Abstract
The pathogenesis of bone metastases is a complex and multifaceted process. Often multiple imaging modalities are needed to follow both the structural and functional changes over time during metastatic bone disease. Researchers face extended data sets of one experiment acquired with multiple modalities at multiple points in time. This review gives an overview of an integrated approach for handling these kinds of complex data. It focuses on the analysis of whole-body micro-computerized tomography and optical data handling. We show how researchers can generate side-by-side visualizations of scans taken with one imaging modality at multiple time points and with multiple modalities at one point. Moreover, we highlight methods for normalized volumes of interest selection and quantification of bone volume and thickness.
Introduction
Bone is one of the preferred sites of breast cancer and prostate cancer metastases. Postmortem examination revealed that ∼70% of patients who died of breast or prostate cancer carried bone metastases.1 The attraction of certain types of cancer toward bone was already noted in 1889 by Paget2 and has been well studied ever since.
The clinical manifestations of overt bone metastases are vast, ranging from severe osteolysis to osteoblastic calcifications outside the bone and inside the marrow cavity.3 Bone metastases are a clinical predictor of poor survival for both breast and prostate cancer patients. The pathology of this so called 'metastatic bone disease' is a result of complex interactions between the tumor and the bone microenvironment. The cross-talk between the tumor and bone results in the disruption of the delicate balance between osteoclasts and osteoblasts.4,5,6
Key regulators involved in osteolytic bone metastases are parathyroid hormone-related protein produced by the tumor and transforming growth factor-beta released from the bone upon resorption. Parathyroid hormone-related protein stimulates osteoclastogenesis and prolongs osteoclast survival. The released transforming growth factor-beta stimulates parathyroid hormone-related protein production by the tumor. It furthermore stimulates proliferation and apoptosis resistance together with other growth factors that are released from the resorbed bone matrix. This positive feedback loop is known as the vicious cycle of osteolytic bone metastases.4,5,6
Bone metastases are difficult to treat because of the strong positive feedback loop between the tumor and the bone microenvironment. Research toward treatment of bone metastases is in need of reliable methods to quantify bone-specific effects of experimental treatments in vivo. Imaging can be an informative tool for that purpose, providing qualitative as well as quantitative results. Here, we give an overview of both the structural and functional whole-body imaging modalities that can be used in research on metastatic bone disease as well as on other bone-related diseases. We will focus on recent advances in normalized comparison of data sets of one animal made at different points in time and combining data generated with multiple imaging modalities.
Structural imaging
Preclinical imaging modalities can be grouped in two classes, structural imaging modalities such as X-ray-based imaging and functional imaging modalities such as bioluminescence imaging (BLI), fluorescence imaging (FLI), positron emission tomography (PET) and single-photon emission computed tomography (SPECT). Structural imaging modalities are used to visualize and quantify structural and architectural features of the skeleton, whereas functional imaging modalities are used to gain insight in cellular, metabolic and molecular processes within and around cancerous bone lesions. For decades, structural imaging of bone and bone lesions has been performed with radiography. The advances in digital computing enabled the development of three-dimensional (3D) computerized tomography (CT) by Godfrey Hounsfield during the 1970s, which was a major revolution in the field of X-ray-based imaging.7,8 Nowadays, specialized micro-CT (μCT) scanners are available for small laboratory animals with resolutions of several micrometers.
Posture correction.
The posture of entire animals varies greatly between scans taken at multiple times. As a result, it can be difficult to identify small osteolytic lesions in longitudinal μCT data sets, even for an experienced observer.9 For some applications, dedicated animal holders can be used to reduce the postural variability,10 but such holders are often not sufficient and limit the throughput, especially for multimodality studies. Therefore, compensation for large posture variations between scans taken at different points in time is needed, to enable the user to identify small lesions within these large, complex whole-body μCT data sets.
