Abstract
Using primary and secondary structure information of an RNA molecule, the program RNA2D3D automatically and rapidly produces a first-order approximation of a 3-dimensional conformation consistent with this information. Applicable to structures of arbitrary branching complexity and pseudoknot content, it features efficient interactive graphical editing for the removal of any overlaps introduced by the initial generating procedure and for making conformational changes favorable to targeted features and subsequent refinement. With emphasis on fast exploration of alternative 3D conformations, one may interactively add or delete base-pairs, adjacent stems can be coaxially stacked or unstacked, single strands can be shaped to accommodate special constraints, and arbitrary subsets can be defined and manipulated as rigid bodies. Compaction, whereby base stacking within stems is optimally extended into connecting single strands, is also available as a means of strategically making the structures more compact and revealing folding motifs. Subsequent refinement of the first-order approximation, of modifications, and for the imposing of tertiary constraints is assisted with standard energy refinement techniques. Previously determined coordinates for any part of the molecule are readily incorporated, and any part of the modeled structure can be output as a PDB or XYZ file. Illustrative applications in the areas of ribozymes, viral kissing loops, viral internal ribosome entry sites, and nanobiology are presented.
Keywords: RNA secondary structure, RNA 3D structure, RNA modeling, molecular modeling, and nanobiology
Introduction
Currently, there is no algorithm for computing the entire 3-dimensional structure of a relatively large (greater than approximately 45 nts) RNA molecule from its primary structure (nucleotide sequence). But its Watson-Crick base-pairing secondary structure can, by a combination of experimental, phylogenetic, and energy-based computational techniques, be determined with reasonable accuracy. Successive base-pairs serve to identify 3D structure motifs in the form of double-stranded helices. These double-stranded helices (hereafter referred to as stems), constitute the secondary structure of the molecule. Base-pairing not occurring in stems relate to the tertiary structure of the molecule. The base-pairing structure of an RNA molecule thus gives a general idea of its 3D conformation in that it determines how some of the coordinates are related to one another. Extrapolating from this partial 3D information to a complete, specific, stereo-chemically correct 3D conformation constitutes the general problem addressed by the program.
Initial motivation for the program was primarily that of testing base-pairing information of large RNAs, however they it may have been derived. Thus, we wished to know if the information is 3D-realizable; that is, whether there is a stereo-chemically correct 3D model of a conformation consistent with this information. For instance, if one considers the proposed base-pairing scheme shown in Figure 6a of the Cricket Paralysis Virus internal ribosome entry site, it is not obvious that it is 3D-realizable. Our 3D model of it, depicted in Figure 6b, proves that it is, and thus provides a basis for detailed, atomic-level studies of structure-function issues.
Figure 6.
CrPV IRES modeling: (a) secondary structure and an associated cryo-EM density map (colored pink) of the IRES bound to the 40S ribosomal subunit of HeLa cells. Copied from Figure 1 of the publication (20); (b) the backbone of our 3D model derived from this secondary structure, colored to match the coloring of the three pseudoknots.
In their review article regarding the modeling of large nucleic acids, Malhorta et al. (1) describe the state-of-the-art computational methodology relevant to this problem. Of the methods described, three are notably relevant to our approach. One is by Major et al. (2) and Gautheret et al. (3), which treat the problem as one of constraint satisfaction using discreet conformational sets to search the conformation space. The method is categorized as an ‘all atom’ one because it retains a representation of each atom of the molecule instead of bundling a set of atoms, such as a nucleotide or entire helix into one unit called a ‘pseudoatom.’ Another of the methods is by Hubbard and Hearst (4, 5), which uses the techniques of distance geometry and molecular mechanics to refine a reduced representation (employing pseudoatoms) of an initially constructed 3D model using interactive graphics. The third is by Malhorta et al. (1), which also uses reduced representations but relies on specially designed energy functions to carry out refinements, such as molecular mechanics. All three produce excellent results, but are not primarily aimed at facilitating interactive exploratory work with large RNAs. (This is especially the case when using distance geometry because the associated computation complexity is of order N3 for structures of N atoms. We, thus, view the use of distance geometry as primarily applicable to small sections of large RNAs.) In addition, Burks et al. (6) describe a system, ERNA-3D, for constructing RNA 3D structures from secondary structures that require manual editing for positioning RNA secondary and 3D structural elements. Massire and Westhof (7) discuss a system, MANIP, that can assemble known RNA fragments into complex structures. Macke et al. (8) describe a system, NAB, that can be used to construct nucleic acid structures in a hierarchical fashion. A recent paper by Das and Baker (9) describes an interesting computationally intense approach (FARNA) for generating RNA 3D structures from sequence that was benchmarked against 20 different structures, most being less than 30 nts in length. The method relies on data derived from the ribosome structure for tri-nucleotide fragments and a simplified energy term. Reasonable results seem to be obtained with this method. Several of these methodologies are reviewed in (10).
Our approach concentrates on the generation of first order 3D approximations that are instantaneously produced from a secondary structure layout that is representative of a 3D unwinding and planar embedding of a 3D structure. Connectivities and structural component orientations are therefore based on this secondary structure 3D embedding; thus, allowing the generation of the first order approximation of a 3D structure layout. The 3D structure, thus defined, can then be interactively modified and refined with special tools. It does not use reduced representations and does not depend on a database of discreet conformation sets.
That we have pursued this kind of an approach is motivated by several considerations. First, complete and accurate base-pairing information for large molecules is more the exception than the rule; and even for exceptions, this information may not be sufficient. The conformation motifs that have been discovered (11, 12) increasingly attest to the need for considering not only non-canonical base-pairing but also base stacking other than that involved with the stacking of successive base-pairs, and even base intercalation. Features such as these do not readily follow from molecular mechanics simulations. Mostly, they need to be incorporated by hand. Part of the exploratory tools of the program is therefore devoted to importing motif coordinates directly into the 3D model being constructed. Additional tools enable interactively adding or deleting Watson-Crick base-pairs and the stacking or unstacking of stems. The latter tool is particularly important because adding or deleting the coaxial stacking of two stems can have a pronounced influence on the 3D conformation even though they may differ but little in free energy, and can thus serve to assess the validity of proposed secondary or tertiary base-pairing. Specific examples of such cases are given in the applications section. Figure 1 depicts a screenshot of an RNA2D3D interactive window. It illustrates both the secondary and 3D structures depictions of a “compacted” (see below) RNA molecule as well as the “picking” of two regions (green) that are about to be coaxially stacked on one another.
