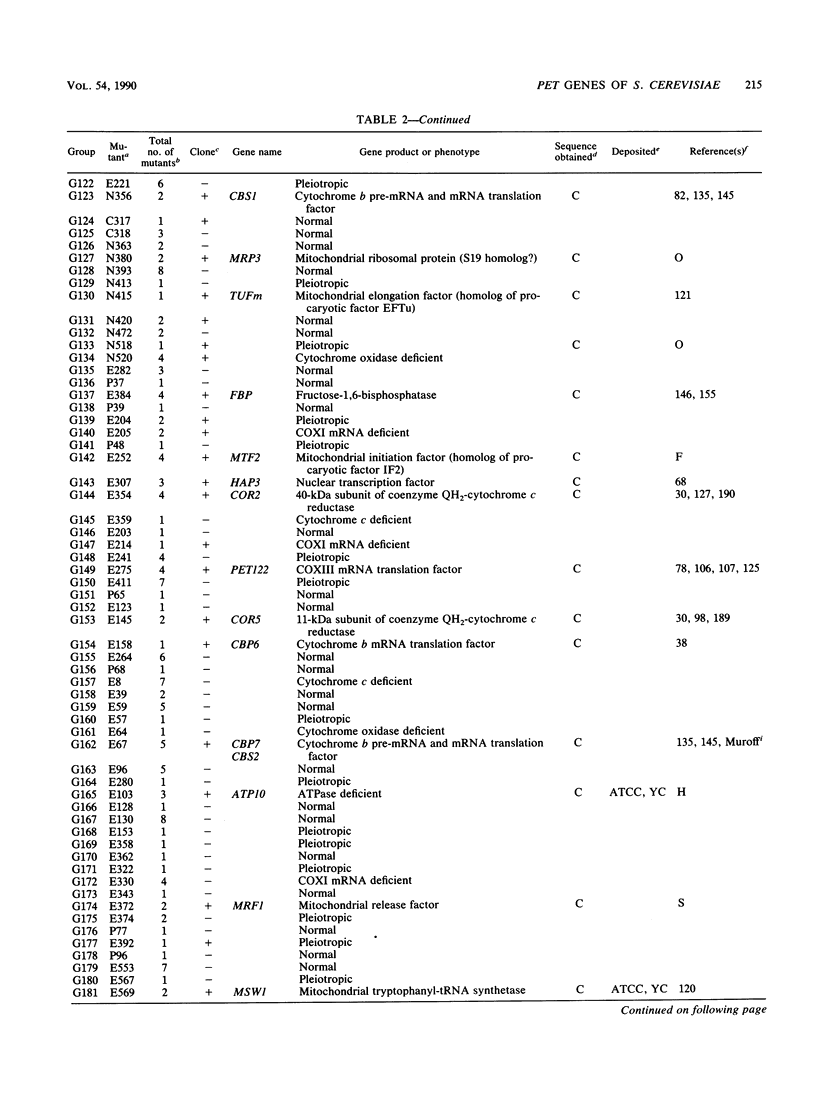
We describe a collection of nuclear respiratory-defective mutants (pet mutants) of Saccharomyces cerevisiae consisting of 215 complementation groups. This set of mutants probably represents a substantial fraction of the total genetic information of the nucleus required for the maintenance of functional mitochondria in S. cerevisiae. The biochemical lesions of mutants in approximately 50 complementation groups have been related to single enzymes or biosynthetic pathways, and the corresponding wild-type genes have been cloned and their structures have been determined. The genes defined by an additional 20 complementation groups were identified by allelism tests with mutants characterized in other laboratories. Mutants representative of the remaining complementation groups have been assigned to one of the following five phenotypic classes: (i) deficiency in cytochrome oxidase, (ii) deficiency in coenzyme QH2-cytochrome c reductase, (iii) deficiency in mitochondrial ATPase, (iv) absence of mitochondrial protein synthesis, and (v) normal composition of respiratory-chain complexes and of oligomycin-sensitive ATPase. In addition to the genes identified through biochemical and genetic analyses of the pet mutants, we have cataloged PET genes not matched to complementation groups in the mutant collection and other genes whose products function in the mitochondria but are not necessary for respiration. Together, this information provides an up-to-date list of the known genes coding for mitochondrial constituents and for proteins whose expression is vital for the respiratory competence of S. cerevisiae.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adrian G. S., McCammon M. T., Montgomery D. L., Douglas M. G. Sequences required for delivery and localization of the ADP/ATP translocator to the mitochondrial inner membrane. Mol Cell Biol. 1986 Feb;6(2):626–634. doi: 10.1128/mcb.6.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher E. B., Groudinsky O., Dujardin G., Altamura N., Kermorgant M., Slonimski P. P. Novel class of nuclear genes involved in both mRNA splicing and protein synthesis in Saccharomyces cerevisiae mitochondria. Mol Gen Genet. 1989 Feb;215(3):517–528. doi: 10.1007/BF00427051. [DOI] [PubMed] [Google Scholar]
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Beck J. C., Mattoon J. R., Hawthorne D. C., Sherman F. Genetic modification of energy-conserving systems in yeast mitochondria. Proc Natl Acad Sci U S A. 1968 May;60(1):186–193. doi: 10.1073/pnas.60.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J. D., Ljungdahl P. O., Lopez J. L., Trumpower B. L. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J Biol Chem. 1987 Jun 25;262(18):8901–8909. [PubMed] [Google Scholar]
- Beltzer J. P., Chang L. F., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast LEU4. The 5' flanking region contains features that predict two modes of control and two productive translation starts. J Biol Chem. 1986 Apr 15;261(11):5160–5167. [PubMed] [Google Scholar]
- Bibus C. R., Lemire B. D., Suda K., Schatz G. Mutations restoring import of a yeast mitochondrial protein with a nonfunctional presequence. J Biol Chem. 1988 Sep 15;263(26):13097–13102. [PubMed] [Google Scholar]
- Bolotin-Fukuhara M. Mitochondrial and nuclear mutations that affect the biogenesis of the mitochondrial ribosomes of yeast. I. Genetics. Mol Gen Genet. 1979;177(1):39–46. doi: 10.1007/BF00267251. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979 Nov;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol Cell Biol. 1983 Oct;3(10):1846–1856. doi: 10.1128/mcb.3.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning K. S., Uhlinger D. J., Reed L. J. Nucleotide sequence for yeast dihydrolipoamide dehydrogenase. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1831–1834. doi: 10.1073/pnas.85.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. F., Cunningham T. S., Gatzek P. R., Chen W. J., Kohlhaw G. B. Cloning and characterization of yeast Leu4, one of two genes responsible for alpha-isopropylmalate synthesis. Genetics. 1984 Sep;108(1):91–106. doi: 10.1093/genetics/108.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B., Walter P., Ebel J. P., Lacroute F., Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem. 1988 Jan 5;263(1):52–57. [PubMed] [Google Scholar]
- Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989 Feb 16;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Cis-dominant regulatory mutations affecting the formation of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1976 Jun 15;145(3):327–333. doi: 10.1007/BF00325831. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Clavilier L., Péré-Aubert G., Somlo M., Slonimski P. P. Réseau d'interactions entre des génes non liés : régulation synergique ou antagoniste de la synthèse de l'iso-1-cytochrome c, de l'iso-2-cytochrome c et du cytochrome b2. Biochimie. 1976;58(1-2):155–172. doi: 10.1016/s0300-9084(76)80366-5. [DOI] [PubMed] [Google Scholar]
- Clavilier L., Péré G., Slonimski P. P. Mise en évidence de plusieurs loci indépendants impliqués dans la synthèse de l'iso-2-cytochrome c chez la levure. Mol Gen Genet. 1969;104(2):195–218. doi: 10.1007/BF00272801. [DOI] [PubMed] [Google Scholar]
- Contamine V., Bolotin-Fukuhara M. A mitochondrial ribosomal RNA mutation and its nuclear suppressors. Mol Gen Genet. 1984;193(2):280–287. doi: 10.1007/BF00330681. [DOI] [PubMed] [Google Scholar]