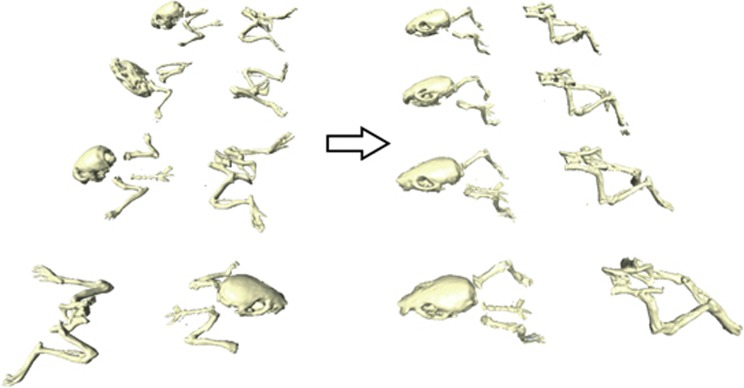
To compensate for the postural differences between scans, a method has been developed based on matching an atlas to whole-body μCT data sets. For this purpose Khmelinskii et al.11 developed kinetic mouse and rat atlases. The skeletons of these atlases have been segmented and each individual skeletal element has been identified. In addition, each joint position has been annotated and the kinetic degrees of freedom have been specified. The position of the bones of such an articulated atlas can be changed to fit the skeleton of an actual animal μCT scan. Also, the positions of major internal organs without μCT contrast are approximated based on the positions of the skeletal elements, the lungs and skin (Figure 1).9 Baiker et al.9 developed a fully automated system for aligning the atlas with μCT scans. The automated system can effectively perform posture corrections in the absence of soft-tissue contrast in in vivo μCT data and without requiring user initialization. The approach is robust with respect to large postural variations and to moderate, pathology-induced structural variations. After aligning the atlas with the scan data, the skeletal elements of the scanned animal can be repositioned to a standardized posture. This articulated atlas-based repositioning of the skeleton facilitates fast data comparison (Figure 2).11
Figure 1.
Aligning the articulated atlas with μCT data. Registration results between the atlas (red) and two different subjects (gray) after coarsely aligning the skeleton (top), after the articulated registration (middle) and after organ approximation (bottom). Adapted with permission from Khmelinskii et al.11
Figure 2.
Demonstration of mapping the skeletons of four different animals to a common reference domain. The large postural differences of the animals (left) are not present anymore (right), enabling a more intuitive comparison of different time points. Adapted with permission from Khmelinskii et al.11
Side-by-side visualizations.
Intracardial injection of bone-specific MDA-MB-231 breast cancer cells is a popular method to induce osteolytic cancer metastases in the skeleton.12 Bone metastases will develop at several sites throughout the skeleton within 1–3 weeks after inoculation with cancer cells. Disease progression of these osteolytic lesions can be followed by comparing whole-body μCT scans taken at different points in time. However, the initial structural disease induced changes in this kind of follow-up data sets can be very small and difficult to identify in complex 3D data sets. Posture normalization of the data sets is helpful, but often not enough. Kok et al.13 developed an approach, which enables side-by-side visualizations of an individual skeletal element at different times. The data used for these detailed visualizations are extracted from longitudinal whole-body μCT data sets.
The generation of side-by-side visualizations individual skeletal elements is a multistep process. First, the μCT data sets taken at each time point are aligned. Next, the scan is divided in normalized subvolumes, which are placed around each of the skeletal elements. These subvolumes are then used to generate an 'exploded view' (Figure 3a) of the skeleton. Finally, the subvolumes are mapped into a common reference frame using articulated planar reformation, where corresponding volumes of interests (VOIs) are visualized side-by-side (Figures 3b and c). The visualizations of the different time points are coupled, in other words, the user selects a section or visualization of a skeletal element of a scan taken at one time and the system will automatically generate the corresponding visualization of the other scans.13
Figure 3.
