Abstract
Severe acute respiratory syndrome (SARS)–coronavirus (CoV) produces a devastating primary viral pneumonia with diffuse alveolar damage and a marked increase in circulating cytokines. One of the major cell types to be infected is the alveolar type II cell. However, the innate immune response of primary human alveolar epithelial cells infected with SARS-CoV has not been defined. Our objectives included developing a culture system permissive for SARS-CoV infection in primary human type II cells and defining their innate immune response. Culturing primary human alveolar type II cells at an air–liquid interface (A/L) improved their differentiation and greatly increased their susceptibility to infection, allowing us to define their primary interferon and chemokine responses. Viral antigens were detected in the cytoplasm of infected type II cells, electron micrographs demonstrated secretory vesicles filled with virions, virus RNA concentrations increased with time, and infectious virions were released by exocytosis from the apical surface of polarized type II cells. A marked increase was evident in the mRNA concentrations of interferon–β and interferon–λ (IL-29) and in a large number of proinflammatory cytokines and chemokines. A surprising finding involved the variability of expression of angiotensin-converting enzyme–2, the SARS-CoV receptor, in type II cells from different donors. In conclusion, the cultivation of alveolar type II cells at an air–liquid interface provides primary cultures in which to study the pulmonary innate immune responses to infection with SARS-CoV, and to explore possible therapeutic approaches to modulating these innate immune responses.
Keywords: lung innate immune response, cytokine responses to SARS coronavirus, lung cell differentiation, air–liquid interface cultures
Clinical Relevance
Severe acute respiratory syndrome–coronavirus (SARS-CoV) produces devastating pneumonia with a marked inflammatory cytokine response and a mortality rate of about 10%. Alveolar epithelial cells are known to be susceptible in vivo, but the extent of infection for submerged primary cultures of human type II cells was too limited to define the cytokine and interferon responses to infection. This report demonstrates that primary human alveolar type II cells can be productively infected with SARS-CoV, and that they can generate a vigorous innate immune response. The critical feature for infecting human type II cells with SARS-CoV involves maintaining and infecting the cells under air–liquid interface conditions. Among our unexpected findings, the expression of angiotensin-converting enzyme–2, the receptor for SARS-CoV, was quite variable among different individuals, and cells from different donors differed in their susceptibility to SARS-CoV infection.
Severe acute respiratory syndrome–associated coronavirus (SARS-CoV) produces devastating viral pneumonia (1, 2). Pathologic changes are most prominent in the lungs, with disruptions of the epithelium in gas exchange areas and conducting airways (2–4). The epithelial cells of the alveoli and the conducting airways are the primary targets of SARS-CoV in the human lung, and they express the SARS receptor, angiotension-converting enzyme–2 (ACE2) (5–8). In autopsies of patients with SARS, coronavirus RNA and proteins have been detected in type II cells via immunocytochemisty and in situ hybridization (7, 9–14). In the aged macaque model of SARS, in which the initial site of infection in the lung can be studied, virus infection was detected in both alveolar type I and type II cells (11, 15). In human SARS autopsy specimens, the infection of alveolar macrophages was also suggested because they contained SARS antigens and formed multinucleated giant cells (2, 11). However, in vitro human alveolar macrophages, monocyte-derived dendritic cells, and monocytes are not readily susceptible to SARS-CoV, and these cell types show only very modest cytokine and interferon responses upon exposure to SARS-CoV (16–21).
One of the major clinical issues in this type of devastating viral pneumonia involves whether SARS is principally a severe primary viral pneumonia, or a systemic disease caused by a cytokine storm out of proportion to the extent of viral pneumonia (22). During the course of SARS-CoV infection, a marked elevation was evident in serum concentrations of proinflammatory cytokines, including CXCL10 (IP-10), CCL2 (MCP-1), IL-6, and CXCL8 (IL-8) (1, 22–26). However, it remains unknown whether these serum cytokines were produced by epithelial cells of the alveoli and conducting airways (where the virus replicates in vivo) or by inflammatory cells recruited to the site of virus infection in the lung. Based on in situ hybridization and immunocytochemistry, He and colleagues suggested that ACE2-positive infected cells in lungs of patients with SARS are the source of proinflammatory cytokines (27).
The primary goal of this research was to define the innate immune response of human alveolar type II cells to SARS-CoV. In our earlier study with submerged human alveolar type II cells on inserts, we detected a very low percentage of infected cells and could not evaluate their innate immune response (18). However, culturing and infecting the type II cell at an air–liquid interface (A/L) allowed for significant infection and characterization of their innate immune response. The robust innate immune response of these cells to SARS-CoV infection consisted of marked elevations in mRNA concentrations of interferon-β, interferon-λ, CXCL10, CXCL11, and IL-6, and these results mirror findings reported in the lungs and sera of patients with SARS (22, 23, 25).
