Abstract
Short palate, lung, and nasal epithelial clone–1 (SPLUNC1) is a protein abundantly expressed by the respiratory epithelium of the proximal lower respiratory tract, a site of great environmental exposure. Previous studies showed that SPLUNC1 exerts antimicrobial effects, regulates airway surface liquid and mucociliary clearance, and suppresses allergic airway inflammation. We studied SPLUNC1 to gain insights into its role in host defense. In the lower respiratory tract, concentrations of SPLUNC1 are high under basal conditions. In models of pneumonia caused by common respiratory pathogens, and in Th1-induced and Th2-induced airway inflammation, SPLUNC1 secretion is markedly reduced. Pathogen-associated molecular patterns and IFN-γ act directly on airway epithelial cells to inhibit SPLUNC1 mRNA expression. Thus, SPLUNC1 is quickly suppressed during infection, in response to an insult on the epithelial surface. These experiments highlight the finely tuned fluctuations of SPLUNC1 in response to exposures in the respiratory tract, and suggest that the loss of SPLUNC1 is a crucial feature of host defense across air-breathing animal species.
Keywords: SPLUNC1, inflammation, innate, immunity, mucosa
Clinical Relevance
Short palate, lung, and nasal epithelial clone–1 (SPLUNC1) is highly expressed in the respiratory tract under basal conditions, and is inhibited by common respiratory pathogens and airway inflammation. The rapid flux of SPLUNC1 suggests that it serves as a sensor of environmental exposure to pathogens and irritants. Understanding the regulation of SPLUNC1 will provide insights into its role during health and disease.
The respiratory tract continuously interacts with environmental irritants and pathogens. These stimuli constitute part of an intricate network of signals that activate host defenses. Airway epithelial cells participate actively in this process by producing antimicrobial and immune mediators that maintain homeostasis during steady-state conditions, and promote inflammation during injury.
The palate, lung, and nasal epithelial clone (PLUNC) genes are members of the bactericidal permeability–increasing protein fold-containing (BPIF)/PLUNC protein family (1, 2). These genes are focally expressed by epithelial cells throughout the respiratory tracts of multiple air-breathing vertebrates (3). In total, 11 human genes and 14 murine genes have been described to date, located within a single locus on chromosome 20 and chromosome 2, respectively (4, 5). These genes code for the PLUNC proteins, which are classified into six short and eight long PLUNCs, based on their lengths.
Short PLUNC 1 (SPLUNC1) is a 25-kD protein secreted by epithelial cells in the upper airway and proximal lower respiratory tract (3, 6–10). SPLUNC1 is readily detected in human saliva and in nasal and bronchial epithelial washings at concentrations ranging from 34.7 ng/ml to 13.8 mg/ml (6, 10–12). In tracheal epithelial-cell supernatants SPLUNC1 constituted 10% of the total soluble protein, making it one of the most highly expressed proteins produced by airway epithelial cells (7).
SPLUNC1 shares structural similarities with the immunomodulatory proteins bactericidal permeability–increasing protein (BPI) and lipopolysaccharide-binding protein (4, 13–15). It was initially hypothesized to express antimicrobial functions, and has been shown to inhibit the growth of multiple pathogens (16–19), inhibit biofilm formation, and possess surfactant properties (20, 21). Although most studies of SPLUNC1 have focused on these antimicrobial effects, SPLUNC1 also regulates the epithelial sodium channel (ENaC) to maintain airway surface liquid depth, affects mucociliary clearance, and regulates inflammation, thereby limiting allergic airway inflammation (10, 22–24).
Here we show that SPLUNC1 is present at high concentrations in murine airways under basal conditions, whereas it is suppressed by low-level environmental pathogen–associated molecular patterns (PAMPs) and inflammatory signals. During acute infection and inflammation, SPLUNC1 is markedly inhibited. These are the first experiments to capitalize on the mechanisms of SPLUNC1 fluctuation. SPLUNC1 is finely tuned by PAMPs and IFN-γ, suggesting that it serves as a sensor of environmental exposures to trigger its multifaceted effects that aid in host defense.
Materials and Methods
Mice
We used 6- to 14-week-old C57BL/6 (National Cancer Institute, Frederick, MD), tlr4−/−, myd88−/− (25–27), ifnγr1−/− (28) (Jackson Laboratories, Bar Harbor, ME), and CC10-rtTA-IL-13 (29) mice in protocols approved by the Animal Care and Use Committee of Yale University.
Bronchoalveolar Lavage
At the time the mice were killed, bronchoalveolar lavage (BAL) was performed with 1 ml of PBS. BAL fluid (BALF) was collected for ELISA, and cells were counted as previously described (30).
Models of Airway Inflammation
LPS (5 μg, Escherichia coli; Sigma-Aldrich, Saint Louis, MO) was administered intranasally, and mice were killed at the time intervals to be indicated. Airway inflammation was induced by the transfer of OT-II (ovalbumin-specific T cell receptor transgenic) CD4 Th1 or Th2 cells into C57BL/6 recipient mice, followed by challenge with inhaled 1% ovalbumin (OVA) for 10 days (30, 31). CC10-rtTA-IL-13 mice, doxycycline-inducible IL-13 transgenic mice, and nontransgenic littermate control mice were treated with doxycycline (Sigma-Aldrich) for 14 days to stimulate IL-13 production in the airway.
Models of Respiratory Infection
Pseudomonas aeruginosa (5 × 104 colony-forming units) and Streptococcus pneumoniae (1 × 105 colony-forming units) were administered intratracheally to C57BL/6 mice that were killed at 4 hours or 3 days, respectively. Influenza A virus (1/20 lethal dose 50%) was administered intranasally, and mice were killed on Day 7. Control mice received PBS intratracheally or intranasally, or underwent no treatment.