Figure 1.
Screenshot of an interactive window of RNA2D3D. Depicted are the “compacted” secondary structure of the RNA molecule being modeled (left side) and its 3D representation (right side). Also shown are two “picked” regions (green) that have been interactively selected for coaxial stacking. Coaxial stacking of the two regions would be realized by clicking on the button labeled “Accept” seen in the right panel.
Because the program is not intended to model folding pathways, it cannot automatically generate a fully satisfactory 3D model from just base-pairing information. An important case in point is that of transfer RNA. Following the generation of the initial 3D approximation, there is required two stem-stacking operations and then a rigid-body rotation of a stacked pair before the tertiary interactions can be invoked to get the characteristic L shape. Hands-on manipulations are required. If the goal were that of checking 3D-realizability, the program is well suited to the task. But it has nothing to say about how tRNA naturally folds to achieve its L-shape. It does not employ a 3D folding algorithm, even though its application may sometimes lead to the exposure of features suggestive of a folding pathway. Instead, emphasis is on efficiently generating a reasonable 3D approximation and then improving it with a variety of tools for conveniently introducing special structural features, which might be relevant to the structural stability or function of the specific RNA being modeled. In short, the program is not of the turn-key type in which an input file of the primary sequence and a secondary structure defined by a list of base-pairs is automatically converted to a stereo chemically correct 3D structure, even though it does have a stand-alone mode that yields an initial, approximate 3D structure in which all bond lengths and angles are correct but does not guarantee the absence of atom overlaps. Built-in examples illustrate the standalone and interactive modes, with special regard to the interactive editing tools, and thus serve as a tutorial for how to best use the program.
In the following there is described how the initial 3D model is generated and how it is subsequently improved. In addition, a few applications are described that further illustrate the use of some of the various tools that have been developed for performing operations conducive to exploratory modeling.
Methodology
Generating the Initial 3D Model
Our method for generating an initial 3D model, regarded as a first-order approximation, starts with an appropriately scaled 2D drawing of the molecule’s secondary structure. The drawing is generated from an input file containing the primary sequence and a list of the base-pairs. (This information is accepted in all the generally used formats). Nodes of the drawing representing successive nucleotides are connected by a line of fixed length L1 and nodes representing a base-pair are connected by a line of fixed length L2. Using a representative backbone atom, such as P, L1 is the distance between successive P atoms in an A-form helix, and L2 is the distance between the two P atoms of a base-pair. A simple example not involving coaxial stem stacking is shown in Figure 2a, and an example involving stem stacking is shown in Figure 4a. The 2D drawing algorithm we have devised automatically detects pseudoknots from the base-pairing list defining the secondary structure and renders the two halves of a pseudoknot as coaxially stacked stems. Additional stem stacking, as might be required for two neighboring stems of a branching loop, is an editing option that can be invoked subsequent to the initial 2D drawing.
Figure 2.
Generation of initial 3D model for the hammerhead molecule: (a) the 2D scaled, secondary-structure drawing. Note that every tenth based is indicated in bold white; (b) the planar 3D embedding of the 2D drawing showing the insertion of the 3D nucleotides; (c) the initial 3D model, obtained by transforming each stem of the planar 3D model into an A-form helical-duplex subject to the constraint of maintaining the planar structure conformation of the single strands. For illustrative convenience the backbone atom used in figures (a) and (b) is C1′ and that in (c) is P.
Figure 4.
Further improvement of 3D model using the stem-stacking, segment-positioning, and energy-refinement tools: (a) 2D redrawing of Figure 3a upon stacking of the red and yellow colored stems. (b) the corresponding stacked 3D model; (c) 3D result of next applying the segment-positioning tool to the white colored segment, and then energy-refinement as described in the text. Nucleotide coloring is: A, magenta; C, cyan; G, green; and U, yellow. Backbone curve passes through the P atoms.
Using this drawing as a backbone template, the corresponding nucleotides are inserted according to the following rule which capitalizes on published x-ray determined 3D coordinates for each of the four nucleotides, (A, C, G, U), and for the base-pairs (A-U), (C-G), and (G-U). Thus, at each node pair representing the backbone atoms of a base-pair we position the corresponding base-pair, as a rigid body, so that the plane determined by its two bases makes a fixed angle X with the template plane. Unpaired nucleotides are then positioned, also as rigid bodies, at their respective nodes such that the orientation of each relative to the template plane is the same as that of a paired nucleotide and such that the plane determined by its base bisects the angle made by the lines connecting the nucleotide to its preceding and succeeding nucleotides (see Figure 2). As a matter of computational convenience we choose the fixed orientation angle X to be 90 degrees. It is not critical.
This 3D insertion of the nucleotides into the planar backbone template gives it a 3D embedding that roughly approximates what one would get by unwinding the stems of the full 3D structure and laying it out flat. Suggestively, a rewinding of the stems while maintaining the template conformation of the inter-connecting single strands might then give a reasonable picture of the original 3D conformation. This simple idea is the basis of our method for generating 3D conformations from secondary structure information. Overall, it consists of the following three steps illustrated in Figures 2a, 2b, and 2c.
Create a scaled, planar backbone template representing the secondary structure information, including that defining pseudoknots.
At each node-pair of the template corresponding to a base-pair, insert a 3D representation of the base-pair; and at each node representing a free nucleotide insert its 3D representation. These insertions are to be made such that the base plane of each nucleotide is perpendicular to the template plane.
Convert the planar form of each stem into a 3D helical A-form subject to the constraint that if a base-pair delimits a loop (inner, hairpin, or branching) the base-pair and the delimited loop be treated as a rigid-body.