- Coruzzi G., Tzagoloff A. Assembly of the mitochondrial membrane system: nuclear suppression of a cytochrome b mutation in yeast mitochondrial DNA. Genetics. 1980 Aug;95(4):891–903. doi: 10.1093/genetics/95.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Mueller P. P., Strick C. A., Fox T. D. Primary structure of wild-type and mutant alleles of the PET494 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 Feb;202(2):294–301. doi: 10.1007/BF00331654. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. At least two nuclear gene products are specifically required for translation of a single yeast mitochondrial mRNA. EMBO J. 1986 Dec 20;5(13):3637–3641. doi: 10.1002/j.1460-2075.1986.tb04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Seaver E. C., Fox T. D. The PET54 gene of Saccharomyces cerevisiae: characterization of a nuclear gene encoding a mitochondrial translational activator and subcellular localization of its product. Genetics. 1989 Jun;122(2):297–305. doi: 10.1093/genetics/122.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Kramer J., Kosic-Smithers J. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4156–4160. doi: 10.1073/pnas.84.12.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E. A., Kramer J., Shilling J., Werner-Washburne M., Holmes S., Kosic-Smithers J., Nicolet C. M. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol Cell Biol. 1989 Jul;9(7):3000–3008. doi: 10.1128/mcb.9.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Verdière J., Gaisne M., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J Mol Biol. 1988 Nov 20;204(2):263–276. doi: 10.1016/0022-2836(88)90574-8. [DOI] [PubMed] [Google Scholar]
- Crivellone M. D., Wu M. A., Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J Biol Chem. 1988 Oct 5;263(28):14323–14333. [PubMed] [Google Scholar]
- Cumsky M. G., Ko C., Trueblood C. E., Poyton R. O. Two nonidentical forms of subunit V are functional in yeast cytochrome c oxidase. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2235–2239. doi: 10.1073/pnas.82.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumsky M. G., Trueblood C. E., Ko C., Poyton R. O. Structural analysis of two genes encoding divergent forms of yeast cytochrome c oxidase subunit V. Mol Cell Biol. 1987 Oct;7(10):3511–3519. doi: 10.1128/mcb.7.10.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan M., van Loon A. P., Kreike J., Vaessen R. T., Grivell L. A. The biosynthesis of the ubiquinol-cytochrome c reductase complex in yeast. DNA sequence analysis of the nuclear gene coding for the 14-kDa subunit. Eur J Biochem. 1984 Jan 2;138(1):169–177. doi: 10.1111/j.1432-1033.1984.tb07896.x. [DOI] [PubMed] [Google Scholar]
- Dickinson J. R., Roy D. J., Dawes I. W. A mutation affecting lipoamide dehydrogenase, pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase activities in Saccharomyces cerevisiae. Mol Gen Genet. 1986 Jul;204(1):103–107. doi: 10.1007/BF00330195. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Homison G., Tzagoloff A. Assembly of the mitochondrial membrane system. Nucleotide sequence of a yeast nuclear gene (CBP1) involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4732–4738. [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Dihanich M. E., Najarian D., Clark R., Gillman E. C., Martin N. C., Hopper A. K. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):177–184. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G., Pajot P., Groudinsky O., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol Gen Genet. 1980;179(3):469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- Dumont M. E., Ernst J. F., Hampsey D. M., Sherman F. Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae. EMBO J. 1987 Jan;6(1):235–241. doi: 10.1002/j.1460-2075.1987.tb04744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner E., Mason T. L., Schatz G. Mitochondrial assembly in respiration-deficient mutants of Saccharomyces cerevisiae. II. Effect of nuclear and extrachromosomal mutations on the formation of cytochrome c oxidase. J Biol Chem. 1973 Aug 10;248(15):5369–5378. [PubMed] [Google Scholar]
- Ebner E., Mennucci L., Schatz G. Mitochondrial assembly in respiration-deficient mutants of Saccharomyces cerevisiae. I. Effect of nuclear mutations on mitochondrial protein synthesis. J Biol Chem. 1973 Aug 10;248(15):5360–5368. [PubMed] [Google Scholar]
- Ellis S. R., Hopper A. K., Martin N. C. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5172–5176. doi: 10.1073/pnas.84.15.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S. R., Morales M. J., Li J. M., Hopper A. K., Martin N. C. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem. 1986 Jul 25;261(21):9703–9709. [PubMed] [Google Scholar]
- Falco S. C., Dumas K. S. Genetic analysis of mutants of Saccharomyces cerevisiae resistant to the herbicide sulfometuron methyl. Genetics. 1985 Jan;109(1):21–35. doi: 10.1093/genetics/109.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco S. C., Dumas K. S., Livak K. J. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 1985 Jun 11;13(11):4011–4027. doi: 10.1093/nar/13.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Simon M. Analysis of a yeast nuclear gene involved in the maturation of mitochondrial pre-messenger RNA of the cytochrome oxidase subunit I. Cell. 1983 Jan;32(1):77–87. doi: 10.1016/0092-8674(83)90498-1. [DOI] [PubMed] [Google Scholar]