Exploded view and side-by-side visualizations of longitudinal μCT data. Osteolytic bone metastases were induced by intracardial injection of 1×106 MDA-MB231-BO2 cells (an osteolytic, bone-specific subclone of the MDA breast carcinoma cell line) in a 6-week-old nude mice. μCT scans were made 3, 5 and 6 weeks after inoculation. The animal was scanned in a different posture per time point, supine (week 3) and prone (week 5 and 6). (a) Longitudinal μCT data have been aligned with the articulated mouse atlas to eliminate differences in posture. Next, the scans were divided over subvolumes per skeletal element. Structural changes between time points have been highlighted. These areas are potentially interesting to the user for further, in depth, inspection. The femur has been selected and is shown side-by-side in detailed views for the three available time points at 3, 5 and 6 weeks into the experiment (b) and (c). Shown for each time are an image plane that can be interactively moved through the volume (b) and a surface rendering of the entire femur (c). At 5 and 6 weeks, bone resorption can clearly be seen near the knee area (indicated by the arrows), even though the animal posture in the original data were highly variable. Note that between the first and second time point, with the subject at 10 and 12 weeks of age, respectively, growth is still taking place, which has to be taken into account when analyzing the images. Figure adapted with permission form Kok et al.13
Methods of mapping data into a common reference frame make the use of animal holders during data acquisition obsolete because postural differences can be compensated for afterwards. The use of subvolumes per skeletal element not only has the advantage of aligning structures of interest between longitudinal scans, but it also enables data analysis at full resolution because only a small part of the whole-body data set is loaded. This is often not possible for whole-body data sets because of the extreme computing requirements owing to the large amount of data.
Quantification.
Treatment and intervention studies on bone metastases often require the quantification of several bone parameters. Currently, the quantification of osteolytic lesions is performed by drawing a region of interest around the lesion on 2D radiographs and measuring the lesion surface.14,15 Lesions projected on top of each other and lesions on the side of bone will be underestimated when quantified owing to the flattening of the 3D structure.16 The large dependency on the manual input results in a strong dependence on experience of the observer and makes the method prone to observer bias. The scoring of such radiographs cannot be automated owing to the variability of shape and gray values between various bones and lesions.
μCT is a 3D modality and therefore data sets provide more accurate information on disease-induced changes like an increase or decrease in bone volume, bone thickness and bone density, both for visual and quantitative assessment. However, the selection of a VOI in 3D data sets is more complex than region of interest selection in 2D data.
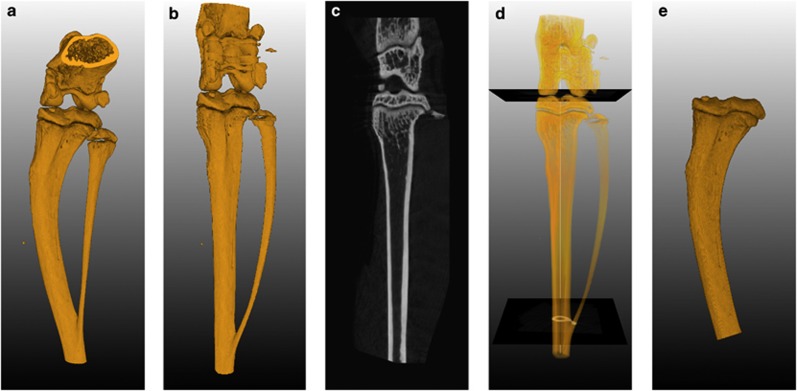
A multiplanar reformation-based approach has been published for normalized selection of VOIs in complex 3D shapes.16 The approach makes use of a centerline, which has been fitted, through an individual bone. This naturally curved centerline can then be 'straightened' to generate a new, normalized volume. The straightened bone is orientated along the z-axis of this new volume. The volume can then be used to extract normalized cross-sections or to define a normalized VOI (Figure 4). The selected VOI is mapped back to the original scan volume, where the actual volume measurements are performed. This way, artefacts that are introduced by the straightening procedure do not influence the volume measurements. The VOIs selected following this method are predefined and relative to the anatomy of the bone. This way, the VOI selection is independent of the observer, reducing the chance of observer bias. Moreover, it becomes possible to select exactly the same part of multiple bones, enabling data comparison between multiple scans.16 This approach has been automated and integrated it in the exploded view visualization system.17
Figure 4.
Selecting a VOI using orthogonal cross-sections. (a) 3D rendering of the original scan volume. (b) 3D rendering of the complete image stack of cross-sections generated perpendicular along the centerline. (c) Longitudinal cross-section generated from the reformatted space. The sectioning plane can be rotated around the longitudinal axis. These sections can be used for side-by-side comparison between different scans. (d) Definition of transversal cutoff planes using the reformatted space to calculate the relative position between the knee and the branch point of the fibula and tibia. (e) 3D rendering of a VOI. The volume was selected in the original space using a region grower limited by the cut off planes, which were mapped back from the reformatted space. Adapted with permission from Snoeks et al.16
In addition to volume measurements, μCT scans can be used to determine the cortical bone thickness. This type of measurements was added to the exploded view workflow. The cortical thickness is measured and projected as a color code on a volume rendering of the bone. Figure 5 shows an example of such thickness maps. In this example, osteolytic breast cancer cells were inoculated in the tibia of nude mice. There is an initial increase in cortical bone thickness around the lesion. After this initial increase, the bone disappears as the local tumor grows and stimulates osteolysis.17 These kind of visualizations are, like volume measurements, of use for the evaluation of both osteolytic and osteosclerotic lesions.