Our procedure for carrying out step 3 is based on using relative coordinates in terms of which rigid-bodies are locally defined. Thus, when the last base-pair of a stem is repositioned to obtain the A form of the stem, the constraint requires that the loop it delimits (hairpin or branching) be also repositioned because they jointly constitute a rigid-body. An initial computation is therefore to compute coordinates of each loop relative to one of the nucleotides of its delimiting base-pair, say the 5’ nucleotide, using the absolute coordinates stored in the template. Any change in the absolute coordinates of the reference nucleotide can therefore be used to compute the required change in the absolute coordinates of the loop nucleotides. Base-pairs in a loop from stems emanating from the loop are thus repositioned; and since such base-pairs are each the first base-pair of a sequence of contiguous base-pairs, there is then the requirement of being able to generate an A-form double-stranded region from the absolute coordinates of its first base-pair. The A-form helical geometry furnishes the required information. Allowing for the general case of a stem being composed of sets of contiguous base-pairs separated by inner loops, we implement its generation by sequentially generating these regions and the delimited loops. We use the 5′ and 3′ nucleotides of a region’s first base-pair to generate its 5′ and 3′ strands, respectively.
In terms of stems as basic units, the secondary structure of an RNA molecule is hierarchically organized; that is, a stem delimits either a hairpin or branching loop, and when the latter is the case, the first stem of each branch is said to be included in its loop. Numerically ordering stems according to the primary sequence position of the 5′ nucleotide of the first base-pair, each stem has associated with it a list of its included stems. The constrained conversion of all the stems of an RNA molecule from its planar 3D template can thus conveniently be programmed as a recursive procedure; that is, the procedure for converting a stem calls upon itself to also convert each of the included stems. Similarly, the 2D drawing of the secondary structure is done recursively.
The numerical calculation of repositioned coordinates is made efficient by taking advantage of the fact that the absolute 3D coordinates of all the atoms of a nucleotide can be obtained from those of any three of its atoms, designated as a reference triad. The triad chosen for nucleotides C and U is (C1′, C4′, N1); for A and G it is (C1′, C4′, N9). Coordinates for the other atoms of the nucleotide relative to the reference triad are then defined as relative to an ortho-normal frame constructed from the triad. The computational advantage of reference triads is that when a nucleotide is moved we need only keep track of its reference triad. In the recursive procedure we use for converting all the stems, only reference triads are moved. Absolute coordinates for all the atoms are updated upon conversion of all the stems. At this point there also is handled the problem of dangling single strands, which occur if the first or the last nucleotide is unpaired. We view such a single strand as a base-stacked extension of the delimiting paired nucleotide and thus provide it with an A-form helical conformation.
The time to complete all three steps of the procedure is essentially instantaneous. On a 1.3 Ghz PC, less than one second is required for the E. Coli 16S ribosomal subunit. It has 1542 nucleotides.
Along with help documentation provided with the program, there is also provided a pull-down menu labeled (2d→3d) mode with the items automatic and movie. The latter option gives the user an interactive view of step 3 being carried out one region or one stem at a time.
Improving the Initial 3D Model
The procedure for generating the initial 3D model insures that all the stems have the correct helical A-form but only an approximate relative positioning. The spatial positioning of the stems is largely dictated by the conformation of the inner and branching loops, which we have chosen to be planar. Partial correction of this deliberate shortcoming can be achieved with molecular mechanics energy refinement techniques or a localized application of the constraint satisfaction methodology (2). But we prefer to be initially guided by the consideration that internal and branching loops have evolved to achieve very specific relative positioning of the stems and that one of the most important folding steps following (or coincident with) stem formation is that of coaxial stacking of stems across branching loops. Accordingly, we have incorporated several features specifically designed to facilitate the generation and assessment of potential stem stacking alternatives within such loops; and with regard to inner loops, there also is provided optional stacking across the loop, that is, the delimiting base-pairs of the loop can be stacked.
Referring to Figure 2c of the initial 3D model for the hammerhead molecule, it is clear that the single strands do not have correct conformations. The imposed planarity condition introduces the artifact of distorted O3′-P bond lengths and O3′-P-O5′ bond angles. This is easily corrected with a locally applied energy-refinement (later described); but, generally, this will only result in a local improvement of the model. What is needed is an adjustment that makes the model more compact in the sense of having fewer unconstrained degrees of freedom. We, therefore, invoke the compacting tool, which automatically makes a pseudo-extension of the stems into the interconnecting single strands by adding pairs of nucleotides to a stem. Pseudo base-pairs are thus created at the expense of the single strand nucleotides. The extension algorithm employed insures maximal recruitment of free nucleotides from inner and branching loops. Figure 3a is a redrawing of Figure 2a, showing the results of the compacting operation, and Figure 3b is the corresponding initial 3D model. The pseudo base-pairs are highlighted by the use of a stippled instead of a solid connecting line. Notable is the significantly increased 3D compactness, the creation of a real base-pair (A-U), the potential non-canonical base-pairs (A-G), and the potential stem stacking in the branching loop whose stems we have colored red, white, and yellow. The red stem is the delimiting stem of the loop and it can potentially coaxially stack with either of the other two. On the basis of known base-pair to base-pair stacking energies, (U-A) with (C-C) of the red to white stem stacking and the (U-A) with (U-A) of the red to yellow stem stacking, it is difficult to assess which of the two cases would be energetically favored. This is because of the unknown influence that the non-canonical base-pairs (A-G) would have on the stacking of the red and yellow stems. On the other hand if one considers opening the (C-C) base-pair in the white stem, there would then be a bulging C nucleotide potentially interfering with the stacking of the red and white stems. We choose the red to yellow stacking case as the preferred conformation to further explore. Using the stem-stacking tool, which only requires graphically picking the two stems to be stacked, we stack the red and yellow stems and, thus, obtain the structures shown in Figure 4a and 4b. What remains to be determined is a stable relative position of the white stem.
Figure 3.
Improving the initial 3D model with the compacting tool: (a) 2D redrawing of Figure 2a; (b) the corresponding improved 3D model. Stems colored red and yellow have potential for stacking. Pseudo base-pairs are distinguished by stippled instead of solid complementary lines.