- Fearon K., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3636–3646. doi: 10.1128/mcb.8.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989 Dec 5;264(34):20552–20560. [PubMed] [Google Scholar]
- Foury F., Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987 May;6(5):1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampel A., Nishikimi M., Tzagoloff A. CBP2 protein promotes in vitro excision of a yeast mitochondrial group I intron. Mol Cell Biol. 1989 Dec;9(12):5424–5433. doi: 10.1128/mcb.9.12.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampel A., Tzagoloff A. Homology of aspartyl- and lysyl-tRNA synthetases. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6023–6027. doi: 10.1073/pnas.86.16.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genga A., Bianchi L., Foury F. A nuclear mutant of Saccharomyces cerevisiae deficient in mitochondrial DNA replication and polymerase activity. J Biol Chem. 1986 Jul 15;261(20):9328–9332. [PubMed] [Google Scholar]
- Goewert R. R., Sippel C. J., Olson R. E. Identification of 3,4-dihydroxy-5-hexaprenylbenzoic acid as an intermediate in the biosynthesis of ubiquinone-6 by Saccharomyces cerevisiae. Biochemistry. 1981 Jul 7;20(14):4217–4223. doi: 10.1021/bi00517a041. [DOI] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Marmur J. Petite mutation in yeast. II. Isolation of mutants containing mitochondrial deoxyribonucleic acid of reduced size. J Bacteriol. 1971 Jul;107(1):377–381. doi: 10.1128/jb.107.1.377-381.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977 May 10;252(9):2846–2854. [PubMed] [Google Scholar]
- Goltz S., Kaput J., Blobel G. Isolation of the yeast nuclear gene encoding the mitochondrial protein, cytochrome c peroxidase. J Biol Chem. 1982 Sep 25;257(18):11186–11190. [PubMed] [Google Scholar]
- Groudinsky O., Dujardin G., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184(3):493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guiard B., Buhler J. M. Yeast cytochrome b2 gene: isolation with antibody probes. Biochimie. 1984 Feb;66(2):151–158. doi: 10.1016/0300-9084(84)90204-9. [DOI] [PubMed] [Google Scholar]
- Guiard B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: the yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985 Dec 1;4(12):3265–3272. doi: 10.1002/j.1460-2075.1985.tb04076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T., Riezman H., Suda K., Schatz G. Import of proteins into mitochondria: nucleotide sequence of the gene for a 70-kd protein of the yeast mitochondrial outer membrane. EMBO J. 1983;2(12):2169–2172. doi: 10.1002/j.1460-2075.1983.tb01718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefta L. J., Lewin A. S., Daignan-Fornier B., Bolotin-Fukuhara M. Nuclear and mitochondrial revertants of a mitochondrial mutant with a defect in the ATP synthetase complex. Mol Gen Genet. 1987 Apr;207(1):106–113. doi: 10.1007/BF00331497. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988 Feb;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordana X., Chatton B., Paz-Weisshaar M., Buhler J. M., Cramer F., Ebel J. P., Fasiolo F. Structure of the yeast valyl-tRNA synthetase gene (VASI) and the homology of its translated amino acid sequence with Escherichia coli isoleucyl-tRNA synthetase. J Biol Chem. 1987 May 25;262(15):7189–7194. [PubMed] [Google Scholar]
- Julou C., Contamine V., Sor F., Bolotin-Fukuhara M. Mitochondrial ribosomal RNA genes of yeast: their mutations and a common nuclear suppressor. Mol Gen Genet. 1984;193(2):275–279. doi: 10.1007/BF00330680. [DOI] [PubMed] [Google Scholar]
- KAKAR S. N., WAGNER R. P. GENETIC AND BIOCHEMICAL ANALYSIS OF ISOLEUCINE-VALINE MUTANTS OF YEAST. Genetics. 1964 Feb;49:213–222. doi: 10.1093/genetics/49.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaput J., Goltz S., Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor. Functional implications of the pre sequence for protein transport into mitochondria. J Biol Chem. 1982 Dec 25;257(24):15054–15058. [PubMed] [Google Scholar]
- Kelly J. L., Greenleaf A. L., Lehman I. R. Isolation of the nuclear gene encoding a subunit of the yeast mitochondrial RNA polymerase. J Biol Chem. 1986 Aug 5;261(22):10348–10351. [PubMed] [Google Scholar]
- Kispal G., Evans C. T., Malloy C., Srere P. A. Metabolic studies on citrate synthase mutants of yeast. A change in phenotype following transformation with an inactive enzyme. J Biol Chem. 1989 Jul 5;264(19):11204–11210. [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen J. E., Poyton R. O. Identification of a third nuclear protein-coding gene required specifically for posttranscriptional expression of the mitochondrial COX3 gene is Saccharomyces cerevisiae. J Bacteriol. 1988 Mar;170(3):1399–1402. doi: 10.1128/jb.170.3.1399-1402.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner T. J., Hill J., Tzagoloff A. Cloning and characterization of the yeast nuclear gene for subunit 5 of cytochrome oxidase. J Biol Chem. 1985 Aug 15;260(17):9513–9515. [PubMed] [Google Scholar]
- Koerner T. J., Homison G., Tzagoloff A. Nuclear mutants of Saccharomyces cerevisiae with altered subunits 4, 5, and 6 of cytochrome oxidase. J Biol Chem. 1985 May 25;260(10):5871–5874. [PubMed] [Google Scholar]