Cell Culture
Murine tracheal epithelial cells (mTECs) were isolated from C57BL/6 mice and cultured on transwell plates at an air–liquid interface (ALI), following previously described methods (32). mTECs were treated with LPS (10–100 ng/ml; Sigma-Aldrich), recombinant murine IFN-γ (rmIFN-γ, 1, 10, or 100 ng/ml; Peprotech, Rocky Hill, NJ), or recombinant murine IL-13 (rmIL-13, 1 or 10 ng/ml; Peprotech) for 24 hours, 48 hours, or 10 days. Two hundred microliters of PBS were placed on the apical membrane surface and collected after 30 minutes to measure protein secretion. NCI-H292 cells were cultured to confluence and treated with recombinant human IFN-γ (1 or 10 ng/ml; eBioscience, San Diego, CA) or human IL-13 (1 or 10 ng/ml; Peprotech). Western blots of H292 cell lysates indicated SPLUNC1 protein production. RNA was extracted from individual wells for quantitative PCR.
ELISA and Lung Immunohistochemistry and Western Blotting
Plates were coated with BALF, followed by polyclonal sheep anti-mouse SPLUNC1 antibody (R&D, Minneapolis, MN), anti-sheep HRP (Millipore, Billerica, MA), 3,3′,5,5′-tetramethylbenzidine substrate (KPL, Gaithersburg, MD), and were measured at optical densities of 450 and 550 nm. Formalin-fixed, paraffin-embedded lung sections were stained with anti-mouse SPLUNC1 (R&D). Trachea, bronchi. and lung parenchyma were dissected from naive C57BL/6 mice, and cell and tissue homogenates were run on nitrocellulose membranes that were probed with anti-mouse SPLUNC1 (R&D).
Quantitative PCR
RNA was prepared from whole lung tissue or tracheal epithelial cells, using the TRIzol reagent (Invitrogen, Carlsbad, CA). Quantitative PCR reactions were performed (SYBR FAST; KAPA Biosystems, Woburn, MA). Murine PCR primers used for amplification included SPLUNC1 (5′-GTCCACCCTTGCCACTGAACCA-3′and 3′-CACCGCTGAGAGCATCTGTGAA-5′) and β-actin (5′-GTCCACACCCGCCACCAGTTCG and 3′-GACCCATTCCCACCATCACACCCT-5′). Human PCR primers used for amplification included SPLUNC1 (5′-TGCTGGAACTTGGCCTTGTGCA-3′ and 3′-ACCAGGGGCGTATTCACTTGGA-5′) and glyceraldehyde 3–phosphate dehydrogenase (GAPDH; 5′-TGGAGAAGGCTGGGGGCTCATTT-3′ and 3′-TGGTGCAGGAGGCATTGCTGAT-5′). For each set of primers, validation experiments showed a linear dependency of threshold cycle values at different RNA concentrations. The data were analyzed after murine genes were normalized to β-actin and human genes were normalized to GAPDH, and P values were obtained using Data Assist software, version 2.0 (Applied Biosystems, Carlsbad, CA).
Statistical Analysis
Results are reported as mean (± SEM) values, unless otherwise specified. Statistical significance was determined by the Student t test.
Results
SPLUNC1 Is Expressed at High Concentrations in the Proximal Airways of Mice
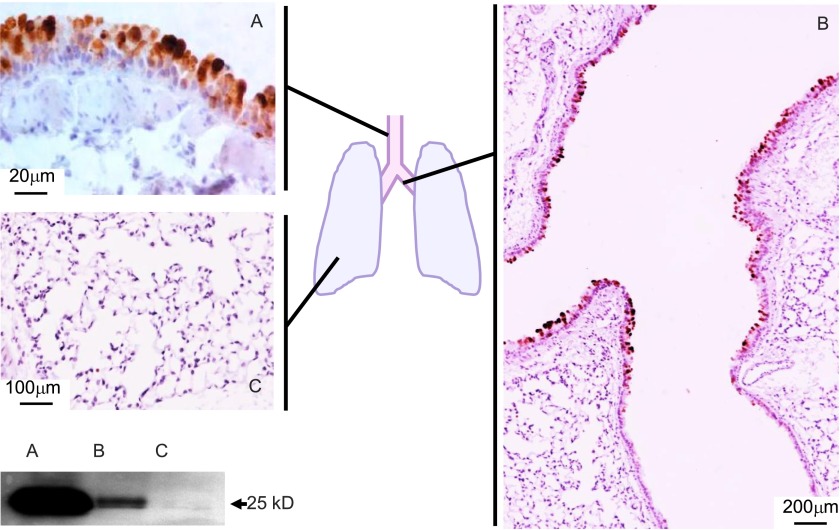
SPLUNC1 is produced by secretory epithelial cells in the trachea and in the proximal large bronchi in the lower respiratory tract (Figure 1A) (3). This is reflected in the high expression and secretion of SPLUNC1 in the large airways, and its minimal expression in the distal airways and lung parenchyma (Figure 1B). The amount of SPLUNC1 protein, measured in naive mice at 2–5 μg/ml in a 1-ml lung lavage sample, is high in the trachea, low in the distal bronchi, and almost absent in the peripheral lung tissue (Figure 1C). SPLUNC1 is age-dependent, gender-dependent, and strain-dependent. SPLUNC1 protein was 2.4-fold higher in BAL fluid from 6-month-old compared with 6-week-old male C57BL/6 mice (4,769 versus 1,997 ng/ml, respectively; P = 0.001), 0.39-fold lower in 6-week-old female compared with male C57BL/6 mice (1,255 versus 1,997 ng/ml, respectively; P = 0.0008), and 2.1-fold higher in 6-week-old BALB/c compared with C57BL/6 mice (4,306 versus 1,997 ng/ml, respectively; P = 0.00013). Based on the high basal level of secretion of SPLUNC1 in the proximal lower airways, the reported multiple functions of SPLUNC1 related to host protection, and the variability of SPLUNC1 under different conditions, we hypothesized that SPLUNC1 is important in the maintenance of airway homeostasis. Thus, we investigated the effects of infection and inflammation on SPLUNC1.