Materials and Methods
Lungs were obtained from deidentified patients whose lungs had been deemed unsuitable for transplantation and were donated for medical research. The Committee for the Protection of Human Subjects at National Jewish Health has deemed that the use of human lung cells from deidentified lung donors does not constitute human subject research, and is therefore exempt from Institutional Review Board approval. (Correspondence to this effect from the Committee for the Protection of Human Subjects at National Jewish Health is available upon request).
Isolation and Culture of Human Alveolar Type II Cells at an Air–Liquid Interface
Cells were isolated as previously described (28, 29). Isolated cells were resuspended in Dulbecco’s Modified Eagle’s Medium supplemented with 10% FBS, and plated at a density of 1 × 106 per cm2 on six-well or 24-well Millicell inserts (Millipore Corp., Bedford, MA) precoated with a mixture of 50% Matrigel (BD Biosciences, San Jose, CA) and 50% rat-tail collagen. After cell adherence for 48 hours, the medium was switched for 2 days to 5% heat-inactivated human serum plus 10 ng/ml keratinocyte growth factor (KGF), 0.1 mM isobutylmethyl xanthine, 0.1 mM 8-bromo–cyclic adenosine monophosphate (cAMP), 10 ng/ml IL-4, and 10 ng/ml IL-13. At this time, cultures were converted to A/L conditions. Cells were incubated for an additional 4 days with medium containing KIA, IL-4, IL-13, and 10 nM dexamethasone (D). After 8 days of culture on the inserts and 6 days under A/L conditions, cells were used for virus inoculation or harvested.
SARS-CoV Infection
The Urbani strain of SARS-CoV was kindly provided by Dr. Bellini at the Centers for Disease Control (Atlanta, GA), and was propagated in Vero E6 cells. All work with infectious SARS-CoV was performed in the Biosafety Level 3 suite at the School of Medicine, University of Colorado (Aurora, CO). type II cells were inoculated on the apical surface with 250 μl per six-well plate insert (or 50 μl per 24-well plate insert) of KIAD with IL-4–supplemented and IL-13–supplemented medium containing SARS-CoV (1 × 107 plaque-forming units (pfu)/ml). After 4 hours of incubation at 37°C, the virus inocula and control media were aspirated from the apical cell surface, and cultures were incubated for a total of 24 or 48 hours after virus inoculation under A/L conditions, after which they were extracted to analyze viral and cellular RNA, or fixed and assayed via electron microscopy or immunofluorescent labeling with antibodies to viral or cell proteins. The yield of infectious virus was titered in washes of apical cell surfaces and basal medium by plaque assays on Vero E6 cells.
The methods for immunocytochemistry, the quantitation of infected cells, virus production, plaque assays, the isolation of RNA, quantitative PCR, flow cytometry, surface biotinylation, and statistical analysis are described in the online supplement.
Results
Culturing Primary Human Alveolar Type II Cells at an Air–Liquid Interface Maintains Their Cellular Differentiation
In our initial studies on SARS-CoV infection with human alveolar type II cells in traditional submerged cultures, we detected only a very low level of infection. Subsequently, we evaluated a variety of changes in the culture system to improve the rate of infection. We found that susceptibility to SARS-CoV infection and the maintenance of differentiation were achieved by culturing cells at an A/L on Millicell inserts coated with a matrix of rat-tail collagen and Matrigel, and by providing a medium that contained IL-13 and IL-4 as well as KGF, isomethylbutyl xanthine, 8 Br-cAMP, dexamethasone, and 5% heat-inactivated human serum (30). Cells contained readily identifiable lamellar bodies according to phase microscopy (Figure 1A). In general, increased type II cell differentiation was observed, as evidenced by the expression of the surfactant proteins (SPs) (Figure 1B). As shown in Figure 1B, the air–liquid interface condition increased the expression of ACE2, SP-A, proSP-B, proSP-C, and ABCA3, but not fatty-acid synthase, compared with the submerged condition in this experiment. However, the magnitude of effect of the A/L condition was more variable with human type II cells than with our experiments on primary rat type II cells (31) (Figure E1 in the online supplement). In addition, we found an increase in ACE2 expression in our cultures after adding IL-4 and IL-13, a finding that differs from results with Vero E6 cells regarding the effect of IL-13 on ACE2 expression (32) (Figure 1B). In general, the level of ACE2 expression, as measured by immunoblotting of whole-cell extracts, was slightly increased in type II cells cultured under A/L conditions, but ACE2 concentrations varied considerably between cells from individual donors (Figure E1). However, according to flow cytometry and surface biotinylation, we were unable to detect any increase in the surface expression of ACE2 in cells maintained under A/L conditions, compared with cells maintained in submerged cultures (data not shown).
Figure 1.
