ESCRT filaments wrap helically around liposomes and assemble into various helical structures in vitro. Dividing Sulfolobus cells further exhibit a thin, dynamic belt coating division furrows. Together these data suggest that spiraling filaments are involved in membrane scission.
Abstract
The endosomal-sorting complex required for transport (ESCRT) is evolutionarily conserved from Archaea to eukaryotes. The complex drives membrane scission events in a range of processes, including cytokinesis in Metazoa and some Archaea. CdvA is the protein in Archaea that recruits ESCRT-III to the membrane. Using electron cryotomography (ECT), we find that CdvA polymerizes into helical filaments wrapped around liposomes. ESCRT-III proteins are responsible for the cinching of membranes and have been shown to assemble into helical tubes in vitro, but here we show that they also can form nested tubes and nested cones, which reveal surprisingly numerous and versatile contacts. To observe the ESCRT–CdvA complex in a physiological context, we used ECT to image the archaeon Sulfolobus acidocaldarius and observed a distinct protein belt at the leading edge of constriction furrows in dividing cells. The known dimensions of ESCRT-III proteins constrain their possible orientations within each of these structures and point to the involvement of spiraling filaments in membrane scission.
INTRODUCTION
The endosomal-sorting complex required for transport (ESCRT) is a set of proteins that bend and separate membranes. ESCRT-mediated membrane scission is a highly conserved process involved in cytokinesis in eukaryotes and Archaea, multivesicular body (MVB) formation, and viral budding (reviewed in Wollert et al., 2009b; Hurley, 2010; McCullough et al., 2013). Based on in vitro models for MVB formation, ESCRT-I and -II deform the membrane to form buds, ESCRT-III cleaves the buds, and Vps4 (written “Vps4” in yeast and “VPS4” in humans), a AAA+ ATPase and the only enzyme in the system, recycles ESCRT-III for additional rounds of budding (Wollert and Hurley, 2010).
Although the molecular mechanisms that drive membrane deformation and cleavage by the ESCRT machinery are unclear, several models have been proposed. Hanson et al. (2008) used “deep-etch” electron microscopy to visualize the plasma membrane of cells overexpressing ESCRT-III proteins and found circular arrays of filaments. Wollert et al. (2009a) reconstituted ESCRT-III–driven membrane constriction in vitro on giant unilamellar vesicles, showing that ESCRT-III proteins at high concentrations were sufficient to produce intralumenal vesicles. Vps4 was required to recycle ESCRT-III subunits for further rounds of budding but was not essential for the actual budding event (Wollert et al., 2009a). These two studies led to a model of scission known as “spiral constriction” in which ESCRT-III filaments form a flat spiral on a membrane and then coil inward as they polymerize, pinching out a membrane bud (Hanson et al., 2009; Wollert et al., 2009a).
ESCRT-III's ability to form dome-shaped caps in vitro led to the related “dome” model (Lata et al., 2008), which posits that as ESCRT-III spirals pinch membranes, they form a dome rather than a flat coil (Fabrikant et al., 2009). Further support for the dome model came from the observation of hemispherically capped cylindrical helices deforming the membranes of cells overexpressing ESCRT-III (Bodon et al., 2011).
A third model arose from the work of (Saksena et al., 2009), who used fluorescence spectroscopy to determine the order in which yeast ESCRT components assemble on the membrane They found that ESCRT-III binds to the ESCRT-II complex and oligomerizes on the membrane into a filament. They reported that other ESCRT-III proteins cap the filament and recruit the AAA+ ATPase Vps4, which disassembles the filament. These observations led to the “purse-string” model in which the ESCRT-III filament forms a ring or spiral that is disassembled by Vps4 from one end. The disassembly causes a decrease in the circumference of the ring, much like pulling the drawstring on a purse closes the opening. In this model, Vps4 drives constriction and is therefore vital to the molecular mechanism.
Recently, a fourth model was proposed based on small-angle x-ray scattering data showing that the ESCRT-I/-II supercomplex has an arched shape (Boura et al., 2012). In this model, a “whorl” of ESCRT-III is created, and Vps4 may promote filament self-association to deform the membrane into a dome. Because of this morphology, the whorl model was described as a modified dome model, but the ESCRT-III filaments were believed to lie within planes including the axis of the budding neck rather than spiraling around it as in other models.
Electron cryotomography (ECT) was used to image HIV-1 budding sites in a near-native, frozen-hydrated state in human cells, but no structure was apparent that could be attributed to the ESCRT machinery (Carlson et al., 2010). Ring-shaped densities were observed, however, in fixed and stained HIV buds arrested in cells lacking some ESCRT-III components (Morita et al., 2011). Guizetti et al. (2011) observed 17-nm-thick helical filaments in the cytokinetic “midbody” of fixed and stained human cells, but the composition of the observed filaments is unclear since 17 nm is much thicker than any filaments formed by ESCRT proteins imaged previously (Hanson et al., 2008; Lata et al., 2008; Bajorek et al., 2009b; Bodon et al., 2011; Morita et al., 2011). The structure of ESCRT complexes in cells and the scission mechanism thus remain unclear.