- Koerner T. J., Myers A. M., Lee S., Tzagoloff A. Isolation and characterization of the yeast gene coding for the alpha subunit of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 1987 Mar 15;262(8):3690–3696. [PubMed] [Google Scholar]
- Kovác L., Lachowicz T. M., Slonimski P. P. Biochemical genetics of oxidative phosphorylation. Science. 1967 Dec 22;158(3808):1564–1567. doi: 10.1126/science.158.3808.1564. [DOI] [PubMed] [Google Scholar]
- Kreike J., Schulze M., Ahne F., Lang B. F. A yeast nuclear gene, MRS1, involved in mitochondrial RNA splicing: nucleotide sequence and mutational analysis of two overlapping open reading frames on opposite strands. EMBO J. 1987 Jul;6(7):2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreike J., Schulze M., Pillar T., Körte A., Rödel G. Cloning of a nuclear gene MRS1 involved in the excision of a single group I intron (bI3) from the mitochondrial COB transcript in S. cerevisiae. Curr Genet. 1986;11(3):185–191. doi: 10.1007/BF00420605. [DOI] [PubMed] [Google Scholar]
- Krzywicki K. A., Brandriss M. C. Primary structure of the nuclear PUT2 gene involved in the mitochondrial pathway for proline utilization in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2837–2842. doi: 10.1128/mcb.4.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körte A., Forsbach V., Gottenöf T., Rödel G. In vitro and in vivo studies on the mitochondrial import of CBS1, a translational activator of cytochrome b in yeast. Mol Gen Genet. 1989 May;217(1):162–167. doi: 10.1007/BF00330956. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Dujardin G., Slonimski P. P. The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell. 1985 May;41(1):133–143. doi: 10.1016/0092-8674(85)90068-6. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Herbert C. J., Dujardin G., Slonimski P. P. Three suppressor mutations which cure a mitochondrial RNA maturase deficiency occur at the same codon in the open reading frame of the nuclear NAM2 gene. EMBO J. 1987 Mar;6(3):713–721. doi: 10.1002/j.1460-2075.1987.tb04812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langgut W., Entrup R., Schweizer E. Isolation of a nuclear yeast gene involved in the mitochondrial import of cytoplasmically synthesized precursor proteins. Curr Genet. 1986;11(3):177–184. doi: 10.1007/BF00420604. [DOI] [PubMed] [Google Scholar]
- Laten H., Gorman J., Bock R. M. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res. 1978 Nov;5(11):4329–4342. doi: 10.1093/nar/5.11.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Jones D., Mueller D. M. The sequence of the yeast ATP5 gene. Nucleic Acids Res. 1988 Aug 25;16(16):8181–8181. doi: 10.1093/nar/16.16.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P., Slonimski P. P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowsky T., Michaelis G. A nuclear gene essential for mitochondrial replication suppresses a defect of mitochondrial transcription in Saccharomyces cerevisiae. Mol Gen Genet. 1988 Oct;214(2):218–223. doi: 10.1007/BF00337714. [DOI] [PubMed] [Google Scholar]
- Lisowsky T. Molecular analysis of the mitochondrial transcription factor mtf2 of Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jan;220(2):186–190. doi: 10.1007/BF00260480. [DOI] [PubMed] [Google Scholar]
- Lombardo A., Scheffler I. E. Isolation and characterization of a Saccharomyces cerevisiae mutant with a disrupted gene for the IP subunit of succinate dehydrogenase. J Biol Chem. 1989 Nov 15;264(32):18874–18877. [PubMed] [Google Scholar]
- Maarse A. C., Grivell L. A. Nucleotide sequence of the gene encoding the 11-kDa subunit of the ubiquinol-cytochrome-c oxidoreductase in Saccharomyces cerevisiae. Eur J Biochem. 1987 Jun 1;165(2):419–425. doi: 10.1111/j.1432-1033.1987.tb11455.x. [DOI] [PubMed] [Google Scholar]
- Maarse A. C., Van Loon A. P., Riezman H., Gregor I., Schatz G., Grivell L. A. Subunit IV of yeast cytochrome c oxidase: cloning and nucleotide sequencing of the gene and partial amino acid sequencing of the mature protein. EMBO J. 1984 Dec 1;3(12):2831–2837. doi: 10.1002/j.1460-2075.1984.tb02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee P. T., Robichon-Szulmajster H. The regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. 2. Identification and characterization of mutants lacking the acetohydroxyacid synthetase. Eur J Biochem. 1968 Feb;3(4):502–506. doi: 10.1111/j.1432-1033.1967.tb19559.x. [DOI] [PubMed] [Google Scholar]
- Marres C. A., Van Loon A. P., Oudshoorn P., Van Steeg H., Grivell L. A., Slater E. C. Nucleotide sequence analysis of the nuclear gene coding for manganese superoxide dismutase of yeast mitochondria, a gene previously assumed to code for the Rieske iron-sulphur protein. Eur J Biochem. 1985 Feb 15;147(1):153–161. doi: 10.1111/j.1432-1033.1985.tb08731.x. [DOI] [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita Y., Kitakawa M., Isono K. Cloning and analysis of the nuclear genes for two mitochondrial ribosomal proteins in yeast. Mol Gen Genet. 1989 Oct;219(1-2):119–124. doi: 10.1007/BF00261166. [DOI] [PubMed] [Google Scholar]
- McAlister-Henn L., Thompson L. M. Isolation and expression of the gene encoding yeast mitochondrial malate dehydrogenase. J Bacteriol. 1987 Nov;169(11):5157–5166. doi: 10.1128/jb.169.11.5157-5166.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. E., Cameron V. L., Poyton R. O. Rapid method for isolation and screening of cytochrome c oxidase-deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1985 Mar;161(3):831–835. doi: 10.1128/jb.161.3.831-835.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J. E., Ko C., Kloeckner-Gruissem B., Poyton R. O. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem. 1986 Sep 5;261(25):11872–11879. [PubMed] [Google Scholar]