Figure 1.
Short palate, lung, and nasal epithelial clone–1 (SPLUNC1) is produced by epithelial cells in the main airways. SPLUNC1 expression was assessed by immunostaining and Western blotting on lung sections and tissue homogenates from naive C57BL/6 mice. The trachea and proximal bronchi show that SPLUNC1 is abundantly expressed in the airway epithelium (A and B), whereas the expression of SPLUNC1 is decreased in the distal portion of the mainstem bronchi (B), and becomes undetectable in peripheral lung tissue (C).
Infection with Respiratory Pathogens and LPS Inhibits SPLUNC1
SPLUNC1 was significantly reduced in the BALF of mice with pneumonia induced by infection with Pseudomonas aeruginosa, Streptococcus pneumoniae, and influenza A (Figure 2). To study the effects of infection on SPLUNC1 over time, we administered the Gram-negative endotoxin LPS intranasally to C57BL/6 mice, and measured SPLUNC1. SPLUNC1 increased 3 hours after LPS administration, then declined, with a nadir at 24 hours, and it remained well below the basal concentration through 48 hours (Figure 3A). SPLUNC1 RNA declined steadily after LPS administration (Figure 3B). This suggests that stored SPLUNC1 is released early after infection, possibly serving to limit the pathogen burden and increase mucociliary clearance. The subsequent decline in SPLUNC1 after pathogen exposure and its persistent low concentrations in infection are not consistent with an antimicrobial function. Moreover, the loss of SPLUNC1 may serve another function in host protection.
Figure 2.
SPLUNC1 is reduced in pneumonia. Pneumonia was induced in C57BL/6 mice with Pseudomonas aeruginosa (4 hours, n = 10 mice/group), Streptococcus pneumoniae (3 days, n = 4–5 mice/group), and influenza A virus (7 days, n = 4 mice/group). Control mice received PBS (S. pneumoniae and influenza A) or no treatment (P. aeruginosa). SPLUNC1 was measured in bronchoalveolar lavage fluid (BALF) by ELISA. Mean values are representative of 2–3 experiments. *P < 0.05.
Figure 3.
SPLUNC1 is inhibited by LPS. LPS (5 μg) was administered intranasally to C57BL/6 mice, and the mice were killed at different time points. (A) SPLUNC1 concentrations, as measured in BALF by ELISA, are compared with total cell counts in BALF at intervals of 3, 8, 24, and 48 hours. (B) SPLUNC1 expression was assessed in RNA from lung tissue by quantitative PCR at intervals of 3, 8, 24, and 48 hours. Mean values are shown (n = 4 mice/group), and are representative of three experiments. RQ, relative quantitation.
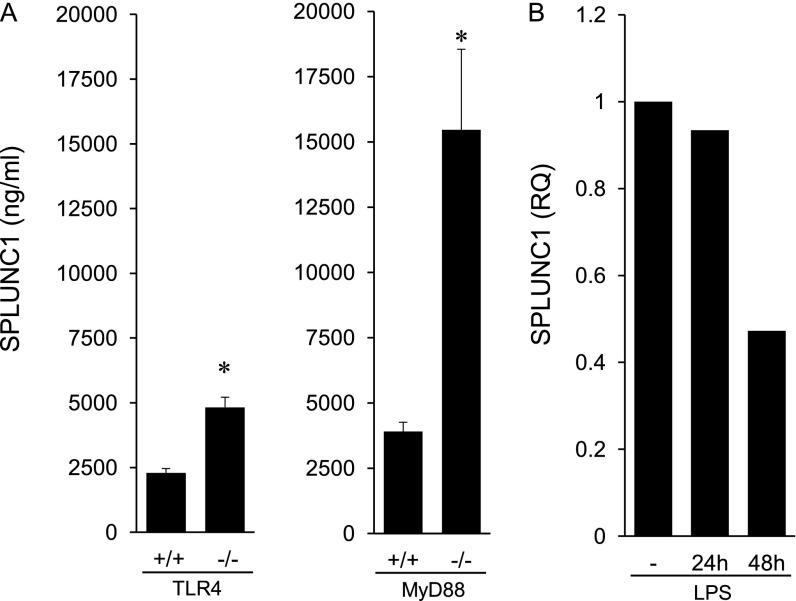
As SPLUNC1 declined in response to LPS, an associated increase in neutrophilic inflammation was evident, suggesting an interrelationship between SPLUNC1 and inflammation. To study the effects of LPS in the regulation of SPLUNC1 independent of inflammation, we investigated SPLUNC1 in mice lacking the LPS signaling receptor Toll-like receptor–4 (TLR4). We hypothesized that in naive mice, exposure to low concentrations of PAMPs from the air and from bacteria colonizing the respiratory tract (33) would be observed, although no airway inflammation would be evident. In the BALF of naive tlr4−/− mice, SPLUNC1 was twofold higher compared with tlr4+/+ mice (Figure 4A), indicating that the basal activation of TLR4 inhibits SPLUNC1. To determine whether the TLR4-mediated effects on SPLUNC1 were transduced through the common TLR adaptor molecule myeloid differentiation primary response gene 88 (MyD88), we measured SPLUNC1 in naive mice deficient in MyD88. SPLUNC1 was also strikingly elevated in the BALF from myd88−/− compared with myd88+/+ mice, indicating that SPLUNC1 is regulated by TLR4 ligands downstream from MyD88 signals (Figure 4A). The amount of SPLUNC1 was higher in myd88−/− compared with tlr4−/− mice, suggesting that the activation of other MyD88-dependent innate immune receptors, in addition to TLR4, suppresses SPLUNC1 under basal conditions. Therefore, these data suggest that PAMPs encountered in the lower respiratory tract under steady-state conditions tonically suppress SPLUNC1 and maintain its normal concentrations.
Figure 4.