Human alveolar type II cells cultured at an air–liquid interface (A/L). Human alveolar type II cells were cultured under A/L conditions, as described for infection with SARS-CoV. (A) Phase micrograph of the epithelial monolayer with inclusions (lamellar bodies), indicated by arrows, clearly visible within the cytoplasm of type II cells. (B) The effect of A/L culture conditions and treatment with IL-4 and IL-13 on selected proteins by immunoblotting. Lane 1 is a extract of freshly isolated type II cells, lane 2 is empty, lanes 3 and 4 contain cells cultured under A/L conditions with KIAD alone, lanes 5 and 6 contain cells cultured under A/L conditions with KIAD, IL-4, and IL-13, lanes 7 and 8 contain cells cultured under submerged conditions with KIAD, and lanes 9 and 10 contain cells cultured under submerged conditions with KIAD, IL-4, and IL-13. A/L conditions increased the expression of angiotensin-converting enzyme–2 (ACE2), surfactant protein (SP)–A, proSP-B, proSP-C, and ATP-binding cassette sub-family A member 3 (ABCA3), but not fatty acid synthase (FAS). IL-4 and IL-13 significantly increased the expression of ACE2. GAPDH, glyceraldehyde 3–phosphate dehydrogenase.
SARS-CoV Readily Infects Primary Human Alveolar Type II Cells Cultured at the Air–Liquid Interface
To test whether type II cells cultured at the A/L were more susceptible to SARS-CoV infection than type II cells in submerged cultures (18), human type II cells cultured at an A/L on six-well inserts were inoculated on the apical surface with 250 μl of medium containing SARS-CoV at an estimated multiplicity of 2 pfu/cell, and allowed to adsorb for 4 hours at 37°C. Then the apical fluid and inoculum were aspirated, and cells were maintained under A/L conditions. SARS-CoV reproducibly caused productive infection of the primary human alveolar type II cells cultured at the A/L. We found little evidence of infection at 4 hours after virus inoculation, but by 24 hours after virus inoculation, infected cells containing viral antigens in the cytoplasm were readily detected. SARS-CoV–infected cells were readily observed in all primary alveolar type II cultures on Millicell inserts at the A/L, although the percentages of infected cells varied considerably in different areas of the same insert and between different human donors. In four donors, the percentage of cells infected ranged from 3.4–34.2% at 24 hours, and from 2.4–27.2% at 48 hours.
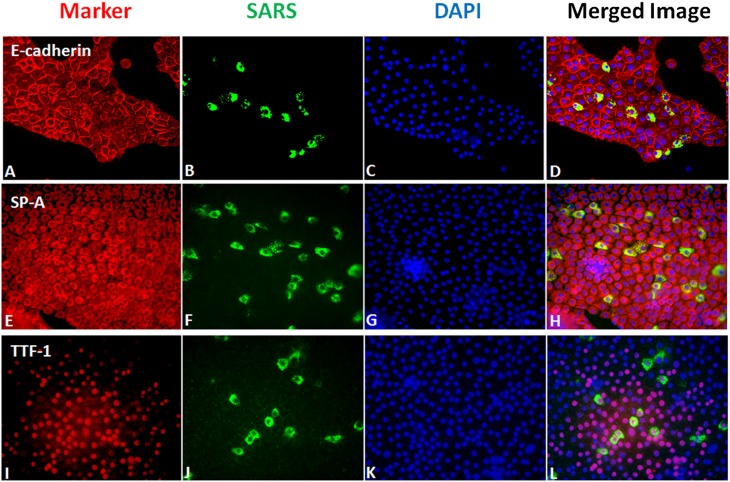
To demonstrate that the SARS-CoV–infected cells were type II cells, we used antibodies to several type II cell differentiation markers, namely, thyroid transcription factor-1 (TTF-1), SP-A, and E-cadherin. Nearly 100% of the cells expressed SP-A. Dual labeling showed that the infected cells were epithelial and expressed TTF-1 and SP-A (Figure 2). These markers were chosen because they could be readily detected in the methanol-fixed cells required for identifying the viral antigens, and could be used with the rabbit antibody to the SARS-CoV nucleocapsid protein. In all experiments, only a portion of the cells were infected, and it was not clear why some cells were infected and others were not. We were not certain if this discrepancy was related to the level of ACE2 expression.
Figure 2.
Immunofluorescent staining for SARS-CoV nucleocapsid protein and selected alveolar type II cell markers. Cells were grown under A/L conditions as described in Materials and Methods, inoculated with SARS-CoV at an estimated multiplicity of infection of 2, and fixed 24 hours after inoculation. (A–D) Staining for E-cadherin (A), SARS-CoV (B), DAPI (C), and merged (D). (E–H) Staining for SP-A (E), SARS-CoV (F), DAPI (G), and merged (H). (I–L) Staining for TTF-1 (I), SARS-CoV (J), DAPI (K), and merged (L). Cells that are infected with SARS-CoV stain for type II cell markers.
Coronavirus RNA is synthesized in virus-induced double-membrane vesicles in the cytoplasm of infected cells. The virions bud intracellularly from membranes of the ER–Golgi-intermediate compartment, are transported through the cytoplasm in secretory vesicles, and are released from cells by an exocytic process. Some coronaviruses are released from polarized epithelial cells by exocytosis at the apical plasma membrane, whereas others are released at basolateral membranes (33). Virions do not bud from the plasma membrane, although released virions often remain adherent to the plasma membrane of infected cells.