- McGraw P., Tzagoloff A. Assembly of the mitochondrial membrane system. Characterization of a yeast nuclear gene involved in the processing of the cytochrome b pre-mRNA. J Biol Chem. 1983 Aug 10;258(15):9459–9468. [PubMed] [Google Scholar]
- Mihara K., Sato R. Molecular cloning and sequencing of cDNA for yeast porin, an outer mitochondrial membrane protein: a search for targeting signal in the primary structure. EMBO J. 1985 Mar;4(3):769–774. doi: 10.1002/j.1460-2075.1985.tb03695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery D. L., Hall B. D., Gillam S., Smith M. Identification and isolation of the yeast cytochrome c gene. Cell. 1978 Jul;14(3):673–680. doi: 10.1016/0092-8674(78)90250-7. [DOI] [PubMed] [Google Scholar]
- Montgomery D. L., Leung D. W., Smith M., Shalit P., Faye G., Hall B. D. Isolation and sequence of the gene for iso-2-cytochrome c in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jan;77(1):541–545. doi: 10.1073/pnas.77.1.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounolou J. C., Jakob H., Slonimski P. P. Mitochondrial DNA from yeast "petite" mutants: specific changes in buoyant density corresponding to different cytoplasmic mutations. Biochem Biophys Res Commun. 1966 Jul 20;24(2):218–224. doi: 10.1016/0006-291x(66)90723-6. [DOI] [PubMed] [Google Scholar]
- Mueller D. M., Biswas T. K., Backer J., Edwards J. C., Rabinowitz M., Getz G. S. Temperature sensitive pet mutants in yeast Saccharomyces cerevisiae that lose mitochondrial RNA. Curr Genet. 1987;11(5):359–367. doi: 10.1007/BF00378178. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Koerner T. J., Tzagoloff A. Characterization of the yeast HEM2 gene and transcriptional regulation of COX5 and COR1 by heme. J Biol Chem. 1987 Dec 15;262(35):16822–16829. [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Tzagoloff A. Assembly of the mitochondrial membrane system. MRP1 and MRP2, two yeast nuclear genes coding for mitochondrial ribosomal proteins. J Biol Chem. 1987 Mar 5;262(7):3388–3397. [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A. MSW, a yeast gene coding for mitochondrial tryptophanyl-tRNA synthetase. J Biol Chem. 1985 Dec 5;260(28):15371–15377. [PubMed] [Google Scholar]
- Müller P. P., Fox T. D. Molecular cloning and genetic mapping of the PET494 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(1-2):275–280. doi: 10.1007/BF00332759. [DOI] [PubMed] [Google Scholar]
- Nagata S., Tsunetsugu-Yokota Y., Naito A., Kaziro Y. Molecular cloning and sequence determination of the nuclear gene coding for mitochondrial elongation factor Tu of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6192–6196. doi: 10.1073/pnas.80.20.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarian D., Dihanich M. E., Martin N. C., Hopper A. K. DNA sequence and transcript mapping of MOD5: features of the 5' region which suggest two translational starts. Mol Cell Biol. 1987 Jan;7(1):185–191. doi: 10.1128/mcb.7.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsoulis G., Hilger F., Fink G. R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986 Jul 18;46(2):235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- Niu X. D., Browning K. S., Behal R. H., Reed L. J. Cloning and nucleotide sequence of the gene for dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7546–7550. doi: 10.1073/pnas.85.20.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley K., Pratt P., Robertson J., Lilly M., Douglas M. G. Selection of the nuclear gene for the mitochondrial adenine nucleotide translocator by genetic complementation of the op1 mutation in yeast. J Biol Chem. 1982 Feb 25;257(4):2097–2103. [PubMed] [Google Scholar]
- Ohmen J. D., Kloeckener-Gruissem B., McEwen J. E. Molecular cloning and nucleotide sequence of the nuclear PET122 gene required for expression of the mitochondrial COX3 gene in S. cerevisiae. Nucleic Acids Res. 1988 Nov 25;16(22):10783–10802. doi: 10.1093/nar/16.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn P., Van Steeg H., Swinkels B. W., Schoppink P., Grivell L. A. Subunit II of yeast QH2:cytochrome-c oxidoreductase. Nucleotide sequence of the gene and features of the protein. Eur J Biochem. 1987 Feb 16;163(1):97–103. doi: 10.1111/j.1432-1033.1987.tb10741.x. [DOI] [PubMed] [Google Scholar]
- Pape L. K., Koerner T. J., Tzagoloff A. Characterization of a yeast nuclear gene (MST1) coding for the mitochondrial threonyl-tRNA1 synthetase. J Biol Chem. 1985 Dec 5;260(28):15362–15370. [PubMed] [Google Scholar]
- Parker J. H., Mattoon J. R. Mutants of yeast with altered oxidative energy metabolism: selection and genetic characterization. J Bacteriol. 1969 Nov;100(2):647–657. doi: 10.1128/jb.100.2.647-657.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partaledis J. A., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3647–3660. doi: 10.1128/mcb.8.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson T. E., Poyton R. O. COX8, the structural gene for yeast cytochrome c oxidase subunit VIII. DNA sequence and gene disruption indicate that subunit VIII is required for maximal levels of cellular respiration and is derived from a precursor which is extended at both its NH2 and COOH termini. J Biol Chem. 1986 Dec 25;261(36):17192–17197. [PubMed] [Google Scholar]