Figure 5.
Cortical thickness maps. MDA-231-B/luc+ cells (2.5×105 cells) were inoculated directly into the right tibia of a 6-week-old female nude mouse. Whole-body μCT scans were made before inoculation (T0), 3 weeks after inoculation (T1) and 6 weeks after inoculation (T2). The measured cortical thickness is mapped to surface representations of tibia, by means of a colormap. The initial osteolytic lesion is surrounded by an area of increased bone thickness at T1. The increased thickness is mainly a result of the inoculation procedure. Most of the cortical bone has disappeared at T2. This method of evaluating bone thickness can only be used for cortical bone and not at the distal part of the femur or the proximal part of the tibia because of distortion of the measurement due to the presence of trabecular bone. Adapted with permission from Baiker et al.17
Altogether, the automated workflow for whole-body μCT scans described above enables the user to select and zoom in on each individual skeletal elements, visually assess structural changes over time of the selected bone and quantify the volumes of pre-defined VOIs within that bone. μCT provides excellent structural and volumetric information and can be used to track changes over time, however, this modality is limited to tissues with a high X-ray contrast.
Functional imaging
Functional imaging modalities are used to gain an insight in cellular, metabolic and molecular processes. Functional imaging includes not only optical imaging modalities such as BLI and FLI but also magnetic resonance imaging (MRI) and radioactive imaging methods such as PET and SPECT.
Optical imaging.
Optical imaging modalities are based on capturing photons in the visible and near infrared part of the spectrum originating from cells and tissues. These photons can be produced by either fluorescence or bioluminescent enzymatic reactions. The choice whether or not to use fluorescence or bioluminescence greatly depends on the research questions and other considerations such as immunological background, skin color, imaging depth and so on.
BLI is most commonly used to follow in vivo tumor growth of luciferase-expressing tumor cells over time.18 In addition, there are a number of transgenic animal models expressing luciferase under tissue-specific promoters such as the prostate-specific PSA-Luc mouse19 and transgenic animals that luminesce upon upregulation or activation of certain signaling cascades such as the vascular endothelial growth factor-receptor 2 luciferase mouse (FVB/N-Tg-(vascular endothelial growth factorR2-Luc)-Xen).20
FLI can be used for tracking of cells expressing certain fluorescent proteins. Other approaches to FLI make use of targeted fluorescent dyes or probes, which fluorescence upon enzymatic activation such as the matrix metalloproteinase-specific MMPSense (PerkinElmer, Waltham, MA, USA)21 and various cysteine proteinase activity-based probes.22,23,24,25,26 The fluorophore of proteinase activity-based probes binds specifically and covalently to the active domain proteases. The fluorescence can be monitored in vivo, but can also be detected in histology can sections or on western blots because of the covalent bond. This enables the exact ex vivo validation and quantification of the signal after the in vivo study. Two other fluorescent probes of interest to the cancer and bone field are OsteoSense (PerkinElmer)27 and BoneTag (LI-COR, Lincoln, NE, USA),28,29 both commercially available dyes, which bind specifically to bone at areas of high turnover such as osteolytic and osteosclerotic lesion sites.
BLI and FLI are essentially 2D modalities. However, recent developments in imaging equipment enabled 3D data capturing and reconstruction. The technological advancements of optical imaging and the use of optical imaging in the field of bone research have been reviewed in detail by Snoeks et al.29
PET, SPECT and MRI.
PET and SPECT are two closely related radioisotope-imaging modalities. Most modern animal PET and SPECT scanners are combined with a CT scanner to provide more structural information. There is a multitude of radiotracers that can be used to image and quantify various targets in metastatic bone. Most common are the clinically used radioactive-labeled bisphosphonates to assess local hotspots in bone turnover,30,31 αvβ3 integrin-targeted tracers to target osteoclasts and activation of pro-inflammatory cells32 and radiolabeled deoxyglucose to image areas of high metabolic activity such as tumors and inflammation.30,33,34,35 Both PET and SPECT offer excellent quantification possibilities.