As imposed by the 2D drawing algorithm, this stem is approximately in the same plane as the stacked stem pair and oriented anti-parallel to it. But since experience with similar cases favors a parallel orientation, we enforce it with use of the segment positioning tool. With this tool a segment can be defined by picking its end nucleotides and then arbitrarily translating and rotating it relative to a moving frame having a fixed orientation. In this particular case, the segment we define is colored white and it is rotated 180 degrees about an axis roughly perpendicular to the stacked stem-pair. A local, small repositioning of this rotated segment is next performed to fix any overlapping of the atoms at the junction of the three stems that may (and usually does) result from a large repositioning operation. Global energy minimization is now employed to determine a stable conformation of the segment relative to the stacked pair. Computational efficiency is obtained by first doing the minimization subject to the constraint that only the predefined segment is allowed to move. This provides a correct conformation to the segment and thus a reasonable optimization of the position of the third stem relative to the stacked pair. Local energy refinement corrects the hairpin loop of the stacked stem pair; and using other built-in tools the potential non-canonical A-G base-pairs are given a sheared hydrogen-bonding conformation. Unconstrained global energy minimization is the final step. The overall result is shown in Figure 4c. Relative to the catalytic pocket, its features are nearly identical to those of the x-ray determined ground state of the hammerhead ribozyme (13, 14).
The energy-refinement tool, which utilizes the Tinker package (15), offers both minimization and dynamics options that primarily are intended to provide reasonably good conformations as starting points for more serious refinement efforts. Thus, only Na+ counter-ions are used to neutralize the phosphates, and the environment dielectric is the simple one of distance-dependency.
Energy minimization is offered in two modes (using the ff99 force field): local and global. Either of these modes can be run using two default convergence values. A coarse-grained minimization terminates when the gradient rms is less than 1.0. A fine grained minimization terminates when the gradient rms is less than .1. In the local mode, any segment or group of non-overlapping segments can be energy-minimized with the end-nucleotides held fixed and treated as an isolated entity; in the global mode the rest of the molecule is brought into play. The local mode is especially useful for rapidly correcting the single-strand distortions introduced by our use of a planar 3D template. It is implemented for interactively controlling the automatic or selective correction of all the single-strands. In the global mode the special single-strand procedure treats them all at once, allowing them to interact with one another and the rest of the molecule that is held fixed.
Some Applications
As reported in (16), an early application of the program was to the modeling of the kissing loop RNA structure that occurs in the genome of the Aids virus and plays an essential role in its replication. The objective was to identify atomic-level features responsible for its stability. Another early, but still on-going investigation, is the modeling of internal ribosome entry sites (IRES) that are used by certain RNA viruses to sequester a host cell’s translation machinery for their own replication. Here, the objective is to identify atomic-level features essential to the sequestering mechanism. In the following we outline these two modeling efforts in order to further illustrate the use of some tools of the program that were specifically developed to deal with these kinds of problems and that are readily adapted to others. For instance, our technique for readily constructing kissing loops has found use in the modeling of nano-structures that use kissing loops as a means for joining together RNA monomers to obtain higher order building blocks (17). And with regard to IRES modeling, which has required tools for efficiently generating diverse pseudoknots, these have found use in the atomic-level modeling of telomerases (18, 19).
In addition to the examples to be described, most of our applications have benefited from specially designed features relating to molecule complexity, visualization, and interactivity. For instance, the branching and pseudoknot complexity of some of the RNAs prompted the need for three different levels of modeling: MOLECULE, BRANCH, and SUBSET. The latter two are conveniently defined in terms of the notion of a segment, which is simply a collection of nucleotides whose primary sequence positions constitute a closed interval; that is, segment [x,y] consists of all the nucleotides whose sequence positions lie between and include positions x and y. Noting that a stem’s base-pairs are each distinguished by the positions of their 5′ and 3′ nucleotides, the first and last base-pairs of a stem are, respectively, the ones having the smallest and largest 5′ positions. Accordingly, a BRANCH is then defined as a segment [x,y] in which x is the 5′ position and y is the 3′ position of the first base-pair of a stem; and a SUBSET is defined as a collection of segments no two of which overlap. Nearly all the operations that can be performed at the MOLECULE level can also be performed at the BRANCH and SUBSET levels.
Of the features designed to enhance visualization and interactivity, a basic one is the simultaneous display of the 2D and the initial 3D structure in separate windows, each of which has its own rendering and manipulation tools. The selection of two stems to be stacked can be done by picking them in either the 2D or 3D window (see Figure 1), as is the selection of segments and subsets. Either window can be keyed into occupying the full screen for enhancing viewing and selection. And to facilitate the comparison of two independent models there is provided two storage arrays, designated as A and B models, either of which can be selected for storing the generated 2D and 3D data of a molecule. Any A-model can then be simultaneously viewed with any B-model at either the 2D or 3D level. Selection of a subset from each model is thereby also facilitated for purposes such as local alignment, subset substitution, and merging. To be later described, the model arrays also provide the means for dealing with multiple 2D structures of the same primary sequence. The corresponding initial 3D models are automatically generated, and these can selectively be viewed, as frames, in an interactive movie.