- Paul M. F., Velours J., Arselin de Chateaubodeau G., Aigle M., Guerin B. The role of subunit 4, a nuclear-encoded protein of the F0 sector of yeast mitochondrial ATP synthase, in the assembly of the whole complex. Eur J Biochem. 1989 Oct 20;185(1):163–171. doi: 10.1111/j.1432-1033.1989.tb15098.x. [DOI] [PubMed] [Google Scholar]
- Petersen J. G., Holmberg S. The ILV5 gene of Saccharomyces cerevisiae is highly expressed. Nucleic Acids Res. 1986 Dec 22;14(24):9631–9651. doi: 10.1093/nar/14.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. H., Kjellin-Stråby K. Studies on microbial ribonucleic acid. IV. Two mutants of Saccharomyces cerevisiae lacking N-2-dimethylguanine in soluble ribonucleic acid. J Mol Biol. 1967 Jun 28;26(3):509–518. doi: 10.1016/0022-2836(67)90318-x. [DOI] [PubMed] [Google Scholar]
- Pillar T., Lang B. F., Steinberger I., Vogt B., Kaudewitz F. Expression of the "split gene" cob in yeast mtDNA. Nuclear mutations specifically block the excision of different introns from its primary transcript. J Biol Chem. 1983 Jul 10;258(13):7954–7959. [PubMed] [Google Scholar]
- Pinkham J. L., Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R. A., Hartl F. U., Cheng M. Y., Ostermann J., Horwich A., Neupert W. The processing peptidase of yeast mitochondria: the two co-operating components MPP and PEP are structurally related. EMBO J. 1988 Nov;7(11):3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poutre C. G., Fox T. D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987 Apr;115(4):637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratje E., Guiard B. One nuclear gene controls the removal of transient pre-sequences from two yeast proteins: one encoded by the nuclear the other by the mitochondrial genome. EMBO J. 1986 Jun;5(6):1313–1317. doi: 10.1002/j.1460-2075.1986.tb04361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading D. S., Hallberg R. L., Myers A. M. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature. 1989 Feb 16;337(6208):655–659. doi: 10.1038/337655a0. [DOI] [PubMed] [Google Scholar]
- Repetto B., Tzagoloff A. Structure and regulation of KGD1, the structural gene for yeast alpha-ketoglutarate dehydrogenase. Mol Cell Biol. 1989 Jun;9(6):2695–2705. doi: 10.1128/mcb.9.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H., Hase T., van Loon A. P., Grivell L. A., Suda K., Schatz G. Import of proteins into mitochondria: a 70 kilodalton outer membrane protein with a large carboxy-terminal deletion is still transported to the outer membrane. EMBO J. 1983;2(12):2161–2168. doi: 10.1002/j.1460-2075.1983.tb01717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D. T., Hiller E., Mitsock L., Orr E. Characterization of the gene for fructose-1,6-bisphosphatase from Saccharomyces cerevisiae and Schizosaccharomyces pombe. Sequence, protein homology, and expression during growth on glucose. J Biol Chem. 1988 May 5;263(13):6051–6057. [PubMed] [Google Scholar]
- Ross J., Reid G. A., Dawes I. W. The nucleotide sequence of the LPD1 gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae: comparison between eukaryotic and prokaryotic sequences for related enzymes and identification of potential upstream control sites. J Gen Microbiol. 1988 May;134(5):1131–1139. doi: 10.1099/00221287-134-5-1131. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980 Apr;94(4):871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D. J., Dawes I. W. Cloning and characterization of the gene encoding lipoamide dehydrogenase in Saccharomyces cerevisiae. J Gen Microbiol. 1987 Apr;133(4):925–933. doi: 10.1099/00221287-133-4-925. [DOI] [PubMed] [Google Scholar]
- Rödel G., Michaelis U., Forsbach V., Kreike J., Kaudewitz F. Molecular cloning of the yeast nuclear genes CBS1 and CBS2. Curr Genet. 1986;11(1):47–53. doi: 10.1007/BF00389425. [DOI] [PubMed] [Google Scholar]
- SHERMAN F. MUTANTS OF YEAST DEFICIENT IN CYTOCHROME C. Genetics. 1964 Jan;49:39–48. doi: 10.1093/genetics/49.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F. Respiration-deficient mutants of yeast. I. Genetics. Genetics. 1963 Mar;48:375–385. doi: 10.1093/genetics/48.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F., SLONIMSKI P. P. RESPIRATION-DEFICIENT MUTANTS OF YEAST. II. BIOCHEMISTRY. Biochim Biophys Acta. 1964 Jul 15;90:1–15. doi: 10.1016/0304-4165(64)90113-8. [DOI] [PubMed] [Google Scholar]
- Sadler I., Suda K., Schatz G., Kaudewitz F., Haid A. Sequencing of the nuclear gene for the yeast cytochrome c1 precursor reveals an unusually complex amino-terminal presequence. EMBO J. 1984 Sep;3(9):2137–2143. doi: 10.1002/j.1460-2075.1984.tb02103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzgaber-Muller J., Kunapuli S. P., Douglas M. G. Nuclear genes coding the yeast mitochondrial adenosine triphosphatase complex. Isolation of ATP2 coding the F1-ATPase beta subunit. J Biol Chem. 1983 Oct 10;258(19):11465–11470. [PubMed] [Google Scholar]
- Schmidt C., Söllner T., Schweyen R. J. Nuclear suppression of a mitochondrial RNA splice defect: nucleotide sequence and disruption of the MRS3 gene. Mol Gen Genet. 1987 Nov;210(1):145–152. doi: 10.1007/BF00337771. [DOI] [PubMed] [Google Scholar]