Of the functional imaging approaches, MRI is the modality that gives the best anatomical and soft-tissue contrast without the use of ionizing radiation. It was first used to image and quantify diffuse bone metastases with and without osteolysis.36 Later measurements of cortical bone thickness were included in MRI analysis to be able to quantify osteolysis in addition to the tumor size.37 Recently, Bauerle et al.38 described a more integrated approach. They combined MRI quantification of the tumor size with volumetric measurements performed with μCT. In addition, they performed vessel size imaging, blood volume measurements and measurements regarding the cellularity of the tumor lesions. This combination of μCT with various MRI-based measurements provides complementary information on tumor growth, angiogenesis and vascularization, bone destruction and the morphological state of the tumor, and it can be used to follow changes over time and treatment response.38
Integrated approaches
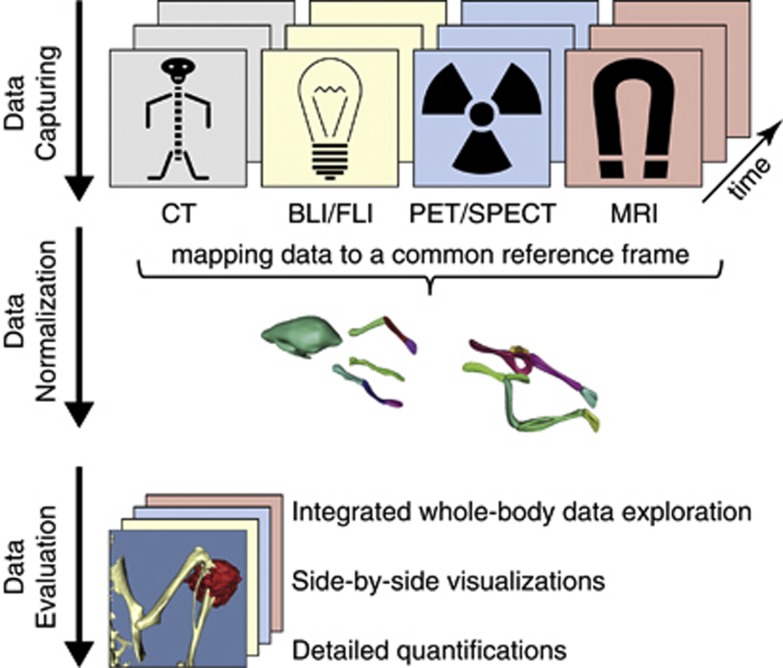
Imaging can be used to visualize and quantify both structural and functional characteristics of tumors and tissues in vivo. Every modality has its own strengths and weaknesses. It is key to combine the data acquired with each modality in order to come to a complete understanding of the disease process or treatment effects. In order to do that, an integrated approach is needed, which maps all the data generated with various modalities throughout an experiment and align this data in space and time (Figure 6).
Figure 6.
Overview of the integrated data workflow. The data generated at different points in time using different modalities are mapped to a common reference frame based on an articulated animal atlas. The atlas is then used to reposition the animal and divide the large data sets in smaller normalized subvolumes. This enables normalized whole-body data evaluation, side-by-side visualizations of a VOI with data generated using multiple modalities at one time, side-by-side visualizations of a VOI with data generated using one modality at different time points and various quantifications per modality.