Regulation of SPLUNC1 by LPS. (A) SPLUNC1 was measured by ELISA in BALF from naive tlr4+/+, tlr4−/−, myd88+/+, and myd88−/− mice (n = 6 mice/group). (B) Tracheal epithelial cells from C57BL/6 mice were grown in culture and treated with LPS (100 μg) for 24 and 48 hours. Control cells (−) were untreated and cultured for 48 hours. SPLUNC1 expression was measured by quantitative PCR. Mean values are shown, and are representative of three experiments. TLR4, Toll-like receptor–4. *P < 0.05.
LPS Inhibits SPLUNC1 through Effects on the Airway Epithelium
To determine whether LPS inhibits SPLUNC1 through TLR4 in airway epithelial cells, we treated cultured primary mTECs with LPS. LPS inhibited SPLUNC1 expression by 48 hours (Figure 4B), which shows that LPS can inhibit SPLUNC1 via direct effects on the tracheal epithelium. However, SPLUNC1 expression remained high in these cultured epithelial cells 24 hours after treatment with LPS (Figure 4B), in contrast with observations of mice in which SPLUNC1 RNA was inhibited by 24 hours. This suggests that other signals, in addition to the action of LPS on airway epithelial cells, may modulate SPLUNC1 in vivo.
Inflammation Inhibits SPLUNC1
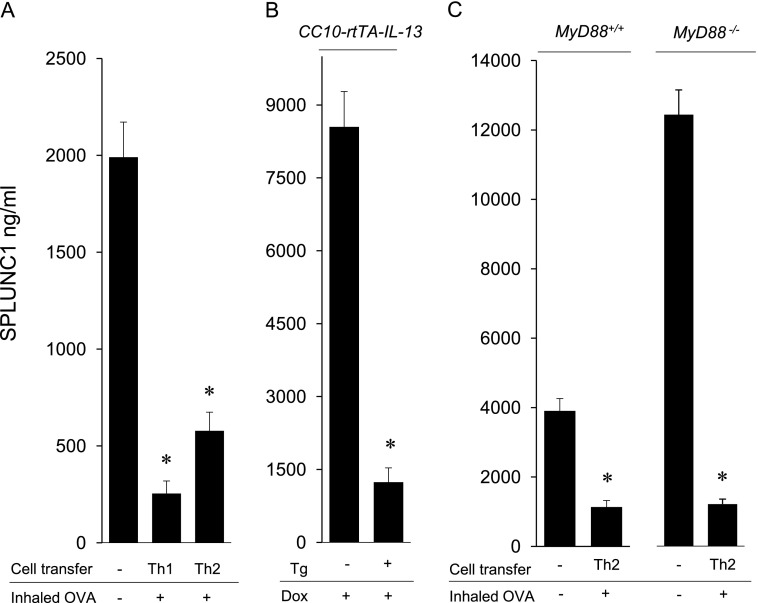
As depicted in Figure 3A, SPLUNC1 concentrations declined in response to a pathogenic stimulus as inflammation increased. To test whether inflammation inhibits SPLUNC1, we measured SPLUNC1 in the BALF of mice with lymphocyte-driven airway inflammation. SPLUNC1 was suppressed in mice with Th1-induced, neutrophil-predominant, and Th2-induced eosinophilic airway inflammation, indicating that the activation of the adaptive immune response also inhibits SPLUNC1 (Figure 5A). Because the induction of inflammation in these models requires an inhaled challenge with OVA that contains LPS (34), we investigated SPLUNC1 in a model of allergic inflammation that does not require inhaled stimuli for the induction of inflammation. Inducible, airway-specific IL-13 transgenic mice (CC10-rtTA-IL-13) were treated with doxycycline for 14 days to stimulate airway inflammation (35). Doxycycline-treated CC10-rtTA-IL-13 mice exhibited a marked reduction in SPLUNC1, compared with doxycycline-treated, nontransgenic littermate control mice (Figure 5B). Note that these models of allergic airway inflammation, although driven by Th2 cytokines, also involve the activation of many inflammatory pathways (31, 34, 36, 37). SPLUNC1 was also reduced in myd88−/− mice after the transfer of Th2 cells, indicating that MyD88 signals are not essential for the inhibition of SPLUNC1 during inflammation (Figure 5C). These studies show that inflammation inhibits SPLUNC1, independent of innate immune signals.
Figure 5.
Th1 and Th2 inflammation inhibit SPLUNC1. (A) SPLUNC1 was measured by ELISA in BALF from C57BL/6 (wild-type) mice with inflammation induced by Th1 or Th2 cells and inhaled ovalbumin (OVA). Mean values are shown (n = 5 mice/group), and are representative of three experiments. (B) SPLUNC1 was measured in BALF from CC10-rtTA-IL-13 doxycycline-inducible transgenic mice (Tg+) and littermate control mice (Tg−) treated with doxycycline (Dox) in drinking water for 14 days to induce allergic inflammation. Mice were 18 weeks of age, and higher SPLUNC1 concentrations were observed in Tg− mice, as expected. (C) SPLUNC1 was measured by ELISA in BALF from myd88+/+ and myd88−/− mice after the transfer of Th2 cells and inhaled OVA. Control mice did not receive a cell transfer or inhaled OVA. Mean values are shown (n = 5 mice/group), and are representative of three experiments. *P < 0.05.