To document definitively that the SARS-CoV–infected cells in these alveolar type II cultures at the A/L were type II cells, we inoculated the cell cultures at the A/L with SARS-CoV, fixed and embedded them in plastic, sectioned them perpendicular to the insert, and observed them with the electron microscope. Coronavirus virions, approximately 100 nm in diameter, were observed within numerous secretory vesicles in cells that also contained lamellar bodies, the definitive morphologic feature of type II cells (Figure 3). The characteristic double-membrane vesicles that comprise the sites of viral RNA replication are depicted in Figure 3B (34, 35), and virions were observed budding into vesicles (Figure 3A) and adsorbed to the apical membrane (Figure 3B).
Figure 3.
Ultrastructure of alveolar type II cells infected with SARS-CoV. Cells were cultured as described in Materials and Methods under A/L conditions, inoculated with SARS-CoV on the apical surface, and then fixed and processed for electron microscopy 48 hours after inoculation. (A) A differentiated alveolar type II cell with its characteristic lamellar bodies (LB) and SARS-CoV virions are found in expanded, smooth-walled secretory vesicles in the cytoplasm of the infected cell. Several budding virions are indicated by arrows. (B) Several smooth-walled, double-membrane vesicles (V), which are the site of replication for SARS-CoV RNA, virions in secretory vesicles near the plasma membrane, and virions adsorbed to the apical plasma membrane. The inset depicts a higher magnification of one of these double-membrane vesicles.
Infected Alveolar Type II Cells Cultured at the Air–Liquid Interface Produce Infectious SARS-CoV
To analyze SARS-CoV infection in type II cells at an A/L, we used both real-time quantitative PCR and plaque assays (Figure 4). Quantitative real-time PCR revealed a consistent 1.5 log increase in viral mRNA at 24 hours after inoculation, as shown for six donors (Figure 4A). Each donor was deidentified and received a number. Most donors were used for only one type of experiment because of the limited number cells isolated from each donor. Cells were isolated from more than 20 donors in the course of these experiments.
Figure 4.
SARS-CoV production by alveolar type II cells. Type II cells were cultured at an A/L and inoculated with SARS-CoV, as described in Materials and Methods. (A) Cells were processed for the extraction of RNA and real-time quantitative RT-PCR at 6 and 24 hours after virus inoculation to measure SARS-CoV–specific viral RNA. For this series of experiments, six different donors were used, and all showed a significant increase in SARS-CoV mRNA at 24 hours after inoculation. (B) The production of infectious virus is shown. In this series of experiments, the yields of virus released from the apical (red) or basolateral (black) surfaces or from cell extracts (blue) at 4, 24, and 48 hours after inoculation were titered by plaque assay in Vero E6 cells and expressed as the total virus yield from each insert. The points represent the means and standard errors from four independent experiments.
Because no medium was recoverable from the cells at the A/L, we washed the apical surface at designated times after virus inoculation, and titered the released virus in the apical wash as well as in the basolateral medium by a plaque assay. Equally high yields of released virus from the apical wash and cell-associated virus from the frozen and thawed samples were detected at 24 hours, and the virus titers in specimens harvested at 48 hours were lower but similar. Very little virus was detected in the basolateral medium (Figure 4B).
Alveolar Type II Cell Cultures at the A/L Display a Vigorous Innate Immune Response to SARS-CoV Infection
Concentrations of multiple proinflammatory cytokines and chemokines have been reported to be increased in the lungs and sera of patients with SARS (22, 23, 25, 26). Because alveolar type II cells at the A/L were readily infected by SARS-CoV, we were able to measure changes in expression of selected genes of the innate immune system in these cells in response to SARS-CoV infection (Figure 5). For technical and biosafety reasons, we did not attempt to measure concentrations of interferon or cytokine proteins from infected cultures. The SARS-CoV infection of type II cells induced markedly increased concentrations of the mRNAs encoding type I and type III interferons, as well as the interferon-responsive genes ISG56, OAS1, Mx1, CXCL10, and CXCL11 (Figure 5). The SARS-CoV infection of type II cultures also elicited large increases in the concentrations of chemokines that recruit inflammatory cells (CXCL8, CCL5, CXCL10, and CXCL11). Thus, human alveolar type II cells are capable of initiating robust innate immune responses that could contribute to the cytokines and chemokines measured in the lungs and sera of patients with SARS. In addition, chemoattractants released by epithelial cells would be expected to recruit inflammatory cells in vivo, and to initiate the innate immune response.
Figure 5.