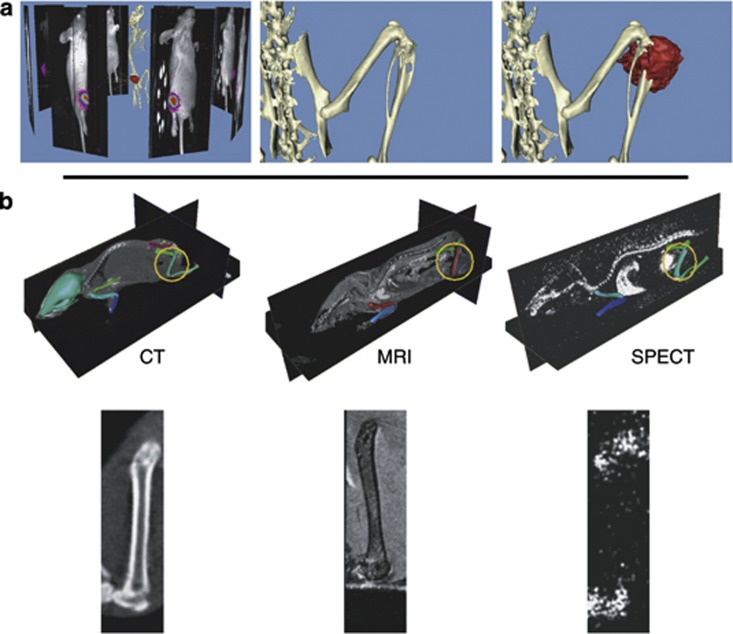
The newly developed 3D optical data capturing techniques and subsequent optical 3D data sets allow for projection of these data sets on more structural modalities such as μCT (Figures 7a–c).18,29 Optical modalities such as BLI completely lack structural information other than the skin. This is the reason why it is important to acquire the optical data together with the structural data such as μCT in the same posture to assure a good estimation of the signal source location. The optical data, coupled to the μCT data, can then be handled in the same workflow as the other modalities.
Figure 7.
Multimodality visualization of bone metastasis. (a) MDA-231-B/luc+ cells (2.5×105 cells) were inoculated directly into the right tibia of a 6-week-old female nude mouse. After 3 weeks tumor cell inoculation, bone metastases where analyzed with an IVIS 3D BLI Imaging system (Caliper Life Sciences, Alameda, CA, USA). The animal was subsequently scanned in a SkyScan 1076μCT scanner (SkyScan, Kontich, Belgium) in the same position as during the BLI measurement. Left: the bioluminescent data captured from eight positions around the animal were reconstructed and projected back onto a CT reconstruction of the animal. Middle: detail of the CT volume visualization of the right hind limb. Right: the tumor-induced osteolytic lesion is clearly visible. Detail of the BLI tumor volume estimation projected onto the CT reconstruction. The source of the bioluminescent signal co-localizes with the osteolytic lesion site. Adapted with permission from Snoeks et al.29 (b) Side-by-side visualizations of CT, MRI and SPECT. SPECT data were obtained using 99mTc-methylene diphosphonate. The articulated atlas has been fit to the scan data (middle row) and side-by-side visualizations of one specific segment have been generated using the atlas-based planar reformation approach. Adapted with permission from Khmelinskii et al.39
MRI and combined PET-CT and SPECT-CT data sets contain enough anatomical data to be able to fit the articulated atlas.11,39 These data sets can then be handled in a similar manner as the μCT data. This enables not only side-by-side visualizations of data generated with one modality at different points in time (as is the case in Figure 3) but also side-by-side visualizations of data generated at one time with different modalities (Figures 7d–f).39,40 The resulting data sets are semi interactive; the user can select a certain field of view in one data set and the corresponding field of view will be automatically generated in the data sets of the other modalities.
Conclusions
The integrated data handling in which all data sets from an experiment are mapped into one common reference frame, as summarized in Figure 6, enables the user to explore the data in an intuitive way. The user can assess disease-induced lesions by generating side-by-side views of changes over time in one skeletal element. A greater understanding of what is happening in a diseased area of the animal is achieved by combining functional and structural data acquired with various imaging modalities.
Recently, a start has been made to implement detailed quantifications, such as bone volume and thickness, into this workflow. This facilitates the objective monitoring of disease-induced lesions and the effectiveness of treatment interventions. The detailed quantitative assessment of optical data is still a challenge. The development of whole-body articulated atlases containing information on optical properties of various tissues might facilitate the development of better signal quantification methods. The overall logic and workflow of integrated data handling can serve as a vehicle to implement this kind of future developments.
Acknowledgments
This work has been supported by the Dutch Cancer Society Koningin Wilhelmina Fonds (grant UL2007-3801; TS).
Footnotes
The authors declare no conflict of interest.