Inflammatory Cytokines Exert Different Effects on SPLUNC1
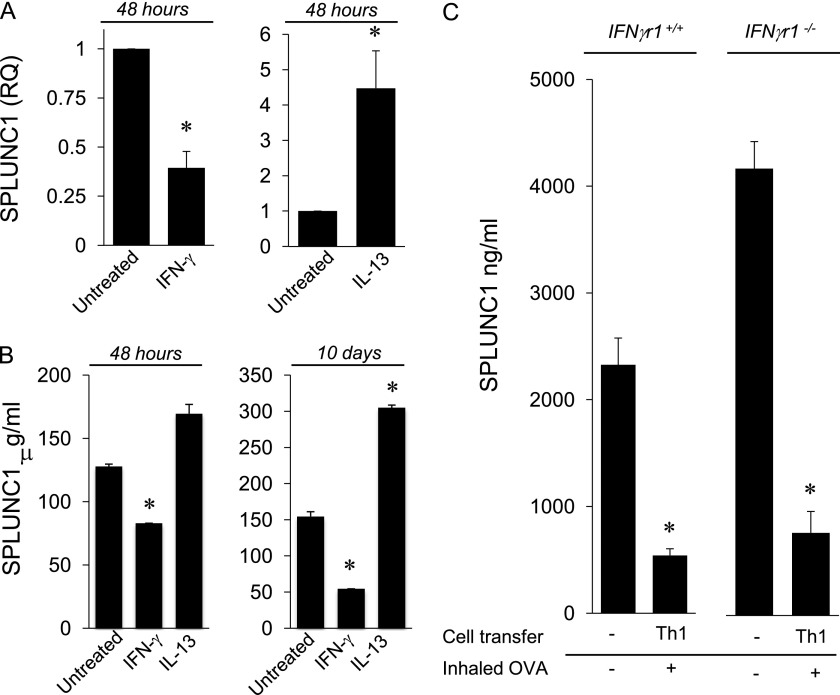
To begin defining the specific inflammatory mediators that regulate SPLUNC1 and to avoid the complex mix of inflammatory cytokines present in vivo in models of Th1 and allergic airway inflammation, we cultured mTECs in vitro with the prototypical Th1 and Th2 cytokines, IFN-γ and IL-13, for 24 and 48 hours. IFN-γ potently inhibited SPLUNC1 expression by 24 hours (not shown), and SPLUNC1 remained suppressed at 48 hours (Figure 6A). IFN-γ inhibited SPLUNC1 RNA over a range of doses (1–100 ng/ml). IFN-γ also reduced SPLUNC1 protein secretion (Figure 6B). In contrast, IL-13 increased SPLUNC1 gene expression at both 24 hours (not shown) and 48 hours (Figure 6A), and this effect was independent of IL-13 dose, because the effect was observed at both 1 ng/ml (not shown) and 10 ng/ml of IL-13. SPLUNC1 secretion was also increased by IL-13 after 48 hours, although this result was not statistically significant. These experiments contrast with previous work showing that IL-13 inhibited SPLUNC1 expression and secretion in airway epithelial cells after longer periods of culture (16, 37). We next treated mTECs with IL-13 for 10 days. Again, IL-13 stimulated SPLUNC1 secretion (Figure 6B) and expression (not shown), whereas 10-day treatment with IFN-γ inhibited SPLUNC1. Thus, longer periods of cytokine treatment did not change the effects on SPLUNC1 production. These findings also indicate that the reduction of SPLUNC1 observed in allergic inflammation induced by Th2 cells or in IL-13 transgenic mice (Figure 5) is not attributable to the effects of IL-13, but rather to the dominant effects of other inflammatory mediators induced by Th2 cytokines that are present in the complex inflammatory response in these animals. Clearly, SPLUNC1 is modulated differently by cytokines in the murine airway epithelium.
Figure 6.
IFN-γ and IL-13 exert different effects on SPLUNC1 in mice. Tracheal epithelial cells were cultured with IFN-γ (10 ng/ml) or IL-13 (10 ng/ml) for 48 hours. (A) SPLUNC1 mRNA expression was measured by quantitative PCR. (B) SPLUNC1 protein secretion was measured by ELISA at 48 hours and at 10 days. (C) Airway inflammation was induced in ifnγr1+/+ or ifnγr1−/− mice after the transfer of Th1 cells and inhaled OVA. Naive mice served as controls. SPLUNC1 was measured in BALF by ELISA. Mean values are shown (n = 5 mice/group). In each experiment, the data are representative of three experiments. IFNγr1, interferon-γ receptor–1. *P < 0.05.
To determine whether IFN-γ is an exclusive regulator of SPLUNC1 in inflammation, we measured SPLUNC1 in IFN-γ receptor (R)–deficient mice with airway inflammation. Th1 airway inflammation inhibited SPLUNC1 in both ifngr1+/+ and ifngr1−/− mice compared with naive mice (Figure 6C). This shows that other factors, in addition to IFN-γ, inhibit SPLUNC1 in inflammation. Yet, in naive ifngr1−/− mice, SPLUNC1 was increased in comparison with ifngr1+/+ mice, indicating that very low concentrations of IFN-γ, not detectable in BALF by ELISA, inhibit SPLUNC1 under steady-state conditions. These experiments show that IFN-γ is a potent inhibitor of SPLUNC1, although other inflammatory mediators also inhibit SPLUNC1.
SPLUNC1 Regulation in Human Airway Epithelial Cells Mirrors Murine Responses
Given the homology of SPLUNC1 in mice and humans and their similar pattern of expression in the lower respiratory tract, we investigated SPLUNC1 regulation in human bronchial epithelial cells to determine whether regulation was also comparable among species. After treatment with IFN-γ, the human mucoepidermoid carcinoma epithelial cell line NCI-H292 exhibited a marked reduction in SPLUNC1, whereas IL-13 stimulated SPLUNC1 expression after 48 hours (Figure 7). These data show that in mice and humans, a parallel regulation of SPLUNC1 by IFN-γ and IL-13 occurs.
Figure 7.
IFN-γ and IL-13 exert different effects on SPLUNC1 in human airway epithelial cells. NCI-H292 cells were cultured with IFN-γ (10 ng/ml) or IL-13 (10 ng/ml) for 48 hours. SPLUNC1 mRNA expression was measured by quantitative PCR. Data are representative of three experiments. *P < 0.05.