In 2008, it was shown that the archaeal species Sulfolobus acidocaldarius uses ESCRT homologues for cell division (Samson et al., 2008), and it has since been shown that these homologues are conserved throughout the crenarchaeal kingdom (Ellen et al., 2009; Lindås et al., 2008; Makarova et al., 2010). Archaea like S. acidocaldarius average about 1 μm in diameter and are therefore more suitable for ECT. Crenarchaea contain ESCRT-III and Vps4 homologues (Lindås et al., 2008; Samson et al., 2008) and also have a unique cell division protein, CdvA, which, like ESCRT-I and -II in eukaryotes, recruits ESCRT-III to the membrane (Samson et al., 2011). Again like other ESCRT proteins, CdvA also contains a membrane-binding domain that is predicted to be largely α-helical in nature (Samson et al., 2011). To withstand extreme conditions, the membranes of thermoacidophilic Archaea like S. acidocaldarius contain bipolar, tetraether-linked lipids (Zhai et al., 2012). A major component of these lipids, called polar lipid fraction E, can form liposomes of varying sizes. Previous negative-stain electron microscopy (EM) revealed that CdvA gives rise to a lattice-like structure on the surface of the liposomes. ESCRT-III alone does not bind to liposomes, but once recruited by CdvA, leads to substantial deformation (Samson et al., 2011). Here we show cryotomograms of CdvA polymerized on membranes. Like human ESCRT-III, CdvA forms filaments of similar dimensions that wrap helically around liposomes.
Additional information comes from imaging ESCRT proteins in vitro. Archaeal ESCRT proteins are insoluble and difficult to purify, but variations of human CHMP1A, CHMP1B, CHMP2A, CHMP3, CHMP4B, and IST1 assemble into helical cylinders and cones in vitro (in humans, ESCRT-III proteins are called charged MVB proteins (CHMPs) and increased sodium tolerance-1 (IST1) (Lata et al., 2008; Bajorek et al., 2009b; Pires et al., 2009). X-ray crystal structures reveal protein packing in numerous ways (Muziol et al., 2006; Bajorek et al., 2009b; Xiao et al., 2009). One of the observed crystal structures is fit into one set of helical tubes observed by ECT, suggesting that monomers interact roughly lengthwise to form the filaments that coil into a one-start helix (Lata et al., 2008).
To further explore the assembly properties of ESCRT proteins, we imaged assemblies of CHMP1B and the IST1 N-terminal domain (IST1NTD) again, but this time with ECT. The IST1NTD is structurally similar to other CHMPs (Xiao et al., 2009; Bajorek et al., 2009b) and has been suggested to play a dual role in regulating Vps4 recycling of ESCRT-III: it recruits Vps4 to the ESCRT machinery for depolymerization, but it also negatively regulates Vps4 by forming a heterodimer with it in the cytoplasm to block binding with ESCRT proteins (Dimaano et al., 2008). It also plays a vital role in cytokinesis (Agromayor et al., 2009; Bajorek et al., 2009a). CHMP1B also recruits VPS4 to depolymerize the complex (Stuchell-Brereton et al., 2007; Bajorek et al., 2009a). We also imaged mixtures of CHMP2A and CHMP3. In addition to confirming previous work showing that ESCRT proteins form cylinders and cones, we show here that at high concentration, these structures also have a propensity to nest. The thick tubes of concentric cylinders and stacks of cones with dome and bell shapes reveal numerous and surprisingly versatile protein–protein contacts.
We also sought to image the ESCRT complex in vivo. Previous immunofluorescence studies showed that all three archaeal components (ESCRT-III, CdvA, and Vps4) localize to the midcell during cell division and form a cytokinetic ring (Samson et al., 2008, 2011). Here imaging plunge-frozen, dividing S. acidocaldarius cells with ECT revealed a thin, flexible protein belt at the leading edge of the ingression furrow throughout cell division. On the basis of the numerous and versatile binding properties of ESCRT proteins seen here and elsewhere, we interpret the belts as flexible helical structures containing cylindrical, conical, spiral, and bell-shaped regions.
RESULTS
CdvA-coated liposomes
In the archaeon S. acidocaldarius, CdvA recruits ESCRT-III to the midcell membrane for constriction (Samson et al., 2011). Although full-length CdvA purified from a different species assembles on DNA (Moriscot et al., 2011), purified S. acidocaldarius CdvA did not polymerize by itself in our hands, but in the presence of lipids it formed filaments (Figure 1 and Supplemental Movie S1). Liposomes were made of a major component of tetraether lipids called polar lipid fraction E that was isolated from S. acidocaldarius. Slender CdvA filaments wrapped around liposomes in helices with a variable spacing of ∼8 nm. As a control, plain liposomes without CdvA were imaged, but no wrapping filaments were ever seen, supporting the interpretation that the filaments were in fact CdvA. It was previously shown that the addition of ESCRT-III protein to CdvA-coated liposomes causes rapid degradation of the liposomes, so the two proteins in complex could not be observed on the liposomes using ECT (Samson et al., 2011).