References
- Abrams HL, Spiro R, Goldstein N. Metastases in carcinoma; analysis of 1000 autopsied cases. Cancer 1950;3:74–85. [DOI] [PubMed] [Google Scholar]
- Paget S. The distribution of secondary growths in cancer of the breast. Lancet 1889;1:571–573. [PubMed] [Google Scholar]
- Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002;2:584–593. [DOI] [PubMed] [Google Scholar]
- Guise TA, Mundy GR. Cancer and bone. Endocr Rev 1998;19:18–54. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer 2011;11:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise TA, Mohammad KS, Clines G, Stebbins EG, Wong DH, Higgins LS et al. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clin Cancer Res 2006;12:6213s–6216s. [DOI] [PubMed] [Google Scholar]
- Ambrose J, Hounsfield G. Computerized transverse axial tomography. Br J Radiol 1973;46:148–149. [PubMed] [Google Scholar]
- Hounsfield GN. Computerized transverse axial scanning (tomography). 1. Description of system. Br J Radiol 1973;46:1016–1022. [DOI] [PubMed] [Google Scholar]
- Baiker M, Milles J, Dijkstra J, Henning TD, Weber AW, Que I et al. Atlas-based whole-body segmentation of mice from low-contrast Micro-CT data. Med Image Anal 2010;14:723–737. [DOI] [PubMed] [Google Scholar]
- Kovacevic N, Hamarneh G, Henkelman M. Anatomically guided registration of whole body mouse MR images. Proc Med Image Comput Comput Assist Intervent 2003;2879 (Pt 2):870–877 (10.1007/978-3-540-39903-2_106). [Google Scholar]
- Khmelinskii A, Baiker M, Kaijzel EL, Chen J, Reiber JH, Lelieveldt BP. Articulated whole-body atlases for small animal image analysis: construction and applications. Mol Imaging Biol 2011;13:898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterwald A, van der Pluijm G, Que I, Sijmons B, Buijs J, Karperien M et al. Optical imaging of cancer metastasis to bone marrow: a mouse model of minimal residual disease. Am J Pathol 2002;160:1143–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok P, Baiker M, Hendriks EA, Post FH, Dijkstra J, Lowik CW et al. Articulated planar reformation for change visualization in small animal imaging. IEEE Trans Vis Comput Graph 2010;16:1396–1404. [DOI] [PubMed] [Google Scholar]
- Nakai M, Mundy GR, Williams PJ, Boyce B, Yoneda T. A synthetic antagonist to laminin inhibits the formation of osteolytic metastases by human melanoma cells in nude mice. Cancer Res 1992;52:5395–5399. [PubMed] [Google Scholar]
- Sasaki A, Boyce BF, Story B, Wright KR, Chapman M, Boyce R et al. Bisphosphonate risedronate reduces metastatic human breast cancer burden in bone in nude mice. Cancer Res 1995;55:3551–3557. [PubMed] [Google Scholar]
- Snoeks TJA, Kaijzel EL, Que I, Mol IM, Löwik CWGM, Dijkstra J. Normalized volume of interest selection and measurement of bone volume in MicroCT scans. Bone 2011;49:1264–1269. [DOI] [PubMed] [Google Scholar]
- Baiker M, Snoeks TJ, Kaijzel EL, Que I, Dijkstra J, Lelieveldt BP et al. Automated bone volume and thickness measurements in small animal whole-body MicroCT data. Mol Imaging Biol (e-pub ahead of print 13 October 2011; 10.1007/s11307-011-0522-2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaijzel EL, Snoeks TJ, Buijs JT, van der Pluijm G, Lowik CW. Multimodal imaging and treatment of bone metastasis. Clin Exp Metastasis 2009;26:371–379. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Xie Z, Liu ZY, Green JE, Martin WD, Datta MW et al. A luciferase transgenic mouse model: visualization of prostate development and its androgen responsiveness in live animals. J Mol Endocrinol 2005;35:293–304. [DOI] [PubMed] [Google Scholar]
- Zhang N, Fang Z, Contag PR, Purchio AF, West DB. Tracking angiogenesis induced by skin wounding and contact hypersensitivity using a Vegfr2-luciferase transgenic mouse. Blood 2004;103:617–626. [DOI] [PubMed] [Google Scholar]