Discussion
SPLUNC1 is heavily secreted into the trachea in healthy humans and other air-breathing animals. Its antimicrobial effects in conjunction with its high expression in the proximal lower airways, where pathogen exposure in the lung is the greatest, support the role of SPLUNC1 as a host-defense protein (33). Here we show that SPLUNC1 is exquisitely controlled by pathogens and inflammatory mediators. Under basal conditions and very early during infection, the amount of SPLUNC1 is high. The rapid down-modulation and loss of SPLUNC1, as these data show, imply that the fluctuation of SPLUNC1 also plays a role in host defense. In addition to its antimicrobial functions, SPLUNC1 suppresses allergic airway inflammation, has surfactant properties, regulates the epithelial sodium channel ENaC that modulates the airway surface liquid, and enhances mucociliary clearance (10, 23, 24, 38). These studies strongly suggested that the loss of SPLUNC1 during inflammation and infection is important in host defense. SPLUNC1 is a multifunctional, airway-specific protein whose production may be coordinated to maintain airway homeostasis in both health and disease.
SPLUNC1 has well-defined antimicrobial properties (16–19). It blocks microbial invasion in the respiratory tract by inhibiting pathogen growth, and through its surfactant properties inhibits biofilm development (16, 19, 20, 24). Already high in the airways of healthy animals, the amount of SPLUNC1 is further increased early after exposure to Gram-negative endotoxin. A boost in SPLUNC1 in response to a pathogen may further inhibit microbial invasion, as suggested by studies of SPLUNC1 transgenic mice in which the overproduction of SPLUNC1 inhibited Pseudomonas infection (18). In addition to its effects on typical Gram-negative pathogens, SPLUNC1 has been shown to inhibit pulmonary infection with Mycoplasma pneumoniae (16). SPLUNC1 blocks the growth of Pseudomonas, Mycoplasma, Haemophilus, and Epstein-Barr virus in vitro (16, 19, 24). Therefore, under steady-state conditions, high concentrations of SPLUNC1 will inhibit many pulmonary pathogens.
High concentrations of SPLUNC1 in the healthy lung may also maintain extracellular sodium and thus sustain liquid on the airway surface, and in conjunction with its surfactant effects, will promote the removal of pathogens through the mucociliary escalator. Under steady-state conditions, SPLUNC1 may also suppress airway inflammation (23). Maintaining an airway free of infection and inflammation through effective airway mucus clearance will ensure the primary function of the lungs, namely, effective gas exchange.
SPLUNC1 drops precipitously after LPS exposure, and was reduced 70% after 24 hours. In models of acute pulmonary infection with both Gram-negative and Gram-positive bacteria or virus, SPLUNC1 was also markedly reduced. Low concentrations of SPLUNC1 in infection contrast with observations of elevated SPLUNC1 in a model of Mycoplasma pneumonia (16). The increase of SPLUNC1 in Mycoplasma infection is an effect downstream from TLR2 (39). Therefore, SPLUNC1 may be differentially regulated in response to the activation of different innate immune pathways. Importantly, the suppression of SPLUNC1 in response to infection appears to be the prototypical response to common pulmonary infections.
Both PAMPs and cytokines inhibit SPLUNC1. Our experiments on naive tlr4−/− and ifngr1−/− mice suggest that amounts of SPLUNC1 would be even higher if environmental irritants and pathogens were absent. Thus, SPLUNC1 is suppressed in the basal state, when very low concentrations of environmental pathogens and inflammatory mediators are present in the lung. SPLUNC1 is more dramatically inhibited by infection and inflammation. These findings suggest that SPLUNC1 serves as a sensor of environmental risk. The greater the exposure, and the higher the risk to the host, the more SPLUNC1 is inhibited. Again, this fine-tuning suggests that the fluctuation of SPLUNC1 aids in host defense.
We show that IFN-γ rapidly and potently inhibits SPLUNC1 by its direct effects on epithelial cells. In contrast, the inhibition of SPLUNC1 by PAMPs was slower and resulted in less suppression of SPLUNC1. This raises the possibility that the pattern recognition receptor–induced inhibition of SPLUNC1 is mediated by cytokines. TNF-α and IL-1β, common cytokines induced downstream from TLR4, did not inhibit SPLUNC1 in vitro when tested on cultured human tracheal epithelial cells (9), indicating that SPLUNC1 inhibition is not a universal response to inflammatory cytokines.
Whereas others showed that IL-13–treated epithelial cells in culture demonstrate reduced SPLUNC1 expression (16, 40), our experiments showed the opposite. These divergent findings may reflect different culture or treatment conditions, because primary epithelial cell cultures require many cell-derived additives, and any material containing LPS may affect SPLUNC1. In contrast to the in vitro effects of a single Th2 cytokine (IL-13), Th2 inflammation in mice inhibits SPLUNC1. Thus, in the complex in vivo allergic environment with many activated inflammatory cells and cytokines, factors other than IL-13 appear to inhibit SPLUNC1. Further defining the inflammatory conditions that modulate SPLUNC1 will strengthen our understanding of the role SPLUNC1 plays in host defense.
Our observations of SPLUNC1 regulation in human airway epithelial cells paralleled our observations in mice. These data conform to observations of humans in whom SPLUNC1 was modulated in response to irritants. Epithelial cells from the lower respiratory tracts of five nonsmokers expressed high concentrations of SPLUNC1 protein, whereas five subjects who smoked expressed markedly lower concentrations of SPLUNC1 (41). Nasal epithelial SPLUNC1 was also reduced in cigarette smoke–exposed and irritant-exposed subjects, compared with control subjects (42). In patients with allergic rhinitis, SPLUNC1 concentrations in nasal lavage fluid were lower during the allergy season, compared with concentrations before allergy season (43). These studies show that irritants and airway inflammation in humans are associated with reduced SPLUNC1. In contrast, SPLUNC1 was increased in the airways of a limited number of subjects with chronic airway diseases such as chronic obstructive pulmonary disease (COPD) and cystic fibrosis (6, 9, 44). Given that these subjects often exhibit chronic infection and impaired mucociliary clearance, such conditions may result in the dysregulation of SPLUNC1.