- Schulze M., Rödel G. Accumulation of the cytochrome c oxidase subunits I and II in yeast requires a mitochondrial membrane-associated protein, encoded by the nuclear SCO1 gene. Mol Gen Genet. 1989 Mar;216(1):37–43. doi: 10.1007/BF00332228. [DOI] [PubMed] [Google Scholar]
- Sedivy J. M., Fraenkel D. G. Fructose bisphosphatase of Saccharomyces cerevisiae. Cloning, disruption and regulation of the FBP1 structural gene. J Mol Biol. 1985 Nov 20;186(2):307–319. doi: 10.1016/0022-2836(85)90107-x. [DOI] [PubMed] [Google Scholar]
- Shannon K. W., Rabinowitz J. C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem. 1988 Jun 5;263(16):7717–7725. [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Jackson M., Gilmore R. A., Parker J. H. Mutants of yeast defective in iso-1-cytochrome c. Genetics. 1974 Jun;77(2):255–284. doi: 10.1093/genetics/77.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Parker J. H., Inhaber E., Shipman N. A., Putterman G. J., Gardisky R. L., Margoliash E. The mutational alteration of the primary structure of yeast iso-1-cytochrome c. J Biol Chem. 1968 Oct 25;243(20):5446–5456. [PubMed] [Google Scholar]
- Sippel C. J., Goewert R. R., Slachman F. N., Olson R. E. The regulation of ubiquinone-6 biosynthesis by Saccharomyces cerevisiae. J Biol Chem. 1983 Jan 25;258(2):1057–1061. [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Sor F., Faye G. Mitochondrial and nuclear mutations that affect the biogenesis of the mitochondrial ribosomes of yeast. II. Biochemistry. Mol Gen Genet. 1979;177(1):47–56. doi: 10.1007/BF00267252. [DOI] [PubMed] [Google Scholar]
- Strick C. A., Fox T. D. Saccharomyces cerevisiae positive regulatory gene PET111 encodes a mitochondrial protein that is translated from an mRNA with a long 5' leader. Mol Cell Biol. 1987 Aug;7(8):2728–2734. doi: 10.1128/mcb.7.8.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa M., Suda K., Schatz G. Isolation of the nuclear yeast genes for citrate synthase and fifteen other mitochondrial proteins by a new screening method. EMBO J. 1984 Aug;3(8):1773–1781. doi: 10.1002/j.1460-2075.1984.tb02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Boulet A., Simon M., Faye G. Construction of a yeast strain devoid of mitochondrial introns and its use to screen nuclear genes involved in mitochondrial splicing. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6810–6814. doi: 10.1073/pnas.84.19.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Boulet A., Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989 Jan 5;337(6202):84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Faye G. MSS18, a yeast nuclear gene involved in the splicing of intron aI5 beta of the mitochondrial cox1 transcript. EMBO J. 1988 May;7(5):1455–1464. doi: 10.1002/j.1460-2075.1988.tb02963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Simon M., Faye G. Primary structure of a gene for subunit V of the cytochrome c oxidase from Saccharomyces cerevisiae. Curr Genet. 1985;9(6):435–439. doi: 10.1007/BF00434047. [DOI] [PubMed] [Google Scholar]
- Takeda M., Chen W. J., Saltzgaber J., Douglas M. G. Nuclear genes encoding the yeast mitochondrial ATPase complex. Analysis of ATP1 coding the F1-ATPase alpha-subunit and its assembly. J Biol Chem. 1986 Nov 15;261(32):15126–15133. [PubMed] [Google Scholar]
- Takeda M., Vassarotti A., Douglas M. G. Nuclear genes coding the yeast mitochondrial adenosine triphosphatase complex. Primary sequence analysis of ATP2 encoding the F1-ATPase beta-subunit precursor. J Biol Chem. 1985 Dec 15;260(29):15458–15465. [PubMed] [Google Scholar]
- Thompson L. M., Sutherland P., Steffan J. S., McAlister-Henn L. Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 1988 Nov 1;27(22):8393–8400. doi: 10.1021/bi00422a015. [DOI] [PubMed] [Google Scholar]
- Thonart P., Bechet J., Hilger F., Burny A. Thermosensitive mutations affecting ribonucleic acid polymerases in Saccharomyces cerevisiae. J Bacteriol. 1976 Jan;125(1):25–32. doi: 10.1128/jb.125.1.25-32.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Apr;7(4):1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Foury F. Assembly of the mitochondrial membrane system XVI. Modified form of the ATPase proteolipid in oligomycin-resistant mutants of Saccharomyces cerevisiae. FEBS Lett. 1976 Jun 15;65(3):391–395. doi: 10.1016/0014-5793(76)80154-8. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Kurkulos M., Repetto B. Homology of yeast mitochondrial leucyl-tRNA synthetase and isoleucyl- and methionyl-tRNA synthetases of Escherichia coli. J Biol Chem. 1988 Jan 15;263(2):850–856. [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R. B. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975 Oct 25;250(20):8228–8235. [PubMed] [Google Scholar]
- Tzagoloff A., Akai A., Needleman R. B. Assembly of the mitochondrial membrane system: isolation of nuclear and cytoplasmic mutants of Saccharomyces cerevisiae with specific defects in mitochondrial functions. J Bacteriol. 1975 Jun;122(3):826–831. doi: 10.1128/jb.122.3.826-831.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Vambutas A., Akai A. Characterization of MSM1, the structural gene for yeast mitochondrial methionyl-tRNA synthetase. Eur J Biochem. 1989 Feb 1;179(2):365–371. doi: 10.1111/j.1432-1033.1989.tb14562.x. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Wu M. A., Crivellone M. Assembly of the mitochondrial membrane system. Characterization of COR1, the structural gene for the 44-kilodalton core protein of yeast coenzyme QH2-cytochrome c reductase. J Biol Chem. 1986 Dec 25;261(36):17163–17169. [PubMed] [Google Scholar]