FIGURE 1:
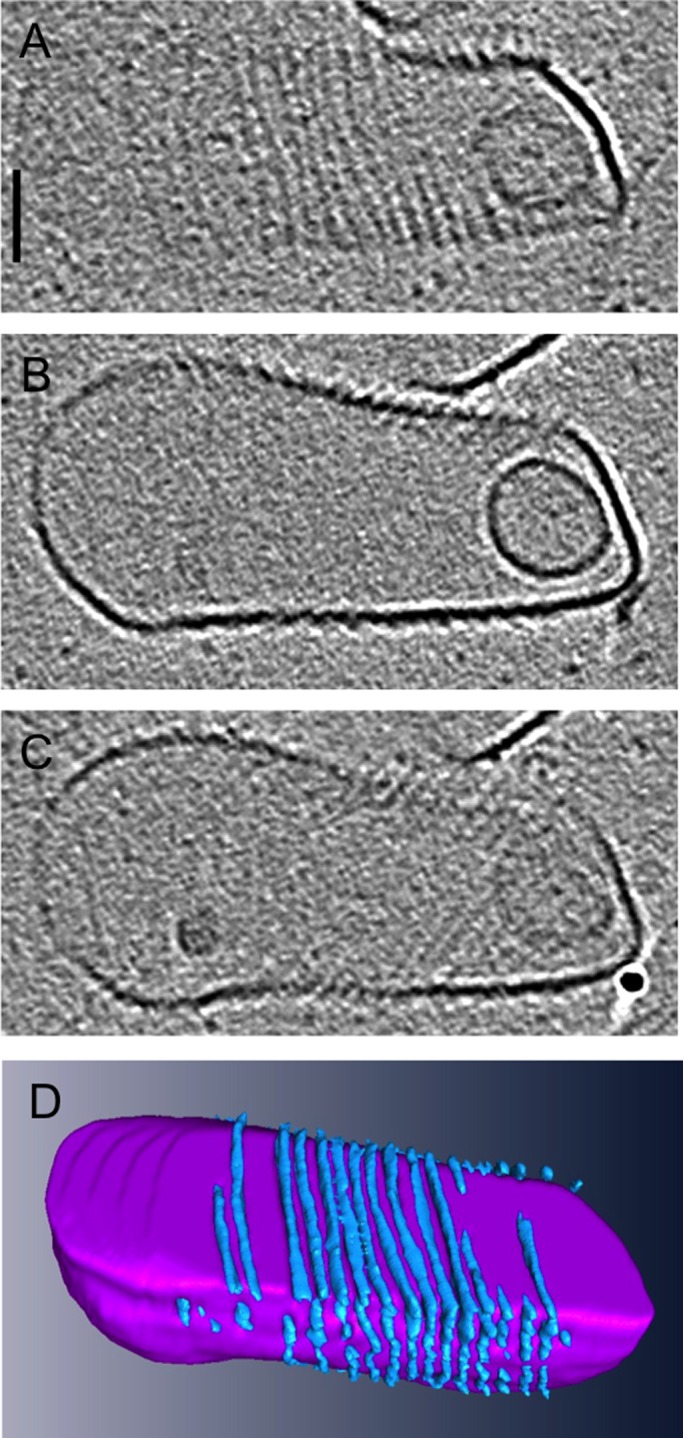
CdvA-coated liposomes. (A–C) Top, middle, and bottom slices through a 3D cryotomogram of a CdvA-decorated tetraether liposome (scale bar, 50 nm). (D) Three-dimensional segmentation, showing CdvA filaments (blue) wrapping around the liposome (purple).
In vitro assemblies of ESCRT-III
Next we sought to image ESCRT-III assemblies, which cause membrane cinching. Archaeal ESCRT-III proteins cannot be purified for in vitro assembly due to their insolubility. Because archaeal and human ESCRT-III proteins are believed to be very structurally similar (Lindås et al., 2008; Supplemental Figure S1), human ESCRT-III proteins were used to observe possible assembly capabilities of the proteins in vitro. ESCRT-III proteins were purified, either negatively stained or plunge-frozen, and imaged with ECT.
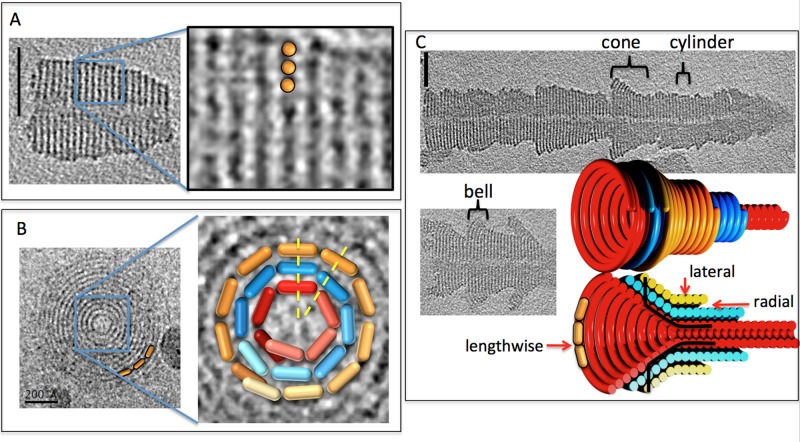
CHMP1B formed thick tubes with variable outer diameters but a consistent 12-nm-diameter hollow inner channel, with clear striations 4.7 nm apart along the long axis (Figure 2A; all measurements reported are from density “peak” to “peak” since this is reliable in EM images, but the exact position of the edges of densities is not). Careful segmentation of a tube segment that froze with its axis parallel to the grid revealed that the structures were one-start helical ramps (e.g., as in a parking garage) rather than stacks of independent disks (Supplemental Movie S2). Several other, short tubular segments froze with their long axis perpendicular to the EM grid. Projections along that view exhibited “bull's-eye” patterns of concentric rings 3 nm apart radially (Figure 2B). The thick tubes are therefore concentrically stacked, thin helices. The smallest-diameter CHMP1B helix had a circumference of 38 nm, and, of interest, the subsequent rings increased in circumference by 19 nm, half the circumference of the smallest ring. The dimensions of CHMP3 are ∼2.5 × 3.5 × 7.1 nm (Muziol et al., 2006; Bajorek et al., 2009b). Because the structures of all CHMP family members are predicted to be very similar and the lateral and radial striations in the tubes are <7.1 nm, CHMP1B monomers must bind each other at least roughly lengthwise to form the filaments that then wrap to form the helices.