Cytokines and other environmental stimuli are known to modulate airway epithelial cell phenotypes, leading, in some cases, to increased secretory and/or goblet cells (45). Goblet cells do not produce SPLUNC1 (46). Other changes in the subsets of proximal, lower respiratory tract airway epithelial cells will likely affect SPLUNC1 production. In Th2 inflammation with low amounts of SPLUNC1 in the airways, numbers of immunoreactive SPLUNC1 cells in the trachea were comparable to those in our observations of naive mice (not shown). However, we have not investigated alterations in SPLUNC1-producing cells among MyD88-deficient mice that exhibited increases in submucosal gland abundance and mucus cells (47). Chronic changes in the airway epithelial cell composition in conjunction with, or as a result of, inflammation may explain elevations in SPLUNC1 in disease states such as COPD and cystic fibrosis (6, 9, 44).
Studies of SPLUNC1 have primarily focused on its antimicrobial properties. We now show that the down-regulation of SPLUNC1 occurs in a range of infections, and these effects appear to be common to mouse and man. Fluctuations of SPLUNC1 may regulate inflammation and mucociliary clearance, thus aiding in host-protective functions that extend beyond its antimicrobial effects. SPLUNC1 may serve as a sensor of exposure to environmental PAMPs and irritants, thus clarifying its high variability in health and disease. The tight control of SPLUNC1 suggests that its flux is important to aid in the rapid activation of innate immune responses and to initiate inflammatory responses, thus serving as a gatekeeper or integrator of innate and adaptive immunity.
Footnotes
This work was supported by National Institutes of Health grants RO1-HL081160 and R21-AI083475 (L.C.) and T32-HL007778 (C.J.B.).
Originally Published in Press as DOI: 10.1165/rcmb.2012-0072OC on March 7, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, Ma L, Tuan RS, Greene RM. Differential display identification of PLUNC, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J Biol Chem. 1999;274:13698–13703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- 2.Bingle CD, Seal RL, Craven CJ. Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold–containing superfamily. Biochem Soc Trans. 2011;39:977–983. doi: 10.1042/BST0390977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingle CD, Bingle L. Characterisation of the human PLUNC gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta. 2000;1493:363–367. doi: 10.1016/s0167-4781(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 4.Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum Mol Genet. 2002;11:937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- 5.Bingle CD, Bingle L, Craven CJ. Distant cousins: genomic and sequence diversity within the BPI fold–containing (BPIF)/PLUNC protein family. Biochem Soc Trans. 2011;39:961–965. doi: 10.1042/BST0390961. [DOI] [PubMed] [Google Scholar]
- 6.Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. Molecular cloning and characterization of SPURT, a human novel gene that is retinoic acid–inducible and encodes a secretory protein specific in upper respiratory tracts. J Biol Chem. 2003;278:1165–1173. doi: 10.1074/jbc.M210523200. [DOI] [PubMed] [Google Scholar]
- 7.Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. Purification and characterization of PLUNC from human tracheobronchial secretions. Am J Respir Cell Mol Biol. 2004;30:184–192. doi: 10.1165/rcmb.2003-0142OC. [DOI] [PubMed] [Google Scholar]
- 8.Bingle L, Cross SS, High AS, Wallace WA, Devine DA, Havard S, Campos MA, Bingle CD. SPLUNC1 (PLUNC) is expressed in glandular tissues of the respiratory tract and in lung tumours with a glandular phenotype. J Pathol. 2005;205:491–497. doi: 10.1002/path.1726. [DOI] [PubMed] [Google Scholar]
- 9.Bingle L, Barnes FA, Cross SS, Rassl D, Wallace WA, Campos MA, Bingle CD. Differential epithelial expression of the putative innate immune molecule SPLUNC1 in cystic fibrosis. Respir Res. 2007;8:79. doi: 10.1186/1465-9921-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, Stutts MJ, Tarran R. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci USA. 2009;106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlgraf KG, Ackermann AR, Burnell KK, Srikantha RN, Joly SA, Bartlett JA, Gakhar L, Johnson GK, McCray PB, Jr, Guthmiller JM, et al. Quantitation of SPLUNC1 in saliva with an xMAP particle–based antibody capture and detection immunoassay. Arch Oral Biol. 2012;57:197–204. doi: 10.1016/j.archoralbio.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Identification of human whole saliva protein components using proteomics. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- 13.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem Soc Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 14.LeClair EE, Nomellini V, Bahena M, Singleton V, Bingle L, Craven CJ, Bingle CD. Cloning and expression of a mouse member of the PLUNC protein family exclusively expressed in tongue epithelium. Genomics. 2004;83:658–666. doi: 10.1016/j.ygeno.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu HW, Thaikoottathil J, Rino JG, Zhang G, Wu Q, Moss T, Refaeli Y, Bowler R, Wenzel SE, Chen Z, et al. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol. 2007;179:3995–4002. doi: 10.4049/jimmunol.179.6.3995. [DOI] [PubMed] [Google Scholar]
- 17.Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, Frasch SC, Michels NM, Case SR, Chu HW. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol. 2011;178:2159–2167. doi: 10.1016/j.ajpath.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukinskiene L, Liu Y, Reynolds SD, Steele C, Stripp BR, Leikauf GD, Kolls JK, Di YP. Antimicrobial activity of PLUNC protects against Pseudomonas aeruginosa infection. J Immunol. 2011;187:382–390. doi: 10.4049/jimmunol.1001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng T, Ma J, Sheng SR. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol Cell Biochem. 2008;309:191–197. doi: 10.1007/s11010-007-9659-3. [DOI] [PubMed] [Google Scholar]