Kissing Loops
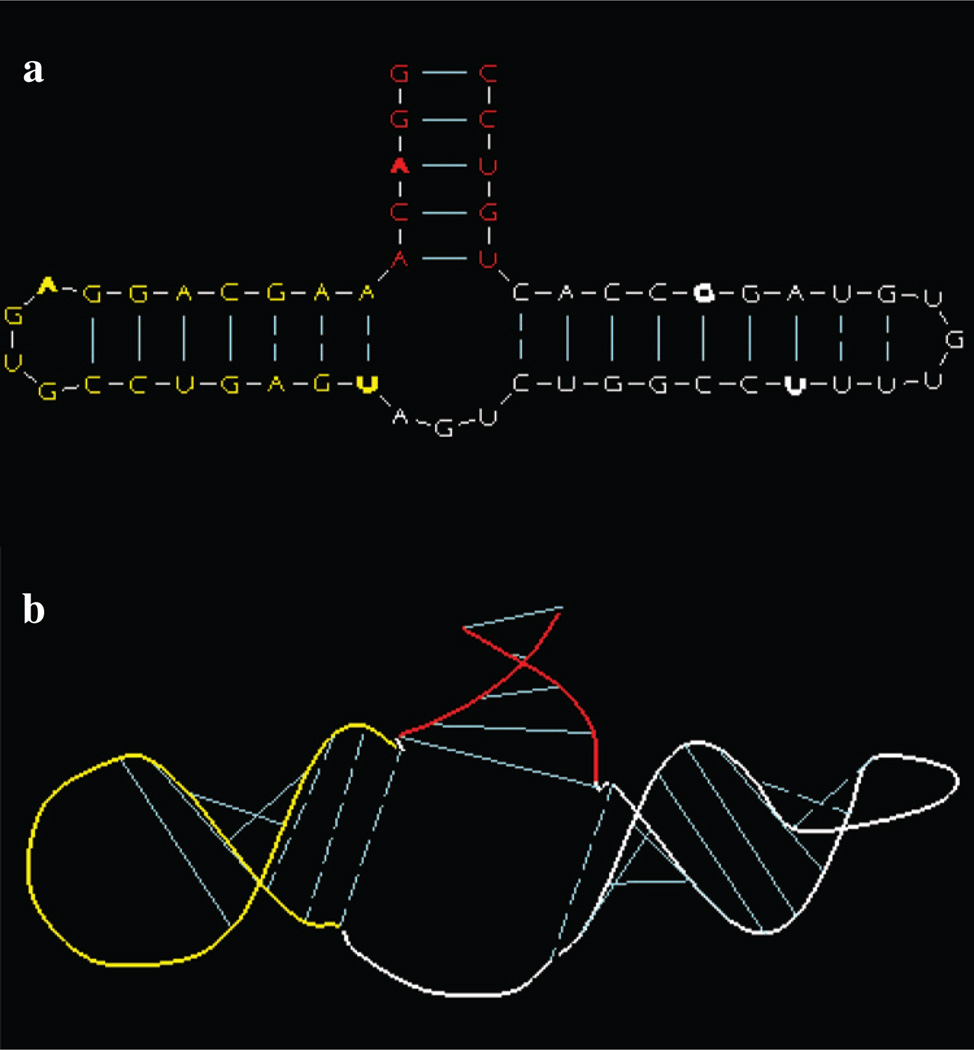
The dimerization of two copies of the HIV-1 genomic RNA is thought to be involved in several steps of the retroviral life cycle, and it has been shown that the dimerization is initiated by a structure termed a kissing loop. As shown in Figure 5, the 9nt kissing loop contains a palindromic 6nt sequence that forms Watson-Crick base-pairs at the kissing site in HIV-1. Deriving a 3D model of the kissing structure involves two major steps: (i) build two copies of the stem-loop structure and (ii) piece them together so that the two palindromic sequences form a stable double stranded helix.
Figure 5.
Kissing-loop modeling: (a) 2D scaled drawing of stem-loop structure. Green colored base indicates nucleotide picked to form kissing loop complex (see d); (b) initial 3D model; (c) improved 3D model obtained by giving the loop palindrome an A-form helix structure with the helix-extending tool and then applying energy-refinement; (d) the kissing complex obtained by duplicating the 3D stem-loop model and then connecting the two 3D models with the base-pair sharing tool.
In Figure 5a there is shown our 2D rendering of the stem-loop secondary structure and Figure 5b is the corresponding initial model. Simple energy-refinement of the hairpin loop does not induce the palindrome to assume a helical conformation as might occur with a more elaborate refinement consisting of both energy minimization and molecular dynamics. We, therefore, invoke the helix-extension tool whereby any nucleotide segment can be given an A-form conformation with either the 5′ or 3′ nucleotide used as the reference nucleotide. After a few attempts aimed at determining the segment that both includes the palindrome and yields a stable conformation of the loop, we find that the segment GUGCACAC with the 3′C paired nucleotide as the reference nucleotide is a suitable one. The helix-extension is followed by an energy minimization to fix the positioning of the two free nucleotides A and G that precede the segment. The resulting structure is shown in Figure 5c. With a favorable 3D conformation for one half of the kissing complex at hand we now duplicate it with the 3D-copying tool to obtain two structures to be joined to form the palindrome duplex. For this we use the base-pairing tool.
Given two separate RNA models, the program provides three alignment options: global rms superposition, local rms superposition, and relative positioning so that a specified base-pair is shared. The base-pairing tool implements this third option. Choosing any one of the presumptive base-pairs in the loop by picking the corresponding two nucleotides from the two copies that were generated, causes it to be formed. And because the two helical strands of the presumptive helical duplex were already constructed, all of the other base-pairs will also be formed. The result is shown in Figure 5d. Notable is the perfect coaxial stacking of the two copies.
Internal Ribosome Entry Sites
Certain RNA viruses sequester a host ribosome to initiate the translation of their genome. The sequestering is done in a manner independent of the 5′ mRNA signal normally used by the host to initiate translation. It is accomplished by means of a special folded RNA structure in the 5′ un-translated region of the virus genome, called the Internal Ribosome Entry Site (IRES). It is used to bind to the 40S ribosomal subunit and to insure that the start codon of its genome becomes strategically placed at the P site of the 40S. Of the known IRESs, the Cricket Paralysis Virus IRES is exceptional because it does not require any factors, such as eIF3, to initiate its translation. A cryo-EM study (20) of its binding to the 40S ribosomal subunit has enabled associating regions of the secondary structure to the density map of the IRES, as shown in our Figure 6a; but there is still required an atomic-level model of the interaction. To this end we constructed the 3D model shown in Figure 6b that could be used as the starting structure for docking studies relevant to identifying atomic-level interactions. The modeling was particularly interesting because of the three pseudoknots involved. The program readily detected them and produced an initial 3D model, which only required a minor segment repositioning prior to a global energy minimization that in turn resulted in a stereo-chemically correct 3D structure.
By way of contrast in methodologies, the recently reported cryo-EM study (21), which now includes a detailed docking effort, features obtaining independent models of the three pseudoknots and of the remaining stem-loops and helices, and then piecing them together so that they individually fit select portions of the IRES cryo-EM electron density map and are connected by stereo-chemically correct single strands. As reported in Supplementary Methods of (21), “Atomic models for helices and stem-loops of the CrPV IRES were generated using the eRNA-3D software and subsequently docked into the cryo-EM density map using the O program. Based on this placement of the secondary structure elements, structural modeling of the CrPV IRES was done using the MANIP package.” The structural modeling referred to was primarily that of using crystal structures of analogous pseudoknots for determining the 3D representations of the CrPV pseudoknots. This is detailed in the Supplementary Materials of (21). The 3D coordinates of the bound CrPV that were, thus obtained are reported as PDB ID 2NOQ. Comparing this structure to ours, we find that although the overall shapes are similar, the relative positioning of two of the pseudoknots (PKII and PKIII) are quite different. This is not surprising because our modeling was not constrained to fit a density map. It is, therefore, reasonable to attribute the difference to the flexibility of the IRES. From the methodology point of view, it is important to note that our approach starts off by first generating a 3D model of the IRES that is consistent with the given secondary structure, including pseudoknots. Only then would it be used to fit a density map. That our approach would give comparable results in the revealing of points of interaction with the ribosome is not predictable. It may indeed require using auxiliary experimental information, such as crystal structures applicable to select portions. If so, they can usually be readily incorporated.