FIGURE 2:
CHMP1B assemblies in vitro. (A) Central slice through a 3D cryotomogram of a CHMP1B assembly (left; scale bar, 50 nm), with enlargement (right) showing a 3D isosurface and segmentation. (B) Projection image of a short CHMP1B tube down its axis (left), with enlarged central region (right) and hypothetical monomers (orange, blue, and red 3D pill shapes) superimposed to illustrate the fact that because each helix has a different circumference, the radial contacts between them (e.g., along the two yellow dotted lines) must be either highly flexible or nonspecific. (C) Central slices through 3D cryotomograms of two longer CHMP1B assemblies (grayscale images; scale bar, 50 nm) and our interpretation (colored 3D objects) highlighting the lengthwise, lateral, and radial contacts present between monomers and the shapes (cone, cylinder, and bell, traversed by black lines) made by ESCRT helices.
The outer diameters of longer tubes changed gradually, resulting in “barbs” (Figure 2C). Such changes in diameter could be due to outer helices terminating, revealing smaller helices within, but because there did not appear to be uniform radial steps, and in analogy to the cones and domes seen in previous studies of other ESCRT-III proteins (Lata et al., 2008; Bajorek et al., 2009b), the barbs more likely represent helical cones. The cone angle varied within and between barbs, however, resulting in regions with outer shapes resembling “cones” and “bells.” In addition to these variations, the bull's-eye patterns of concentric rings and the segments of the tubes with constant diameter proved that there were also regions where the helices remained cylindrical (projections down the axis of a cone do not exhibit distinct circular rings when viewed from the top because each new turn of the helix overlaps with the previous turn). Our interpretation is that individual filaments wrap to form cylinders, cones, and bells in different regions, transitioning between these shapes along the length of the filament, and then these structures often stack, as illustrated by the colored three dimensional (3D) model in Figure 2. CHMP1B can therefore bind neighboring monomers lengthwise (to form a filament), laterally (between the coils of each helix), and radially (to the helix of just smaller or just larger radius).
Because each helix within a multilayered cylindrical tube has a different circumference, each must contain different numbers of monomers per turn, regardless of the length or orientation of the monomer (but assuming that its dimensions are constant throughout the tube). Radially adjacent monomers in the tubes cannot therefore all be in a single consistent register, so the radial contacts must be either highly flexible or nonspecific (Figure 2B). The cones must have a similar property: because each turn of a cone has a different circumference, monomers in adjacent turns cannot all be in a single consistent register, so there must be a similar flexibility/nonspecificity in the lateral (diagonal) contacts from one turn to the next. The nested cones must possess such flexible/nonspecific contacts both radially and diagonally.
IST1NTD (Bajorek et al., 2009b) and CHMP2A/3 assemblies were also imaged (Lata et al., 2008). IST1NTD formed hollow, striated tubes with different inner and outer diameters (Figure 3A). Side views of tubes in negative stain showed striations spaced 5 nm apart and oriented ∼13° from perpendicular to the long axis of the tubes. Tomographic slices perpendicular to the long axis of tubes exhibited concentric rings spaced 3.5 nm apart radially. (Note that because of the complex point-spread function, which includes effects of both defocus and the “missing wedge” of data—reviewed in Pilhofer et al., 2010—densities are surrounded by fringes, which have the effect here of emphasizing the outermost and innermost rings and suppressing the contrast of the middle rings. In addition, the “missing wedge” of data reduces the resolution in the direction parallel to the beam, obscuring the rings across the “top” and “bottom” of the tubes in the view shown.) The features exhibited by the IST1NTD tubes were very similar to those of the CHMP1B tubes, including a hollow central channel. Similarly, negatively stained CHMP2A/3 formed striated cones and tubes (Figure 3B) as seen before (Lata et al., 2008), but here stacked cones were also seen, providing evidence of nonspecific radial contacts. Thus all three human ESCRT-III proteins demonstrated interactions in all three directions, including nonspecific interactions radially and laterally.
FIGURE 3:
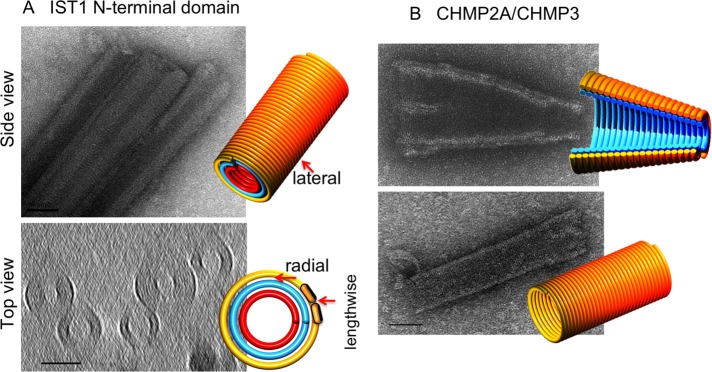
IST1 and CHMP2A/CHMP3 assemblies in vitro. (A) Projection image of three negatively stained IST1NTD tubes (top) and a central slice through a 3D cryotomogram of five plunge-frozen tubes (bottom; scale bar, 50 nm). The 3D models highlight the lengthwise, lateral, and radial contacts present between monomers (orange pills) as in Figure 1. (B) Projection images of negatively stained, full-length CHMP2A/CHMP3 tubes, with 3D models illustrating the nested cones (top) or tube (bottom; scale bar, 50 nm).