Innate immune responses of alveolar type II cells to SARS-CoV infection. type II cells at an A/L were cultured and inoculated with SARS-CoV, as described in Materials and Methods. After the 4-hour inoculation period, the inoculum was aspirated, and cultures were harvested 6 and 24 hours later. The specific mRNA concentrations of chemokines and cytokines were measured by quantitative RT-PCR, and normalized to the constitutive probe 36B4 (acidic ribosomal phosphoprotein). The white bars represent data from mock-inoculated inserts, and the black bars represent data from SARS-CoV–infected inserts. (A) At 6 hours after virus inoculation, the only significant increase in detected mRNA concentrations involved the interferon-responsive gene ISG56. (B) At 24 hours after virus inoculation, a marked increase in the expression of all selected genes was evident. *P < 0.05. **P < 0.01. ***P < 0.001. The results were derived from six independent donors, with duplicate inserts for each condition and time.
Some Alveolar Type I–Like Cells Are Susceptible to SARS-CoV Infection in A/L Cultures
The focus of these studies involved differentiated human type II cells, but we also performed a limited number of experiments to determine whether type I–like cells transdifferentiated from type II cells grown under A/L conditions were also susceptible to SARS-CoV. In our previous study (18), we detected no SARS-CoV infection of human alveolar type I–like cells under submerged conditions, and ACE2 protein expression was below the level of detection. In pilot experiments with unselected donors, we detected very little infection of type I–like cells under A/L conditions at 24 hours after inoculation. However, when we undertook screening to select a donor (i.e., patient 101) who expressed a high concentration of ACE2 in the freshly isolated cells, we observed that transdifferentiated type I–like cells could be infected with SARS-CoV (Figures E1 and E2). Hence, these alveolar type I–like cells at the A/L were also susceptible to SARS-CoV, although they were much less susceptible to infection than differentiated human type II cells. These type I–like cells derived from type II cells in vitro may differ from type I cells in vivo (36).
Discussion
The goal of this study involved developing a primary culture system with human alveolar type II cells that was permissive to SARS-CoV infection, so that their innate immune response to infection could be defined. Human alveolar type II cells maintained at an A/L were well differentiated (Figures 1–3) and permissive for SARS-CoV infection, as shown by immunocytochemistry, by increases in the synthesis of viral RNA and protein, and by the production and release of infectious virions. Although the air–liquid interface culture conditions greatly increased the susceptibility of type II cells to SARS-CoV, the mechanism for this increased susceptibility to infection is not yet established. In the lung, very little fluid is present on the apical surface of the alveolar epithelium (37, 38). Hence, the A/L conditions approximate conditions in vivo. A/L culture conditions improved the differentiation state of type II alveolar epithelial cells in vitro, and greatly enhanced their susceptibility to SARS-CoV infection.
Innate immunity serves as the first response to protect host cells from viral infection, and the characteristics of the innate immune response to a pathogen can significantly shape subsequent adaptive immune responses and affect the outcome of infection (39). When the alveolar epithelium becomes infected, infected cells alert other cells and organs to the infection by secreting cytokines and chemokines. The infected epithelial cells are likely the first responders, and initiate the innate immune response. The earliest response of human alveolar type II cells to influenza virus infection involves the expression of interferon and interferon-responsive genes (40). Presumably this interferon response increases the resistance of neighboring cells to infection. The next phase of the innate immune response involves the expression of cytokines, which can both attract and activate immune cells and stimulate neighboring cells to amplify the defensive response greatly by secreting additional cytokines and chemokines. The innate response also signals the bone marrow to release additional inflammatory cells and initiates the adaptive immune response to recruit antigen-specific lymphocytes. In this study, we measured the mRNAs encoding interferon-β and interferon-λ, and both were markedly elevated in response to SARS-CoV infection. Both of these interferons have been reported to be elevated in experimental SARS-CoV infections in primates and in cell lines infected with SARS-CoV (15, 24, 41). IL-29 is important in the innate immune response and may help control SARS-CoV infection (29, 41, 42).
The cytokine responses of human type II alveolar epithelial cells at the A/L were similar to those detected in the sera and lungs of patients and in nonhuman primates infected with SARS-CoV (15, 24, 43). He and colleagues suggested that resident ACE2-expressing lung epithelial cells, and not the subsequently recruited inflammatory cells, constitute the primary source of elevated cytokines in patients infected with SARS-CoV, and that these epithelial-derived cytokines play an important role in the acute lung injury and pathogenesis of SARS (27). Our data show that type II alveolar cells likely contribute to the high concentrations of cytokines and chemokines detected in the gas-exchange portion of the lungs in patients with SARS. In the sera of patients with SARS, the cytokine that increases most rapidly is CXCL10 (IP-10) (22, 23). Similarly, in our type II cells at the A/L, the level of CXCL10 mRNA expression increased more than 1,000 fold by 24 hours after virus inoculation (Figure 5). The other cytokine that appears to predict the clinical course and outcome of SARS is IL-6 (43). The concentration of mRNA encoding IL-6 was elevated approximately 10-fold in our SARS-CoV–infected alveolar type II cultures at the A/L (Figure 5). In these particular experiments, we did not evaluate ultraviolet light–inactivated virus, but we knew from previous studies that type II cells do not present a cytokine or interferon response to ultraviolet light–inactivated influenza (40). Additional studies will be required to define the cell–cell interactions whereby infected epithelial cells signal to nearby uninfected parenchymal cells or inflammatory cells. We found that rat alveolar type I–like cells infected with rat coronavirus stimulated noninfected cells in the culture to secrete cytokines and chemokines through an IL-1–dependent mechanism (30). Paracrine signaling by IL-1, which is released from infected type II cells during the activation of the inflammasome, may also play important roles, both in protecting type II cells from infection and in amplifying the innate immune response.