Alternative Secondary Structures
Several secondary structure generating programs produce more than one solution for a given primary structure (10, 22). For instance, MFOLD (23), which generates secondary structures using a dynamic programming algorithm, furnishes sub-optimal solutions in addition to the optimal one. And MPGAfold (24–28), which generates secondary structures using a genetic algorithm, furnishes multiple solutions potentially significant as intermediates to a final conformation that may be analyzed by STRUCTURELAB (29–32). In order to facilitate comparative analysis, as well as that of corresponding 3D structures, the input secondary structure file allows the listing of more than one secondary structure for the same primary structure. Our program automatically generates an initial 3D structure for all members of the list and stores them as frames, with each frame consisting of both the secondary and its corresponding initial 3D structure. These frames may then be displayed as an interactive movie. And since the frames may be generated in either the A or B model array, generating them in both gives the capability of simultaneously displaying frame pairs for visually comparing their 2D or 3D members.
Multiple 3D Structures and Nanobiology
As noted, the foregoing kissing loop application is an example of modeling the joining of two 3D monomers, via base-pairing, to obtain a 3D dimer. Implementation of this feature is a special case of 3D alignment whereby two 3D monomers, one from the model A array and the other from the model B array, can be superimposed relative to an arbitrary common subset or made to be connected by base-pairing. Providing this base-pairing capability to n-mers with n > 2, as required for some nanobiology applications, is achieved with an additional model array termed the C array. Each of its frames is used to represent a monomer, and intermonomer connectivity via base-pairing is specified with an input file. As next described, we use this array to model the RNA structures reported in (17), in which the monomers have three base-pairing connection points, two of which are used to construct 4-mers called tectosquares and the third is used to join tectosquares to obtain higher order structures.
The experimental results reported in (17) strongly indicate that tectosquares can be constructed under suitable experimental conditions. Labeling its monomers as A, B, C, and D, there are no restraints to the formation of a tetramer A-B-C-D, but for the formation of a tectosquare it is required that the tetramer have a conformation conducive to the stable formation of the cyclic bond D-A. The extent to which this is satisfied should, therefore, be revealed by our modeling of the monomers and of the subsequent tetramer A-B-C-D.
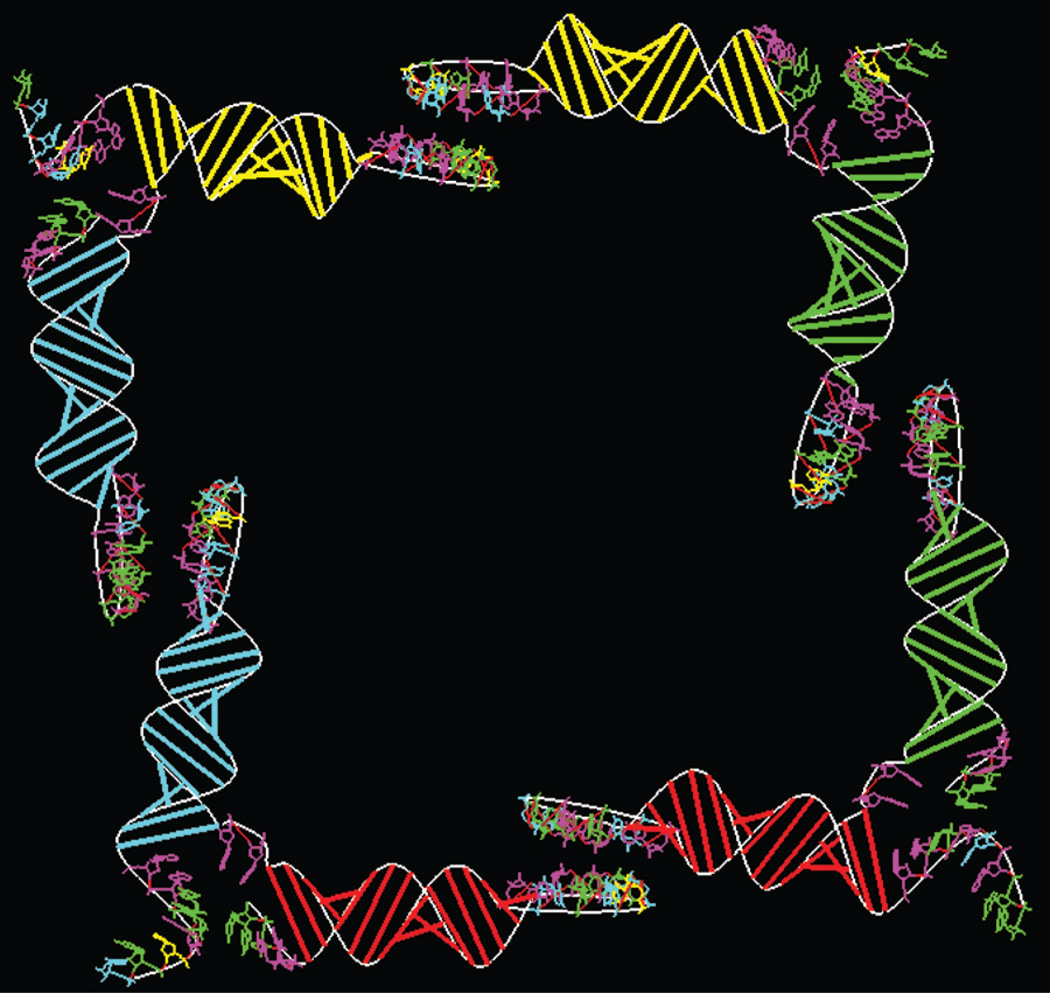
Starting with the primary structures of the tectoRNAs A2′s, B6′s, C4′s, and D1′s defined in (17) as the monomers for tectosquare LT20, we generated corresponding secondary structures with the MPGAfold program (24–28) and then used RNA2D3D to obtain initial 3D structures for them. Except for the 3′ single strand of 8 nucleotides, termed the tail of the monomer, and for the kissing loops, all four monomers are identical. All the initial 3D structures will, therefore, be identical. They are displayed in Figure 7, colored and spatially disposed to describe how they would form the four corners of a tectosquare once they become connected via base-pairing of the stem-loops having the same color. Notable is that even in the initial 3D structure of the monomer its two stems seem to already form a right angle as would be required of a corner of a square. This is partly due to how the secondary structure is initially laid out; but it is also the case when our initial 3D structure is subjected to extensive molecular dynamics and energy minimization that insure a stable conformation of the junction connecting its two stems (data not shown). The junction conformation, thus, obtained differs significantly from that for which its primary structure was specifically designed, namely that of an 11 nucleotide structural motif called “the right angle (RA) motif,” which connects adjacent helices in the ribosome (33, 34).