Dividing Sulfolobus
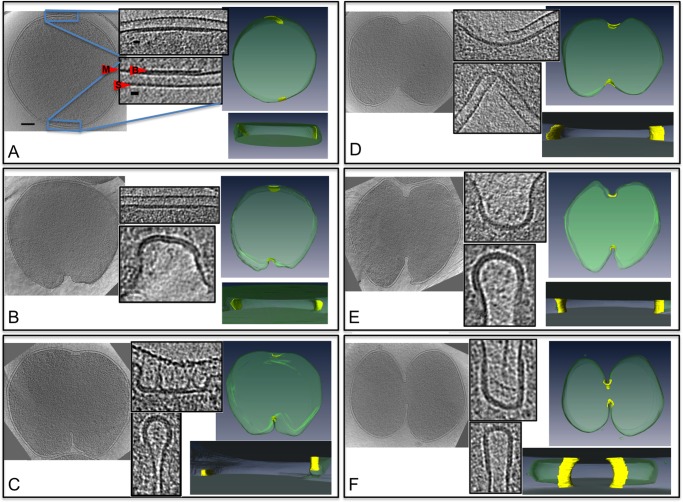
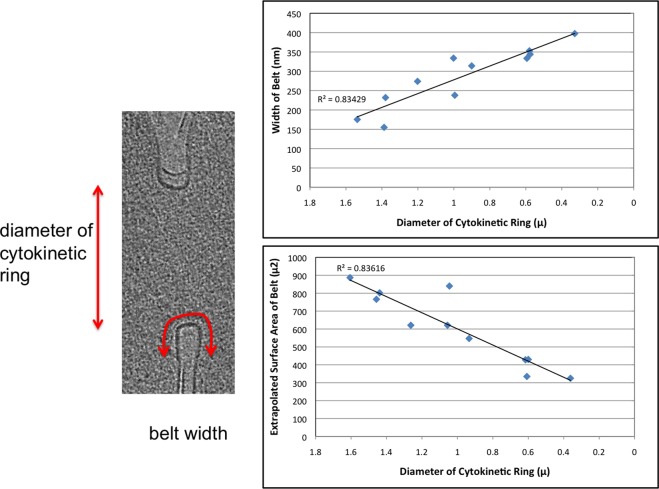
To visualize the ESCRT-III and CdvA complex in a physiologically relevant setting, the constriction rings of S. acidocaldarius cells were observed with ECT. S. acidocaldarius cells flattened somewhat when placed on EM grids and were therefore small enough to be imaged intact. Cells were synchronized for division as previously described (Duggin et al., 2008) and plunge-frozen at the time of division (as determined by flow cytometry). Tilt series were collected of 12 dividing cells and 159 nondividing cells (example tilt series are shown in Supplemental Movie S3 and a tomogram in Supplemental Movie S4). The dividing cells exhibited different degrees of ingression, ranging from 1.6 μm remaining to divide (unconstricted) to 362 nm (examples in Figure 4). In each dividing cell, the ingressing membrane was lined with an apparently proteinaceous belt ∼3.5 nm thick, 150−400 nm wide, and 6 nm away from the membrane (center-to-center) into the cytoplasm (Figure 4 and Supplemental Movie S5). As with the in vitro helices, although the belts likely wrapped around the entire circumference of the cells, they were obscured across the tops and bottoms of the cells due to the well-known “missing-wedge” artifact inherent in ECT. As cell division progressed (measured by the diameter of the cytokinetic ring), the width of the belt increased, but its thickness remained constant (Figure 5). The total surface area of the belt decreased, however, indicating a net loss of material. The curvature across the width of the belt also increased as the cell progressed through division and the membrane folded more deeply. Cross sections of the belt progressed from initially flat at the beginning of division to teardrop shaped later. Of interest, the cleavage furrow was always more advanced and differently shaped on one side of the cell than the other, suggesting further flexibility within individual belts. This was the case in cells prepared by traditional methods (fixed, embedded, and sectioned) as well, so it is not likely an artifact of flattening during plunge-freezing. The S-layer was often incomplete at the site of ingression, and many budding vesicles were observed on the side of the cells, adjacent to the ingression site, where the membrane is at maximum curvature.
FIGURE 4:
Protein belt in dividing S. acidocaldarius cells. (A–F) Left to right, central slices through cryotomograms; insets of magnified division furrows revealing protein belt on the membrane; and top and side views of corresponding segmentations of the cell membrane (green) and protein belt (yellow). Scale bars, 200 nm for the whole cells, 20 nm for the insets. (A) A nondividing cell exhibiting a flat protein belt. The three layers of the insets shown are the belt (B), membrane (M), and S-layer (S). (B) A cell that has started ingressing on one side only. (C) A cell that shows one side ingressing deeper into the cell with a small radius of curvature and the other side still very shallow and flat. (D–F) Cells farther along in the division process with both sides ingressing and becoming narrower.
FIGURE 5:
Width and surface area of the belt. As division progressed (measured as the diameter of the cytokinetic ring), the protein belt widened, but the total surface area of the belt decreased. The surface area was calculated as described in Materials and Methods.
DISCUSSION
Although purified ESCRT proteins were already known to assemble into helical tubes, cones, spirals, and domes (Lata et al., 2008; Bajorek et al., 2009b; Pires et al., 2009), here we showed that they can also form nested cylinders and stacked cones and bells and even transition between a range of shapes, apparently within the length of a single helical filament. Two factors challenge, however, the relevance of the in vitro structures. First, they were formed at very high concentration. Second, they were homopolymers, and it is known that heteromeric complexes act in human cells (Bajorek et al., 2009b). The structures nevertheless reveal a general property of ESCRT that has not been highlighted in the literature: because each helix within a thick-walled tube or turn within a cone has a different circumference, the radial and lateral/diagonal contacts must be either highly flexible or nonspecific.