- Urban-Grimal D., Labbe-Bois R. Genetic and biochemical characterization of mutants of Saccharomyces cerevisiae blocked in six different steps of heme biosynthesis. Mol Gen Genet. 1981;183(1):85–92. doi: 10.1007/BF00270144. [DOI] [PubMed] [Google Scholar]
- Urban-Grimal D., Volland C., Garnier T., Dehoux P., Labbe-Bois R. The nucleotide sequence of the HEM1 gene and evidence for a precursor form of the mitochondrial 5-aminolevulinate synthase in Saccharomyces cerevisiae. Eur J Biochem. 1986 May 2;156(3):511–519. doi: 10.1111/j.1432-1033.1986.tb09610.x. [DOI] [PubMed] [Google Scholar]
- Valencik M. L., Kloeckener-Gruissem B., Poyton R. O., McEwen J. E. Disruption of the yeast nuclear PET54 gene blocks excision of mitochondrial intron aI5 beta from pre-mRNA for cytochrome c oxidase subunit I. EMBO J. 1989 Dec 1;8(12):3899–3904. doi: 10.1002/j.1460-2075.1989.tb08569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon A. P., De Groot R. J., De Haan M., Dekker A., Grivell L. A. The DNA sequence of the nuclear gene coding for the 17-kd subunit VI of the yeast ubiquinol-cytochrome c reductase: a protein with an extremely high content of acidic amino acids. EMBO J. 1984 May;3(5):1039–1043. doi: 10.1002/j.1460-2075.1984.tb01924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velours J., Durrens P., Aigle M., Guérin B. ATP4, the structural gene for yeast F0F1 ATPase subunit 4. Eur J Biochem. 1988 Jan 4;170(3):637–642. doi: 10.1111/j.1432-1033.1988.tb13745.x. [DOI] [PubMed] [Google Scholar]
- Verdière J., Creusot F., Guérineau M. Regulation of the expression of iso 2-cytochrome c gene in S. cerevisiae: cloning of the positive regulatory gene CYP1 and identification of the region of its target sequence on the structural gene CYP3. Mol Gen Genet. 1985;199(3):524–533. doi: 10.1007/BF00330769. [DOI] [PubMed] [Google Scholar]
- Verdière J., Gaisne M., Guiard B., Defranoux N., Slonimski P. P. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. II. Missense mutation suggests alternative Zn fingers as discriminating agents of gene control. J Mol Biol. 1988 Nov 20;204(2):277–282. doi: 10.1016/0022-2836(88)90575-x. [DOI] [PubMed] [Google Scholar]
- Vincent R. D., Hofmann T. J., Zassenhaus H. P. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Apr 25;16(8):3297–3312. doi: 10.1093/nar/16.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol. 1986 Jul;6(7):2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C., Jensen R. E., Yaffe M. P., Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988 May;7(5):1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. M., Dircks L. K., Poyton R. O. Characterization of COX9, the nuclear gene encoding the yeast mitochondrial protein cytochrome c oxidase subunit VIIa. Subunit VIIa lacks a leader peptide and is an essential component of the holoenzyme. J Biol Chem. 1986 Dec 25;261(36):17183–17191. [PubMed] [Google Scholar]
- Wright R. M., Ko C., Cumsky M. G., Poyton R. O. Isolation and sequence of the structural gene for cytochrome c oxidase subunit VI from Saccharomyces cerevisiae. J Biol Chem. 1984 Dec 25;259(24):15401–15407. [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. Identification and characterization of a new gene (CBP3) required for the expression of yeast coenzyme QH2-cytochrome c reductase. J Biol Chem. 1989 Jul 5;264(19):11122–11130. [PubMed] [Google Scholar]
- Wu M., Tzagoloff A. Mitochondrial and cytoplasmic fumarases in Saccharomyces cerevisiae are encoded by a single nuclear gene FUM1. J Biol Chem. 1987 Sep 5;262(25):12275–12282. [PubMed] [Google Scholar]
- Yaffe M. P., Jensen R. E., Guido E. C. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J Biol Chem. 1989 Dec 15;264(35):21091–21096. [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E. T., Pilgrim D. Isolation and DNA sequence of ADH3, a nuclear gene encoding the mitochondrial isozyme of alcohol dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3024–3034. doi: 10.1128/mcb.5.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorec M., Buhler J. M., Treich I., Keng T., Guarente L., Labbe-Bois R. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J Biol Chem. 1988 Jul 15;263(20):9718–9724. [PubMed] [Google Scholar]
- Zhu H., Conrad-Webb H., Liao X. S., Perlman P. S., Butow R. A. Functional expression of a yeast mitochondrial intron-encoded protein requires RNA processing at a conserved dodecamer sequence at the 3' end of the gene. Mol Cell Biol. 1989 Apr;9(4):1507–1512. doi: 10.1128/mcb.9.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon A. P., Maarse A. C., Riezman H., Grivell L. A. Isolation, characterization and regulation of expression of the nuclear genes for the core II and Rieske iron-sulphur proteins of the yeast ubiquinol-cytochrome c reductase. Gene. 1983 Dec;26(2-3):261–272. doi: 10.1016/0378-1119(83)90196-8. [DOI] [PubMed] [Google Scholar]
- van Loon A. P., de Groot R. J., van Eyk E., van der Horst G. T., Grivell L. A. Isolation and characterization of nuclear genes coding for subunits of the yeast ubiquinol-cytochrome c reductase complex. Gene. 1982 Dec;20(3):323–337. doi: 10.1016/0378-1119(82)90201-3. [DOI] [PubMed] [Google Scholar]