Figure 7.
Layout of the four initially modeled tectoRNAs (A, B, C, D) to be interconnected via base-pairing of their stem-loops having the same color. No refinement of the stem junctions within a monomer or of the kissing loops has been implemented. The nucleotides shown are colored: A is magenta, G is green, U is yellow and C is cyan.
Incorporation of the ribosome RA motif into the initial 3D monomer was done with our tool for importing 3D coordinates whereby a subset in one 3D structure can be replaced by a homologous (same primary sequence) subset of another 3D structure. In this case the subset is GCAAGU. Next, the putative kissing loops were shaped with the helix-extension tool in preparation for their mutual base-pairing. The resulting monomers are redrawn in Figure 8.
Figure 8.
Layout of the four refined modeled tectoRNAs. The kissing loops have been shaped to accommodate base-pairing and the stem junctions of each monomer have been given the ribosome right-angle motif.
Adopting the terminology that A, B, C, and D are, respectively, the monomers A2′s, B6′s, C4′s, and D1′s, we use the kissing procedure to first obtain the dimer A-B. In this operation the absolute atom coordinates of monomer B become changed to correspond to those of monomer A. We then join C to B to obtain the trimer A-B-C, and then D to C to obtain the tetramer A-B-C-D. The result is shown in Figure 9a, which clearly indicates that closure of the tetramer was not obtained even though coaxial stacking of the kissing stems was rigorously implemented. The large distance between the D and A kissing loops argues that their coming together in experimental conditions must be mitigated by structural flexibility or by distortions in the motifs. As a potential remedy we, therefore, sought an alternative conformation of the right-angle junction that would give a significantly better approximation to a cyclic conformation.
Figure 9.
Resulting 3D conformation of the interconnected complex A-B-C-D. (a) Notable is the significant departure from a closed (cyclic) conformation even though the ribosome right-angle motif has been incorporated and the kissing stems are perfectly coaxially stacked; (b) the conformation of (a) corrected by changing the conformation of the right-angle motif. The 5′ stem was rotated by 26 degrees about its cylindrical axis in the four monomers and also slightly translated while maintaining the right-angle feature. See text for details. Achieved is full closure.
Global molecular dynamics and energy minimization, suitably constrained to insure maintenance of the kissing conformations was one possible approach. But since there are about 12,000 atoms in a tectosquare, considerable computation effort is involved. Instead, we concentrated on finding parameters which determine the A-B-C-D conformation that results from the four successive kissing operations and whose variation does not affect the right angle feature of the corners. Rotation of either stem of a monomer relative to its cylindrical axis fits this requirement. Fortuitously, the segment positioning tool can provide rotation of a stem about its cylindrical axis. As the segment we chose nucleotides 1–45. It includes all of the 5′ stem and the two free adenine nucleotides connecting the two stems. If Q is the midpoint of the line connecting the P atoms of nucleotides 1 and 45 and COM is the center-of-mass of the segment, then the line connecting Q and COM happens to nearly match the 5′ stem cylindrical axis. Interactive rotation is applied to each of the four monomers, A, B, C, and D. The kissing loop conformations were then restored. In this way, it was determined that a rotation of about 26 degrees applied to each monomer provided significant improvement. And when aided by mild, rigid translations of the segment, which preserve the right angle relative conformation of the two stems, an exact cyclic conformation was obtained. It is shown in Figure 9b.
Our modeling, thus, proved that there exists a stereo-chemically correct model of the cyclic conformation for this particular set of tectoRNAs. Unfortunately, the same sort of adjustment does not work well when tried for some of the other tectosquares proposed in (17). As suggested by conversations with Luc Jaeger, we therefore, next concentrated on actual design parameters such as stemlength. By editing the file defining the primary and secondary structure, we experimented by adding and subtracting base-pairs to the 3′ stem of the LT20 RNAs. The addition of exactly one base-pair yielded a significantly improved model that appears to be conducive to cyclic closure. It is shown in Figure 10.
Figure 10.
Depiction of the significant improvement towards cyclization of the tectosquare by the insertion of an extra base-pair in each of the 3′ stems of the four RNA tectosquare building blocks.
A further effort was then directed at the modeling of tectosquare patterns, such as the pattern LT17–20 described in (17). It consists of four tectosquares linked sequentially via base-pairing of their tails. Automating the kissing required in the tectrosquares and in the linking tails, this pattern is efficiently generated from the sixteen preformed monomers contained in the C model array. The result is shown in Figure 11a, which depicts a highly non-planar construct that is far from being cyclic, - contrary to what seems to be experimentally observed. Automating stem-length and tail-length adjustment applicable to the entire structure quickly revealed that the optimal adjustment consisted of increasing the monomer 3′ stem-length by one base-pair and decreasing tail-length by one base-pair. The result of this adjustment is shown in Figure 11b. The planarity is considerably improved as is the favoring of closure in both the tectosquares and in the pattern. This attests to how 3D modeling that is coupled to the efficient adjustment of key structure parameters can contribute to the design of tecto RNA structures. However, it should be noted that because the structure parameters of stem and tail length are very coarse grained, the modeling of complete cyclic closure, if required, will require the use of more finely grained parameters, such as that of stem rotation about its helical axis that we first employed in the initial modeling effort. It is not guaranteed to yield complete closure in all cases, but does seem to improve the model over what can be achieved with just the stem-length and tail-length tools.