These properties became of even greater interest when we imaged intact S. acidocaldarius cells in a near-native (plunge-frozen) state in 3D by ECT and observed a distinct protein belt lining the ingression furrow of all dividing cells. Although ECT can be used to image wild-type cells in native states, identifying specific protein densities within cryotomograms can be a challenge. In previous work we identified densities by perturbing their abundance or stability (e.g., Li et al., 2007; Ingerson-Mahar et al., 2010), through correlated light and electron microscopies (Briegel et al., 2008; Schlimpert et al., 2012), or by their structural signatures when purified or heterologously expressed (Pilhofer et al., 2010; Basler et al., 2012). Unfortunately in this case no stable ESCRT-III or CdvA genetic modifications with penetrance suitably high for ECT studies are available in Sulfolobus, so we could not perturb the abundance of ESCRT system components, and lysed cells were not amenable to immunogold labeling. Immunofluorescence studies have shown, however, that the ESCRT system localizes to a ring at the midcell region specifically throughout division (including when the constriction ring had the diameters seen here by ECT; Samson et al., 2011), and the belt was present here in every dividing cell imaged but only one nondividing cell (out of 159). Because the one nondividing cell containing this protein density was most likely preparing to divide, the correlation between light and electron microscopies is strong. The belt is further structurally consistent with previous crystallography and in vitro imaging of ESCRT (see later discussion), and other species of Archaea and bacteria that do not divide using the ESCRT system have not exhibited this structure when dividing (Li et al., 2007; Tocheva et al., 2011). Thus, although we have been unable to confirm that the protein belt seen here is ESCRT, our working hypothesis is that it is, and we present a structural interpretation to stimulate further experiments and discussion. Future superresolution light microscopy, or ECT of ESCRT assemblies in other contexts, such as other dividing Archaea, eukaryotic midbodies, or viral budding sites, should confirm or refute our hypothesis that the belt is ESCRT.
On the assumption that the belt seen in dividing Sulfolobus cells is the ESCRT machinery, the long axis of the ESCRT monomers (∼7 nm) must lie parallel to the membrane, since the belt's thickness is only ∼3.5 nm. The smooth edges of the belt further suggest that ESCRT filaments wrap around the circumference of the cytokinetic ring rather than perpendicular to the ring, and the different belt widths likely reflect different numbers of coils. The data are therefore consistent with the model that the belt consists of helical ESCRT filaments wrapping around the circumference of the cytokinetic ring (Figure 6). It remains unclear how ESCRT-III and CdvA are oriented with respect to each other and the membrane, but CdvA is not likely to be in the belt, since CdvA would not then be in direct contact with the membrane. Instead, since CdvA recruits ESCRT-III to the membrane, one likely scenario compatible with the observed structures is that it lies between the two (Figure 6). This structural arrangement would be consistent with the finding that although every ESCRT-III ring colocalizes with CdvA, not every CdvA ring has an ESCRT-III structure decorating it (Samson et al., 2011). If CdvA forms a layer between the membrane and the belt, however, it is clear from the low density in the tomograms that the layer is much less closely packed than the ESCRT belt. This is reasonable because it is not necessary that every ESCRT-III be bound to a CdvA: ESCRT-III will form a coherent belt by itself and may only be sparsely linked to the membrane by CdvA.
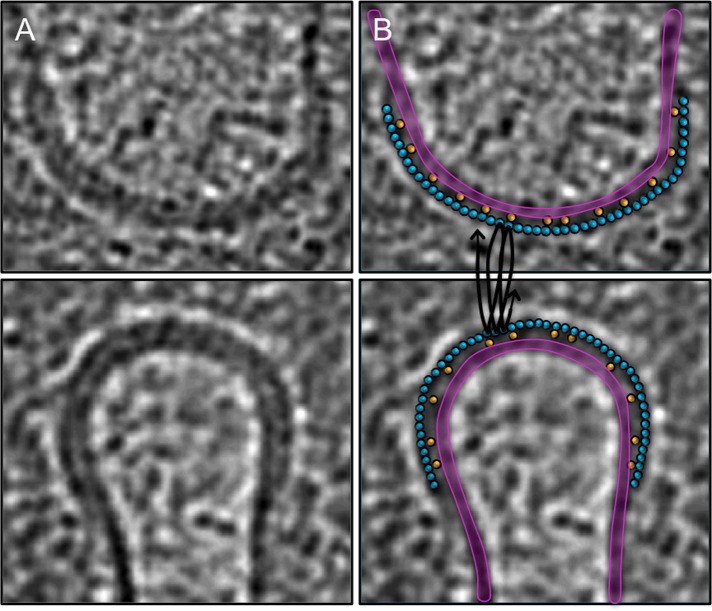
FIGURE 6:
Interpretation of the protein belt as ESCRT helices. (A) Central slices through opposing (top and bottom) edges of the division furrow in a S. acidocaldarius cell (as also shown in Figure 4E). (B) Same slices with the membrane (purple) and our model for how ESCRT (blue) filaments wrap helically (black lines) around the cytokinetic ring to form the protein belt. The ESCRT filaments appear as circles because they are cut in cross section, but their diameter matches the width of the belt. CdvA filaments (yellow, also in cross section) are supposed to lie between the membrane and the ESCRT belt, linking them together.