We explored the hypothesis that the increased rate of SARS-CoV infection in type II cells under A/L conditions was attributable to an increased concentration of ACE2, the SARS-CoV receptor. We observed that type II cells from donors who expressed higher concentrations of ACE2 and from culture conditions that increased ACE2 expression generally correlated with a higher susceptibility to SARS-CoV infection. However, we did not measure ACE2 concentrations in cells from enough different donors who were also inoculated with SARS-CoV, and we did not measure the extent of virus infection to prove this quantitatively. Although immunoblotting showed a modest but variable increase in ACE2 expression by type II cells when cultured at the A/L compared with submerged conditons, we did not detect a consistent increase in the surface expression of ACE2 by either flow cytometry or surface biotinylation (data not shown). Similarly, when we transdifferentiated type II cells at the A/L into type I–like cells that expressed very low levels of ACE2, we observed very little SARS-CoV infection. However, in type I–like cells transdifferentiated from a type II cell donor selected to express high concentrations of ACE2, we did observe the infection of type I–like cells (Figures E1 and E2). Because we did not demonstrate a clear increase in the surface expression of ACE2 under A/L conditions, we cannot conclude that the enhanced role of infection of type II cells at the A/L was attributable solely to the concentration of ACE2.
We did not focus on the potential of adding IL-4 and IL-13 to these cultures. De Lang and colleagues reported that IL-4 and IL-13 decreased ACE2 expression in Vero E6 cells (32). However, as shown in Figure 1, we found that IL-4/IL-13 actually increased ACE2 expression in our primary human alveolar type II cells. Hence, we added IL-4/IL-13 to increase ACE2 expression and thereby increase SARS-CoV infection. We previously reported on the effects of IL-13 on human type II cells, and noted that IL-13 decreases SP-C and SP-D but exerts little effect on SP-A or SP-B (44). The slight decrease in SP-D could enhance the SARS infection, although this is unlikely, based on our experiments with 229E-6V (another coronavirus), in which SP-D did not make a major difference in the 229E-6V infectability of human alveolar macrophages (45). Nevertheless, SP-D has been reported to interact with the SARS spike glycoprotein, the attachment protein (46).
Alternatively, A/L culture conditions may have increased the level of expression in some other coreceptor for SARS-CoV, such as CD209L (47). In pilot experiments, we did not find a consistent increase in CD209L according to flow cytometry under A/L conditions (data not shown). Another consideration involves the details of the inoculation and the volume of apical fluid. In the A/L cultures, the inoculum was aspirated and cells were not washed, so some of the inoculum persisted in the surface fluid for the duration of the experiment. Perhaps the A/L conditions facilitated multiple cycles of virus infection. For example, after the first cycle of virus replication, the newly synthesized virus would be released into the scant amount of fluid on the apical surface, and this concentrated virus might accelerate secondary rounds of infection. In pilot studies, we observed that an increased susceptibility of type II cells to SARS-CoV infection required A/L culture conditions both before and after virus inoculation.
Other investigators identified Oct-4–positive lung cells that could be infected with SARS-CoV (43, 48). In neonatal mice, these cells expressed club cell secretory protein but not alveolar epithelial cell markers (48). In human lung tissue, these cells did not express cytokeratin or surfactant proteins (43). Our cells were different and did not express Oct-4 or the club cell secretory protein, but they expressed surfactant proteins and cytokeratin, and contained characteristic lamellar bodies.
In conclusion, culturing at the A/L renders primary human alveolar type II cells susceptible to productive SARS-CoV infection, and the cells mount a robust innate immune response, similar to that observed in the lungs and sera of patients with SARS. type II alveolar epithelial cells cultured at an A/L offer an excellent in vitro system in which to isolate and propagate emerging respiratory viruses that present with fastidious requirements for growth in highly differentiated human lung cells. These cultures also provide excellent in vitro models in which to evaluate the effects of candidate therapeutics that target the innate immune responses to respiratory virus infection.
Acknowledgments
Acknowledgments
The authors thank Beata Kosmider, Taylor Afford, Elise Messier, and Piruz Nahreini for assistance with the type II cell isolations; Dorothy Dill for assistance with electron microscopy; Boyd Jacobson for the medical illustrations; and Lydia Orth, Sarah Murrell, and Teneke Warren at National Jewish Health for help in preparing the manuscript.
Footnotes
This work was supported by National Institutes of Health grants AI05976 and R01-HL-29891, the ExxonMobil Foundation (R.J.M.), and the Parker B. Francis Foundation (J.W.).