- 20.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PB., Jr PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS ONE. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett JA, Gakhar L, Penterman J, Singh PK, Mallampalli RK, Porter E, McCray PB., Jr PLUNC: a multifunctional surfactant of the airways. Biochem Soc Trans. 2011;39:1012–1016. doi: 10.1042/BST0391012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaillard EA, Kota P, Gentzsch M, Dokholyan NV, Stutts MJ, Tarran R. Regulation of the epithelial Na+ channel and airway surface liquid volume by serine proteases. Pflugers Arch. 2010;460:1–17. doi: 10.1007/s00424-010-0827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright PL, Yu J, Di YP, Homer RJ, Chupp G, Elias JA, Cohn L, Sessa WC. Epithelial reticulon 4B (Nogo-B) is an endogenous regulator of Th2-driven lung inflammation. J Exp Med. 2010;207:2595–2607. doi: 10.1084/jem.20100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGillivary G, Bakaletz LO. The multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PLoS ONE. 2010;5:e13224. doi: 10.1371/journal.pone.0013224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1– and IL-18–mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)–deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the LPS gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 27.Zhang X, Shan P, Qureshi S, Homer R, Medzhitov R, Noble PW, Lee PJ. Cutting edge: TLR4 deficiency confers susceptibility to lethal oxidant lung injury. J Immunol. 2005;175:4834–4838. doi: 10.4049/jimmunol.175.8.4834. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel RM, Aguet M. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z, Lee CG, Zheng T, Chupp G, Wang J, Homer RJ, Noble PW, Hamid Q, Elias JA. Airway inflammation and remodeling in asthma: lessons from interleukin 11 and interleukin 13 transgenic mice. Am J Respir Crit Care Med. 2001;164:S67–S70. doi: 10.1164/ajrccm.164.supplement_2.2106070. [DOI] [PubMed] [Google Scholar]
- 30.Niu N, Laufer T, Homer RJ, Cohn L. Cutting edge: limiting MHC Class II expression to dendritic cells alters the ability to develop Th2-dependent allergic airway inflammation. J Immunol. 2009;183:1523–1527. doi: 10.4049/jimmunol.0901349. [DOI] [PubMed] [Google Scholar]
- 31.Cohn L, Homer RJ, Marinov A, Rankin J, Bottomly K. Induction of airway mucus production by T helper 2 (Th2) cells: a critical role for interleukin 4 in cell recruitment but not mucus production. J Exp Med. 1997;186:1737–1747. doi: 10.1084/jem.186.10.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 33.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, Toll-like receptor 4–dependent T helper cell Type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ, Jr, Chapman HA, Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase– and cathepsin-dependent emphysema. J Clin Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohn L, Homer RJ, Niu N, Bottomly K. T helper 1 cells and interferon gamma regulate allergic airway inflammation and mucus production. J Exp Med. 1999;190:1309–1318. doi: 10.1084/jem.190.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai XF, Zhu J, Zhang GX, Kaponides G, Hojeberg B, van der Meide PH, Link H. IL-10 suppresses experimental autoimmune neuritis and down-regulates Th1-type immune responses. Clin Immunol Immunopathol. 1997;83:117–126. doi: 10.1006/clin.1997.4331. [DOI] [PubMed] [Google Scholar]
- 38.Rollins BM, Garcia-Caballero A, Stutts MJ, Tarran R. SPLUNC1 expression reduces surface levels of the epithelial sodium channel (ENaC) in Xenopus laevis oocytes. Channels (Austin) 2010;4:255–259. doi: 10.4161/chan.4.4.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu HW, Gally F, Thaikoottathil J, Janssen-Heininger YM, Wu Q, Zhang G, Reisdorph N, Case S, Minor M, Smith S, et al. SPLUNC1 regulation in airway epithelial cells: role of Toll-like receptor 2 signaling. Respir Res. 2010;11:155. doi: 10.1186/1465-9921-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh TH, Lee SY, Hsu WC. Expression of SPLUNC1 protein in nasal polyp epithelial cells in air–liquid interface culture treated with IL-13. Am J Rhinol Allergy. 2010;24:17–20. doi: 10.2500/ajra.2010.24.3381. [DOI] [PubMed] [Google Scholar]
- 41.Steiling K, Kadar AY, Bergerat A, Flanigon J, Sridhar S, Shah V, Ahmad QR, Brody JS, Lenburg ME, Steffen M, et al. Comparison of proteomic and transcriptomic profiles in the bronchial airway epithelium of current and never smokers. PLoS ONE. 2009;4:e5043. doi: 10.1371/journal.pone.0005043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghafouri B, Kihlstrom E, Stahlbom B, Tagesson C, Lindahl M. PLUNC (palate, lung and nasal epithelial clone) proteins in human nasal lavage fluid. Biochem Soc Trans. 2003;31:810–814. doi: 10.1042/bst0310810. [DOI] [PubMed] [Google Scholar]
- 43.Ghafouri B, Irander K, Lindbom J, Tagesson C, Lindahl M. Comparative proteomics of nasal fluid in seasonal allergic rhinitis. J Proteome Res. 2006;5:330–338. doi: 10.1021/pr050341h. [DOI] [PubMed] [Google Scholar]
- 44.Roxo-Rosa M, da Costa G, Luider TM, Scholte BJ, Coelho AV, Amaral MD, Penque D. Proteomic analysis of nasal cells from cystic fibrosis patients and non–cystic fibrosis control individuals: search for novel biomarkers of cystic fibrosis lung disease. Proteomics. 2006;6:2314–2325. doi: 10.1002/pmic.200500273. [DOI] [PubMed] [Google Scholar]
- 45.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8:432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 46.Bingle L, Wilson K, Musa M, Araujo B, Rassl D, Wallace WA, LeClair EE, Mauad T, Zhou Z, Mall MA, et al. BPIFB1 (LPLUNC1) is upregulated in cystic fibrosis lung disease. Histochem Cell Biol. 2012;138:749–758. doi: 10.1007/s00418-012-0990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giangreco A, Lu L, Mazzatti DJ, Spencer-Dene B, Nye E, Teixeira VH, Janes SM. MyD88 deficiency influences murine tracheal epithelial metaplasia and submucosal gland abundance. J Pathol. 2011;224:190–202. doi: 10.1002/path.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]