The in vitro assemblies imaged here and elsewhere showed that ESCRT filaments can form cylinders, domes, cones, spirals, bells, nested cylinders, and stacked cones and transition between them. This supports our interpretation of the protein belt as ESCRT spirals wrapping around the cytokinetic ring, since the belt exhibited all these shapes. In early-dividing cells, the protein belt is flat in cross section, which would be similar to the lateral interaction of the monomers in the cylindrical helices in vitro. Cells farther into division exhibit curved cross sections similar to those exhibited by the cones, domes, and bells observed in vitro, indicating flexible/nonspecific diagonal interactions. Later, washer-shaped regions at the rising edges of the ingression furrow have the same morphology as the flat spirals seen in vivo (Hanson et al., 2008) as well as cross sections through the nested cylinders here. In fact, all the interactions required to sustain the dynamic belt can be seen in the barbed, thick tubes of purified CHMP1B (black lines in Figure 2C).
Similar structures are likely to be involved in other contexts in which the ESCRT machinery acts, including, for instance, cytokinesis in eukaryotes. ESCRT proteins do not localize to the intercellular bridge in eukaryotes until the membranes are ∼1 μm apart (Guizetti and Gerlich, 2012), a diameter similar to the cytokinetic belt observed here in S. acidocaldarius. Thus the actin/myosin contractile ring may have evolved to constrict very large eukaryotic cells sufficiently for the ESCRT machinery to take over. Although there are known differences in protein sequences between the human, yeast, and archaeal ESCRTs, the functions performed are very similar. Archaea do not have sequence homologues to ESCRT-0, -I, or –II, but CdvA performs the same function of recruiting ESCRT-III to the membrane. Guizetti et al. (2011) also visualized helical filaments in dividing eukaryotic cells, but, puzzlingly, they were 17 nm in diameter, substantially larger than any reported ESCRT filament, and each turn was separated from adjacent turns, unlike the apparently tight coil seen here in Sulfolobus. The filaments in the Guizetti study were identified as ESCRT-III because CHMP2A RNA interference–treated cells did not exhibit similar filaments, but because of their size perhaps the filaments are composed of, or consist largely of, other proteins that depend on CHMP2A for assembly.
Of interest, no belts or spiraling filaments have been observed by ECT at sites of viral budding (Carlson et al., 2010). Because 1) eukaryotic ESCRT-III can bind to membranes directly (no requirement for ESCRT-I, II/CdvA), 2) individual ESCRT-III proteins are only ∼3 nm wide, and 3) there may only be a few turns of the filament needed to constrict the small neck of viral or MVB buds, the density may have simply been too small or transient (Jouvenet et al., 2011) to resolve against the membrane with ECT. Fixation and dehydration may have further obscured the filaments in earlier conventional EM studies (Gelderblom et al., 1989). It is known, for example, that several bacterial cytoskeletal filaments are not preserved by these methods (Pilhofer et al., 2010). The CdvA spacer in Sulfolobus may have made the ESCRT-III belt uniquely visible here by distancing it from the membrane.
How spiraling ESCRT filaments drive membrane scission remains unclear. Our images of dividing Sulfolobus cells fail to support the “purse-string” and “whorl” models, since corresponding structures were not seen, but the images are consistent with the “spiral” and “dome” models (Fabrikant et al., 2009). Filament growth in a spiraling pattern toward smaller radius may therefore drive constriction.
MATERIALS AND METHODS
In vitro ESCRT-III proteins: purification, assembly, and imaging
CHMP1B, CHMP2A, CHMP3, CHMP31-150, and IST1NTD were purified as previously described (Bajorek et al., 2009b). CHMP2A and CHMP3 coassemblies were formed by concentrating both proteins to 20 mg/ml (CHMP2A, 830 μM; CHMP3, 790 μM), mixing (1:1 vol/vol) and incubating at room temperature for 30 min. Monomeric IST1NTD was assembled by dialyzing concentrated protein (265 μM for negative stain image and 240 μM for cryo image) against 50 mM Tris, pH 7.0, 100 mM NaCl, 5% (vol/vol) glycerol, and 5 mM beta-mercaptoethanol (BME) for 12 h at 4°C. Monomeric CHMP1B was assembled by concentrating the protein to 440 μM in 10 mM Tris, pH 8.0, 100 mM NaCl, and 5 mM BME and incubating for 24 h at 4°C.
CHMP1B tomograms were collected with 4- to 6-μm defocus, 150 e−/Å2, and pixel size of 1 nm. CHMP2A/3 tubes were negatively stained on glow-discharged carbon/Formvar grids with 2% uranyl acetate and imaged at 52,000× magnification. IST1NTD tubes were similarly stained and imaged, as well as plunge-frozen and imaged using electron cryotomography. The parameters of tomographic data collection were 41,000× magnification, −3.5-μm defocus, tilt ± 60°, 2° tilt increments, a dose ranging between 120 and 240 e−/Å2, and pixel size of 0.5 nm. Tomograms were reconstructed using eTomo or Raptor and were binned by two. Software packages Amira and IMOD were used for making supplemental movies.
S. acidocaldarius
Growth conditions and synchronization.
S. acidocaldarius DSM639 was grown in Brock's medium, pH 3.2, at 78°C and synchronized as described in Duggin et al. (2008). Specifically, 75 ml of culture at approximately OD600 = 0.1 was applied to a poly-d-lysine–coated membrane within the baby machine apparatus. After pumping of Brock's medium through the apparatus for 3.5 h at ∼0.8 ml/min, newly divided cells were collected on ice over a period of 30 min. Synchronized grow-out was initiated by transferring the vessel containing the cells from ice to a water bath heated to 78°C.
Purification of protein, polar lipid fraction E lipids, and antibodies.