Author Contributions: Z.Q. performed most of the experiments with severe acute respiratory syndrome (SARS) infections and helped write the manuscript. E.A.T. helped isolate the type II cells, performed the mRNA experiments to define innate immune responses, performed the flow cytometry, and helped write the manuscript. L.O. oversaw the initial SARS infections. K.E. performed all the type II cell cultures and some of the immunocytochemistry. A.B. performed some of the immunocytochemistry. J.W. helped isolate human type II cells, and advised on the manuscript and on innate immune responses. Y.I. helped with type II cell isolations and performed additional experiments to document the state of differentiation in type II cells. K.V.H. advised on all aspects of the project, and edited the manuscript. R.J.M. advised on all aspects of the project, including type II cell isolation, and wrote most of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2012-0339OC on February 15, 2013
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Chen J, Subbarao K. The immunobiology of SARS. Annu Rev Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 2.Franks TJ, Chong PY, Chui P, Galvin JR, Lourens RM, Reid AH, Selbs E, McEvoy CP, Hayden CD, Fukuoka J, et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol. 2003;34:743–748. doi: 10.1016/S0046-8177(03)00367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang DM, Chamberlain DW, Poutanen SM, Low DE, Asa SL, Butany J. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol. 2005;18:1–10. doi: 10.1038/modpathol.3800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2) J Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, Farzan M, Wohlford-Lenane C, Perlman S, McCray PB., Jr ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims AC, Burkett SE, Yount B, Pickles RJ. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mossel EC, Huang C, Narayanan K, Makino S, Tesh RB, Peters CJ. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J Virol. 2005;79:3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X, Tan L, Tschernig T, Deng H, Naim HY, et al. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome–associated coronavirus. J Gen Virol. 2006;87:1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- 11.Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shieh WJ, Hsiao CH, Paddock CD, Guarner J, Goldsmith CS, Tatti K, Packard M, Mueller L, Wu MZ, Rollin P, et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36:303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao CH, Chang MF, Hsueh PR, Su IJ. Immunohistochemical study of severe acute respiratory syndrome–associated coronavirus in tissue sections of patients. J Formos Med Assoc. 2005;104:150–156. [PubMed] [Google Scholar]
- 14.Tse GM, To KF, Chan PK, Lo AW, Ng KC, Wu A, Lee N, Wong HC, Mak SM, Chan KF, et al. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS) J Clin Pathol. 2004;57:260–265. doi: 10.1136/jcp.2003.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smits SL, de Lang A, van den Brand JM, Leijten LM, van IWF, Eijkemans MJ, van Amerongen G, Kuiken T, Andeweg AC, Osterhaus AD, et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 2010;6:e1000756. doi: 10.1371/journal.ppat.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scagnolari C, Trombetti S, Cicetti S, Antonelli S, Selvaggi C, Perrone L, Visca M, Romano S, Antonelli G. Severe acute respiratory syndrome coronavirus elicits a weak interferon response compared to traditional interferon-inducing viruses. Intervirology. 2008;51:217–223. doi: 10.1159/000154258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yilla M, Harcourt BH, Hickman CJ, McGrew M, Tamin A, Goldsmith CS, Bellini WJ, Anderson LJ. SARS–coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107:93–101. doi: 10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mossel EC, Wang J, Jeffers S, Edeen KE, Wang S, Cosgrove GP, Funk CJ, Manzer R, Miura TA, Pearson LD, et al. Sars-CoV replicates in primary human alveolar Type II cell cultures but not in Type I–like cells. Virology. 2008;372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler T, Matikainen S, Ronkko E, Osterlund P, Sillanpaa M, Siren J, Fagerlund R, Immonen M, Melen K, Julkunen I. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte–derived dendritic cells. J Virol. 2005;79:13800–13805. doi: 10.1128/JVI.79.21.13800-13805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL. Chemokine up-regulation in SARS–coronavirus–infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, Chan KH, Yuen KY, Gordon S, Guan Y, et al. Cytokine responses in severe acute respiratory syndrome coronavirus–infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang NL, Chan PK, Wong CK, To KF, Wu AK, Sung YM, Hui DS, Sung JJ, Lam CW. Early enhanced expression of interferon-inducible protein–10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin Chem. 2005;51:2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Xu J, Zhou C, Wu Z, Zhong S, Liu J, Luo W, Chen T, Qin Q, Deng P. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am J Respir Crit Care Med. 2005;171:850–857. doi: 10.1164/rccm.200407-857OC. [DOI] [PubMed] [Google Scholar]