Figure 11.
Modeling of the tectosquare pattern LT17–20 described in reference (17). It consists of four tectosquares sequentially coupled by their tails to form a cycle: (a) the initial model without stem-length and tail-length adjustments; (b) the modified model with the optimal adjustment of one base-pair added to the 3′ stem of each monomer, and of one base-pair deleted from the coupling tails.
Under development is an extension of all our RNA modeling to that of DNA so that the design of DNA nano-structures might also benefit by an efficient search of optimal 3D geometries.
Implementation Specifics and Availability
The program is written in the C language. It uses OpenGL for graphics, and the TINKER molecular modeling program for the energy refinement computations. It runs on PCs using the Red Hat Linux operating system and on SGI platforms using the IRIX operating system. It is available upon request, separately or as a component of STRUCTURELAB. A comparable version that will include a DNA modeling component is also under development.
Acknowledgements
Initial computational assistance was provided by the Molecular Graphics Laboratory of the University of California, San Francisco. Use of the molecular graphics program MIDAS greatly aided initial stages of the development. Of the same institute, we would also like to acknowledge the early help of Dr. Peter Kollman’s staff in the use of AMBER for energy minimization and molecular dynamics computations.
Dr. Shu-Yen Le of this laboratory generously provided early secondary structure data for the initial modeling of internal ribosome entry sites and also assisted in assessing the interactive utility of the program. Dr. Danielle A. Konings provided invaluable advice concerning initial design considerations, especially with regard to the modeling of pseudoknots. Dr. Yaroslava Yingling and Wojciech Kasprzak of this laboratory tested the program extensively and applied it to specific applications.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Also to be gratefully acknowledged is the Advanced Biomedical Computing Center, NCI, Frederick for computer time and staff support.
Footnotes
Publisher's Disclaimer: For full terms and conditions of use, see: http://www.tandfonline.com/page/terms-and-conditions esp. Part II. Intellectual property and access and license types, § 11. (c) Open Access Content
The use of Taylor & Francis Open articles and Taylor & Francis Open Select articles for commercial purposes is strictly prohibited.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae, and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand, or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
References
- 1.Malhotra A, Gabb HA, Harvey SC. Current Opinion In Structural Biology. 1993;3:241–246. [Google Scholar]
- 2.Major F, Turcotte M, Gautheret D, Lapalme G, Fillion E, Cedergren R. Science. 1991;253:1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- 3.Gautheret D, Major F, Cedergren R. J Mol Biol. 1993;229:1049–1064. doi: 10.1006/jmbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- 4.Hubbard JM, Hearst JE. J Mol Biol. 1991;221:889–907. doi: 10.1016/0022-2836(91)80182-t. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard JM, Hearst JE. Biochemistry. 1991;30:5458–5465. doi: 10.1021/bi00236a019. [DOI] [PubMed] [Google Scholar]
- 6.Burks J, Zwieb C, Muller F, Wower I, Wower J. BMC Mol Biol. 2005;6:14–31. doi: 10.1186/1471-2199-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massire C, Westhof E. J Mol Graph Model. 1998;16:197–205. doi: 10.1016/s1093-3263(98)80004-1. 255-197. [DOI] [PubMed] [Google Scholar]
- 8.Macke T, Svrcek-Seiler W, Brown R, Case D. The NAB molecular manipulation language. Scripps Research Institute; 2006. [Google Scholar]
- 9.Das R, Baker D. Proc Natl Acad Sci USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Noller HF. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 12.Leontis NB, Westhof E. Curr Opin Struct Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 13.Scott WG, Finch JT, Klug A. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 14.Hermann T, Auffinger P, Westhof E. Eur Biophys J. 1998;27:153–165. doi: 10.1007/s002490050121. [DOI] [PubMed] [Google Scholar]
- 15.Ponder J. Tinker - Software tools for molecular design - Version 4.2. 2006. http://dasher.wustl.edu/tinker/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattabiraman N, Martinez HM, Shapiro BA. J Biomol Struct Dyn. 2002;20:397–412. doi: 10.1080/07391102.2002.10506858. [DOI] [PubMed] [Google Scholar]
- 17.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 18.Yingling YG, Shapiro BA. J Mol Graph Model. 2006;25:261–274. doi: 10.1016/j.jmgm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Yingling YG, Shapiro BA. J Biomol Struct Dyn. 2007;24:303–320. doi: 10.1080/07391102.2007.10531238. [DOI] [PubMed] [Google Scholar]
- 20.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 22.Mathews DH, Turner DH. Curr Opin Struct Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro BA, Bengali D, Kasprzak W, Wu JC. J Mol Biol. 2001;312:27–44. doi: 10.1006/jmbi.2001.4931. [DOI] [PubMed] [Google Scholar]
- 25.Shapiro BA, Navetta J. J Supercomputing. 1994;8:195–207. [Google Scholar]
- 26.Shapiro BA, Wu JC. CABIOS. 1996;12:171–180. doi: 10.1093/bioinformatics/12.3.171. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro BA, Wu JC. CABIOS. 1997;13:459–471. doi: 10.1093/bioinformatics/13.4.459. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro BA, Wu JC, Bengali D, Potts MJ. Bioinformatics. 2001;17:137–148. doi: 10.1093/bioinformatics/17.2.137. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro BA, Kasprzak W. J Mol Graph. 1996;14:194–205. doi: 10.1016/s0263-7855(96)00063-x. 222-194. [DOI] [PubMed] [Google Scholar]
- 30.Kasprzak W, Shapiro B. Bioinformatics. 1999;15:16–31. doi: 10.1093/bioinformatics/15.1.16. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro BA, Kasprzak W, Grunewald C, Aman J. J Mol Graph Model. 2006;25:514–531. doi: 10.1016/j.jmgm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Linnstaedt SD, Kasprzak WK, Shapiro BA, Casey JL. Rna. 2006;12:1521–1533. doi: 10.1261/rna.89306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 34.Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]