S. acidocaldarius CdvA (residues 69–238) was purified as described in Samson et al. (2011). Polar lipid fraction E (PLFE) tetraether lipids were isolated from S. acidocaldarius cells, and PLFE liposomes were prepared as previously described (Samson et al., 2011). Affinity columns were prepared and antisera were purified as described (Samson et al., 2008).
EM sample preparation, data collection, tomographic reconstruction, and analysis.
EM R2/2 copper Quantifoil grids were made hydrophilic by using a Harrick plasma cleaner for 30 s. The synchronized cells were mixed 3:1 with 10-μm colloidal gold particles and 5% bovine serum albumin. A 3-μl droplet of the cell mixture was applied to each grid and then blotted and plunge-frozen into liquid ethane using a Gatan Cryoplunge 3 (Gatan, Pleasanton, CA). The grids were stored in a liquid nitrogen–cooled dry shipper and shipped to Caltech for imaging.
EM images were collected using a Polara 300-kV FEG transmission electron microscope (FEI, Hillsboro, OR) equipped with an energy filter and a lens-coupled 4k × 4k Ultracam (Gatan). Tilt series were recorded from −65 to +65° with an increment of 1° at −15 μm under focus using the Leginon software package (Suloway et al., 2005). Cumulative doses of up to 200 e−/Å2 were used.
The tilt series were binned by two, and 3D tomograms were calculated using the IMOD software package (Kremer et al., 1996). Dividing cells were 3D rendered using Amira (Visage Imaging, San Diego, CA). The surface area was calculated for each cell using the average of the belt width × the average diameter of the belt ring × 2π for the surface area of a cylinder. It was assumed that the ring was complete around the cell, even though not all of it could be measured due to the missing wedge, and that it was circular, even though the cell flattened on the grid. This surface area calculation was performed in the same way for all cells and is used to compare cells rather than for accurate independent values.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Heath Grant P50 GM082545 to G.J.J., National Science Foundation Grant DMR1105277 to P.L.C., and a gift to Caltech from the Gordon and Betty Moore Foundation. We thank Wesley I. Sundquist for advice and critical reading of the manuscript, Kay Grunewald for use of laboratory equipment, and Morgan Beeby, Jason Porath, and Jean Choi for their help with illustrations.
Abbreviations used:
- CHMP
charged multivesicular body protein
- ECT
electron cryotomography
- EM
electron microscopy
- ESCRT
endosomal-sorting complex required for transport
- HIV
human immunodeficiency virus
- IST1
increased sodium tolerance-1
- MVB
multivesicular body
- NTD
N-terminal domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0785) on June 12, 2013.
REFERENCES
- Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009a;20:1360−1373. doi: 10.1091/mbc.E08-05-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009b;16:754−762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodon G, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276−40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, Eaton WA, Hummer G, Hurley JH. Solution structure of the ESCRT-I and –II supercomplex: Implications for membrane budding and scission. Structure. 2012;20:874−886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L-A, De Marco A, Oberwinkler H, Habermann A, Briggs JAG, Kräusslich H-G, Grünewald K. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog. 2010;6:1−11. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C, Jones CB, Hanono A, Curtiss M, Babst M. Ist1 regulates Vps4 localization and assembly. Mol Biol Cell. 2008;19:465−474. doi: 10.1091/mbc.E07-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105:16737−16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellen AF, et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles. 2009;13:67−79. doi: 10.1007/s00792-008-0199-x. [DOI] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JAG, Weissenhorn W, Kozlov MK. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comp Biol. 2009;5:1−11. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelderblom HR, Ozel MC, Pauli G. Morphogenesis and morphology of HIV. Structure-function relations. Arch Virol. 1989;106:1−13. doi: 10.1007/BF01311033. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133−140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, Müller-Reichert T, Gerlich DW. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616−1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389−402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Opin Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463−487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;8:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Matronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71−76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger H, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354−1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindås A-C, Karlsson EA, Lindgren MT, Ettema TJG, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci USA. 2008;105:18942−18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen G. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694−4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731−741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:24.1–24.30. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriscot C, Gribaldo S, Jault J-M, Krupovic M, Arnaud J, Jamin M, Schoehn G, Forterre P, Weissenhorn W, Renesto P. Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS One. 2011;6:1–9. doi: 10.1371/journal.pone.0021921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III Factor CHMP3. Dev Cell. 2006;10:821−830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Ladinsky MS, McDowall AW, Jensen G. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21−45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- Pires R, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843−856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson A, Emr S. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97−109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in Archaea. Science. 2008;322:1710−1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL, Williams RL, Bell SD. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell. 2011;41:186−196. doi: 10.1016/j.molcel.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlimpert S, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270−1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740−744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41−60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tocheva EI, Matson EG, Morris DM, Moussavi F, Leadbetter JR, Jensen G. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799−812. doi: 10.1016/j.cell.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley J. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864−869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley J. Membrane scission by the ESCRT-III complex. Nature. 2009a;458:172−177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Yang D, Ren X, Lee HH, Im YJ, Hurley J. The ESCRT machinery at a glance. J Cell Sci. 2009b;122:2163−2166. doi: 10.1242/jcs.029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514−3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Chong PL, Taylor LJ, Erlkamp M, Grobelny S, Czeslik C, Watkins E, Winter R. Physical properties of archaeal tetraether lipid membranes as revealed by differential scanning and pressure perturbation calorimetry, molecular acoustics, and neutron reflectometry: effects of pressure and cell growth temperature. Langmuir. 2012;28:5211−5217. doi: 10.1021/la300142r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.