- 24.de Lang A, Baas T, Teal T, Leijten LM, Rain B, Osterhaus AD, Haagmans BL, Katze MG. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV–infected macaques. PLoS Pathog. 2007;3:e112. doi: 10.1371/journal.ppat.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao CH, Chang MF, Hsueh PR, Su IJ. Immunohistochemical study of severe acute respiratory syndrome–associated coronavirus in tissue sections of patients. J Formos Med Assoc. 2005;104:150–156. [PubMed] [Google Scholar]
- 27.He L, Ding Y, Zhang Q, Che X, He Y, Shen H, Wang H, Li Z, Zhao L, Geng J, et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV–infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Edeen K, Manzer R, Chang Y, Wang S, Chen X, Funk CJ, Cosgrove GP, Fang X, Mason RJ. Differentiated human alveolar epithelial cells and reversibility of their phenotype in vitro. Am J Respir Cell Mol Biol. 2007;36:661–668. doi: 10.1165/rcmb.2006-0410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Oberley-Deegan R, Wang S, Nikrad M, Funk CJ, Hartshorn KL, Mason RJ. Differentiated human alveolar Type II cells secrete antiviral IL-29 (IFN–lambda 1) in response to influenza A infection. J Immunol. 2009;182:1296–1304. doi: 10.4049/jimmunol.182.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura TA, Wang J, Holmes KV, Mason RJ. Rat coronaviruses infect rat alveolar Type I epithelial cells and induce expression of CXC chemokines. Virology. 2007;369:288–298. doi: 10.1016/j.virol.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y, Ahmad A, Kewley E, Mason RJ. Hypoxia-inducible factor regulates expression of surfactant protein in alveolar Type II cells in vitro. Am J Respir Cell Mol Biol. 2011;45:938–945. doi: 10.1165/rcmb.2011-0052OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lang A, Osterhaus AD, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. 2006;353:474–481. doi: 10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossen JW, Bekker CP, Strous GJ, Horzinek MC, Dveksler GS, Holmes KV, Rottier PJ. A murine and a porcine coronavirus are released from opposite surfaces of the same epithelial cells. Virology. 1996;224:345–351. doi: 10.1006/viro.1996.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stertz S, Reichelt M, Spiegel M, Kuri T, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. The intracellular sites of early replication and budding of SARS–coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulasli M, Verheije MH, de Haan CA, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez R, Yang YH, Griffin C, Allen L, Tigue Z, Dobbs L. Freshly isolated rat alveolar Type I cells, Type II cells, and cultured Type II cells have distinct molecular phenotypes. Am J Physiol Lung Cell Mol Physiol. 2005;288:L179–L189. doi: 10.1152/ajplung.00272.2004. [DOI] [PubMed] [Google Scholar]
- 37.Mason RJ, Williams MC, Widdicombe JH, Sanders MJ, Misfeldt DS, Berry LC., Jr Transepithelial transport by pulmonary alveolar Type II cells in primary culture. Proc Natl Acad Sci USA. 1982;79:6033–6037. doi: 10.1073/pnas.79.19.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fehrenbach H. Alveolar epithelial Type II cell: defender of the alveolus revisited. Respir Res. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouse BT, Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol. 2010;10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, et al. Innate immune response to influenza A virus in differentiated human alveolar Type II cells. Am J Respir Cell Mol Biol. 2011;45:582–591. doi: 10.1165/rcmb.2010-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshikawa T, Hill TE, Yoshikawa N, Popov VL, Galindo CL, Garner HR, Peters CJ, Tseng CT. Dynamic innate immune responses of human bronchial epithelial cells to severe acute respiratory syndrome–associated coronavirus infection. PLoS ONE. 2010;5:e8729. doi: 10.1371/journal.pone.0008729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Chan VS, Zheng B, Chan KY, Xu X, To LY, Huang FP, Khoo US, Lin CL. A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J Exp Med. 2007;204:2529–2536. doi: 10.1084/jem.20070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ito Y, Mason RJ. The effect of interleukin-13 (IL-13) and interferon-gamma (IFN-gamma) on expression of surfactant proteins in adult human alveolar Type II cells in vitro. Respir Res. 2010;11:157. doi: 10.1186/1465-9921-11-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funk CJ, Wang J, Ito Y, Travanty EA, Voelker DR, Holmes KV, Mason RJ. Infection of human alveolar macrophages by human coronavirus strain 229E. J Gen Virol. 2012;93:494–503. doi: 10.1099/vir.0.038414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leth-Larsen R, Zhong F, Chow VT, Holmskov U, Lu J. The SARS coronavirus spike glycoprotein is selectively recognized by lung surfactant protein D and activates macrophages. Immunobiology. 2007;212:201–211. doi: 10.1016/j.imbio.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Jr, Thackray LB, Young MD, Mason RJ, et al. CD209l (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling TY, Kuo MD, Li CL, Yu AL, Huang YH, Wu TJ, Lin YC, Chen SH, Yu J. Identification of pulmonary Oct-4+ stem/progenitor cells and demonstration of their susceptibility to SARS coronavirus (SARS-CoV) infection in vitro. Proc Natl Acad Sci USA. 2006;103:9530–9535. doi: 10.1073/pnas.0510232103. [DOI] [PMC free article] [PubMed] [Google Scholar]