- Bremer C, Tung CH, Weissleder R. In vivo molecular target assessment of matrix metallo-proteinase inhibition. Nat Med 2001;7:743–748. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C et al. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol 2005;1:33–38. [DOI] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ et al. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol 2005;1:203–209. [DOI] [PubMed] [Google Scholar]
- Blum G, Weimer RM, Edgington LE, Adams W, Bogyo M. Comparative assessment of substrates and activity based probes as tools for non-invasive optical imaging of cysteine protease activity. PLoS ONE 2009;4:e6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N et al. Non- invasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med 2009;15:967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol 2007;3:668–677. [DOI] [PubMed] [Google Scholar]
- Kozloff KM, Weissleder R, Mahmood U. Noninvasive optical detection of bone mineral. J Bone Miner Res 2007;22:1208–1216. [DOI] [PubMed] [Google Scholar]
- Kovar J, Xu X, Simpson M, Olive D. Effective bone labelling for in vivo nir noninvasive imaging in nude mice. Joint Molecular Imaging Conference, Providence, RI 2007. [Google Scholar]
- Snoeks TJ, Khmelinskii A, Lelieveldt BP, Kaijzel EL, Lowik CW. Optical advances in skeletal imaging applied to bone metastases. Bone 2011;48:106–114. [DOI] [PubMed] [Google Scholar]
- Chua S, Gnanasegaran G, Cook GJ. Miscellaneous cancers (lung, thyroid, renal cancer, myeloma, and neuroendocrine tumors): role of SPECT and PET in imaging bone metastases. Semin Nucl Med 2009;39:416–430. [DOI] [PubMed] [Google Scholar]
- Ben-Haim S, Israel O. Breast cancer: role of SPECT and PET in imaging bone metastases. Semin Nucl Med 2009;39:408–415. [DOI] [PubMed] [Google Scholar]
- Wadas TJ, Deng H, Sprague JE, Zheleznyak A, Weilbaecher KN, Anderson CJ. Targeting the alphavbeta3 integrin for small-animal PET/CT of osteolytic bone metastases. J Nucl Med 2009;50:1873–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Kuge Y, Yi M, Zhao Y, Hatano T, Magota K et al. Dynamic (11)C-methionine PET analysis has an additional value for differentiating malignant tumors from granulomas: an experimental study using small animal PET. Eur J Nucl Med Mol Imaging 2011;38:1876–1886. [DOI] [PubMed] [Google Scholar]
- Wang TC, Hsiao IT, Cheng YK, Wey SP, Yen TC, Lin KJ. Noninvasive monitoring of tumor growth in a rat glioma model: comparison between neurological assessment and animal imaging. J Neurooncol 2011;104:669–678. [DOI] [PubMed] [Google Scholar]
- Rogers IS, Tawakol A. Imaging of coronary inflammation with FDG-PET: feasibility and clinical hurdles. Curr Cardiol Rep 2011;13:138–144. [DOI] [PubMed] [Google Scholar]
- Gauvain KM, Garbow JR, Song SK, Hirbe AC, Weilbaecher K. MRI detection of early bone metastases in b16 mouse melanoma models. Clin Exp Metastasis 2005;22:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MH, Sharp JC, Latta P, Hassard TH, Orr FW. Early detection and quantification of murine melanoma bone metastases with magnetic resonance imaging. Skeletal Radiol 2007;36:659–666. [DOI] [PubMed] [Google Scholar]
- Bauerle T, Merz M, Komljenovic D, Zwick S, Semmler W. Drug-induced vessel remodeling in bone metastases as assessed by dynamic contrast enhanced magnetic resonance imaging and vessel size imaging: a longitudinal in vivo study. Clin Cancer Res 2010;16:3215–3225. [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Baiker M, Kok P, de Swart J, Reiber JHC, de Jong M et al. Atlas-based articulated skeleton segmentation of spect mouse data. In: 2011 IEEE International Symponsium on Biomedical Imaging: From Nano to Macro; pp 437–440 (10.1109/ISBI.2011.5872440).
- Lelieveldt BP, Botha CP, Kaijzel EL, Hendriks EA, Reiber JH, Lowik CW et al. Towards integrated analysis of longitudinal whole-body small animal imaging studies. In: 2011 IEEE International Conference on Aucostics, Speech and Signal Processing: pp 5768–5771 (10.1109/ICASSP.2011.5947671).